Abstract
The polyubiquitin signal is post-translationally attached to a large number of proteins, often directing formation of macromolecular complexes resulting in the translocation, assembly or degradation of the attached protein. Recent structural and functional studies reveal general mechanisms by which different architectures and length of the signal are distinguished.
Introduction
Post-translational modification of proteins is a widespread mechanism to fine tune the structure, function and location of proteins encoded by the surprisingly small number of genes in most organisms. Among these modifications, one of the most versatile and intriguing modifications is the covalent attachment of ubiquitin or ubiquitin-like proteins to target proteins [1-5]. In this commentary we will argue that, in contrast to modifications such as phosphorylation, methylation, acetylation, etc., the attachment of one or more ubiquitins provides a large interaction surface by which the modification can be decoded [6] and results in a vast number of potential signals by virtue of the varied architectures linking the multiple ubiquitin molecules. Ubiquitination of proteins directs the modified proteins to different cellular fates such as translocation, assembly or degradation, depending on the type of tagging. Recent studies have begun to define the roles of these different polyubiquitin signals in physiology and disease [7-11], and it is obvious that the manipulation of these signals and their recognition will be important in understanding and treating human disease. For example, defects in the pathway by which Ubiquitination tags proteins for proteolysis in the proteosome have been linked to neurodegenerative disorders such as Alzheimer's [7] and Parkinson's disease [8].
Diversity of polyubiquitin chains and linkages
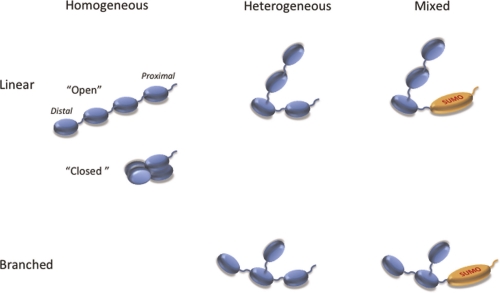
Polyubiquitin chains consist of ubiquitin monomers linked to each other covalently by an isopeptide bond between the C-terminus of one ubiquitin and an amine from one of the seven lysines or the N-terminal methionine of the next ubiquitin. A variety of polyubiquitin chains have been detected in cellular lysates [12], varying in length and linkage (Figure 1). Moreover, the chains can also be mixed, i.e. containing other heterologous proteins from the ubiquitin family such as SUMO (small ubiquitin-like modifier) [13].
Figure 1. Architecture of polyubiquitin chains.
Unanchored chains are shown here. These are often found attached to a target protein via an isopeptide bond between the free C-terminus of the proximal ubiquitin and an amino group on the target protein. Chains can be “linear” (no more than one amino group of each ubiquitin is linked to another ubiquitin) or “branched” (at least one ubiquitin is attached to other ubiquitins via two or more different amino groups). The ubiquitin containing the free C-terminus is the “proximal” subunit and the ubiquitin(s) lacking any amino group linkage is a “distal subunit”. The polymers are referred to as “homogeneous” if all linkages use the same amino group on each ubiquitin. They are termed “heterogeneous” if ubiquitin linkages involve different amino groups on different subunits. Chains containing other ubiquitin-like proteins are called “heterologous” or “mixed”. The type of linkage is designated by the identity of the amino group used, i.e. Met1-, Lys6-, Lys11-, Lys27-, Lys29-, Lys33-, Lys48-, Lys63-linked. Note that Met1-linked ubiquitin is identical to the proprotein product of the polyubiquitin gene and has often been referred to as linear polyubiquitin. We propose the Met1-linked nomenclature to avoid the confusion with unbranched chains described above. In the “open” conformations the hydrophobic patch of ubiquitin is exposed, while in the “closed” conformations this patch is buried by ubiquitin-ubiquitin interactions. Abbreviations: SUMO, small ubiquitin-like modifier.
Polyubiquitin can signal in the cell as unanchored chains [14, 15], with one free C-terminus on the proximal subunit, or as a molecular tag attached through this C-terminus to a lysine in a target protein. As the latter might have several lysines, the same protein can carry several different polyubiquitin tags associated with it [16].
Assembly and disassembly of different polyubiquitin chains
Some unanchored polyubiquitin chains are synthesised by the sequential reactions of two enzymes: E1 (the ubiquitin activating enzyme) and E2 (the ubiquitin conjugating enzyme). Pioneering work from Cecile Pickart showed that E1 and E2-25K synthesize unanchored homogeneous Lys48-linked chains, while ligation of similar chains linked through Lys63 requires the UBC (ubiquitin C)13/MMS2 heterodimer as the E2 [17]. Many other isolated E2s synthesize multiple types of linkages. The length and linkage type of polyubiquitin chains can also be modulated by a third enzyme, the E3 ubiquitin ligase. In most cases, this E3 ligase is required for assembly of a polyubiquitin chain on a protein acceptor.
Chains can also be remodelled or disassembled by various deubiquitinating enzymes, proteases that cleave the (iso)peptide bond between ubiquitins or ubiquitin and a target protein. Deubiquitinating enzymes can show considerable specificity. For instance, isopeptidase T (USP5 [ubiquitin-specific protease 5]) only acts efficiently on free polyubiquitin chains but shows little linkage selectivity [18]. Proteasomal deubiquitinating enzymes require association with the proteasome for maximal efficiency although the precise chain specificity is still unclear. Structural analysis of several deubiquitinases explains why TRABID cleaves the Lys29 linkage 40-fold more efficiently than the Lys63 linkage, Cezanne is specific for Lys11 linked chains [19], CYLD prefers Lys63-linked chains [20], and OTU1/Otubain is selective for long Lys48-linked chains [21, 22]. In many other cases the deubiquitinating enzyme's specificity is either minimal or has not been determined. Thus, the type of polyubiquitin chain that is accumulated will depend on the specificity of the enzymes that synthesize the chain (E2/E3) and disassemble it (deubiquitinating enzymes).
Indeed, because of the sequential action of various deubiquitinases (deubiquitinating enzymes) and/or conjugating enzymes/ligases, the length and linkage composition of a given chain can be dynamic [23]. For instance, the promiscuous UBCH5 family of E2s combines with either of two E3 ligases, CIAP (a cellular inhibitor of apoptosis) or APC (the anaphase promoting complex), to monoubiquitinate their respective substrates [24-27]. Subsequently Lys11-linked chains are elaborated by these E3s using the E2 enzyme UBE (ubiquitin-activating enzyme)2S. BRCA1/BARD1 autoubiquitination also shows this progression of priming (monoubiquitination) followed by elongation (polyubiquitination). Six different E2s can monoubiquitinate BRCA1, while different E2s are then used to elongate the chains with Lys63 (UBC13/MMS2), Lys48 (UBE2K), or Lys6 (UBCH5) linkages [28]. Certain pairs of E2 and E3 can also synthesize branched chains [16]. In another classic example of chain remodelling, A20 (a large enzyme with both deubiquitinating enzymes and E3 ligase activity) is thought to first trim K63-linked substrates of the NF-κB (nuclear transcription factor kappa B) pathway and then elaborate a K48-linked chain that results in the degradation of the substrate.
