Abstract
Purpose
High-throughput profiling experiments have linked altered expression of microRNAs (miRNA) to different types of cancer. Tumor tissues are a heterogeneous mixture of not only cancer cells, but also supportive and reactive tumor microenvironment elements. To clarify the clinical significance of altered miRNA expression in solid tumors, we developed a sensitive fluorescence-based in situ hybridization (ISH) method to visualize miRNA accumulation within individual cells in formalin-fixed, paraffin-embedded tissue specimens. This ISH method was implemented to be compatible with routine clinical immunohistochemical (IHC) assays to enable the detection of miRNAs and protein markers in the same tissue section for colocalization and functional studies.
Experimental Design
We used this combined ISH/IHC assay to study a subset of cancer-associated miRNAs, including miRNAs frequently detected at low (miR-34a and miR-126) and high (miR-21 and miR-155) levels, in a panel of breast, colorectal, lung, pancreas, and prostate carcinomas.
Results
Despite the distinct histopathologic alterations of each particular cancer type, general trends emerged that pinpointed distinct source cells of altered miRNA expression. Although altered expressions of miR-21 and miR-34a were manifested within cancer cells, those of miR-126 and miR-155 were predominantly confined to endothelial cells and immune cells, respectively. These results suggest a heterogeneous participation of miRNAs in carcinogenesis by intrinsically affecting cancer cell biology or by modulating stromal, vascular, and immune responses.
Conclusions
We described a rapid and sensitive multicolor ISH/IHC assay and showed that it could be broadly applied as an investigational tool to better understand the etiologic relevance of altered miRNA expression in cancer.
MicroRNAs (miRNA) are a class of short noncoding regulatory RNA genes which act as posttranscriptional regulators of gene expression (1–3). By binding to the 3′-untranslated region of target mRNAs, the ~21 to 23 nucleotide–long miRNAs could trigger translational downregulation and/or increased degradation of mRNA from target genes (4). The recent explosion of miRNA research in biomedical sciences, and particularly in cancer biology, attests to their perceived importance to human disease (5, 6). High-throughput expression profiling of RNA extracted from whole tissue biopsies has provided a short list of miRNAs that could serve as useful biomarkers for the early detection, diagnosis, and/or prognosis of different types of cancer (7). Low levels of let-7, miR-34, miR-126, and miR-145 and high levels of miR-21, miR-155, and miR-221 have been frequently reported in association with breast, colorectal, gastrointestinal, lung, pancreas, prostate, and/or thyroid cancer (7, 8). These high-throughput profiling results have been technically confirmed by miRNA-specific quantitative reverse transcription-PCR (RT-PCR) analysis and several studies based on RT-PCR analysis of miRNA expression have further supported the clinical application of miRNAs as informative biomarkers. However, these detection assays cannot directly determine whether these expression changes occur specifically within cancer cells, reactive stroma, and/or infiltrating immune cells recruited to the cancerous lesion. Moreover, tissue heterogeneity among specimens and unequal representation of source cells (cancer cell and/or other cell types) with altered miRNA expression might confound the interpretation of RT-PCR analyses, unless they are done on samples highly enriched for the source cells. Visualization of miRNA expression within individual cells by in situ hybridization (ISH) provides an independent tool to clinically validate miRNAs that have been highlighted by expression profiling analysis and also to more closely assess the etiologic relevance and clinical significance of altered miRNA expression. Moreover, a refined understanding of the source cell(s) of miRNA deregulation in cancer could shed light onto the molecular mechanisms at work.
Locked nucleic acids (LNA), a class of bicyclical high-affinity RNA analogues (9), made the detection of miRNAs by ISH possible. LNA-modified DNA probes have overcome the technical limitations of achieving specific and avid hybridization to the short RNA sequence of mature miRNAs as it was first shown in zebrafish and mouse embryos by whole-mount ISH using chromogenic staining (10, 11). Subsequently, we and others implemented ISH methods to detect miRNA expression in formalin-fixed, paraffin-embedded (FFPE) brain (12), breast (13), colon (14), lung (15), and pancreatic tissue sections (16, 17). These methods followed a similar general strategy in which DNA probes modified with LNAs were terminally tagged with a hapten molecule, either digoxigenin (DIG) or fluorescein (FITC or FAM), and a single-step antibody conjugated to alkaline phosphatase or horseradish peroxidase (HRP) was used for probe recognition and signal staining. Although other groups used alkaline phosphatase–mediated (i.e., 5-bromo-4-chloro-3-indolyl phosphate/nitroblue tetrazolium) or HRP-mediated (i.e., 3,3′-diaminobenzidine) chromogenic staining of tissue sections (12, 14, 16, 17), we used HRP-mediated (i.e., FITC) fluorescent staining (13, 15). These recent advancements showed the feasibility of detecting cancer-associated miRNAs by ISH in FFPE tissue specimens, but further technical improvements in signal quantification, reproducibility, and sensitivity are still required for the development of miRNA-based clinical assays.
Most diagnostic and prognostic decisions are based on the examination of FFPE tissue sections in which overall histology is revealed by H&E staining and specific protein markers are revealed by immunohistochemical (IHC) staining. Here, we sought to develop a rapid and sensitive fluorescence-based ISH assay with similar and compatible experimental and instrumental requirements for clinical IHC assays to allow for seamless adoption of miRNA bio-markers into routine clinical practice. Codetection of multiple miRNAs, other noncoding RNAs, and/or proteins on the same FFPE tissue section was achieved with custom-designed LNA-modified DNA probes and in-house synthesized fluorescent substrates. To show the general clinical application of this combined ISH/IHC assay, we characterized the expression of a panel of cancer-associated miRNAs in archived FFPE specimens of breast, colorectal, lung, pancreatic, and prostate carcinomas, which collectively are the leading causes of cancer-related mortality in the United States. Our results uncovered a complex and distinct contribution of different cell types to altered expression of individual miRNAs in tumor tissues. These findings underscore the necessity for spatial characterization of miRNA expression to determine whether the cancer cells, supportive, and/or reactive microenvironment elements are the principal sources of miRNA deregulation in solid tumors. This information will be essential to understand the role of miRNAs in cancer initiation and progression as well as to interpret the effect of miRNA changes in a diagnostic test.
Materials and Methods
Processing and procurement of tissue specimens
Tissue specimens were obtained through the research pathology services from the Tissue Bank at Dartmouth-Hitchcock Medical Center, Lebanon, NH. These surgical specimens were processed in the surgical pathology laboratory using an institutional standardized protocol. Briefly, surgical specimens were sectioned in 2-mm slices and fixed in 10% formalin for up to 24 hours and then were paraffin-embedded in a fully automated Shandon Path-centre instrument using a standard overnight procedure (2 × 10% formalin for 80 minutes, ethanol series 75–100% in six sequential 45-minute steps, 2× xylene for 45 minutes, and 4× paraplast for 45 minutes).
