Abstract
Phosphatase and tensin homologue (PTEN) loss and activation of the Akt-mammalian target of rapamycin (mTOR) pathway increases mRNA translation, increases levels of the antiapoptotic protein FLIPS, and confers resistance to tumor necrosis factor–related apoptosis-inducing ligand (TRAIL)–induced apoptosis in glioblastoma multiforme (GBM). In PTEN-deficient GBM cells, however, the FLIPS protein also exhibited a longer half-life than in PTEN mutant GBM cells, and this longer half-life correlated with decreased FLIPS polyubiquitination. FLIPS half-life in PTEN mutant GBM cells was reduced by exposure to an Akt inhibitor, but not to rapamycin, suggesting the existence of a previously undescribed, mTOR-independent linkage between PTEN and the ubiquitin-dependent control of protein stability. Total levels of the candidate FLIPS E3 ubiquitin ligase atrophin-interacting protein 4 (AIP4) were comparable in PTEN wild-type (WT) and PTEN mutant GBM cells, although in PTEN-deficient cells, AIP4 was maintained in a stable polyubiquitinated state that was less able to associate with FLIPS or with the FLIPS-containing death inducing signal complex. Small interfering RNA–mediated suppression of AIP4 levels in PTEN WT cells decreased FLIPS ubiquitination, prolonged FLIPS half-life, and increased TRAIL resistance. Similarly, the Akt activation that was previously shown to increase TRAIL resistance did not alter AIP4 levels, but increased AIP4 ubiquitination, increased FLIPS steady-state levels, and suppressed FLIPS ubiquitination. These results define the PTEN-Akt-AIP4 pathway as a key regulator of FLIPS ubiquitination, FLIPS stability, and TRAIL sensitivity and also define a novel link between PTEN and the ubiquitin-mediated control of protein stability.
Introduction
Tumor necrosis factor–related apoptosis-inducing ligand (TRAIL) is a proapoptotic peptide that binds to the death receptors DR4/DR5 and induces formation of the death-inducing signaling complex and engagement of the type I extrinsic apoptotic pathway (1, 2). Although many cancer cells are preferentially sensitive to TRAIL-induced apoptosis, the sensitivity of glioblastoma multiforme (GBM), the most aggressive form of brain cancer, is variable, and most short-term primary GBM cultures are TRAIL insensitive (3, 4). Although many factors contribute to TRAIL resistance in GBM, the levels of FLIPS, a truncated splice variant of FLIP, have been shown to be of particular importance (5, 6). Levels of FLIPS in TRAIL-resistant cells have in turn been shown to be regulated by the phosphatase and tensin homologue (PTEN)-Akt-mammalian target of rapamycin (mTOR) pathway, and PTEN loss and Akt activation correlate in vitro, in human GBM xenografts, and in primary human GBM samples with increased FLIPS mRNA translation, high levels of FLIPS expression, and TRAIL resistance (5).
In the process of completing studies related to the PTEN-dependent translational regulation of FLIPS, we noted that the high amounts of FLIPS in PTEN-defective TRAIL resistant GBM cells were also associated with a greatly prolonged half-life of the protein. Protein stability is frequently regulated by ubiquitination, a process by which the small protein ubiquitin is covalently attached to lysine residues in target proteins by E3 ubiquitin ligases (7, 8). Whereas the ligation of a single ubiquitin molecule at one or multiple lysines in the target protein (monoubiquitination) can change target protein activity and cellular location, chain-like addition of multiple ubiquitin molecules to the sites of monoubiquitination (at lysines 48 or 63 in ubiquitin itself; polyubiquitination) leads to alterations in protein sorting and activity (K63 polyubiquitination) or, more critically, to degradation of the targeted protein (K48 polyubiquitination; refs. 9, 10). Although PTEN has not, to date, been reported to regulate the ubiquitination process, the association between PTEN status and FLIPS stability suggested that PTEN may use regulation of ubiquitination, in addition to regulation of protein production, to control the levels of FLIPS and TRAIL sensitivity. In the present study, we explored this possibility.
