Abstract
A method based on the Carr-Purcell-Meiboom-Gill relaxation dispersion experiment is presented for measuring the temperature coefficients of amide proton chemical shifts of low populated ‘invisible’ protein states that exchange with a ‘visible’ ground state on the millisecond time-scale. The utility of the approach is demonstrated with an application to an I58D mutant of the Pfl6 Cro protein that undergoes exchange between the native, folded state and a cold denatured, unfolded conformational ensemble that is populated at a level of 6% at 2.5°C. A wide distribution of amide temperature coefficients is measured for the unfolded state. The distribution is centered about –5.6 ppb/K, consistent with an absence of intra-molecular hydrogen bonds, on average. However, the large range of values (-standard deviation of 2.1 ppb/K) strongly supports the notion that the unfolded state of the protein is not a true random coil polypeptide chain.
Keywords: CPMG Relaxation Dispersion, Temperature Coefficients, Amide Protons, Cold Denaturation
Proteins are not rigid molecules. Often they undergo significant conformational fluctuations leading to transiently populated sub-states that may be critically important for function (Boehr et al., 2006; Henzler-Wildman et al., 2007; Ishima et al., 1999; Karplus & Kuriyan, 2005). These dynamics, involving the inter-conversion of a ground (G), highly populated state and at least one low-populated, excited state (E) are invisible to most standard structural biology techniques (Palmer et al., 2001). Yet so long as the exchange processes involve states with lifetimes on the order of 0.5–5 ms, a highly populated ground state and excited state fractional populations in excess of approximately 0.5% they can be studied in some detail (Korzhnev & Kay, 2008; Palmer et al., 2001) using Carr-Purcell-Meiboom-Gill (CPMG) relaxation dispersion NMR spectroscopy (Carr & Purcell, 1954; Meiboom & Gill, 1958). These experiments quantify transverse relaxation rates of NMR probes attached to the visible ground state conformer as a function of the spacing between successive refocusing pulses applied as a pulse train. Using suitably labeled samples and recently developed pulse schemes it is possible to measure 15N, 13Cα, 13Cβ, 13CO, 1HN, 1Hα chemical shifts (Hansen et al., 2008; Ishima et al., 2004; Loria et al., 1999; Lundstrom et al., 2008; Lundstrom et al., 2009; Lundstrom & Kay, 2009; Tollinger et al., 2001), 1H and 13C methyl shifts (Baldwin et al., 2010; Lundstrom et al., 2007) as well as anisotropic interactions such as residual dipolar couplings (Vallurupalli et al., 2007) and chemical shift anisotropies (Vallurupalli et al., 2008a) of excited state conformers that together form the basis for structure determination of these excited states (Korzhnev et al., 2010; Vallurupalli et al., 2008b).
One parameter that has not being exploited to date in studies of excited states is the amide proton temperature coefficient, Δδ̃HN/ΔT, that is sensitive to hydrogen bonding in proteins (Baxter & Williamson, 1997; Dyson et al., 1988). Studies of folded proteins for which high resolution crystal structures are available establish that in over 90% of cases for which Δδ̃HN/ΔT > −4.6 ppb/K the amide in question participates in an intra-molecular hydrogen bond (Cierpicki & Otlewski, 2001; Cierpicki et al., 2002). Conversely, for the majority of amide protons for which Δδ̃HN/ΔT < −4.6 ppb/K the amide proton was found to be hydrogen bonded to solvent water. Although the temperature coefficient is a qualitative parameter at best, it nevertheless provides some insight into structure (hydrogen bond) formation even at an early stage in data analysis when only 15N and 1HN chemical shifts are generally available. Herein we describe a straightforward approach for obtaining accurate Δδ̃HN/ΔT values of excited protein states using relaxation dispersion NMR spectroscopy and provide an illustrative example involving a protein folding system in which a folded (ground) state exchanges with an unfolded, invisible (excited) conformer.
The Pfl6 Cro I58D protein studied here (Pfl6 I58D) is a member of the Cro family of bacteriophage transcription factors with a mixed α+β fold similar to the λ Cro repressor, with the mutation at position 58 ensuring a monomer structure in solution (LeFevre & Cordes, 2003; Roessler et al., 2008). Initial 15N and 1HN CPMG relaxation dispersion experiments established that the protein exchanges between two states, , with values for kex = kEG+kGE and the fractional population of the excited state, pE, of (875±9)s−1 and (6.3±0.1)%, respectively (2.5°C). Interestingly, as the temperature is increased pE decreases - to (3.9±0.1)% at 15°C. The chemical shifts of the excited state that are extracted from fits of the dispersion experiment compare vary favorably with shifts obtained from small ‘random coil peptides’ published by Wishart and coworkers (Wishart et al., 1995), indicating that this state is unfolded, Figure 1. The exchange reaction studied is thus the result of the cold denaturation of Pfl6 I58D with ‘G’ the native conformation of the protein and ‘E’ an ensemble of disordered states that interconvert very rapidly on the NMR chemical shift time-scale. In order to assess whether the cold denatured state of Pfl6 I58D is completely ‘unfolded-like’, similar to small, unstructured peptides in solution or whether there are ‘pockets’ of structure one could, of course, measure additional dispersion profiles using 13Cα or 1Hα probes. Alternatively, we are interested here in evaluating whether insight can be obtained from Δδ̃HN/ΔT values that are available from the temperature dependent relaxation dispersion series that has already been performed.