How does chain architecture determine the fate of polyubiquitinated proteins?
Importantly, all these polyubiquitin forms appear to serve different functions (targeting/localization, complex assembly, modulation of function or stability). The underlying structure-function hypothesis is that the chain linkage (and possibly chain length) defines the ensemble of structures/conformations that a particular chain can adopt [10, 24, 29-31]. In turn, this structure defines the chain's ability to interact with specific receptors in a linkage specific manner [22, 32-39]. The critical question is how signalling specificity is achieved despite the vast variety of polyubiquitin chain structures/conformations.
Two features of ubiquitin chains are essential for this signalling: the hydrophobic patch on one face of the monomers [40, 41], and a highly flexible C-terminus, resulting in significant flexibility of each ubiquitin-ubiquitin linker in the chain [42]. In fact, Lys48-linked di- and tetra-ubiquitin chains under physiological conditions are in equilibrium between a predominantly “closed” conformation (where the hydrophobic patches are sequestered at the ubiquitin/ubiquitin interface) and one or more open conformations [42-44], whereas Lys63-linked chains predominantly adopt an extended structure exposing the hydrophobic patches and making them readily available for interactions with receptors [38, 45]. The emerging structural data and computer modelling indicate an even greater structural variability for chains connected through other lysines [19, 32, 46, 47]. We anticipate that chain branching might result in structures and intra-chain contacts unavailable for linear-topology chains, thus potentially further enhancing the signalling capability of polyubiquitin.
Polyubiquitin receptors
Specific recognition of polyubiquitin is accomplished by proteins containing one or more ubiquitin-binding domains. There are at least 20 families of ubiquitin-binding domains (Table 1) and many polyubiquitin-binding proteins contain multiple copies, often consisting of two or three different ubiquitin-binding domains connected by flexible linkers [48-53]. The most common of these domains, including the UBA (ubiquitin-associated) domain and UIM (ubiquitin-interacting motif), bind to a conserved hydrophobic patch on ubiquitin consisting of Leu8, Ile44, and Val70 [48-53]. The affinity of all these domains for ubiquitin is modest (µM) but tight binding is achieved by avidity; the immobilization of polyubiquitin by binding to one ubiquitin-binding domain lowers the entropic barrier for binding of an adjacent ubiquitin to another ubiquitin-binding domain, or for other protein-protein interactions between the ligand and the intact receptor [33, 48, 54].
Table 1. Ubiquitin binding domains (See text and [48-53] for details).
| Structure | Domain | Comments |
|---|---|---|
| Single Helices | UIM | Binds to groove on ubiquitin containing hydrophobic patch |
| DUIM | Double UIM | |
| MIU | Binds to groove on ubiquitin but in opposite orientation | |
| Three helix bundles | UBA | Widespread, often combined with other ubiquitin-binding domains |
| CUE | UBA variant involved in ERAD | |
| VHS | Seven helices in folded domain, endosomal sorting | |
| GAT | Found in endosomal sorting pathways, UBA+2 ZnF | |
| Zinc Fingers | NZF | Found in Npl4, TAB2, and NZF2 of ESCRT II |
| ZnF UBP | Found in deubiquitinating enzymes, BRAP2 ligase and HDAC6 | |
| A20 ZnF | Variant of ZnF ubiquitin-specific protease domain, found in A20 deubiquitinating enzyme and Rabex 5 | |
| UBZ | Found in Y-family polymerases involved in DNA repair | |
| Catalytic Domains | UBC | Catalytic domain of E2 ubiquitin-conjugating enzymes |
| UEV | Variant UBC lacking enzymatic Cys residue, inactive | |
| SENP | SUMO-specific protease domain | |
| USP | Ubiquitin-specific protease deubiquitinating enzyme domain | |
| UCH | Ubiquitin C-terminal hydrolase deubiquitinating enzyme domain | |
| OTU | Ovarian Tumor-related deubiquitinating enzyme domain | |
| Jab1/MPN | Metalloprotease deubiquitinating enzymes | |
| Other Domains | GLUE | GRAM-like ubiquitin-binding domain in EAP45 |
| PRU | PH domain of RPN13 | |
| SH3 | SH3 domain in many endocytic proteins | |
| UBM | Ubiquitin-binding domain in Y-family DNA polymerases | |
| PFU | Ubiquitin-binding domain of PLAA family proteins | |
| WD40 | WD40 domain of UFD2/DOA1/PLAA | |
| UBAN | Coiled Coil, binds to Met1-linked polyubiquitin |
Abbreviations: DOA1, degradation of alpha2 1; ERAD, endoplasmic reticulum-associated protein degradation; GLUE, GRAM-like ubiquitin-binding domain in EAP45; MIU, ubiquitin interacting domain; NZF, Npl4 zinc fingers; OTU, Ovarian Tumor; PFU, PLAA family ubiquitin binding domain; PH, pleckstin homology; PLAA, phospholipase A2 activating protein; SUMO, small ubiquitin-like modifier; UBA, ubiquitin associated; UBAN, ubiquitin binding in ABIN and NEMO proteins; UBC, ubiquitin C; UBM, ubiquitin-binding motif; UBP, ubiquitin-specific protease domain; UIM, ubiquitin interacting motif; UBZ, ubiquitin-binding zinc finger; UCH, ubiquitin C-terminal hydrolase; UEV, ubiquitin E2 variant; UFD2, ubiquitin fusion degradation 2; USP, ubiquitin-specific protease; ZnF zinc fingers.
Shuttling receptors
Moving ubiquitinated proteins within the cell is facilitated by shuttling receptors [55-59]. These ubiquitin-binding proteins contain one or more ubiquitin-binding domains. One type of shuttling receptor, the UBX (ubiquitin regulatory X domain) family of p97 cofactors, contains both a ubiquitin-binding domain (of the UBA family) and a ubiquitin-like domain (of the UBX family) and acts as an adapter localizing polyubiquitinated proteins to the p97 AAA ATPase (an ATPase associated with a variety of cellular activities) [60, 61]. Subsequent delivery of the ubiquitinated proteins to the proteasome could be direct or the substrate could be handed off to other shuttling proteins [62]. Ubiquitin-like domains can act as autoinhibitory sequences by binding to the ubiquitin-binding domain, competing with ubiquitin binding [55] and/or directing the ubiquitin-binding protein to another receptor or the proteasome. Networks of ubiquitin binding domain /ubiquitin-like domain containing proteins could assemble into oligomers or a lattice that would provide a highly selective array of ubiquitin-binding sites that exhibit specificity for particular polyubiquitin chain linkages. Indeed, studies on artificial oligomeric UBA domains such as GST (glutathione S-transferase)-UBA fusions or TUBES (tandem ubiquitin-binding entities) have emphasized that the observed specificity is determined more by the oligomeric state of these domains than it is by the weak specificity inherent in the isolated domains [37, 48].