Materials and Methods
Cell culture and manipulation
Flank xenografts of individual human GBM were established in mice as previously described (5). Freshly resected xenograft material was then obtained from the UCSF Brain Tumor Research Tissue Bank, dissected into small (<1 mm diameter) pieces, passed through a 100 µm pore size tissue culture sieve, and grown on reduced Matrigel-coated dishes (Fisher Scientific). Each resultant culture (e.g., 5, 10, 14) is therefore derived from a unique patient tumor. Transformed mouse astrocytes (TMA) and GBM xenograft cells (5) were cultured in DMEM (H-21) supplemented with 10% fetal bovine serum at 37°C in a 5% CO2 atmosphere. Cells were incubated with vehicle (DMSO), MG132 (10 µmol/L, 24 h), Akt inhibitor III (50 µmol/L, 24 h; Calbiochem), rapamycin (100 nmol/L, 24 h), or TRAIL (800 ng/mL, 24 h; Genentech) and, where indicated, with cycloheximide (100 µg/mL) or vehicle for an additional 24 to 72 h. Pools of retrovirally infected or transfected (Lipofectamine) cells (11) were obtained by selection with neomycin (1 mg/mL, 7 d) or hygromycin B (400 µg/mL, 7 d). Atrophin-interacting protein 4 (AIP4)–targeted small interfering RNA (siRNA; 300 nmol/L; Ambion, identification no. 120674) or scramble control (300nmol/L; silencer negative control #1, Ambion) was transfected, and target protein levels were analyzed 1 to 3 d after exposure to vehicle or cycloheximide.
Immunoprecipitation and Western blot analysis
Control or hemagglutinin (HA)-ubiquitin–expressing cells were lysed, and AIP4, FLIPS, or death receptor 5 (DR5) was immunoprecipitated from lysates by incubation (1 h, 4°C) with the appropriate antibody pre-conjugated to protein-G beads (Santa Cruz Biotechnology). Levels of proteins in the cell lysate and the eluted immunoprecipitates were assessed by Western blot using the appropriate antibody. Levels of ubiquitinated protein were assessed by Western blot using a goat polyclonal antibody against FLIPS (Santa Cruz Biotechnology) or a rabbit polyclonal antibody against AIP4 or HA (Cell Signaling Technology), followed by detection with antigoat IgG or antirabbit IgG (Santa Cruz Biotechnology) using enhanced chemiluminescence. Densitometric measurements were acquired using an AlphaImager 2200 (Alpha Innotech Corporation). Immunoprecipitations carried out using a nonspecific normal rabbit IgG antibody (for AIP4 immunoprecipitations) or a goat IgG antibody (for FLIPS immunoprecipitations) were included as negative controls. Analysis of TRAIL-induced apoptosis was done as previously described (12).