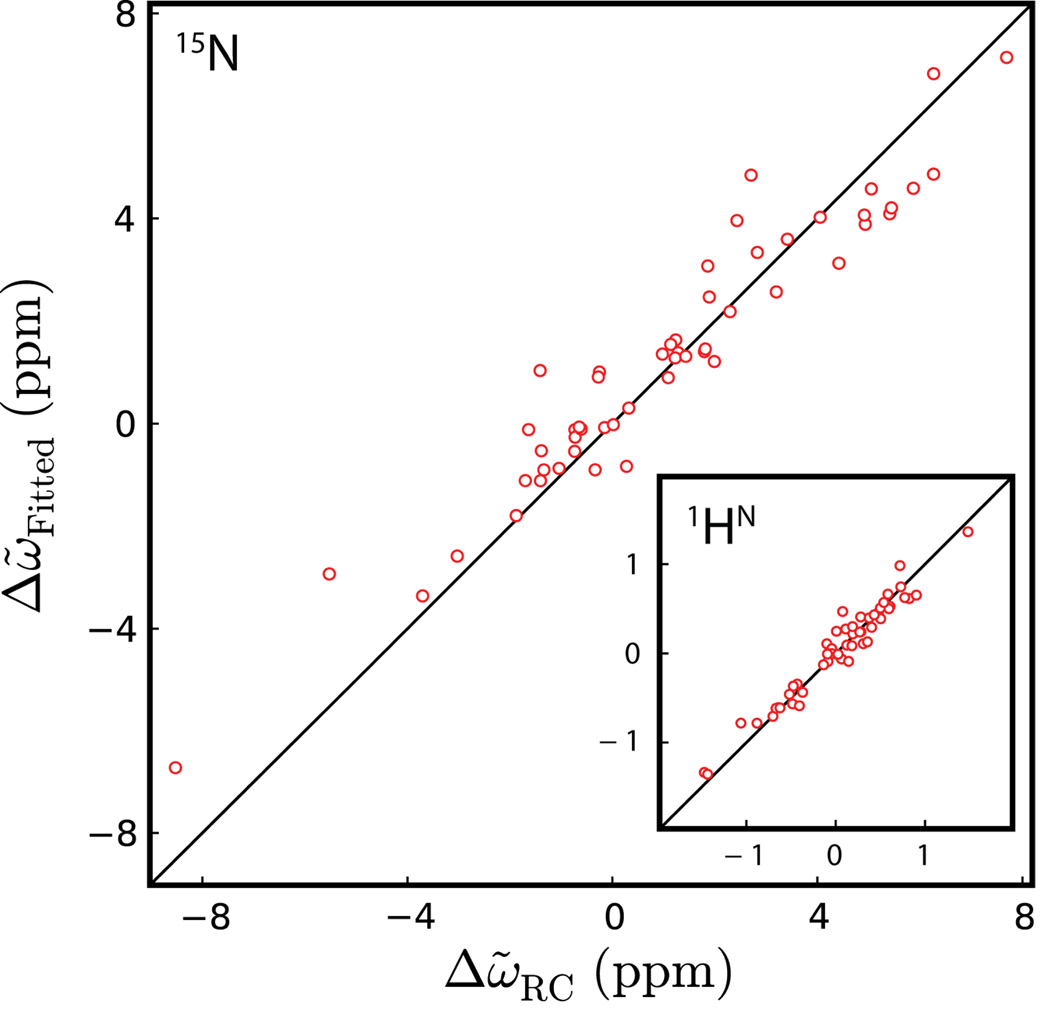
Figure 1.
Correlation plot of 15N (1HN, inset) chemical shift differences between ground and excited states of Pfl6 I58D (Δω̃Fitted) measured by relaxation dispersion experiments (5°C) and predicted shift differences assuming that the excited state is unfolded (Δω̃RC) calculated as described by Wishart and coworkers (Wishart et al., 1995). The RMSDs of the 15N and 1HN correlations are 0.95 and 0.13 ppm, respectively.
Figure 2 shows 1HN dispersion profiles recorded at a static magnetic field of 18.8 T, R2,eff(υCPMG), as a function of temperature for Asn 26 and Leu 44 of Pfl6 I58D. Profiles for all of the residues obtained at 11.7 and 18.8 T were fitted together, with separate kex and pE for each temperature and with the difference in chemical shifts between an amide proton in states E and G, , assumed to vary linearly with temperature. The fits so obtained for Asn 26 and Leu 44 are shown with solid lines in the Figure. Only |Δω̃HN| values are obtained from analysis of relaxation dispersion data and the sign of the shift difference is critical for calculation of excited state temperature coefficients (see below). Signs of Δω̃HN were obtained from (i) a comparison of the amide proton resonance positions measured from (ground state) peaks in 15N-1HN HSQC spectra recorded at 11.7 and 18.8 T (Bouvignies et al., 2010) and (ii) from a comparison of 1HN-15N zero- and double quantum CPMG relaxation dispersion profiles in cases where the signs of 15N Δω̃ values were available (Orekhov et al., 2004).