Specific recognition of chain linkage
Specificity of chain binding is a required attribute of receptors that sort proteins carrying differently linked ubiquitins to different cellular fates. For instance Lys6-, Lys11-, Lys29- and Lys48-linked substrates can be directed to the proteasome [9, 63], while Lys63-linked and Met1-linked chains serve non-proteolytic roles by assembling protein complexes in the DNA-damage response and NF-κB signalling pathways [15, 64, 65].
In general, it appears that proteins consisting of flexibly linked ubiquitin-binding domains show little chain specificity, in part because the polyubiquitin chain itself is also very flexible [43, 44, 47]. If this conformational flexibility is restricted in a physiological complex or lattice, even these binding proteins could show modest specificity. In contrast, receptors that recognize either the isopeptide linkage or adjacent faces of ubiquitin that are juxtaposed only in certain linkages can show much higher chain linkage specificity [35, 53, 54, 66, 67] (see Figure 2).
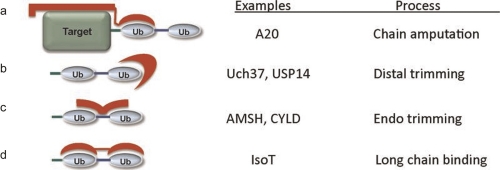
Figure 2. Specific recognition of polyubiquitin chains by deubiquitinating enzymes.
Modes of recognition by deubiquitinating enzymes involve: a) binding to the proximal ubiquitin and the ubiquitinated protein resulting in removal (amputation) of the chain; b) binding to the distal copy of ubiquitin only resulting in shortening (distal trimming) of the chain; c) binding to the specific linkage and/or faces of ubiquitin uniquely juxtaposed by that linkage resulting in specific cleavage of one type of linkage; or d) binding of adjacent ubiquitins in a chain by a deubiquitinating enzyme containing multiple ubiquitin binding domains, each bound to a single ubiquitin (pan specific). Abbreviations: Ub, ubiquitin. Reprinted with permission from Reyes-Turcu FE, Wilkinson KD. Polyubiquitin binding and disassembly by deubiquitinating enzymes. Chem Rev. 2009 Apr;109(4):1495-508. Copyright 2009, American Chemical Society.
Importance of chain length
Since the initial observation of Pickart's group that K48-linked tetraubiquitin was the minimal effective length to deliver proteins to the proteasome [68], the question of how chain length affects the fate of a ubiquitinated protein has been debated. Chains must achieve a length that provides sufficient binding affinity through avidity, but long chains may be so tightly bound as to prevent dissociation of catalytic intermediates or “hand off” from one receptor to the next [69]. In part, chain length can be controlled by the degree of processiveness (repetitive rounds of catalysis without releasing the substrate) exhibited by the ubiquitin ligases [70-72] or by ubiquitin chain editing catalyzed by deubiquitinating enzymes. The modular nature of receptors containing multiple ubiquitin-binding domains and the ability of longer polyubiquitin chains to bind multiple receptors could serve to act as length sensors (Figure 3). For instance, the deubiquitinating enzyme USP5 selectively binds tetraubiquitin with sub-nanomolar affinity using an ensemble of four ubiquitin-binding sites [18]. Alternatively, a lattice consisting of ubiquitin-binding domain/ubiquitin-like domain-containing proteins that bind each other and/or compete for ubiquitin and ubiquitin-like domain binding can show a marked length dependence on the resulting complexes (as for example, Dsk2 and Rpn10 [56]). Longer polyubiquitin chains could be “handed off” from one receptor to another in such a lattice, as exemplified by the trafficking of ubiquitinated proteins through the ESCRT (endosomal sorting complex required for transport) complex [69]).
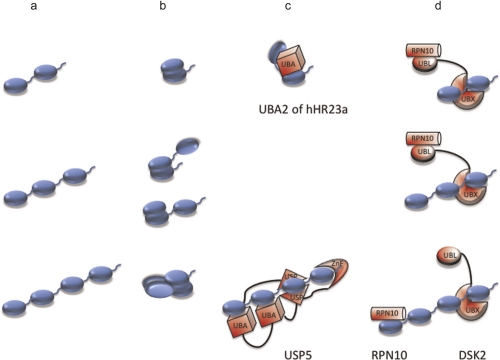
Figure 3. Figure 3. Length dependence of linear chain structure and recognition.
Each row represents a different length of linear ubiquitin: a) extended chains with exposed hydrophobicity such as Met1- and Lys63-linked; b) closed conformations where some of the ubiquitins contain intramolecularly buried hydrophobic patches as seen in Lys48-linked chains [67, 88, 89]; c) length selection by binding to a single receptor polypeptide (UBA [ubiquitin associated domain]2 of hHR23a and USP5 [ubiquitin-specific protease 5] are shown [18, 39]); and d) combinatorial length selection by two different ubiquitin receptors (RPN10 and DSK2 are shown [56]). Abbreviations: UBA, ubiquitin-associated domain; UBL, ubiquitin-like; UBX, ubiquitin regulatory X domain; USP, ubiquitin-specific protease; ZnF, zinc finger.
Localization of the polyubiquitin signal
Met1-linked chains (both unanchored and attached to NEMO – an NF-κB modifier) can activate signalling in the NF-κB pathway and directly activate some kinases [15, 29, 73, 74]. Unanchored Met1-linked ubiquitin is also the primary gene product of several ubiquitin genes and is post-translationally assembled in response to genotoxic stress and TNF-α by the E3 ligase LUBAC (linear ubiquitin chain assembly complex) [75, 76]. Levels of M1-linked ubiquitin chains in cells are normally very low [77], in part because the primary gene product is co-translationally cleaved to monomeric ubiquitin at the ribosome and in part because of the presence of large amounts of USP5 deubiquitinating enzyme, the enzyme responsible for disassembling polyubiquitin intermediates that might otherwise accumulate in the cell [18]. Thus, it is unlikely that Met1-linked chains are widely distributed in the cell, and more likely that they are locally generated at the site(s) of signalling when required. A similar argument can be made in the case of ubiquitinated proteins that accumulate during signalling. A great deal of cellular specificity in the ubiquitin pathway seems to depend on the use of adaptors and scaffolds that colocalize polyubiquitin and the enzymes that metabolize it [78]. For instance, deubiquitinating enzymes are very often found in the same protein complexes as the ubiquitin ligases that synthesize polyubiquitin [78, 79]. This suggests that if a polyubiquitin or polyubiquitinated protein is not properly channelled to its target by ubiquitin receptors it can be disassembled before it leaves the site of synthesis.