Statistical analysis
Data presented are representative of at least three independent experiments. All statistical analyses were done using the Student t test, with significance defined as P < 0.05 (*).
Results and Discussion
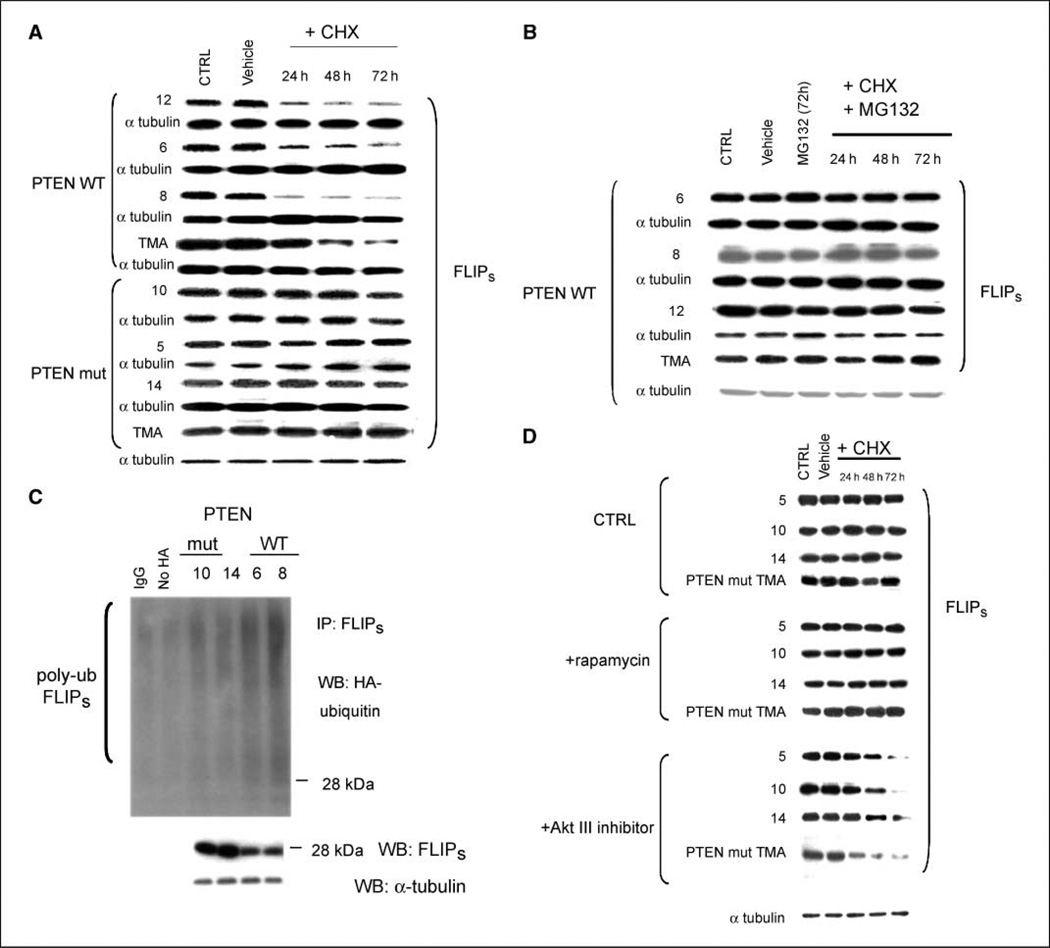
We previously showed that PTEN loss causes a mTOR-dependent increase in the translation of the FLIPS mRNA levels, increased levels of the antiapoptotic protein FLIPS, and increased TRAIL resistance (5). In cycloheximide-treated PTEN-deficient GBM xenograft cells and in cycloheximide-treated TMA derived from PTEN knockout mouse embryos, however, the half-life of preexisting FLIPS was also significantly longer than in corresponding PTEN wild-type (WT) cells (Fig. 1A). Differences in FLIPS protein stability were not due to effects of cycloheximide on FLIPS mRNA levels (Supplementary Fig. S1), nor were they an artifact of the low initial levels of FLIPS in PTEN WT cells because overexpression of FLIPS did not alter FLIPS half-life (Supplementary Fig. S2). MG132-mediated suppression of the proteasome, however, greatly prolonged the half-life of the FLIPS protein in PTENWT human GBM cells and PTEN WT TMA (Fig. 1B), suggesting that FLIPS half-life was regulated by protein degradation in a PTEN-dependent manner. FLIPS immunoprecipitates from PTEN WT cells transiently transfected with a construct encoding HA-ubiquitin and subjected to Western blot analysis using a HA-targeted antibody also contained more >28-kDa FLIPS than FLIPS immunoprecipitates from PTEN mutant cells (Fig. 1C; each ubiquitin added adds 7 kDa of mass), indicative of increased FLIPS polyubiquitination and suggesting that the increased FLIPS stability in PTEN mutant cells resulted from the inability of these cells to mark FLIPS for proteasomal degradation. Because the PTEN-Akt-mTOR pathway regulates translation of FLIPS mRNA (5), we questioned whether the PTEN-associated effects on FLIPS protein stability were mediated by the same pathway. Incubation of PTEN-deficient GBM and TMA cells with an Akt inhibitor suppressed Akt phosphorylation (Supplementary Fig. S3) and significantly shortened FLIPS half-life, although exposure of the cells to concentrations of rapamycin that suppressed S6 phosphorylation (Supplementary Fig. S3) did not (Fig. 1D). These results suggest that the PTEN-Akt pathway is linked to the control of FLIPS stability in a novel, Akt-dependent but mTOR-independent manner.