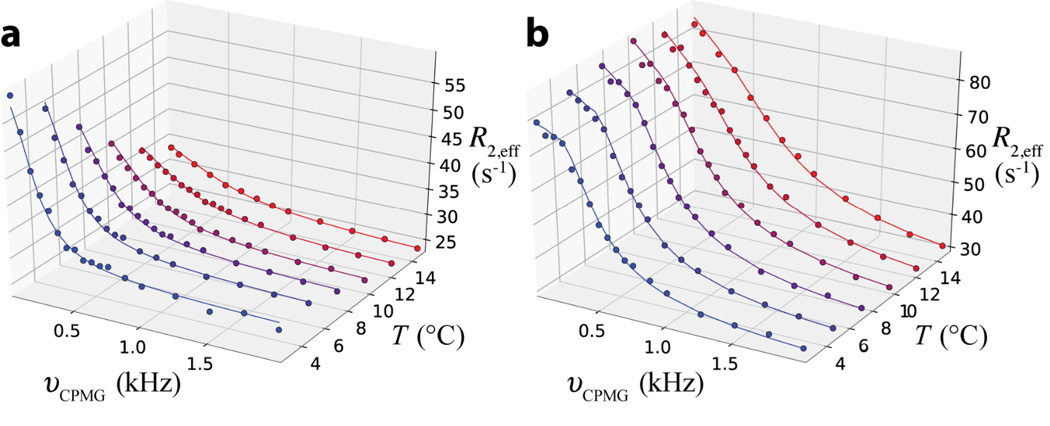
Figure 2.
Experimental 1HN dispersion profiles (circles) for Asn 26 (a) and Leu 44 (b) of Pfl6 I58D measured at temperatures (T) ranging from 2.5°C (blue) to 15.0°C (red), along with the best fit dispersion profiles (solid line). Dispersion profiles were fit to models assuming (i) ΔR2 = 0 or (ii) . The slope of the linear correlation of the resulting temperature coefficients of the excited state obtained using both approaches is 0.98 with a pair-wise rmsd of 0.2 ppb/K, indicating that for this application the results are not sensitive to the values of ΔR2.
Figure 3 shows the temperature dependence of the ground state amide chemical shifts for Asn 26 and Leu 44 of Pfl6 I58D (green circles) obtained from 15N-1HN correlation spectra (insets). For a system that is not in exchange the temperature coefficient (of the ground state amide proton) would be calculated simply as the slope of the shift vs. temperature profile. However, exchange leads to a small perturbation in the position of the peaks derived from exchanging spins, δ̃ex. In the case where pG≫pE it has been shown that , where ξHN = Δω̃HN/kEG, ρ = ΔR2/kEG and is the difference in intrinsic transverse relaxation rates between nuclei in the two states (Anet & Basus, 1978; Skrynnikov et al., 2002). Values of kEG vary from approximately 900 s−1 (2.5°C) to 3000 s−1 (15°C), with s−1 (11.7 T) so that ρ can be neglected with little error. Using the relation for δ̃ex along with the exchange parameters that are isolated from fits of dispersion profiles (Figure 2) the position of the ground state in the absence of exchange is calculated from the measured chemical shift . These values are plotted in Figure 3 (blue circles) along with the best-fit lines from which the amide temperature coefficients in the ground state are calculated, . Once values for are obtained the corresponding chemical shift values in the excited state, are calculated from the relation and the excited state temperature coefficients subsequently generated from the slope of vs. temperature, Figure 4.
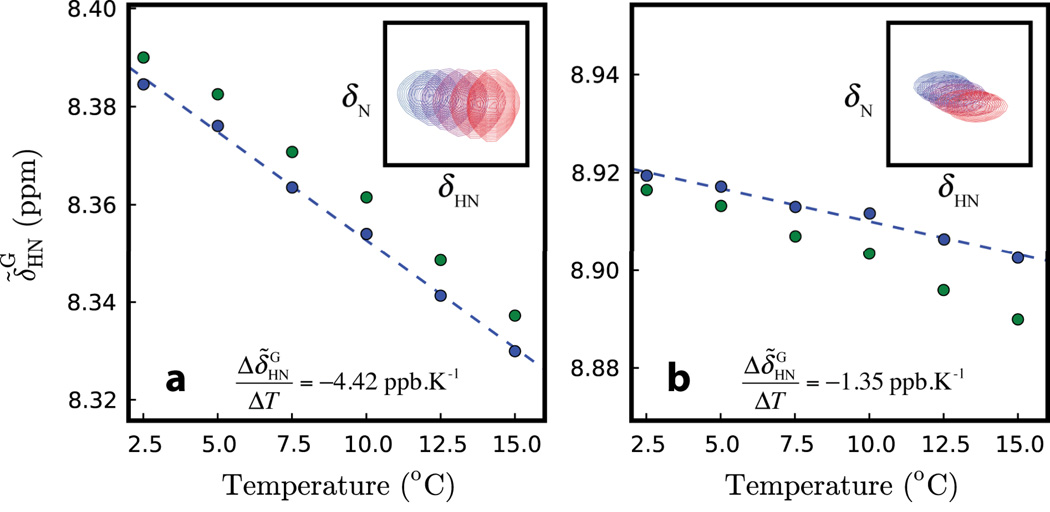
Figure 3.