Future Directions
The incredible diversity of polyubiquitin chains observed in vivo suggests that there is a similar complexity in the receptors that recognize chains. It also seems likely that additional ubiquitin-binding motifs and domains remain to be discovered. In spite of the limited specificity of isolated ubiquitin binding domains different chain architectures effectively signal different cellular fates, suggesting that a deeper understanding will be gained from studies on binding specificity in the context of the full-length receptors. As engineered and synthetic polyubiquitins become available [46, 80-87], these will aid structure determination of polyubiquitin-receptor complexes that will be vital to understand the decoding of the polyubiquitin signals. Finally, we need a more sophisticated understanding of the “hand off” of a receptor bound polyubiquitin from one receptor to the next in a pathway. For instance it is still a mystery how shuttling receptors hand off ubiquitin from the ligases and chaperone complexes to the proteasome, or how sequential ESCRT complexes can direct endocytic cargos carrying the ubiquitin signal [69]. A detailed understanding of these issues may pay dividends in treating diseases involving errors in the ubiquitination pathway.
Acknowledgments
We thank Robert Cohen for sharing his manuscript and helpful comments.
Abbreviations
- AAA ATPase
ATPases associated with a variety of cellular activities;
- ESCRT
endosomal sorting complex required for transport
- GST
glutathione S-transferase
- LUBAC
linear ubiquitin chain assembly complex
- NF-κB
nuclear transcription factor kappa B
- SUMO
small ubiquitin-like modifier
- TUBE
tandem ubiquitin-binding entity
- UBA
ubiquitin associated
- UBC
ubiquitin C
- UBE
ubiquitin-activating enzyme
- UBX
ubiquitin regulatory X domain
- UIM
ubiquitin interacting motif
- USP5
ubiquitin-specific protease 5
Competing interests
The authors declare that they have no competing interests.
The electronic version of this article is the complete one and can be found at: http://f1000.com/reports/b/3/26
References
- 1.Bedford L, Lowe J, Dick LR, Mayer RJ, Brownell JE. Ubiquitin-like protein conjugation and the ubiquitin-proteasome system as drug targets. Nat Rev Drug Discov. 2011;10:29–46. doi: 10.1038/nrd3321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Geng J, Klionsky DJ. The Atg8 and Atg12 ubiquitin-like conjugation systems in macroautophagy. 'Protein modifications: beyond the usual suspects' review series. EMBO Rep. 2008;9:859–64. doi: 10.1038/embor.2008.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hochstrasser M. Origin and function of ubiquitin-like proteins. Nature. 2009;458:422–9. doi: 10.1038/nature07958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wilkinson KA, Henley JM. Mechanisms, regulation and consequences of protein SUMOylation. Biochem J. 2010;428:133–45. doi: 10.1042/BJ20100158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang D, Zhang DE. Interferon-stimulated gene 15 and the protein ISGylation system. J Interferon Cytokine Res. 2011;31:119–30. doi: 10.1089/jir.2010.0110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Casado-Vela J, Cebrian A, del Pulgar MT, Sanchez-Lopez E, Vilaseca M, Menchen L, Diema C, Selles-Marchart S, Martinez-Esteso MJ, Yubero N, Bru-Martínez R, Lacal JC. Lights and shadows of proteomic technologies for the study of protein species including isoforms, splicing variants and protein post-translational modifications. Proteomics. 2011;11:590–603. doi: 10.1002/pmic.201000287. [DOI] [PubMed] [Google Scholar]
- 7.Dammer EB, Na CH, Xu P, Seyfried NT, Duong DM, Cheng D, Gearing M, Rees H, Lah JJ, Levey AI, Rush J, Peng J. Polyubiquitin linkage profiles in three models of proteolytic stress suggest the etiology of Alzheimer disease. J Biol Chem. 2011;286:10457–65. doi: 10.1074/jbc.M110.149633. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 6Evaluated by Keith Wilkinson 17 Nov 2011
- 8.Lee FK, Wong AK, Lee YW, Wan OW, Chan HY, Chung KK. The role of ubiquitin linkages on alpha-synuclein induced-toxicity in a Drosophila model of Parkinson's disease. J Neurochem. 2009;110:208–19. doi: 10.1111/j.1471-4159.2009.06124.x. [DOI] [PubMed] [Google Scholar]; F1000 Factor 6Evaluated by Keith Wilkinson 17 Nov 2011
- 9.Xu P, Duong DM, Seyfried NT, Cheng D, Xie Y, Robert J, Rush J, Hochstrasser M, Finley D, Peng J. Quantitative proteomics reveals the function of unconventional ubiquitin chains in proteasomal degradation. Cell. 2009;137:133–45. doi: 10.1016/j.cell.2009.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 8Evaluated by Karin Romisch 22 Apr 2009, Jane Endicott 13 May 2009, Keith Wilkinson 17 Nov 2011
- 10.Li W, Ye Y. Polyubiquitin chains: functions, structures, and mechanisms. Cell Mol Life Sci. 2008;65:2397–406. doi: 10.1007/s00018-008-8090-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ciechanover A, Iwai K. The ubiquitin system: from basic mechanisms to the patient bed. IUBMB Life. 2004;56:193–201. doi: 10.1080/1521654042000223616. [DOI] [PubMed] [Google Scholar]
- 12.Peng J, Schwartz D, Elias JE, Thoreen CC, Cheng D, Marsischky G, Roelofs J, Finley D, Gygi SP. A proteomics approach to understanding protein ubiquitination. Nat Biotechnol. 2003;21:921–6. doi: 10.1038/nbt849. [DOI] [PubMed] [Google Scholar]; F1000 Factor 10Evaluated by John Yates 05 Aug 2003, Gourisankar Ghosh 29 Aug 2003, Visith Thongboonkerd 08 Sep 2003, Andy Golden 23 Oct 2003, Keith Wilkinson 17 Nov 2011