Figure 1.
The PTEN-Akt pathway controls FLIPS protein stability. Mouse PTEN knockout or WT TMA or PTEN WT or mutant human xenografted GBM cells, or the same cells infected with a blank vector or construct encoding HA-ubiquitin (C), were incubated with vehicle or MG132 (10 µmol/L, 24 h; B), rapamycin (100 nmol/L, 24 h; D), or Akt III inhibitor (50 µmol/L, 24 h; D), after which cells were incubated with either vehicle or cycloheximide (CHX; 100 µg/mL; A, B, and D), lysed at the indicated time points, and analyzed either for levels of FLIPS and α-tubulin (A, B, D) or for the extent of HA ubiquitination in FLIPS immunoprecipitates (C). The α-tubulin blot shown in D is representative of those for all experimental groups.
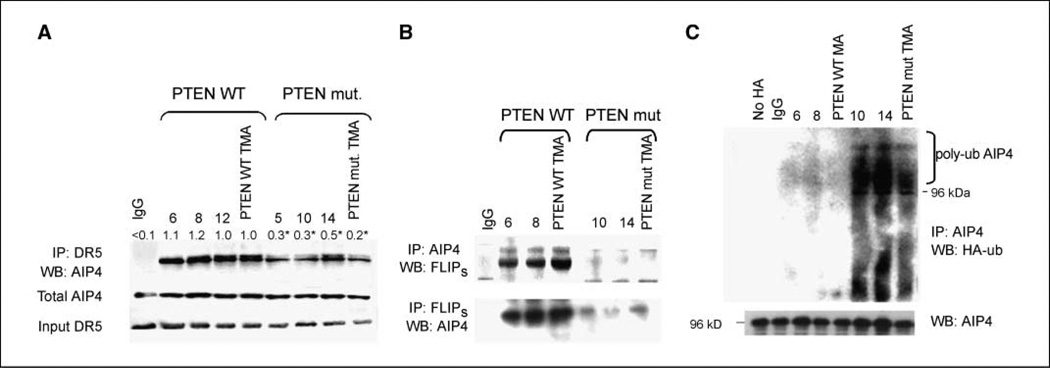
To better define the linkage between PTEN loss, Akt activation, and ubiquitin-mediated regulation of FLIPS stability, we took a candidate approach and ultimately focused on the E3 ubiquitin ligase AIP4 (13). Levels of AIP4 were comparable in PTEN WT and PTEN-deficient cells, although less AIP4 was found in FLIPS immunoprecipitates from PTEN-deficient cells than from PTEN-deficient cells (Fig. 2A). Similarly, DR5 immunoprecipitates of the death-inducing signaling complex (which contains both DR5 and FLIPS; ref. 14) from TRAIL-treated PTEN mutant GBM cells also contained less AIP4 than those from PTEN WT cells (Fig. 2B). The immunoprecipitated AIP4 from PTEN-deficient cells was also maintained in a more highly polyubiquitinated form than that in PTEN WT cells (Fig. 2C), suggesting that PTEN loss and Akt activation lead to the generation of a (perhaps K63-) ubiquitinated AIP4, which, although not less stable, is decreased in its ability to interact with and ubiquitinate FLIPS.
Figure 2.
AIP4 interacts with FLIPS in a PTEN- and ubiquitin-dependent manner. PTEN WT or mutant cells were transfected with a blank vector or a construct encoding HA-ubiquitin (C) or exposed to vehicle (B) or TRAIL (800 ng/mL, 24 h; A). Cells were then lysed and immunoprecipitated using antibodies specific for IgG or for DR5 (A), AIP4 or FLIPS (B), or AIP4 (C). Lysates and immunoprecipitates were then subjected to Western blot analysis of AIP4 and DR5 (A), FLIPS and AIP4 (B), and AIP4 and HA-AIP4 (C) levels.