Probe design and hybridization conditions
LNA-modified DNA probes were designed to have a predicted melting temperature (Tm) of between ~75°C and 78°C (see Supplementary Table S1 for details) following the general strategy as previously described (18). Briefly, LNA-modified nucleotides were intercalated at every third nucleotide among DNA nucleotides. When this strategy rendered a probe with a Tm lower than 70°C, we introduced more LNA-modified nucleotides, which were intercalated at every second or third position. We preferentially chose C or G for LNA modifications. The Tm of the probe against complementary miRNA sequences was calculated using LNA design SciTools (Integrated DNA Technologies, Inc.; http://www.idtdna.com/analyzer/Applications/lna/), with default variables, except for [Na+], which was set to 100 mmol/L. When ordering probes from IDT (http://www.idtdna.com/), these are the codes for the hapten addition onto a terminal 5′ and/or 3′ extra T nucleotides (these Ts are not part of the complementary miRNA sequence:/5biosg/t,/55Br-dU/tt,/5DigN/t,/56-FAM/t,/5Biosg/t[…]t/3Bio/,/55Br-dU/tt[…]tt/i5Br-dU/t,/5DigN/t[…]t/3Dig_N/, and/56-FAM/t[…]t/36-FAM/). When ordering from Exiqon (http://www.exiqon.com/), these haptens could be added onto the 5′ and/or 3′ end of the probe.
Combined ISH/IHC assay
Four-micrometer-thick tissue sections were mounted on positively charged barrier frame slides, dewaxed in xylenes, and rehydrated through an ethanol dilution series (100% to 25%). Tissue sections were digested with 5 μg/mL of proteinase K for 20 minutes at 37°C to facilitate probe penetration and exposure of miRNA species. To minimize nonspecific binding based on charge interactions, tissues were subjected to a brief acetylation reaction [66 mmol/L HCl, 0.66% acetic anhydride (v/v) and 1.5% triethanolamine (v/v) in RNase-free water]. Then, tissue sections were prehybridized at the hybridization temperature (see Supplementary Table S1 for details) for 30 minutes in prehybridization solution which consisted of 50% deionized formamide, 5× sodium chloride/sodium citrate buffer, 1× Denhardt’s solution, 500 μg/mL of yeast tRNA, and 0.01% Tween. The prehybridization solution was replaced with 200 μL of hybridization solution containing 10 pmol of the hapten-labeled LNA probe and tissues were incubated for 90 minutes at the hybridization temperature and washed thrice for 10 minutes in sodium chloride/sodium citrate buffer at the established stringency of sodium chloride/sodium citrate (see Supplementary Table S1 for details). At this point, tissue slides were loaded onto the Biogenex i6000 staining machine (BioGenex Laboratories, Inc.), which was programmed to dispense 400 μL of the appropriate reagent per step. Slides were treated with 3% H2O2 to inactivate endogenous peroxidase and block with 5% bovine serum albumin in PBS (w/v). Followed by primary and secondary antibody incubation in PBT [1% bovine serum albumin (w/v), 0.1% Tween 20 (v/v) in PBS] and washes in PBST [0.01% Tween 20 (v/v) in PBS], tyramine-conjugated fluorochrome was applied to the slide and the tyramide signal amplification (TSA) reaction was allowed to proceed for 10 to 30 minutes. Sequential TSA rounds for the detection of other miRNAs, noncoding RNAs, or proteins followed the same protocol. Finally, slides were washed extensively with PBST and mounted with antifading ProLong Gold Solution (Invitrogen) with or without 4′,6-diamidino-2-phenylindole (for nuclear counterstaining). Please see Supplementary Table S2 for a list of antibodies used and Supplementary Table S3 for the preparation of tyramide-conjugated fluorescent substrates.
Microscopy and image analysis
Fluorescent images were captured with an F-view II monochrome camera (Olympus, U-CMAD3) mounted on an Olympus BX60 microscope with filter cubes for AMCA/Dylight405/DAPI (Chroma Filter Set 31000), fluorescein (Olympus Filter Set U-MNIBA), rhodamine/Cy3 (Chroma Filter Set SP102V1), Dylight594/TR (Chroma Filter Set SP107), Alexa647/Dylight649/Cy5 (Chroma Filter Set SP104V2), and Dylight680/Cy5.5 (Chroma Filter Set SP105) using the cellSens software package (Olympus). Gray scale .TIF files were later colorized as displayed in Figs. 1–4. Gray scale .TIF files were converted to RGB .TIF files for image analysis with intensity measurement tools of Image-Pro Plus software package (Media Cybernetics).
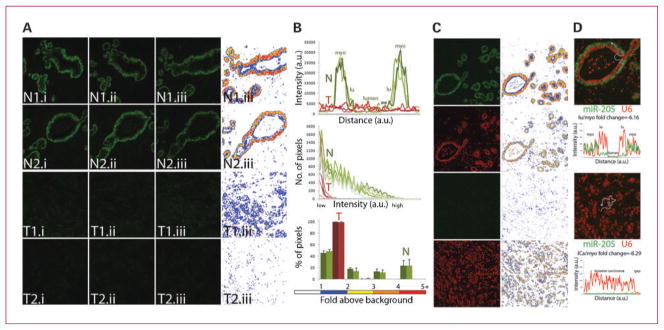
Fig. 1.
Reproducibility of the ISH method and signal analysis. Matched normal and tumor FFPE breast tissue sections were used to codetect miR-205 and U6 snRNA using FAM2×- and Bio2×-tagged probes, respectively. miR-205 and U6 signals were revealed by sequential TSA reactions with FITC-tyramine (green for miR-205 probe) and rhodamine-tyramine (red for U6 probe) substrates. A, consecutive (i, ii, iii) matched normal (N) and tumor (T) tissue sections were assayed on separate days (1 and 2). Left, raw fluorescent image of the same representative field for each intraexperimental replica. Right, a heat map rendition of intensity classes of miR-205 signal above background noise (see color scale in B). B, the signal intensity of miR-205 was measured across the mammary duct in N1 tissue and across invasive carcinoma lesion in T1 (top). Line profile of N1.iii was slightly shifted to the right (gap) to match the signal from the myoepithelial (myo) and luminal (lu) cellular structures of N1.i and N1.ii. Distribution of pixel intensity of the whole images in A indicate concordant readings for each intraexperimental and interexperimental replica (middle). Columns, mean percentage of pixels within each intensity class for each set of intraexperimental replica; bars, SD (bottom). C, raw images of miR-205 and snRNA U6 were captured with the same exposure and gain setting in normal and tumor tissue (left), which are displayed as a heat map graph (right). D, signal intensity of miR-205 and U6 expression was measured using a line profile tool (merged images are displayed as a 2× magnification of those in C). Background intensity was subtracted from the recorded intensity value and these corrected intensity values were used to generate line graphs. Square dot indicates the beginning of the line profile reading and corresponds with the left-most value of the graph. Average intensity of miR-205 and U6 signal in the indicated structures was used to calculate relative fold decrease of miR-205 expression in luminal and cancer cells compared with myoepithelial cells (lu/myo and ICa/myo, respectively).