Ground state 1HN chemical shifts vs. temperature for Asn 26 (a) and Leu 44 (b) of Pfl6 I58D. Shifts measured directly from spectra are plotted in green; they are then corrected for exchange that ‘moves’ the ground and excited state correlations towards each other and subsequently replotted in blue (see text). The dashed lines are the best linear fits to the ‘corrected’ peak positions from which the ground state 1HN temperature coefficients are extracted. Insets show the corresponding 1HN-15N correlation peaks from spectra recorded at temperatures ranging from 2.5°C (blue) to 15.0°C (red).
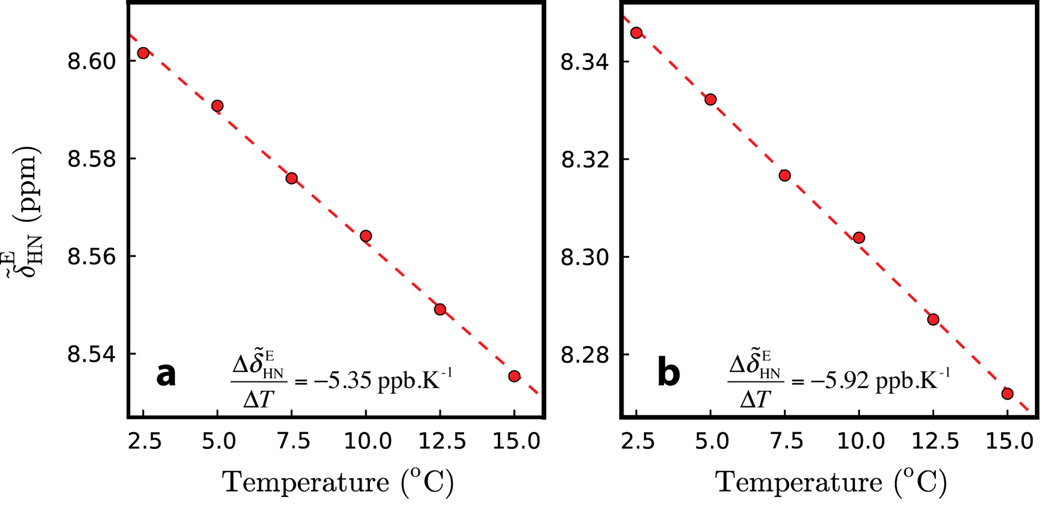
Figure 4.
Excited state 1HN chemical shifts vs. temperature for Asn 26 (a) and Leu 44 (b) of Pfl6 I58D. Note that Asn 26 and Leu 44 have different temperature coefficients in the ground state yet similar values in the excited state.
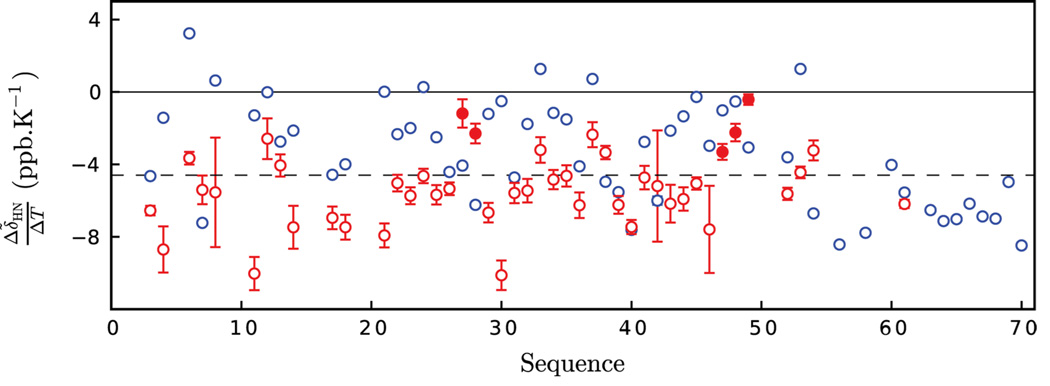
Figure 5 plots both the ground, folded (blue) and the excited, unfolded (red) state temperature coefficients calculated as described above. It is noteworthy that the dispersion methodology allows for measurements to be made on both states under identical conditions so that Δδ̃HN/ΔT values can be properly compared. Not surprisingly a range of values are noted for the ground state with the majority of values greater than −4.6 ppb/K, the cutoff for intra-molecular hydrogen bonding (Cierpicki & Otlewski, 2001; Cierpicki et al., 2002). For residues at the C-terminus, extending from Arg 56 and beyond the temperature coefficients are centered about −7.5 ppb/K, much lower than for the remaining protein. This provides strong evidence that these residues are unstructured, as expected on the basis of 15N and 1HN chemical shifts. An NMR study of a closely related protein, an A33W/F58D/Y26Q triple mutant of the λ Cro repressor that is monomeric in solution, shows that the C-terminal 10 residues of the native state are highly disordered, with no medium or long range NOEs observed in this region and with a very high level of backbone dynamics based on 15N relaxation studies (Newlove et al., 2006). In contrast to residues in the structured region of the folded state (residues 3–55), for which the majority of Δδ̃HN/ΔT values exceed −4.6 ppb/K, temperature coefficients for the excited state are in general smaller, consistent with the absence of intra-molecular hydrogen bonding. This is in keeping with expectations for a denatured ensemble of conformers. Interestingly, however there are a (small) number of residues with larger temperature coefficients, including Gln 27, Ser 28 and Asp 47, Gly 48 and Arg 49 that potentially indicate some level of structure even in the denatured state. These ‘anomalous’ Δδ̃HN/ΔT values are not artifactual; high quality fits of the