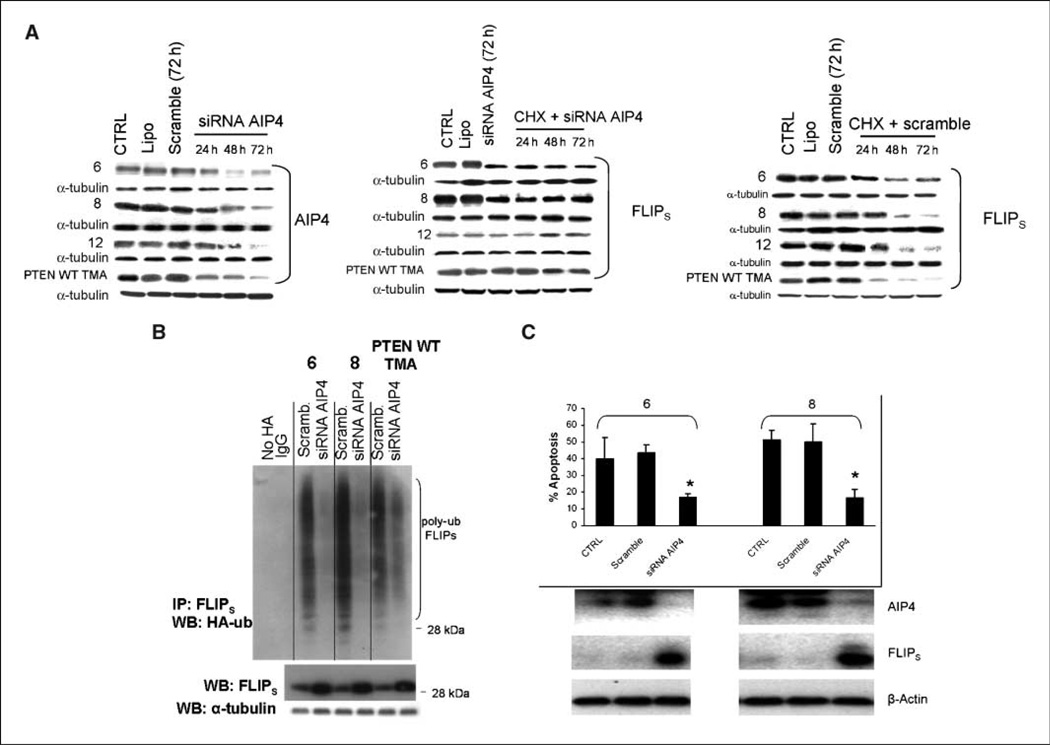
To more definitively link AIP4 to the control of FLIPS stability and TRAIL sensitivity, PTEN WT cells were exposed to siRNA targeting AIP4, after which effects on FLIPS levels, FLIPS ubiquitination, FLIPS stability, and TRAIL sensitivity were monitored. The siRNA-mediated suppression of AIP4 levels in PTEN WT cells (Fig. 3A, left) significantly increased the half-life of FLIPS (Fig. 3A, middle) relative to that noted in cells receiving a nontargeted siRNA (Fig. 3A, right). increased steady-state levels of FLIPS (Fig. 3B, bottom), decreased levels of polyubiquitinated FLIPS (Fig. 3B, top), and increased resistance to TRAIL-induced apoptosis (Fig. 3C). These results show that AIP4 is directly linked to the control of FLIPS ubiquitination and stability, and that AIP4 targets FLIPS for ubiquitination and proteasomal destruction.
Figure 3.
AIP4 is linked to the control of FLIPS ubiquitination and stability and TRAIL sensitivity. PTEN WT or mutant cells were transfected with a scrambled siRNA or siRNA targeting AIP4. Cells were then incubated with vehicle or cycloheximide (A), transfected with a blank vector or a construct encoding HA-ubiquitin (B), or exposed to TRAIL (800 ng/mL, 24 h; C). Cells were then collected at the indicated time points and subjected to Western blot analysis of AIP4, FLIPS, and α-tubulin levels (A and B); immunoprecipitated using antibodies specific for FLIPS and subjected to analysis of extent of HA-ubiquitination of FLIPS (B); or analyzed for extent of TRAIL-induced apoptosis.