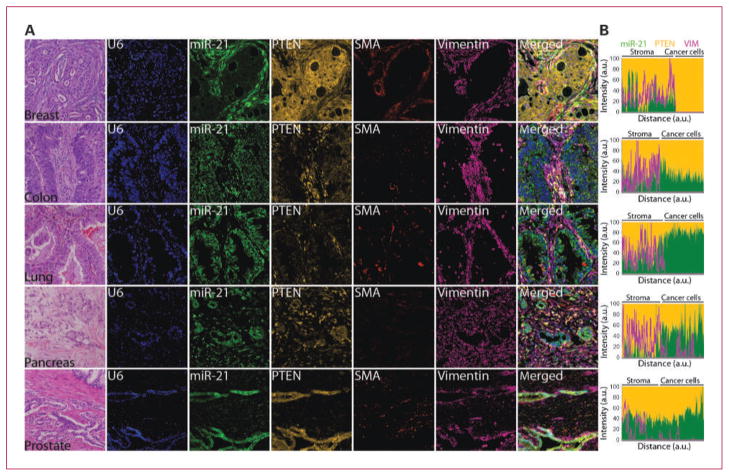
Fig. 4.
Codetection of miR-21 with PTEN in clinical specimens. A, serial 4-μm FFPE tumor tissue sections of the indicated organs were stained with H&E to reveal histologic features or subjected to standard ISH assays using FAM2×-tagged probe against miR-21 and Bio2×-tagged probe against snRNA U6. The miR-21 and snRNA U6 signals were revealed by sequential TSA reactions with FITC-tyramine (green) and AMCA-tyramine (blue) substrates. Smooth muscle actin expression (SMA) was revealed by TSA reaction with Dylight594 (red). After a HIER with citrate, PTEN and vimentin expression were revealed by TSA reaction with rhodamine-tyramine (orange) and Dylight680 (magenta), respectively. B, line profile analysis was used to quantitate the intensity of miR-21, PTEN, and vimentin expression. Background intensity was subtracted and intensity values were normalized setting the point with maximum intensity to 100 and calculating other values in relation to this reference. Vimentin expression pattern was plotted as a line; independently, relative expression of miR-21 and PTEN for each data point was plotted as stacked areas. Displayed images were modified by optimizing the contrast with the process enhancement function from cellSens software package (Olympus), please see Supplementary Fig. S5 for raw fluorescent images and details on signal analysis.
Results and Discussion
Optimization of experimental variables for miRNA detection in FFPE specimens
We first systematically tested and optimized the conditions of our ISH method (Supplementary Fig. S1 and Supplementary Table S1; data not shown). We determined that digestion with Proteinase K at 5 μg/mL for 20 minutes at 37°C provided optimal results for archived FFPE human breast, colon, lung, pancreas, and prostate tissues. To further improve the sensitivity of detection, we designed probes with a hapten at both the 5′ and 3′ ends on extra T nucleotides that were not part of the miRNA complementary sequence, and we introduced a secondary antibody step (antibody sandwich amplification) using HRP-conjugated antibody against the primary anti-hapten antibody (Supplementary Table S2). By using double hapten–tagged (5′ and 3′ ends; hapten 2×) versus single hapten–tagged (5′ end; hapten 1×) probes, we observed a >3-fold increase in the sensitivity of signal detection of miRNAs and other noncoding RNA species (Supplementary Figs. S1 and S2). Importantly, this improved protocol allowed us to visualize the expression of miRNAs that had previously fallen below the detection limit of the ISH assay.
Assessing experimental reproducibility and signal analysis using continuous measurements
Given the importance of reproducibility in the clinical setting, we assessed the intraexperimental and interexperimental variation of the ISH method. Consecutive sections of matched normal and tumor breast tissue were mounted on the same slide and groups of three of these slides were independently subjected to the ISH method on separate days for the detection of miR-205 (Fig. 1A). To minimize experimental variations due to the operator, experiments were done in a Food and Drug Administration–approved automated staining station (Biogenex i6000 machine) for all steps after probe washes. The signal intensity and overall staining pattern of miR-205 expression was highly concordant as determined by visual examination and by computer-assisted image analysis. We used heat map plots and line profile analysis tools to measure the intensity of miR-205 signal as an effort to objectivize, standardize, and eventually, automate a scoring system for miRNA expression (Fig. 1B-D). Below, we describe the use of similar analytic tools for signal quantification and colocalization studies of miRNA and protein expression (Figs. 2–4)
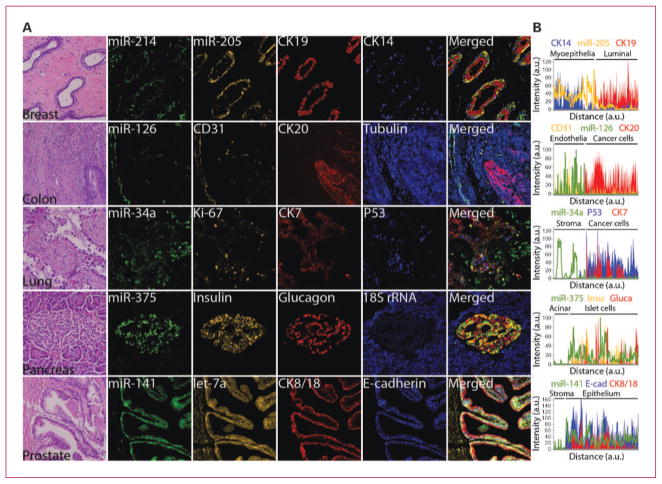
Fig. 2.