- 13.Geoffroy MC, Hay RT. An additional role for SUMO in ubiquitin-mediated proteolysis. Nat Rev Mol Cell Biol. 2009;10:564–8. doi: 10.1038/nrm2707. [DOI] [PubMed] [Google Scholar]; F1000 Factor 6Evaluated by Keith Wilkinson 17 Nov 2011
- 14.Parvatiyar K, Harhaj EW. Cell signaling. Anchors away for ubiquitin chains. Science. 2010;328:1244–5. doi: 10.1126/science.1192296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xia Z, Sun L, Chen X, Pineda G, Jiang X, Adhikari A, Zeng W, Chen Z. Direct activation of protein kinases by unanchored polyubiquitin chains. Nature. 2009;461:114–9. doi: 10.1038/nature08247. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 19Evaluated by Mark Bothwell 07 Sep 2009, John Kyriakis 09 Sep 2009, Adolfo Garcia-Sastre 18 Sep 2009, David Wotton 02 Oct 2009, Michael Rape 29 Oct 2009, William Hahn 04 Feb 2010, Keith Wilkinson 17 Nov 2011
- 16.Kim HT, Kim KP, Lledias F, Kisselev AF, Scaglione KM, Skowyra D, Gygi SP, Goldberg AL. Certain pairs of ubiquitin-conjugating enzymes (E2s) and ubiquitin-protein ligases (E3s) synthesize nondegradable forked ubiquitin chains containing all possible isopeptide linkages. J Biol Chem. 2007;282:17375–86. doi: 10.1074/jbc.M609659200. [DOI] [PubMed] [Google Scholar]; F1000 Factor 7Evaluated by Titia Sixma 28 Jan 2008, Keith Wilkinson 17 Nov 2011
- 17.Pickart CM, Raasi S. Controlled synthesis of polyubiquitin chains. Methods Enzymol. 2005;399:21–36. doi: 10.1016/S0076-6879(05)99002-2. [DOI] [PubMed] [Google Scholar]
- 18.Reyes-Turcu FE, Shanks JR, Komander D, Wilkinson KD. Recognition of polyubiquitin isoforms by the multiple ubiquitin binding modules of isopeptidase T. J Biol Chem. 2008;283:19581–92. doi: 10.1074/jbc.M800947200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bremm A, Freund SM, Komander D. Lys11-linked ubiquitin chains adopt compact conformations and are preferentially hydrolyzed by the deubiquitinase Cezanne. Nat Struct Mol Biol. 2010;17:939–47. doi: 10.1038/nsmb.1873. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 6Evaluated by Keith Wilkinson 17 Nov 2011
- 20.Komander D, Lord CJ, Scheel H, Swift S, Hofmann K, Ashworth A, Barford D. The structure of the CYLD USP domain explains its specificity for Lys63-linked polyubiquitin and reveals a B box module. Mol Cell. 2008;29:451–64. doi: 10.1016/j.molcel.2007.12.018. [DOI] [PubMed] [Google Scholar]; F1000 Factor 6Evaluated by Keith Wilkinson 17 Nov 2011
- 21.Messick TE, Russell NS, Iwata AJ, Sarachan KL, Shiekhattar R, Shanks JR, Reyes-Turcu FE, Wilkinson KD, Marmorstein R. Structural basis for ubiquitin recognition by the Otu1 ovarian tumor domain protein. J Biol Chem. 2008;283:11038–49. doi: 10.1074/jbc.M704398200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang T, Yin L, Cooper EM, Lai MY, Dickey S, Pickart CM, Fushman D, Wilkinson KD, Cohen RE, Wolberger C. Evidence for bidentate substrate binding as the basis for the K48 linkage specificity of otubain 1. J Mol Biol. 2009;386:1011–23. doi: 10.1016/j.jmb.2008.12.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Newton K, Matsumoto ML, Wertz IE, Kirkpatrick DS, Lill JR, Tan J, Dugger D, Gordon N, Sidhu SS, Fellouse FA, Komuves L, French DM, Ferrando RE, Lam C, Compaan D, Yu C, Bosanac I, Hymowitz SG, Kelley RF, Dixit VM. Ubiquitin chain editing revealed by polyubiquitin linkage-specific antibodies. Cell. 2008;134:668–78. doi: 10.1016/j.cell.2008.07.039. [DOI] [PubMed] [Google Scholar]; F1000 Factor 10Evaluated by Ze'ev Ronai 08 Sep 2008, Paolo Rossi 22 Sep 2008, Keith Wilkinson 17 Nov 2011
- 24.Bremm A, Komander D. Emerging roles for Lys11-linked polyubiquitin in cellular regulation. Trends Biochem Sci. 2011;36:355–63. doi: 10.1016/j.tibs.2011.04.004. [DOI] [PubMed] [Google Scholar]
- 25.Dynek JN, Goncharov T, Dueber EC, Fedorova AV, Izrael-Tomasevic A, Phu L, Helgason E, Fairbrother WJ, Deshayes K, Kirkpatrick DS, Vucic D. c-IAP1 and UbcH5 promote K11-linked polyubiquitination of RIP1 in TNF signalling. Embo J. 2010;29:4198–209. doi: 10.1038/emboj.2010.300. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 6Evaluated by Keith Wilkinson 17 Nov 2011
- 26.Garnett MJ, Mansfeld J, Godwin C, Matsusaka T, Wu J, Russell P, Pines J, Venkitaraman AR. UBE2S elongates ubiquitin chains on APC/C substrates to promote mitotic exit. Nat Cell Biol. 2009;11:1363–9. doi: 10.1038/ncb1983. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 6Evaluated by Keith Wilkinson 17 Nov 2011
- 27.Williamson A, Wickliffe KE, Mellone BG, Song L, Karpen GH, Rape M. Identification of a physiological E2 module for the human anaphase-promoting complex. Proc Natl Acad Sci U S A. 2009;106:18213–8. doi: 10.1073/pnas.0907887106. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 6Evaluated by Keith Wilkinson 17 Nov 2011
- 28.Christensen D, Brzovic P, Klevit R. E2-BRCA1 RING interactions dictate synthesis of mono- or specific polyubiquitin chain linkages. Nat Struct Mol Biol. 2007;14:941–8. doi: 10.1038/nsmb1295. [DOI] [PubMed] [Google Scholar]; F1000 Factor 9Evaluated by Brian Kuhlman 02 Oct 2007, Jane Endicott 25 Oct 2007, Thomas Kodadek 02 Nov 2007, Keith Wilkinson 17 Nov 2011
- 29.Zhao S, Ulrich HD. Distinct consequences of posttranslational modification by linear versus K63-linked polyubiquitin chains. Proc Natl Acad Sci U S A. 2010;107:7704–9. doi: 10.1073/pnas.0908764107. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 6Evaluated by Keith Wilkinson 17 Nov 2011