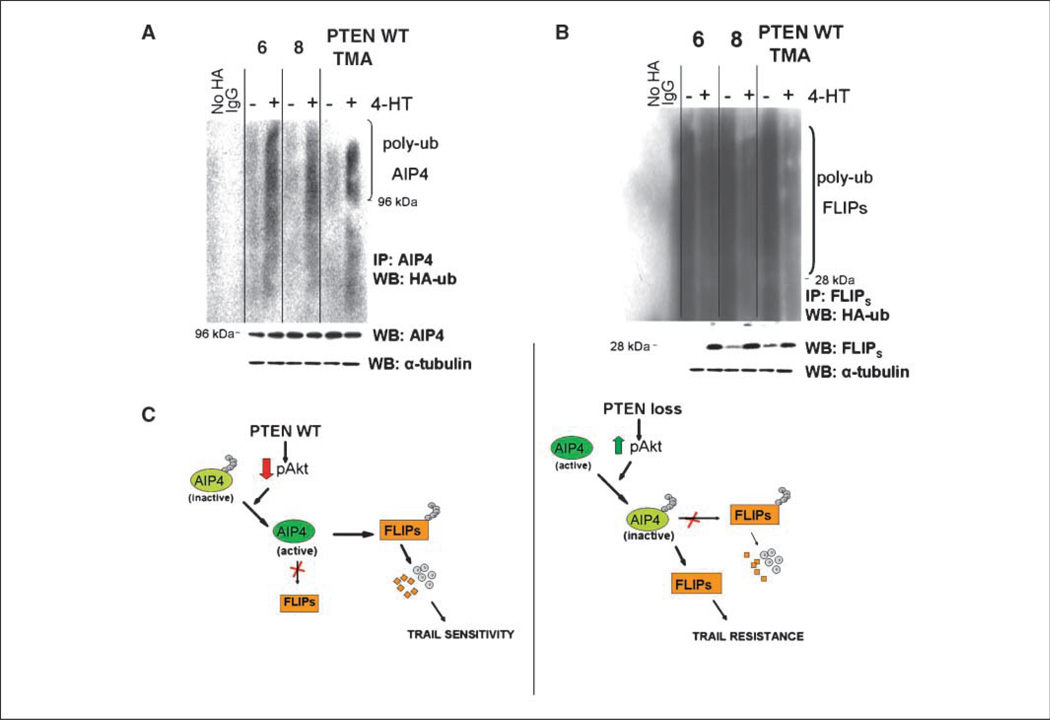
Finally, to formally establish the role of AIP4 in linking the PTEN-Akt pathway to the control of FLIPS stability and TRAIL sensitivity, PTEN WT cells were retrovirally infected with a construct encoding Akt-ER (15) and incubated with vehicle or 4-hydroxytamoxifen (4HT), after which the known TRAIL-desensitizing effects of 4HT-mediated Akt activation were compared with the effects on AIP4 and FLIPS levels and ubiquitination. The 4HT-mediated activation of Akt-ER in PTEN WT cells (Supplementary Fig. S4) that was previously shown to increase resistance to TRAIL-induced apoptosis (5) did not alter AIP4 levels (Fig. 4A, bottom), but significantly increased the extent of AIP4 polyubiquitination (Fig. 4A, top), suppressed levels of FLIPS ubiquitination (Fig. 4B, top), and increased levels of total FLIPS (Fig. 4B, top). Taken as a whole, these results show the existence of a novel pathway that links PTEN to the control of the extrinsic apoptotic pathway.
Figure 4.
AIP4 links the PTEN-Akt pathway to FLIPS ubiquitination and stability. PTEN WT or mutant cells were transfected with a blank construct or a construct encoding a 4HT-inducible Akt-ER (A and B). Cells were then incubated with vehicle or 4HT (100 µmol/L, 24 h), transfected with a blank vector or a construct encoding HA-ubiquitin and lysed, then subjected either to Western blot analysis of FLIPS, AIP4, and α-tubulin or to immunoprecipitation using antibodies specific for AIP4 or FLIPS, and then analyzed for the extent of HA-ubiquitination of AIP4 (A) and FLIPS (B). C, schematic representation of the PTEN-mediated control of FLIPS ubiquitination and TRAIL sensitivity.