dispersion profiles and the resultant vs. temperature curves are obtained in these cases (Figure S1 of Supporting Information). Notably, however, the 15N and 1HN excited state chemical shifts for these residues fall within the range expected for denatured proteins, suggesting that, at least in some cases, temperature coefficients may provide a more sensitive measure of residual structure. In native Pfl6, residues 27 and 28 form the center of a motif (residues 24–29; VNQSAI) that caps helix 3 at its N-terminus. The sequence and structure of this region partially resemble a classic helix capping box (Seale et al., 1994), a stable initiation motif that could retain partial order even in the absence of the global tertiary fold. Similarly, residues 47–49 form a β-turn in the folded state (Newlove et al., 2006). It may well be that such a turn is present – at least partially – in the unfolded state as well, serving to initiate the native state β-sheet that is formed in this region. A more quantitative description must await further relaxation dispersion studies focusing on measurement of 1Hα as well as 13C backbone and 13Cβ chemical shifts that are powerful indicators of secondary structure (Wishart & Case, 2001).
Figure 5.
Ground (blue) and excited (red) state temperature coefficients Δδ̃HN/ΔT for Pfl6 I58D as a function of primary sequence. The dashed line is plotted at −4.6 ppb/K, the cut-off used to establish whether amide protons participate in intra-molecular hydrogen-bonding (Cierpicki & Otlewski, 2001; Cierpicki et al., 2002). Excited state temperature coefficients for residues 27,28 and 47–49 highlighted in the text are indicated by filled circles. Temperature coefficients are reported only in cases where signs of Δω̃ values could be determined.
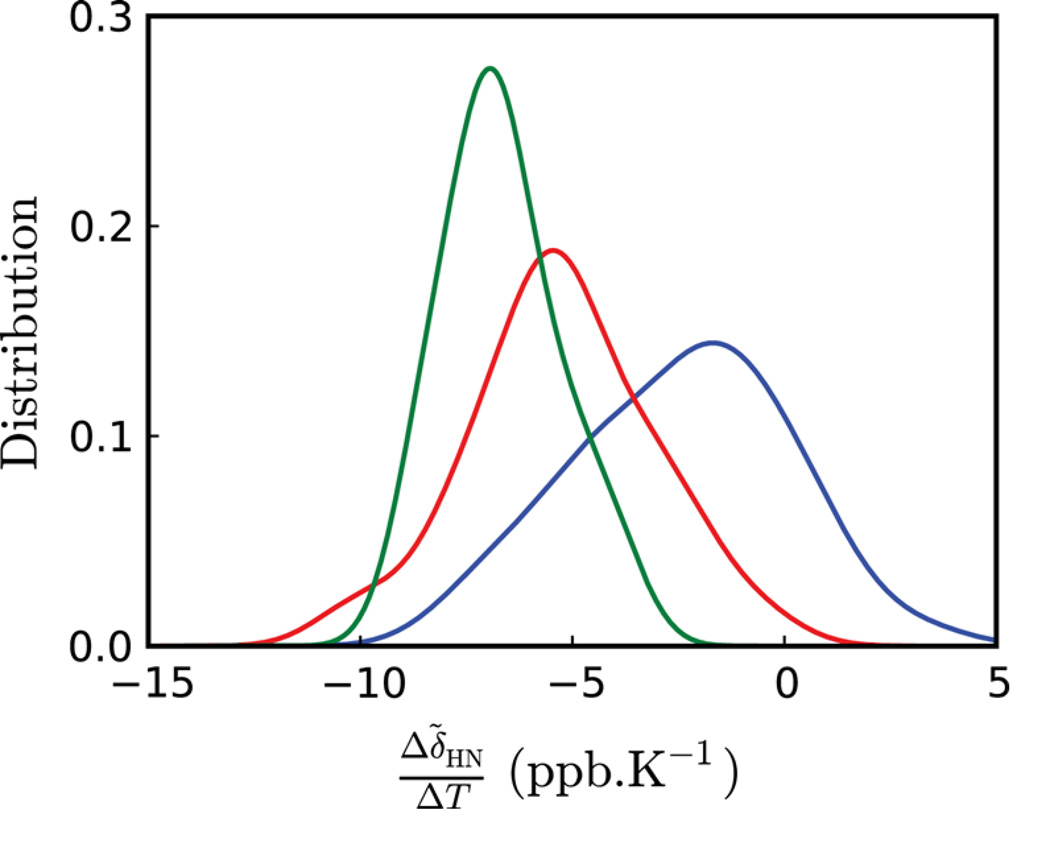
Further insight into the nature of the cold denatured ensemble of Pfl6 I58D can be obtained by comparing the distribution of amide proton temperature coefficients for this state with those obtained for both the folded conformer (minus the C-terminus) and the C-terminal region of the folded state that is highly disordered. Figure 6 plots the three distributions that have been normalized so that each has the same area. Notably Δδ̃HN/ΔT values for the cold denatured state are centered between those of the well-folded native state conformer and a random coil peptide corresponding to the C-terminal end of the folded protein. The wide distribution of temperature coefficients for the excited state suggests that the structure of the cold denatured state does not resemble a highly dynamic, random coil chain. Studies by Merutka et al. (1995) using a disordered small linear peptide model system –GGXGG– showed that values of Δδ̃HN/ΔT ranged from −6.4 (Asp) to −9.3 ppb/K (Tyr) with a median(±std) of −7.65±0.68 ppb/K over all 20 amino acids, X. The cold denatured state of Pfl6 I58D shows a much larger range than for these peptides (Figure 6, red curve). This is consistent with other studies of unfolded protein states, such as the unfolded ensemble of the N-terminal SH3 domain from the protein drk (Crowhurst & Forman-Kay, 2003; Zhang & Forman-Kay, 1997), which clearly shows regions with structure, including non-native interactions, despite the fact that the protein is ‘unfolded’.
Figure 6.
Normalized distributions of 1HN temperature coefficient values for the ground (blue, minus the C-terminus) and excited (red) states of Pfl6 I58D; the disordered C-terminal region of the ground state is shown in green. The distributions were obtained using a Gaussian kernel density estimation (Scott, 1992).
In summary, we have presented a CPMG relaxation dispersion approach for measuring amide proton temperature coefficients of excited protein states. Because both ground and excited conformers are present in solution it becomes possible to measure temperature coefficients of both states under identical conditions so that they can be readily compared. The methodology has been applied to the study of the cold denatured ensemble of Pfl6 I58D where Δδ̃HN/ΔT values are for the most part significantly more negative than for the folded conformer and less than −4.6ppb/K, consistent with the absence of intra-molecular hydrogen bonds, on average. Nevertheless, temperature coefficients are not uniform and span a significant range (5.6±2.1 ppb/K), suggesting that the unfolded state does not approximate a random coil chain, but like other unfolded proteins, samples a distribution of transiently formed conformers. The methodology presented adds to a growing set of experiments for characterizing small transient populations of proteins under ‘native-like’ conditions that can provide a detailed atomic resolution description of conformations that are recalcitrant to study using other techniques.
Supplementary Material
Acknowledgements
G.B. acknowledges the European Molecular Biology Organization and the Canadian Institutes of Health Research (CHIR) for post-doctoral fellowships. D.F.H was supported by a post-doctoral fellowship from the CIHR. This work was supported by grants from the CIHR and the Natural Sciences and Engineering Research Council of Canada (LEK) and the NIH (GM066806 to M.H.J.C.). L.E.K. holds a Canada Research Chair in Biochemistry.
Footnotes
Supporting Information
Details of protein production, NMR measurements and data analysis, 1 figure showing fits of dispersion/chemical shift profiles with temperature (for Q27 and R49) and a table of temperature coefficients for the ground and excited states of Pfl6 I58D is provided.