Codetection of miRNAs with cell type–specific and prognostic protein markers. A, serial FFPE tissue sections of the indicated organs were stained with H&E to reveal histologic features or subjected to ISH assay using FAM2×-tagged probes against miR-34a, miR-126, miR-141, miR-214, and miR-375 mixed with Bio2×-tagged probe against 18S rRNA, BrdU2×-tagged probe against miR-205, or DIG2×-tagged probe against let-7a. miRNA signals were revealed by sequential TSA reactions with FITC-tyramine (green for FAM2× probes), rhodamine-tyramine (orange for BrdU2× and DIG2× probes), and AMCA-tyramine (blue for Bio2× probe) substrates. After HIER as needed (Supplementary Table S2), expression of the indicated proteins was revealed by sequential TSA reactions with Dylight594-tyramine or Dylight680-tyramine (red) and AMCA-tyramine (blue) substrates, each TSA reaction was preceded by incubation with specific antibodies against each protein and appropriate anti-host species/HRP antibody; except for insulin expression, which was revealed by an anti–guinea pig secondary antibody conjugated to Cy3 (orange), obviating the need for the TSA step. B, line profile analysis was used to quantitate the intensity of RNA or protein expression. Background intensity was subtracted and intensity values were normalized setting the point with maximum intensity to 100 and calculating other values in relation to this reference. miRNA expression pattern was plotted as a line; independently, expression patterns of each protein were plotted as stacked areas. Displayed images were modified by optimizing the contrast with the process enhancement function from cellSens software package (Olympus), please see Supplementary Fig. S3 for raw fluorescent images and details on signal analysis.
Codetection of miRNAs and other noncoding RNAs in FFPE specimens
To assess the quality and integrity of RNA preservation in tissue samples and to normalize changes in miRNA expression, we codetected several distinct miRNAs and abundant control noncoding RNAs, either small nuclear RNA U6 (snRNA U6) or 18S ribosomal RNA (18S rRNA; Figs. 1, 2 and 4). Staining with ubiquitously expressed snRNA U6 and 18S rRNA was also used to identify tissue dehydration (dry spots), spurious edge effects, or other technical problems caused by suboptimal sample handling. Moreover, we set out to codetect multiple miRNA species because this could maximize the amount of information obtained per tissue section, which would be especially important (a) if tissue availability were limited such as tissue cores of a tissue microarray, core needle biopsies, and cell preparations obtained by fine-needle aspiration; and (b) if concomitant analysis of miRNAs with opposite expression changes between normal and cancer cells or within different cell types within the tumor lesion yielded a more powerful clinical indicator than either miRNA alone. For codetection of miRNAs and/or control RNAs, we used probes that were 5′ and 3′ terminally tagged (hapten 2×) with biotin (Bio)-, digoxigenin (DIG)-, 5-bromo-2-deoxyuridine (BrdU)-, or 5- (and 6)-carboxy-fluorescein (FAM). The desired FAM2×, Bio2×, BrdU2×, and/or DIG2× probes were mixed together and annealed with their respective complementary RNA sequences in a single hybridization incubation step. After washes to eliminate unbound probes, the codetection of multiple RNA species was achieved by sequential TSA (19) reactions using different in-house synthesized tyramine-conjugated fluorescent substrates (Supplementary Table S3). Following this strategy, up to five independent RNA and/or protein markers (see below) could be codetected on the same tissue section (Fig. 4; Supplementary Fig. S5 and Supplementary Table S3).
Codetection of miRNAs and protein markers in FFPE specimens
To ascribe miRNA expression to a specific cell type and to better understand the physiologic or pathologic status of the cell, we set out to codetect miRNAs and clinically important protein markers on the same tissue section. Numerous protein markers expressed in normal tissues also display specific expression patterns in tumor tissues that could be especially informative: (a) cytokeratin 19 (CK19) is an epithelial-specific marker, and as such, is widely used to identify carcinoma cells; (b) CK5/6 and CK14 are expressed in normal myoepithelial cells and specifically identify the aggressive basal breast cancer subtype (20); (c) the CK7/CK20 signal could be used to discern the organ of origin of certain cancers, for example, most primary and metastatic colorectal carcinoma cells are CK7−/CK20+ and lung carcinoma cells are CK7+/CK20−(21); (d) glucagon and insulin are specifically expressed in α and β endocrine pancreatic cells, respectively, and as such, are useful markers for establishing the type of endocrine pancreatic tumors (22). In addition, miRNAs have been shown to mediate the repression of and/or to be regulated by important oncogenic and tumor suppressor genes (5, 6). Thus, codetection of miRNA and protein markers could have important clinical applications.
The detection of protein markers was generally compatible with PK digestion and other chemical treatments used in the preceding ISH steps (Supplementary Table S2). Some useful epithelial-specific markers such as CK19 and other cytokeratins (e.g., CK7, CK8/18, and CK20) were detected without further tissue processing, whereas other protein markers required heat-induced epitope retrieval (HIER) to be efficiently detected. Although HIER did not quench the fluorescent signal from previous steps, tissue damage and detachment were observed in some slides and the signal was diminished by physically washing off the anchored fluorochrome molecules. Thus, the mildest PK treatment (limited incubation time) and HIER treatment (limited temperature of HIER and/or incubation time) should be used for optimal codetection conditions of miRNAs and proteins. In the following sections, we provide several examples of miRNAs frequently associated with cancer to show the feasibility of codetection of miRNA and protein markers and to illustrate the potential uses of miRNAs as novel biomarkers. In Supplementary Table S1, we also provide a “broad strokes” summary of the expression pattern of the 18 miRNAs that we analyzed in this study.
Colocalization of miRNAs and cell type–specific protein markers reveals distinct source cells for altered miRNA expression
We previously reported that the expression of miR-205 was confined to a subpopulation of mammary epithelial cells and that there was a positive trend of association between miR-205 expression and a favorable clinical outcome in patients of the basal breast cancer subtype (ER−PR−HER2−; ref. 13). Costaining with CK14 (myoepithelial cells) and CK19 (luminal epithelial cells) protein markers confirmed myoepithelia-restricted expression of miR-205 in breast tissue (Fig. 2). Recent reports based on RT-PCR analyses indicated that miR-205 expression could be useful to distinguish squamous and adenocarcinoma subtypes of lung cancer (23, 24). In an independent study, low levels of miR-205 were associated with poor prognosis in head and neck squamous carcinomas (25). Thus, a combined ISH/IHC assay for the detection of miR-205 and basal/squamous epithelial cell marker (e.g., CK14) would increase the interpretative power of miR-205 expression changes and could be further implemented as a diagnostic and/or prognostic tool in solid tumors.