The pathway that links PTEN to the control of FLIPS ubiquitination described in this work is presented in Fig. 4C. In this model, PTEN suppresses levels of pAkt (left), which in turn retains AIP4 in a state in which it can interact with and (likely K48-) polyubiquitinate FLIPS. K48-polyubiquitinated FLIPS then undergoes ubiquitin-mediated degradation, leaving the cell susceptible to TRAIL-induced apoptosis. Loss of PTEN function (Fig. 4D, right), in contrast, increases pAkt levels and retains AIP4 in a (perhaps K63-) polyubiquitinated state in which it can no longer interact with and target FLIPS for destruction, thereby allowing FLIPS to accumulate and suppress TRAIL-induced apoptosis. Although the means by which Akt activation enhances AIP4 ubiquitination are not clear, many E3 ligases including AIP4 regulate their own ubiquitination (16–18), and Akt may directly modulate this process. Alternatively, because ubiquitination is a reversible process, Akt may interact with any of a number of deubiquitinases (19), which may in turn tailor the pattern of AIP4 ubiquitination and serve to regulate AIP4 function. Cell type–specific factors that influence AIP4 ubiquitination and/or protein interactions may also help explain the ability of AIP4 to target FLIPS in GBM cells, but not other cell types (20).
In light of the present work, PTEN seems to exert coordinate control on FLIPS, suppressing FLIPS mRNA translation (5) while at the same time contributing to the destabilization of the protein. This coordinate system could therefore allow both immediate resetting of the apoptotic threshold of cells (via rapid regulation of protein stability) and a more long-term restructuring of apoptotic potential (via changes in protein translation), and on inactivation (as in a significant percentage of PTEN-deficient gliomas; ref. 19), it could allow near-complete inhibition of the extrinsic apoptotic response. Whereas pharmacologic manipulation of either arm of this coordinate pathway could effectively resensitive tumor cells to TRAIL, manipulation of both arms may ultimately prove most effective. In a broader sense, the present work also has implications for PTEN function. It seems unlikely that PTEN has evolved the ability to regulate ubiquitination solely to control AIP4 activity, FLIPS stability, and TRAIL sensitivity. Rather, it seems more likely that PTEN may regulate the activity of many of the more than 1,000 E3 ubiquitin ligases in the human cell (8) and, in doing so, may control the stability of a wide range of proteins critical in regulating the transformed phenotype. These ideas are currently being investigated.
Acknowledgments
Grant support: NIH grants CA115638 and CA136774 (R.O. Pieper) and CA097257 (A.T. Parsa and R.O. Pieper).
We thank L. Bin, K. Carraway, and M. McMahon for various construct; S. Baker and G. Bergers for the PTEN WT and knockout TMA; A. Ashkenazi for TRAIL; and D. James for the help with the human GBM xenografts.
Footnotes
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Boldin MP, Goncharov TM, Goltsev YV, Wallach D. Involvement of MACH, a novel MORT1/FADD-interacting proteinase, in Fas/APO-1- and TNF receptor-induced cell death. Cell. 1996;85:803–815. doi: 10.1016/s0092-8674(00)81265-9. [DOI] [PubMed] [Google Scholar]
- 2.Muzio M, Chinnaiyan AM, Kischel FC, et al. FLICE, a novel FADD-homologous ICE/CED-3-like protease, is recruited to the CD95 (Fas/APO-1) death-inducing signaling complex. Cell. 1996;85:817–827. doi: 10.1016/s0092-8674(00)81266-0. [DOI] [PubMed] [Google Scholar]
- 3.Sheridan JP, Marsters SA, Pitti RM, et al. Control of TRAIL-induced apoptosis by a family of signaling and decoy receptors. Science. 1997;277:818–821. doi: 10.1126/science.277.5327.818. [DOI] [PubMed] [Google Scholar]