- 30.Huang H, Jeon M, Liao L, Yang C, Elly C, Yates J, Liu Y. K33-linked polyubiquitination of T cell receptor-zeta regulates proteolysis-independent T cell signaling. Immunity. 2010;33:60–70. doi: 10.1016/j.immuni.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 10Evaluated by Amnon Altman 13 Oct 2010, Larry Kane 14 Jun 2011, Keith Wilkinson 17 Nov 2011
- 31.Pickart CM, Fushman D. Polyubiquitin chains: polymeric protein signals. Curr Opin Chem Biol. 2004;8:610–6. doi: 10.1016/j.cbpa.2004.09.009. [DOI] [PubMed] [Google Scholar]
- 32.Matsumoto ML, Wickliffe KE, Dong KC, Yu C, Bosanac I, Bustos D, Phu L, Kirkpatrick DS, Hymowitz SG, Rape M, Kelley RF, Dixit VM. K11-linked polyubiquitination in cell cycle control revealed by a K11 linkage-specific antibody. Mol Cell. 2010;39:477–84. doi: 10.1016/j.molcel.2010.07.001. [DOI] [PubMed] [Google Scholar]; F1000 Factor 6Evaluated by Keith Wilkinson 17 Nov 2011
- 33.Sims JJ, Haririnia A, Dickinson BC, Fushman D, Cohen RE. Avid interactions underlie the Lys63-linked polyubiquitin binding specificities observed for UBA domains. Nat Struct Mol Biol. 2009;16:883–9. doi: 10.1038/nsmb.1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang Q, Young P, Walters KJ. Structure of S5a bound to monoubiquitin provides a model for polyubiquitin recognition. J Mol Biol. 2005;348:727–39. doi: 10.1016/j.jmb.2005.03.007. [DOI] [PubMed] [Google Scholar]; F1000 Factor 6Evaluated by Keith Wilkinson 17 Nov 2011
- 35.Sato Y, Yoshikawa A, Mimura H, Yamashita M, Yamagata A, Fukai S. Structural basis for specific recognition of Lys 63-linked polyubiquitin chains by tandem UIMs of RAP80. Embo J. 2009;28:2461–8. doi: 10.1038/emboj.2009.160. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 7Evaluated by Christiane Riedinger and Jane Endicott 12 Aug 2009, Keith Wilkinson 17 Nov 2011
- 36.Raasi S, Orlov I, Fleming KG, Pickart CM. Binding of polyubiquitin chains to ubiquitin-associated (UBA) domains of HHR23A. J Mol Biol. 2004;341:1367–79. doi: 10.1016/j.jmb.2004.06.057. [DOI] [PubMed] [Google Scholar]; F1000 Factor 9Evaluated by Jane Endicott 20 Sep 2004, Keith Wilkinson 17 Nov 2011
- 37.Sims JJ, Cohen RE. Linkage-specific avidity defines the lysine 63-linked polyubiquitin-binding preference of rap80. Mol Cell. 2009;33:775–83. doi: 10.1016/j.molcel.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 7Evaluated by Zhen-Qiang Pan 23 Apr 2009, Keith Wilkinson 17 Nov 2011
- 38.Varadan R, Assfalg M, Haririnia A, Raasi S, Pickart C, Fushman D. Solution conformation of Lys63-linked di-ubiquitin chain provides clues to functional diversity of polyubiquitin signaling. J Biol Chem. 2004;279:7055–63. doi: 10.1074/jbc.M309184200. [DOI] [PubMed] [Google Scholar]; F1000 Facftor 6Evaluated by Jane Endicott 19 Mar 2004
- 39.Varadan R, Assfalg M, Raasi S, Pickart C, Fushman D. Structural determinants for selective recognition of a Lys48-linked polyubiquitin chain by a UBA domain. Mol Cell. 2005;18:687–98. doi: 10.1016/j.molcel.2005.05.013. [DOI] [PubMed] [Google Scholar]; F1000 Factor 9Evaluated by Jane Endicott 27 Jun 2005
- 40.Beal R, Deveraux Q, Xia G, Rechsteiner M, Pickart C. Surface hydrophobic residues of multiubiquitin chains essential for proteolytic targeting. PNAS. 1996;93:861–6. doi: 10.1073/pnas.93.2.861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Beal RE, Toscano-Cantaffa D, Young P, Rechsteiner M, Pickart CM. The Hydrophobic Effect Contributes to Polyubiquitin Chain Recognition. Biochemistry. 1998;37:2925–34. doi: 10.1021/bi972514p. [DOI] [PubMed] [Google Scholar]
- 42.Varadan R, Walker O, Pickart C, Fushman D. Structural properties of polyubiquitin chains in solution. J Mol Biol. 2002;324:637–47. doi: 10.1016/S0022-2836(02)01198-1. [DOI] [PubMed] [Google Scholar]
- 43.Ryabov Y, Fushman D. Interdomain Mobility in Di-Ubiquitin Revealed by NMR. Proteins. 2006;63:787–96. doi: 10.1002/prot.20917. [DOI] [PubMed] [Google Scholar]
- 44.Ryabov YE, Fushman D. A Model of Interdomain Mobility in a Multidomain Protein. J Am Chem Soc. 2007;129:3315–27. doi: 10.1021/ja067667r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Datta AB, Hura GL, Wolberger C. The structure and conformation of Lys63-linked tetraubiquitin. J Mol Biol. 2009;392:1117–24. doi: 10.1016/j.jmb.2009.07.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Virdee S, Ye Y, Nguyen DP, Komander D, Chin JW. Engineered diubiquitin synthesis reveals Lys29-isopeptide specificity of an OTU deubiquitinase. Nat Chem Biol. 2010;6:750–7. doi: 10.1038/nchembio.426. [DOI] [PubMed] [Google Scholar]
- 47.Fushman D, Walker O. Exploring the linkage dependence of polyubiquitin conformations using molecular modeling. J Mol Biol. 2010;395:803–14. doi: 10.1016/j.jmb.2009.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lopitz-Otsoa F, Rodriguez MS, Aillet F. Properties of natural and artificial proteins displaying multiple ubiquitin-binding domains. Biochem Soc Trans. 2010;38:40–5. doi: 10.1042/BST0380040. [DOI] [PubMed] [Google Scholar]; F1000 Factor 6Evaluated by Keith Wilkinson 17 Nov 2011
- 49.Hofmann K. Ubiquitin-binding domains and their role in the DNA damage response. DNA repair. 2009;8:544–56. doi: 10.1016/j.dnarep.2009.01.003. [DOI] [PubMed] [Google Scholar]; F1000 Factor 6Evaluated by Keith Wilkinson 17 Nov 2011
- 50.Dikic I, Wakatsuki S, Walters KJ. Ubiquitin-binding domains - from structures to functions. Nat Rev Mol Cell Biol. 2009;10:659–71. doi: 10.1038/nrm2767. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 6Evaluated by Keith Wilkinson 17 Nov 2011