Expression profiling analyses detected miR-126 at lower levels in colorectal and other carcinomas compared with normal tissue (26–30). Costaining with CD31/PECAM-1 (endothelial cells) and CK20 (epithelial cells) protein markers indicated an endothelial-specific expression of miR-126 in colonic and other tissues (Fig. 2; Supplementary Fig. S6; data not shown). Our ISH results suggest that miR-126 expression changes in tumor tissue could mainly reflect differences in the distribution of blood vessels compared with normal tissue, and perhaps, differences in the integrity and/or structure of the neovasculature (Supplementary Fig. S6; data not shown). Similarly, expression profiling analyses detected miR-375 at lower levels in pancreatic tumor tissues compared with normal pancreas (31). Costaining with insulin and glucagon confirmed endocrine-restricted expression of miR-375 (Fig. 2) in pancreatic tissue. Our ISH results suggest that miR-375 is expressed in cell types that do not contribute to pancreatic ductal adenocarcinoma formation.
Lastly, miR-141 was reported as a potential serum bio-marker for prostate cancer detection (32). Independent studies have shown that members of the miR-141/miR-200 family act to maintain epithelial genetic programs via binding to 3′-untranslated region sites on the mRNAs of ZEB1/2 zinc finger transcriptional repressors (33–37). Costaining with CK8/18 and E-cadherin indicated that the expression of miR-141 was confined to prostatic epithelium (Fig. 2). Our ISH results support the notion that the presence of miR-141 in the peripheral circulation of these patients might closely reflect changes within the neoplastic cells.
Potential value of miRNAs as functional biomarkers
miR-34 family members (miR-34a,b,c) are transcriptionally activated by the p53 tumor suppressor gene in response to DNA damage and mitogenic signals (38). Consistent with a tumor-suppressive role, miR-34s were detected at lower levels in breast, lung, and other solid tumors by expression profiling analyses in whole tissue biopsies (7). Costaining with CK7 (epithelial marker), p53 (tumor suppressor gene), and Ki-67 (proliferation marker) protein markers indicated a lower accumulation of miR-34a levels within malignant cells in lung and breast cancer compared with normal epithelial cells as well as normal and reactive stroma (Fig. 2; data not shown). Detection of p53 expression by IHC is suggestive of discrete or point mutations that interfere with p53 tumor-suppressive functions, negative IHC staining is not informative because it cannot distinguish other mutated forms or chromosomal deletion of p53 from wild-type and functional p53 genes (39). A miR-34–based ISH assay could inform by proxy of p53 activity status and provide a better prognostic indicator than a p53-based IHC assay alone.
miRNAs and the immune response
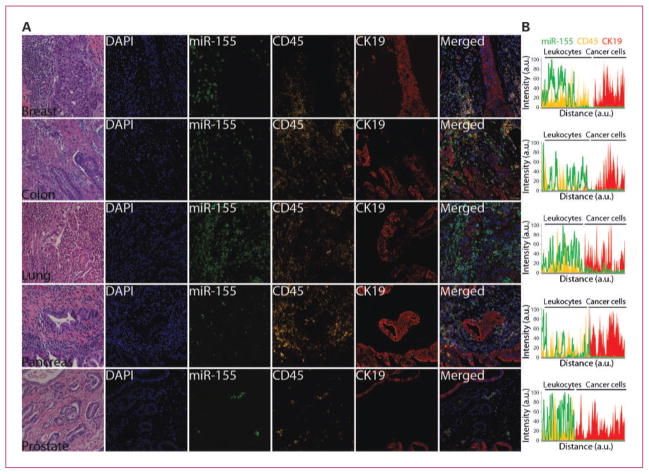
High levels of miR-155 expression have been frequently detected in leukemias, lymphomas as well as breast, lung, pancreas, and thyroid carcinomas (40, 41). High levels of miR-155 correlated with poor outcome in patients with lung and pancreatic cancer (42, 43). In vitro and in vivo studies have unraveled an oncogenic role for miR-155 in hematologic and solid tumors (40, 41, 44, 45). A large body of evidence also attributes an important role for miR-155 in the immune system as a mediator of lymphoid and myeloid cell responses to infection and inflammation (40, 41, 46, 47), which is further supported by immunologic deficiency exhibited by mir-155 knockout mouse models (48, 49). Costaining with CK19 and CD45 (leukocyte marker) indicated that expression of miR-155 was predominantly confined to a subpopulation of infiltrating immune cells in breast, colorectal, lung, pancreas, and prostate tumor lesions (Fig. 3). Further characterization with specific lymphoid and myeloid markers will be required to establish the identity of miR-155–expressing cells and whether a common cell population is present in solid tumors or distinct subpopulations in each tumor type. Nonetheless, these results suggest that the majority of miR-155 signals detected by RT-PCR assays in whole tissue biopsies or blood samples likely emanate from tumor-associated immune cells, and thus, this fact should be taken into consideration when interpreting RT-PCR results. In some tumor specimens, a low level of miR-155 expression was detected within cancer cells (Fig. 3; data not shown). Because ISH is not as sensitive as other detection methods, particularly real-time RT-PCR, undetectable or low levels of expression of miR-155 by ISH might still be functionally important within cancer cells. Recent studies reported the detection of miR-155 by ISH within immune cells, and cancer cells in colorectal adenocarcinoma lesions and predominantly within cancer cells in pancreatic adenocarcinoma lesions (45, 50). Further studies will be needed to elucidate the technical and/or biological reasons behind these discrepancies.
Fig. 3.
Colocalization of miR-155 with CD45 immune cell marker. A, serial 4-μm FFPE tumor tissue sections of the indicated organs were stained with H&E to reveal histologic features or subjected to standard ISH assays using FAM2×-tagged probe against miR-155. The miR-155 signal was revealed by TSA reaction with FITC-tyramine (green). CK19 expression was revealed by TSA reaction with Dylight680-tyramine (red). After a HIER with citrate, CD45 expression was revealed by TSA reaction with rhodamine-tyramine (orange). Tissue sections were counterstained with nuclear marker 4′,6-diamidino-2-phenylindole (blue). B, line profile analysis was used to quantitate the intensity of RNA or protein expression. Background intensity was subtracted and intensity values were normalized setting the point with maximum intensity to 100, and calculating other values in relation to this reference. miRNA expression pattern was plotted as a line; independently, expression patterns of each protein were plotted as stacked areas. Displayed images were modified by optimizing the contrast with the process enhancement function from cellSens software package (Olympus), please see Supplementary Fig. S4 for raw fluorescent images and details on signal analysis.