- 4.Ashley DM, Riffkin CD, Lovric MM, et al. In vitro sensitivity testing of minimally passaged and uncultured gliomas with TRAIL and/or chemotherapy drugs. Br J Cancer. 2008;99:294–304. doi: 10.1038/sj.bjc.6604459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Panner A, James CD, Berger MS, Pieper RO. mTOR controls FLIPS translation and TRAIL sensitivity in glioblastoma multiforme cells. Mol Cell Biol. 2005;25:8809–8823. doi: 10.1128/MCB.25.20.8809-8823.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schultze K, Böck B, Eckert A, et al. Troglitazone sensitizes tumoRcells to TRAIL-induced apoptosis via downregulation of FLIP and survivin. Apoptosis. 2006;11:1503–1512. doi: 10.1007/s10495-006-8896-3. [DOI] [PubMed] [Google Scholar]
- 7.Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 8.Hicke L, Schubert HL, Hill CP. Ubiquitin-binding domains. Nat Rev Mol Cell Biol. 2005;6:610–621. doi: 10.1038/nrm1701. [DOI] [PubMed] [Google Scholar]
- 9.Haglund K, Dikic I. Ubiquitylation and cell signaling. EMBO J. 2005;24:3353–3359. doi: 10.1038/sj.emboj.7600808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hoeller D, Hecker CM, Dikic I. Ubiquitin and ubiquitin-like proteins in cancer pathogenesis. Nat Rev. 2006;6:776–788. doi: 10.1038/nrc1994. [DOI] [PubMed] [Google Scholar]
- 11.Sonoda Y, Ozawa T, Aldape KD, Deen DF, Berger MS, Pieper RO. Akt pathway activation converts anaplastic astrocytoma to glioblastoma multiforme in a human astrocyte model of glioma. Cancer Res. 2001;61:6674–6678. [PubMed] [Google Scholar]
- 12.Panner A, Nakamura JL, Parsa AT, et al. mTOR independent translational control of the extrinsic cell death pathway by RalA. Mol Cell Biol. 2006;26:7345–7357. doi: 10.1128/MCB.00126-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chang L, Kamata H, Solinas G, et al. The E3 ubiquitin ligase itch couples JNK activation to TNFα-induced cell death by inducing c-FLIP(L) turnover. Cell. 2006;124:601–613. doi: 10.1016/j.cell.2006.01.021. [DOI] [PubMed] [Google Scholar]
- 14.Yu JW, Shi Y. FLIP and the death effector domain family. Oncogene. 2008;27:6216–6227. doi: 10.1038/onc.2008.299. [DOI] [PubMed] [Google Scholar]
- 15.Hirose Y, Katayama M, Mirzoeva OK, Berger MS, Pieper RO. Akt activation suppresses Chk2-mediated methylating agent-induced G2 arrest and protects from temozolomide-induced mitotic catastrophe and cellular senescence. Cancer Res. 2005;65:4587–4596. doi: 10.1158/0008-5472.CAN-04-2633. [DOI] [PubMed] [Google Scholar]
- 16.Evans PC, Ovaa H, Hamon M, et al. Zinc-finger protein A20, a regulator of inflammation and cell survival, has de-ubiquitinating activity. Biochem J. 2004;378:727–734. doi: 10.1042/BJ20031377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wilkinson KD. Regulation of ubiquitin-dependent processes by deubiquitinating enzymes. FASEB J. 1997;11:1245–1256. doi: 10.1096/fasebj.11.14.9409543. [DOI] [PubMed] [Google Scholar]
- 18.Scialpi F, Malatesta M, Peschiaroli A, Rossi M, Melino G, Bernassola F. Itch self-polyubiquitylation occurs through lysine-63 linkages. Biochem Pharmacol. 2008;76:1515–1521. doi: 10.1016/j.bcp.2008.07.028. [DOI] [PubMed] [Google Scholar]
- 19.Steck PA, Pershouse MA, Jasser SA, et al. Identification of a candidate tumour suppressor gene, MMAC1, at chromosome 10q23.3 that is mutated in multiple advanced cancers. Nat Genet. 1997;15:356–362. doi: 10.1038/ng0497-356. [DOI] [PubMed] [Google Scholar]
- 20.Chang L, Kamata H, Solinas G, et al. The E3 ubiquitin ligase Itch couples JNK activation to TNFα-induced cell death by inducing c-FLIPL turnover. Cell. 2006;124:601–613. doi: 10.1016/j.cell.2006.01.021. [DOI] [PubMed] [Google Scholar]