- 51.Hurley JH, Lee S, Prag G. Ubiquitin-binding domains. Biochem J. 2006;399:361–72. doi: 10.1042/BJ20061138. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 6Evaluated by Keith Wilkinson 17 Nov 2011
- 52.Hicke L, Schubert HL, Hill CP. Ubiquitin-binding domains. Nat Rev Mol Cell Biol. 2005;6:610–21. doi: 10.1038/nrm1701. [DOI] [PubMed] [Google Scholar]; F1000 Factor 6Evaluated by Keith Wilkinson 17 Nov 2011
- 53.Wu H, Lo YC, Lin SC. Recent advances in polyubiquitin chain recognition. F1000 Biol Rep. 2010;2:1–5. doi: 10.3410/B2-20. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 6Evaluated by Keith Wilkinson 17 Nov 2011
- 54.Markin CJ, Xiao W, Spyracopoulos L. Mechanism for recognition of polyubiquitin chains: balancing affinity through interplay between multivalent binding and dynamics. J Am Chem Soc. 2010;132:11247–58. doi: 10.1021/ja103869x. [DOI] [PubMed] [Google Scholar]; F1000 Factor 6Evaluated by Keith Wilkinson 17 Nov 2011
- 55.Riedinger C, Boehringer J, Trempe JF, Lowe ED, Brown NR, Gehring K, Noble ME, Gordon C, Endicott JA. Structure of Rpn10 and its interactions with polyubiquitin chains and the proteasome subunit Rpn12. J Biol Chem. 2010;285:33992–4003. doi: 10.1074/jbc.M110.134510. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 6Evaluated by Keith Wilkinson 17 Nov 2011
- 56.Zhang D, Chen T, Ziv I, Rosenzweig R, Matiuhin Y, Bronner V, Glickman MH, Fushman D. Together, Rpn10 and Dsk2 can serve as a polyubiquitin chain-length sensor. Mol Cell. 2009;36:1018–33. doi: 10.1016/j.molcel.2009.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wade SL, Auble DT. The Rad23 ubiquitin receptor, the proteasome and functional specificity in transcriptional control. Transcr. 2010;1:22–6. doi: 10.4161/trns.1.1.12201. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 6Evaluated by Keith Wilkinson 17 Nov 2011
- 58.Chen X, Lee BH, Finley D, Walters KJ. Structure of proteasome ubiquitin receptor hRpn13 and its activation by the scaffolding protein hRpn2. Mol Cell. 2010;38:404–15. doi: 10.1016/j.molcel.2010.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 6Evaluated by Keith Wilkinson 17 Nov 2011
- 59.Zhang N, Wang Q, Ehlinger A, Randles L, Lary JW, Kang Y, Haririnia A, Storaska AJ, Cole JL, Fushman D, Walters KJ. Structure of the s5a:k48-linked diubiquitin complex and its interactions with rpn13. Mol Cell. 2009;35:280–90. doi: 10.1016/j.molcel.2009.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hanzelmann P, Buchberger A, Schindelin H. Hierarchical binding of cofactors to the AAA ATPase p97. Structure. 2011;19:833–43. doi: 10.1016/j.str.2011.03.018. [DOI] [PubMed] [Google Scholar]; F1000 Factor 6Evaluated by Keith Wilkinson 17 Nov 2011
- 61.Schuberth C, Buchberger A. UBX domain proteins: major regulators of the AAA ATPase Cdc48/p97. Cell Mol Life Sci. 2008;65:2360–71. doi: 10.1007/s00018-008-8072-8. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 6Evaluated by Keith Wilkinson 17 Nov 2011
- 62.Verma R, Oania R, Fang R, Smith GT, Deshaies RJ. Cdc48/p97 mediates UV-dependent turnover of RNA Pol II. Mol Cell. 2011;41:82–92. doi: 10.1016/j.molcel.2010.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 7Evaluated by Alexandra Segref and Thorsten Hoppe 28 Feb 2011, Keith Wilkinson 17 Nov 2011
- 63.Bedford L, Layfield R, Mayer RJ, Peng J, Xu P. Diverse polyubiquitin chains accumulate following 26S proteasomal dysfunction in mammalian neurones. Neurosci Lett. 2011;491:44–7. doi: 10.1016/j.neulet.2010.12.064. [DOI] [PubMed] [Google Scholar]; F1000 Factor 6Evaluated by Keith Wilkinson 17 Nov 2011
- 64.Zeng W, Sun L, Jiang X, Chen X, Hou F, Adhikari A, Xu M, Chen Z. Reconstitution of the RIG-I pathway reveals a signaling role of unanchored polyubiquitin chains in innate immunity. Cell. 2010;141:315–30. doi: 10.1016/j.cell.2010.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 16Evaluated by Caetano Reis e Sousa 23 Apr 2010, Robert Krug 26 Apr 2010, Danielle Brabant and John Brumell 04 Jun 2010, Carolyn Coyne 18 Jun 2010, Keith Wilkinson 17 Nov 2011
- 65.Chen J, Bozza W, Zhuang Z. Ubiquitination of PCNA and its essential role in eukaryotic translesion synthesis. Cell. Biochem Biophys. 2011;60:47–60. doi: 10.1007/s12013-011-9187-3. [DOI] [PubMed] [Google Scholar]; F1000 Factor 6Evaluated by Keith Wilkinson 17 Nov 2011
- 66.Winget JM, Mayor T. The diversity of ubiquitin recognition: hot spots and varied specificity. Mol Cell. 2010;38:627–35. doi: 10.1016/j.molcel.2010.05.003. [DOI] [PubMed] [Google Scholar]
- 67.Phillips CL, Thrower J, Pickart CM, Hill CP. Structure of a new crystal form of tetraubiquitin. Acta Cryst D. 2000;57:341–4. doi: 10.1107/S090744490001800X. [DOI] [PubMed] [Google Scholar]
- 68.Thrower JS, Hoffman L, Rechtenstein M, Pickart CM. Recognition of the polyubiquitin proteolytic signal. EMBO J. 2000;19:94–102. doi: 10.1093/emboj/19.1.94. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 6Evaluated by Keith Wilkinson 17 Nov 2011
- 69.Hurley JH. The ESCRT complexes. Crit Rev Biochem Mol Biol. 2010;45:463–87. doi: 10.3109/10409238.2010.502516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Maspero E, Mari S, Valentini E, Musacchio A, Fish A, Pasqualato S, Polo S. Structure of the HECT:ubiquitin complex and its role in ubiquitin chain elongation. EMBO Rep. 2011;12:342–9. doi: 10.1038/embor.2011.21. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 6Evaluated by Keith Wilkinson 17 Nov 2011