Correlation of miRNA and target gene expression in clinical samples
Like miR-155, high levels of miR-21 expression have been frequently detected in hematologic and solid tumors, and miR-21 is considered to be an important oncogenic miRNA based on functional studies in cancer cell lines and animal models (6, 51). However, we found that in solid tumors, the source cells of altered expression for miR-21 and miR-155 were strikingly different. We detected higher levels of miR-21 expression within cancer cells and tumor-associated fibroblasts compared with matched normal tissues (Supplementary Fig. S7; data not shown). Generally, miR-21 was predominantly expressed at higher levels within cancer cells in lung, pancreas, and prostate cancer; whereas in breast and colorectal cancer, higher levels of miR-21 expression were more apparent within tumor-associated fibroblasts as supported by costaining with the mesenchymal markers vimentin and smooth muscle actin (Fig. 4; Supplementary Fig. S7). The clinical significance of these distinct patterns of increased miR-21 expression within cancer cells and/or tumor-associated fibroblasts in different types of solid tumors will need to be further evaluated in future studies. These observed patterns of miR-21 accumulation were consistent with our previous studies in breast and lung cancer and in an independent study in colorectal adenocarcinomas (13–15), but was in disagreement with the reported nuclear-enriched staining of miR-21 within cancer cells in pancreatic adenocarcinoma lesions (16).
In vitro studies indicated that miR-21 inhibits the expression of tumor-suppressive PTEN in colon, liver, and pancreatic cancer cell lines (51, 52). To test this interaction in a clinical setting, we analyzed the coexpression of miR-21 and PTEN protein levels in carcinoma lesions of the breast, colon, lung, pancreas, and prostate tissue. In this small sample size, PTEN expression exhibited a partially inverse pattern to that of miR-21 both within cancer cells and in the stromal compartment, providing some support for this regulatory interaction, especially in lung adenocarcinomas (Fig. 4; Supplementary Fig. S7). This example also underscores the feasibility of codetecting the miRNA and protein levels of target gene(s) on the same tissue section. Maspin, PDCD-4, RECK, TIMP3, and TPM1 are among the other miR-21 target genes (6, 51) which could be studied using this combined ISH/IHC assay and whose regulatory interactions could be clinically informative.
Clinical application of combined ISH/IHC assay for disease management
Here, we described a combined multicolor ISH/IHC assay for the codetection of miRNA and protein markers in the same FFPE tissue section. Special emphasis was placed on implementing an ISH method that could be compatible and would meet the reproducibility standards of current clinical IHC assays. Approximately 6 hours of manpower was required for a typical experiment with up to 48 slides, from slide baking to posthybridization washes and preparation of antibodies and reagents for steps carried out on an automated staining station, providing a comparable turnaround time to routine IHC clinical assays. The technical reproducibility and high-throughput capability of this ISH assay was greatly enhanced by using a Food and Drug Administration–approved automated staining station, due to precise and equal incubation periods for time-sensitive TSA reaction, which would be difficult to achieve manually when handling a large number of samples. This automation also permitted the rapid detection of up to five RNA and/or protein markers by sequential rounds of TSA reactions with different fluorescent substrates without the need for any human intervention (unless HIER was needed for protein detection).
Some of the examples above documented, for the first time, miRNA expression in FFPE tissue specimens by ISH. Although the clinical significance of these findings will need to be assessed in a larger cohort of patients, our observations suggest that altered miRNA expression might not be confined to cancer cells to be etiologically relevant (e.g., miR-21 in tumor-associated fibroblasts), whereas other miRNA expression changes might simply reflect tissue heterogeneity (e.g., endocrine cell–expressed miR-375 in pancreatic ductal adenocarcinoma). Coregistration with cell type–specific protein markers indicated a previously unappreciated contribution of tumor micro-environment elements to altered miRNA expression. Thus, the source cell(s) of altered miRNA should be carefully considered when designing and interpreting miRNA-based diagnostic assays. Our results also have implications for miRNA-based therapeutic interventions aimed at restoring basal miRNA activity; because the source cell(s) of miRNA deregulation could be the cancer cells, reactive stroma, infiltrating immune cells, and/or other involved cell types, different targeted delivery strategies might be required for different miRNAs (6, 53).
Large collections of FFPE specimens are archived in tissue banks of medical centers and hospitals, thus this combined ISH/IHC could be a powerful investigational tool to examine miRNA and protein expression in these readily available specimens. If independent retrospective and longitudinal studies validate the informative value of miRNAs as biomarkers, we believe that this combined ISH/IHC method could be seamlessly translated into miRNA-based clinical assays to assist with the management of patient care.
Supplementary Material
Translational Relevance.
The examination of formalin-fixed, paraffin-embedded (FFPE) tissues is the cornerstone for histologic and molecular pathology diagnosis of solid tumors. Micro-RNA (miRNA)–based diagnostics is rapidly becoming a promising area of cancer management. Several methodologies have been applied to detect miRNAs in FFPE specimens. However, an in situ hybridization (ISH) assay has the unique feature of determining the cellular compartment(s) of altered miRNA expression, which, as we show here, is paramount to accurately interpreting the clinical significance of miRNA changes. We implemented a combined ISH and immunohistochemistry (IHC) assay for the detection of miRNAs and clinically important protein markers in FFPE specimens. This ISH/IHC assay could provide a useful investigational tool to assess and validate the clinical value of cancer-associated miRNAs. This fluorescence-based multicolor ISH/IHC assay is rapid, sensitive, and fully compatible with current automated clinical IHC assays. As such, it could be seamlessly adopted for the detection of miRNA biomarkers in routine clinical practice.
Acknowledgments
We thank Carol Hart, Rebecca O’Meara, Mary Schwab, and Maudine Waterman at the Pathological Translation Research Laboratory and Beth Allen, David Beck, Laura Gordon, Eve Kemble, and Eric York at the Research Clinical Pathology Laboratory for technical assistance; Kenneth A. Orndorff at The Herbert C. Englert Cell Analysis Laboratory for technical assistance with imaging and microscopy; Drs. Daniel Longnecker, Vincent Memoli, and Alan Schned for providing and reviewing pancreas, lung, and prostate tissue specimens, respectively; Dr. Todd MacKenzie for advice on data analysis; and Drs. Elena Bryleva and Catherine Carriere for comments and suggestions on this manuscript.
Grant Support
NIH and National Cancer Institute grants R21 CA133715 (M. Korc), R21 RR024411-01A1 (W. Wells), and R03 CA141564-01 (L. Sempere), as well as a postdoctoral fellowship grant from the Susan G. Komen Breast Cancer Foundation (L. Sempere), a Hitchcock Foundation grant (L. Sempere), AACR-Pancreatic Cancer Action Network Award (L. Sempere), and the Danish National Advanced Technology Foundation (S. Kauppinen).