References
- Anet FA, Basus VJ. Limiting equations for exchaning broadening in 2-site NMR systmes with very unequal populations. J. Magn. Reson. 1978;32:339–343. [Google Scholar]
- Baldwin AJ, Religa TL, Hansen DF, Bouvignies G, Kay LE. 13CHD2 methyl group probes of millisecond time scale exchange in proteins by 1H relaxation dispersion: an application to proteasome gating residue dynamics. J Am Chem Soc. 2010;132:10992–10995. doi: 10.1021/ja104578n. [DOI] [PubMed] [Google Scholar]
- Baxter NJ, Williamson MP. Temperature dependence of 1H chemical shifts in proteins. J Biomol NMR. 1997;9:359–369. doi: 10.1023/a:1018334207887. [DOI] [PubMed] [Google Scholar]
- Boehr DD, McElheny D, Dyson HJ, Wright PE. The dynamic energy landscape of dihydrofolate reductase catalysis. Science. 2006;313:1638–1642. doi: 10.1126/science.1130258. [DOI] [PubMed] [Google Scholar]
- Bouvignies G, Korzhnev DM, Neudecker P, Hansen DF, Cordes MH, Kay LE. A simple method for measuring signs of 1H(N) chemical shift differences between ground and excited protein states. J Biomol NMR. 2010;47:135–141. doi: 10.1007/s10858-010-9418-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr HY, Purcell EM. Effects of diffusion on free precession in nuclear magnetic resonance experiments. Phys. Rev. 1954;54:630–638. [Google Scholar]
- Cierpicki T, Otlewski J. Amide proton temperature coefficients as hydrogen bond indicators in proteins. J Biomol NMR. 2001;21:249–261. doi: 10.1023/a:1012911329730. [DOI] [PubMed] [Google Scholar]
- Cierpicki T, Zhukov I, Byrd RA, Otlewski J. Hydrogen bonds in human ubiquitin reflected in temperature coefficients of amide protons. J Magn Reson. 2002;157:178–180. doi: 10.1006/jmre.2002.2597. [DOI] [PubMed] [Google Scholar]
- Crowhurst KA, Forman-Kay JD. Aromatic and methyl NOEs highlight hydrophobic clustering in the unfolded state of an SH3 domain. Biochemistry. 2003;42:8687–8695. doi: 10.1021/bi034601p. [DOI] [PubMed] [Google Scholar]
- Dyson HJ, Rance M, Houghten RA, Lerner RA, Wright PE. Folding of immunogenic peptide fragments of proteins in water solution. I. Sequence requirements for the formation of a reverse turn. J Mol Biol. 1988;201:161–200. doi: 10.1016/0022-2836(88)90446-9. [DOI] [PubMed] [Google Scholar]
- Hansen DF, Vallurupalli P, Lundstrom P, Neudecker P, Kay LE. Probing Chemical Shifts of Invisible States of Proteins with Relaxation Dispersion NMR Spectroscopy: How Well Can We Do? J. Am. Chem. Soc. 2008:2667–2675. doi: 10.1021/ja078337p. [DOI] [PubMed] [Google Scholar]
- Henzler-Wildman KA, Lei M, Thai V, Kerns SJ, Karplus M, Kern D. A hierarchy of timescales in protein dynamics is linked to enzyme catalysis. Nature. 2007;450:913–916. doi: 10.1038/nature06407. [DOI] [PubMed] [Google Scholar]
- Ishima R, Baber J, Louis JM, Torchia DA. Carbonyl carbon transverse relaxation dispersion measurements and ms-micros timescale motion in a protein hydrogen bond network. J. Biomol. NMR. 2004;29:187–198. doi: 10.1023/B:JNMR.0000019249.50306.5d. [DOI] [PubMed] [Google Scholar]
- Ishima R, Freedberg DI, Wang YX, Louis JM, Torchia DA. Flap opening and dimer-interface flexibility in the free and inhibitor- bound HIV protease, and their implications for function. Structure Fold Des. 1999;7:1047–1055. doi: 10.1016/s0969-2126(99)80172-5. [DOI] [PubMed] [Google Scholar]
- Karplus M, Kuriyan J. Molecular dynamics and protein function. Proc. Natl. Acad. Sci. U.S.A. 2005;102:6679–6685. doi: 10.1073/pnas.0408930102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korzhnev DM, Kay LE. Probing invisible, low-populated States of protein molecules by relaxation dispersion NMR spectroscopy: an application to protein folding. Acc Chem Res. 2008;41:442–451. doi: 10.1021/ar700189y. [DOI] [PubMed] [Google Scholar]
- Korzhnev DM, Religa TL, Banachewicz W, Fersht AR, Kay LE. A transient and low-populated protein-folding intermediate at atomic resolution. Science. 2010;329:1312–1316. doi: 10.1126/science.1191723. [DOI] [PubMed] [Google Scholar]
- LeFevre KR, Cordes MH. Retroevolution of lambda Cro toward a stable monomer. Proc Natl Acad Sci U S A. 2003;100:2345–2350. doi: 10.1073/pnas.0537925100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loria JP, Rance M, Palmer AG. A relaxation compensated CPMG sequence for characterizing chemical exchange. J. Am. Chem. Soc. 1999;121:2331–2332. doi: 10.1023/a:1008355631073. [DOI] [PubMed] [Google Scholar]
- Lundstrom P, Hansen DF, Kay LE. Measurement of carbonyl chemical shifts of excited protein states by relaxation dispersion NMR spectroscopy: comparison between uniformly and selectively (13)C labeled samples. J Biomol NMR. 2008;42:35–47. doi: 10.1007/s10858-008-9260-4. [DOI] [PubMed] [Google Scholar]