- 71.Kleiger G, Saha A, Lewis S, Kuhlman B, Deshaies RJ. Rapid E2-E3 assembly and disassembly enable processive ubiquitylation of cullin-RING ubiquitin ligase substrates. Cell. 2009;139:957–68. doi: 10.1016/j.cell.2009.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 11Evaluated by Chih-Cheng Yang and Dieter Wolf 14 Dec 2009, Keith Wilkinson 17 Nov 2011
- 72.Ye Y, Rape M. Building ubiquitin chains: E2 enzymes at work. Nat Rev Mol Cell Biol. 2009;10:755–64. doi: 10.1038/nrm2780. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 6Evaluated by Keith Wilkinson 17 Nov 2011
- 73.Behrends C, Harper JW. Constructing and decoding unconventional ubiquitin chains. Nat Struct Mol Biol. 2011;18:520–8. doi: 10.1038/nsmb.2066. [DOI] [PubMed] [Google Scholar]
- 74.Kirisako T, Kamei K, Murata S, Kato M, Fukumoto H, Kanie M, Sano S, Tokunaga F, Tanaka K, Iwai K. A ubiquitin ligase complex assembles linear polyubiquitin chains. Embo J. 2006;25:4877–87. doi: 10.1038/sj.emboj.7601360. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 6Evaluated by Jane Endicott 23 Nov 2006
- 75.Verhelst K, Verstrepen L, Carpentier I, Beyaert R. Linear ubiquitination in NF-kappaB signaling and inflammation: What we do understand and what we do not. Biochem Pharmacol. 2011;82:1057–65. doi: 10.1016/j.bcp.2011.07.066. [DOI] [PubMed] [Google Scholar]; F1000 Factor 6Evaluated by Keith Wilkinson 17 Nov 2011
- 76.Tokunaga F, Nakagawa T, Nakahara M, Saeki Y, Taniguchi M, Sakata S, Tanaka K, Nakano H, Iwai K. SHARPIN is a component of the NF-kappaB-activating linear ubiquitin chain assembly complex. Nature. 2011;471:633–6. doi: 10.1038/nature09815. [DOI] [PubMed] [Google Scholar]; F1000 Factor 6Evaluated by Keith Wilkinson 17 Nov 2011
- 77.Ikeda F, Deribe YL, Skanland SS, Stieglitz B, Grabbe C, Franz-Wachtel M, van Wijk SJ, Goswami P, Nagy V, Terzic J, Tokunaga F, Androulidaki A, Nakagawa T, Pasparakis M, Iwai K, Sundberg JP, Schaefer L, Rittinger K, Macek B, Dikic I. SHARPIN forms a linear ubiquitin ligase complex regulating NF-kappaB activity and apoptosis. Nature. 2011;471:637–41. doi: 10.1038/nature09814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ventii KH, Wilkinson KD. Protein partners of deubiquitinating enzymes. Biochem J. 2008;414:161–175. doi: 10.1042/BJ20080798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sowa ME, Bennett EJ, Gygi SP, Harper JW. Defining the human deubiquitinating enzyme interaction landscape. Cell. 2009;138:389–403. doi: 10.1016/j.cell.2009.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Castaneda CA, Liu J, Chaturvedi A, Nowicka U, Cropp TA, Fushman D. Non-enzymatic assembly of natural polyubiquitin chains of any linkage composition and isotopic labeling scheme. J Am Chem Soc. 2011;133:17855–68. doi: 10.1021/ja207220g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Virdee S, Kapadnis PB, Elliott T, Lang K, Madrzak J, Nguyen DP, Riechmann L, Chin JW. Traceless and site-specific ubiquitination of recombinant proteins. J Am Chem Soc. 2011;133:10708–11. doi: 10.1021/ja202799r. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 6Evaluated by Keith Wilkinson 17 Nov 2011
- 82.Kumar KS, Bavikar SN, Spasser L, Moyal T, Ohayon S, Brik A. Total chemical synthesis of a 304 amino acid K48-linked tetraubiquitin protein. Angew Chem Int Ed Engl. 2011;50:6137–41. doi: 10.1002/anie.201101920. [DOI] [PubMed] [Google Scholar]; F1000 Factor 6Evaluated by Keith Wilkinson 17 Nov 2011
- 83.El Oualid F, Merkx R, Ekkebus R, Hameed DS, Smit JJ, de Jong A, Hilkmann H, Sixma TK, Ovaa H. Chemical synthesis of ubiquitin, ubiquitin-based probes, and diubiquitin. Angew Chem Int Ed Engl. 2010;49:10149–53. doi: 10.1002/anie.201005995. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 6Evaluated by Keith Wilkinson 17 Nov 2011
- 84.Yang R, Pasunooti KK, Li F, Liu XW, Liu CF. Synthesis of K48-linked diubiquitin using dual native chemical ligation at lysine. Chem Commun (Camb) 2010;46:7199–201. doi: 10.1039/c0cc01382j. [DOI] [PubMed] [Google Scholar]; F1000 Factor 6Evaluated by Keith Wilkinson 17 Nov 2011
- 85.Castaneda CA, Spasser L, Bavikar SN, Brik A, Fushman D. Segmental Isotopic Labeling of Ubiquitin Chains To Unravel Monomer-Specific Molecular Behavior. Angew Chem Int Ed Engl. 2011;50:11210–4. doi: 10.1002/anie.201104649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kumar KS, Spasser L, Erlich LA, Bavikar SN, Brik A. Total chemical synthesis of di-ubiquitin chains. Angew Chem Int Ed Engl. 2010;49:9126–31. doi: 10.1002/anie.201003763. [DOI] [PubMed] [Google Scholar]; F1000 Factor 6Evaluated by Keith Wilkinson 17 Nov 2011
- 87.Castaneda CA, Liu J, Kashyap TR, Singh RK, Fushman D, Cropp TA. Controlled enzymatic synthesis of natural-linkage, defined-length polyubiquitin chains using lysines with removable protecting groups. Chem Commun (Camb) 2011;47:2026–8. doi: 10.1039/c0cc04868b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Cook WJ, Jeffrey LC, Carson M, Chen Z, Pickart CM. Structure of a diubiquitin conjugate and a model for interaction with ubiquitin conjugating enzyme (E2) J Biol Chem. 1992;267:16467–71. doi: 10.2210/pdb1aar/pdb. [DOI] [PubMed] [Google Scholar]
- 89.Eddins MJ, Varadan R, Fushman D, Pickart CM, Wolberger C. Crystal Structure and Solution NMR Studies of Lys48-linked Tetraubiquitin at Neutral pH. J Mol Biol. 2007;367:204–11. doi: 10.1016/j.jmb.2006.12.065. [DOI] [PubMed] [Google Scholar]