Footnotes
Note: Supplementary data for this article are available at Clinical Cancer Research Online (http://clincancerres.aacrjournals.org/).
L.F. Sempere designed and performed experiments, and wrote the paper; M. Preis helped with experimental design and performed experiments; T. Yezefski and H. Ouyang performed experiments; A.A. Suriawinata, J.R. Conejo-Garcia, and W. Wells provided reagents and assisted with data analysis; A. Silahtaroglu and S. Kauppinen provided technical advice during the project; and M. Korc provided advice, technical expertise on pancreatic cancer, and general guidance during the project. All co-authors provided conceptual and/or editorial suggestions to the manuscript.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Lee RC, Ambros V. An extensive class of small RNAs in Caenorhabditis elegans. Science. 2001;294:862–4. doi: 10.1126/science.1065329. [DOI] [PubMed] [Google Scholar]
- 2.Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–62. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- 3.Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–8. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- 4.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–33. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ventura A, Jacks T. MicroRNAs and cancer: short RNAs go a long way. Cell. 2009;136:586–91. doi: 10.1016/j.cell.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sempere LF, Kauppinen S. Translational implications of microRNAs in clinical diagnostics and therapeutics. In: Bradshaw RA, Dennis EA, editors. Handbook of cell signaling. 2. Oxford: Academic Press; 2009. pp. 2965–81. [Google Scholar]
- 7.Barbarotto E, Schmittgen TD, Calin GA. MicroRNAs and cancer: profile, profile, profile. Int J Cancer. 2008;122:969–77. doi: 10.1002/ijc.23343. [DOI] [PubMed] [Google Scholar]
- 8.Volinia S, Calin GA, Liu CG, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006;103:2257–61. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kauppinen S, Vester B, Wengel J. Locked nucleic acid: high-affinity targeting of complementary RNA for RNomics. Handb Exp Pharmacol. 2006:405–22. doi: 10.1007/3-540-27262-3_21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kloosterman WP, Wienholds E, de BE, Kauppinen S, Plasterk RH. In situ detection of miRNAs in animal embryos using LNA-modified oligonucleotide probes. Nat Methods. 2006;3:27–9. doi: 10.1038/nmeth843. [DOI] [PubMed] [Google Scholar]
- 11.Wienholds E, Kloosterman WP, Miska E, et al. MicroRNA expression in zebrafish embryonic development. Science. 2005;309:310–1. doi: 10.1126/science.1114519. [DOI] [PubMed] [Google Scholar]
- 12.Nelson PT, Baldwin DA, Kloosterman WP, Kauppinen S, Plasterk RH, Mourelatos Z. RAKE and LNA-ISH reveal microRNA expression and localization in archival human brain. RNA. 2006;12:187–91. doi: 10.1261/rna.2258506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sempere LF, Christensen M, Silahtaroglu A, et al. Altered microRNA expression confined to specific epithelial cell subpopulations in breast cancer. Cancer Res. 2007;67:11612–20. doi: 10.1158/0008-5472.CAN-07-5019. [DOI] [PubMed] [Google Scholar]
- 14.Yamamichi N, Shimomura R, Inada K, et al. Locked nucleic acid in situ hybridization analysis of miR-21 expression during colorectal cancer development. Clin Cancer Res. 2009;15:4009–16. doi: 10.1158/1078-0432.CCR-08-3257. [DOI] [PubMed] [Google Scholar]
- 15.Liu X, Sempere LF, Ouyang H, et al. MicroRNA-31 functions as an oncogenic microRNA in mouse and human lung cancer cells by repressing specific tumor suppressors. J Clin Invest. 2010;120:1298–309. doi: 10.1172/JCI39566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dillhoff M, Liu J, Frankel W, Croce C, Bloomston M. MicroRNA-21 is overexpressed in pancreatic cancer and a potential predictor of survival. J Gastrointest Surg. 2008;12:2171–6. doi: 10.1007/s11605-008-0584-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Habbe N, Koorstra JB, Mendell JT, et al. MicroRNA miR-155 is a bio-marker of early pancreatic neoplasia. Cancer Biol Ther. 2009;8:340–6. doi: 10.4161/cbt.8.4.7338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Valoczi A, Hornyik C, Varga N, Burgyan J, Kauppinen S, Havelda Z. Sensitive and specific detection of microRNAs by northern blot analysis using LNA-modified oligonucleotide probes. Nucleic Acids Res. 2004;32:e175. doi: 10.1093/nar/gnh171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Speel EJ, Hopman AH, Komminoth P. Tyramide signal amplification for DNA and mRNA in situ hybridization. Methods Mol Biol. 2006;326:33–60. doi: 10.1385/1-59745-007-3:33. [DOI] [PubMed] [Google Scholar]
- 20.Moriya T, Kasajima A, Ishida K, et al. New trends of immunohistochemistry for making differential diagnosis of breast lesions. Med Mol Morphol. 2006;39:8–13. doi: 10.1007/s00795-006-0309-8. [DOI] [PubMed] [Google Scholar]
- 21.Tot T. Cytokeratins 20 and 7 as biomarkers: usefulness in discriminating primary from metastatic adenocarcinoma. Eur J Cancer. 2002;38:758–63. doi: 10.1016/s0959-8049(02)00008-4. [DOI] [PubMed] [Google Scholar]
- 22.Tomita T. New markers for pancreatic islets and islet cell tumors. Pathol Int. 2002;52:425–32. doi: 10.1046/j.1440-1827.2002.01368.x. [DOI] [PubMed] [Google Scholar]
- 23.Bishop JA, Benjamin H, Cholakh H, Chajut A, Clark DP, Westra WH. Accurate classification of non-small cell lung carcinoma using a novel microRNA-based approach. Clin Cancer Res. 2010;16:610–9. doi: 10.1158/1078-0432.CCR-09-2638. [DOI] [PubMed] [Google Scholar]
- 24.Lebanony D, Benjamin H, Gilad S, et al. Diagnostic assay based on hsa-miR-205 expression distinguishes squamous from non-squamous non-small-cell lung carcinoma. J Clin Oncol. 2009;27:2030–7. doi: 10.1200/JCO.2008.19.4134. [DOI] [PubMed] [Google Scholar]