- Lundstrom P, Hansen DF, Vallurupalli P, Kay LE. Accurate measurement of alpha proton chemical shifts of excited protein states by relaxation dispersion NMR spectroscopy. J Am Chem Soc. 2009;131:1915–1926. doi: 10.1021/ja807796a. [DOI] [PubMed] [Google Scholar]
- Lundstrom P, Kay LE. Measuring 13Cβ chemical shifts of invisible excited states in proteins by relaxation dispersion NMR spectroscopy. J. Biomol. NMR. 2009;44:139–155. doi: 10.1007/s10858-009-9321-3. [DOI] [PubMed] [Google Scholar]
- Lundstrom P, Vallurupalli P, Religa TL, Dahlquist FW, Kay LE. A single-quantum methyl 13C-relaxation dispersion experiment with improved sensitivity. J Biomol NMR. 2007;38:79–88. doi: 10.1007/s10858-007-9149-7. [DOI] [PubMed] [Google Scholar]
- Meiboom S, Gill D. Modified spin-echo method for measuring nuclear magnetic relaxation times. Rev. Sci. Instrum. 1958;29:688–691. [Google Scholar]
- Merutka G, Dyson HJ, Wright PE. 'Random coil' 1H chemical shifts obtained as a function of temperature and trifluoroethanol concentration for the peptide series GGXGG. J Biomol NMR. 1995;5:14–24. doi: 10.1007/BF00227466. [DOI] [PubMed] [Google Scholar]
- Newlove T, Atkinson KR, Van Dorn LO, Cordes MH. A trade between similar but nonequivalent intrasubunit and intersubunit contacts in Cro dimer evolution. Biochemistry. 2006;45:6379–6391. doi: 10.1021/bi052541c. [DOI] [PubMed] [Google Scholar]
- Orekhov VY, Korzhnev DM, Kay LE. Double- and zero-quantum NMR relaxation dispersion experiments sampling millisecond time scale dynamics in proteins. J. Am. Chem. Soc. 2004;126:1886–1891. doi: 10.1021/ja038620y. [DOI] [PubMed] [Google Scholar]
- Palmer AG, Kroenke CD, Loria JP. NMR methods for quantifying microsecond-to-millisecond motions in biological macromolecules. Methods in Enzymol. 2001;339:204–238. doi: 10.1016/s0076-6879(01)39315-1. [DOI] [PubMed] [Google Scholar]
- Roessler CG, Hall BM, Anderson WJ, Ingram WM, Roberts SA, Montfort WR, Cordes MH. Transitive homology-guided structural studies lead to discovery of Cro proteins with 40% sequence identity but different folds. Proc Natl Acad Sci U S A. 2008;105:2343–2348. doi: 10.1073/pnas.0711589105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott D. Multivariate density estimation: theory, practice, and visualization. Wiley; 1992. [Google Scholar]
- Seale JW, Srinivasan R, Rose GD. Sequence determinants of the capping box, a stabilizing motif at the N-termini of alpha-helices. Protein Sci. 1994;3:1741–1745. doi: 10.1002/pro.5560031014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skrynnikov NR, Dahlquist FW, Kay LE. Reconstructing NMR spectra of "invisible" excited protein states using HSQC and HMQC experiments. J. Am. Chem. Soc. 2002;124:12352–12360. doi: 10.1021/ja0207089. [DOI] [PubMed] [Google Scholar]
- Tollinger M, Skrynnikov NR, Mulder FAA, Forman-Kay JD, Kay LE. Slow dynamics in folded and unfolded states of an SH3 domain. J. Am. Chem. Soc. 2001;123:11341–11352. doi: 10.1021/ja011300z. [DOI] [PubMed] [Google Scholar]
- Vallurupalli P, Hansen DF, Kay LE. Probing structure in invisible protein states with anisotropic NMR chemical shifts. J. Am. Chem. Soc. 2008a;130:2734–2735. doi: 10.1021/ja710817g. [DOI] [PubMed] [Google Scholar]
- Vallurupalli P, Hansen DF, Kay LE. Structures of invisible, excited protein states by relaxation dispersion NMR spectroscopy. Proc Natl Acad Sci U S A. 2008b;105:11766–11771. doi: 10.1073/pnas.0804221105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallurupalli P, Hansen DF, Stollar EJ, Meirovitch E, Kay LE. Measurement of bond vector orientations in invisible excited states of proteins. Proc. Natl. Acad. Sci. U.S.A. 2007;104:18473–18477. doi: 10.1073/pnas.0708296104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wishart DS, Bigam CG, Holm A, Hodges RS, Sykes BD. 1H, 13C and 15N random coil NMR chemical shifts of the common amino acids. I. Investigations of nearest-neighbor effects. J Biomol NMR. 1995;5:67–81. doi: 10.1007/BF00227471. [DOI] [PubMed] [Google Scholar]
- Wishart DS, Case DA. Use of chemical shifts in macromolecular structure determination. Methods Enzymol. 2001;338:3–34. doi: 10.1016/s0076-6879(02)38214-4. [DOI] [PubMed] [Google Scholar]
- Zhang O, Forman-Kay J. NMR Studies of Unfolded States of an SH3 Domain in Aqueous Solution and Denaturing Conditions. Biochemistry. 1997;36:3959–3970. doi: 10.1021/bi9627626. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.