- 25.Wu H, Zhu S, Mo YY. Suppression of cell growth and invasion by miR-205 in breast cancer. Cell Res. 2009;19:439–48. doi: 10.1038/cr.2009.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tavazoie SF, Alarcon C, Oskarsson T, et al. Endogenous human mi-croRNAs that suppress breast cancer metastasis. Nature. 2008;451:147–52. doi: 10.1038/nature06487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guo C, Sah JF, Beard L, Willson JK, Markowitz SD, Guda K. The noncoding RNA, miR-126, suppresses the growth of neoplastic cells by targeting phosphatidylinositol 3-kinase signaling and is frequently lost in colon cancers. Genes Chromosomes Cancer. 2008;47:939–46. doi: 10.1002/gcc.20596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Diaz R, Silva J, Garcia JM, et al. Deregulated expression of miR-106a predicts survival in human colon cancer patients. Genes Chromosomes Cancer. 2008;47:794–802. doi: 10.1002/gcc.20580. [DOI] [PubMed] [Google Scholar]
- 29.Li X, Zhang Y, Zhang Y, Ding J, Wu K, Fan D. Survival prediction of gastric cancer by a seven-microRNA signature. Gut. 2010;59:579–85. doi: 10.1136/gut.2008.175497. [DOI] [PubMed] [Google Scholar]
- 30.Miko E, Czimmerer Z, Csanky E, et al. Differentially expressed micro-RNAs in small cell lung cancer. Exp Lung Res. 2009;35:646–64. doi: 10.3109/01902140902822312. [DOI] [PubMed] [Google Scholar]
- 31.Mardin WA, Mees ST. MicroRNAs: novel diagnostic and therapeutic tools for pancreatic ductal adenocarcinoma? Ann Surg Oncol. 2009;16:3183–9. doi: 10.1245/s10434-009-0623-1. [DOI] [PubMed] [Google Scholar]
- 32.Mitchell PS, Parkin RK, Kroh EM, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A. 2008;105:10513–8. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Burk U, Schubert J, Wellner U, et al. A reciprocal repression between ZEB1 and members of the miR-200 family promotes EMT and invasion in cancer cells. EMBO Rep. 2008;9:582–9. doi: 10.1038/embor.2008.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gregory PA, Bert AG, Paterson EL, et al. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008;10:593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- 35.Korpal M, Lee ES, Hu G, Kang Y. The miR-200 family inhibits epithelial-mesenchymal transition and cancer cell migration by direct targeting of E-cadherin transcriptional repressors ZEB1 and ZEB2. J Biol Chem. 2008;283:14910–4. doi: 10.1074/jbc.C800074200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Park SM, Gaur AB, Lengyel E, Peter ME. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev. 2008;22:894–907. doi: 10.1101/gad.1640608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hurteau GJ, Carlson JA, Spivack SD, Brock GJ. Overexpression of the microRNA hsa-miR-200c leads to reduced expression of transcription factor 8 and increased expression of E-cadherin. Cancer Res. 2007;67:7972–6. doi: 10.1158/0008-5472.CAN-07-1058. [DOI] [PubMed] [Google Scholar]
- 38.He L, He X, Lowe SW, Hannon GJ. microRNAs join the p53 network—another piece in the tumour-suppression puzzle. Nat Rev Cancer. 2007;7:819–22. doi: 10.1038/nrc2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhu CQ, Shih W, Ling CH, Tsao MS. Immunohistochemical markers of prognosis in non-small cell lung cancer: a review and proposal for a multiphase approach to marker evaluation. J Clin Pathol. 2006;59:790–800. doi: 10.1136/jcp.2005.031351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tili E, Croce CM, Michaille JJ. miR-155: on the crosstalk between inflammation and cancer. Int Rev Immunol. 2009;28:264–84. doi: 10.1080/08830180903093796. [DOI] [PubMed] [Google Scholar]
- 41.Faraoni I, Antonetti FR, Cardone J, Bonmassar E. miR-155 gene: a typical multifunctional microRNA. Biochim Biophys Acta. 2009;1792:497–505. doi: 10.1016/j.bbadis.2009.02.013. [DOI] [PubMed] [Google Scholar]
- 42.Yanaihara N, Caplen N, Bowman E, et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9:189–98. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 43.Greither T, Grochola LF, Udelnow A, Lautenschlager C, Wurl P, Taubert H. Elevated expression of microRNAs 155, 203, 210 and 222 in pancreatic tumors is associated with poorer survival. Int J Cancer. 2010;126:73–80. doi: 10.1002/ijc.24687. [DOI] [PubMed] [Google Scholar]
- 44.Jiang S, Zhang HW, Lu MH, et al. MicroRNA-155 functions as an OncomiR in breast cancer by targeting the suppressor of cytokine signaling 1 gene. Cancer Res. 2010;70:3119–27. doi: 10.1158/0008-5472.CAN-09-4250. [DOI] [PubMed] [Google Scholar]
- 45.Valeri N, Gasparini P, Fabbri M, et al. Modulation of mismatch repair and genomic stability by miR-155. Proc Natl Acad Sci U S A. 2010;107:6982–7. doi: 10.1073/pnas.1002472107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Okada H, Kohanbash G, Lotze MT. MicroRNAs in immune regulation—opportunities for cancer immunotherapy. Int J Biochem Cell Biol. 2010;42:1256–61. doi: 10.1016/j.biocel.2010.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tsitsiou E, Lindsay MA. microRNAs and the immune response. Curr Opin Pharmacol. 2009;9:514–20. doi: 10.1016/j.coph.2009.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thai TH, Calado DP, Casola S, et al. Regulation of the germinal center response by microRNA-155. Science. 2007;316:604–8. doi: 10.1126/science.1141229. [DOI] [PubMed] [Google Scholar]
- 49.Rodriguez A, Vigorito E, Clare S, et al. Requirement of bic/microRNA-155 for normal immune function. Science. 2007;316:608–11. doi: 10.1126/science.1139253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ryu JK, Hong SM, Karikari CA, Hruban RH, Goggins MG, Maitra A. Aberrant MicroRNA-155 expression is an early event in the multistep progression of pancreatic adenocarcinoma. Pancreatology. 2010;10:66–73. doi: 10.1159/000231984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Krichevsky AM, Gabriely G. miR-21: a small multi-faceted RNA. J Cell Mol Med. 2009;13:39–53. doi: 10.1111/j.1582-4934.2008.00556.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Park JK, Lee EJ, Esau C, Schmittgen TD. Antisense inhibition of microRNA-21 or -221 arrests cell cycle, induces apoptosis, and sensitizes the effects of gemcitabine in pancreatic adenocarcinoma. Pancreas. 2009;38:e190–9. doi: 10.1097/MPA.0b013e3181ba82e1. [DOI] [PubMed] [Google Scholar]
- 53.Petri A, Lindow M, Kauppinen S. MicroRNA silencing in primates: towards development of novel therapeutics. Cancer Res. 2009;69:393–5. doi: 10.1158/0008-5472.CAN-08-2749. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.