SUMMARY
IDH1 and IDH2 mutations occur frequently in gliomas and acute myeloid leukemia, leading to simultaneous loss and gain of activities in the production of α-ketoglutarate (α-KG) and 2-hydroxyglutarate (2-HG), respectively. Here we demonstrate that 2-HG is a competitive inhibitor of multiple α-KG-dependent dioxygenases, including histone demethylases and the TET family of 5-methlycytosine (5mC) hydroxylases. 2-HG occupies the same space as α-KG does in the active site of histone demethylases. Ectopic expression of tumor-derived IDH1 and IDH2 mutants inhibits histone demethylation and 5mC hydroxylation. In glioma, IDH1 mutations are associated with increased histone methylation and decreased 5-hydroxylmethylcytosine (5hmC). Hence, tumor-derived IDH1 and IDH2 mutations reduce α-KG and accumulate an α-KG antagonist, 2-HG, leading to genome-wide histone and DNA methylation alterations.
INTRODUCTION
The NADP+-dependent isocitrate dehydrogenase genes IDH1 and IDH2 are mutated in >75% of low grade gliomas and secondary glioblastoma multiforme (GBM) and ~20% of acute myeloid leukemia (AML) (Mardis et al., 2009; Parsons et al., 2008; Yan et al., 2009). IDH1 mutation has rapidly emerged as a reliable diagnostic and prognostic marker for identifying low grade gliomas and for distinguishing secondary and primary GBM (Ducray et al., 2009). In addition to the highly restricted tumor spectrum, IDH1 and IDH2 mutations identified thus far are heterozygous and produce single amino acid substitutions either at arginine 132 (R132) in IDH1 or corresponding arginine 172 (R172) in IDH2 in glioma and leukemia, or at arginine 140 (R140) in IDH2 in leukemia. Tumor-derived mutations targeting R132 in IDH1 nearly completely abolish its normal catalytic activity of oxidizing and decarboxylating isocitrate (ICT) to produce α-KG, resulting in decreased α-KG and α-KG-dependent prolyl hydroxylase (PHD) activity and leading to an increase in a PHD substrate, HIF-1α (Zhao et al., 2009).
In addition to losing its normal catalytic activity, mutant IDH1 and IDH2 also gained the function of catalyzing the reduction of α-KG to produce D-2-HG (also known as R-2-HG), resulting in an accumulation of D-2-HG in IDH1 or IDH2 mutated gliomas and AML (Dang et al., 2009; Gross et al., 2010; Ward et al., 2010). In IDH1 mutated glioma, D-2-HG accumulated to astonishingly high levels of ~5–35 μmol/g of GBM (Dang et al., 2009), which could be equivalent to 5–35 mM assuming the tissue density of 1 g/ml. Accumulation of a different enantiomer, L-2-HG (also known as S-2-HG), has previously been linked to L-2-hydroxyglutaric aciduria (L-2HGA), a rare metabolic disorder that is caused by a defect in L-2-HG dehydrogenase in mitochondria and is associated with psychomotor retardation, progressive ataxia and leukodystrophy (Rzem et al., 2004; Topcu et al., 2004), and in a few cases increased risk of developing brain tumors (Aghili et al., 2009). Although 2-HG has been proposed to be an oncometabolite, its mechanism of action is not known. 2-HG and α-KG are structurally similar except that the oxygen atom linked to C2 in α-KG is replaced by a hydroxyl group in 2-HG. This similarity suggests the possibility that 2-HG may bind to and function as a competitive inhibitor of α-KG-dependent dioxygenases. Mammalian cells express >60 dioxygenases that utilize α-KG as a cosubstrate (Iyer et al., 2009; Loenarz and Schofield, 2008), including the JmjC domain containing histone demethylases (Tsukada et al., 2006) and recently discovered TET family of 5-methylcytosine (5mC) hydroxylases that convert 5mC to 5-hydroxylmethycytosine (5hmC [Tahiliani et al., 2009]). Many of these α-KG-dependent dioxygenases have a Km for α-KG near physiological concentrations (Clifton et al., 2006; Couture et al., 2007; Loenarz and Schofield, 2008; Simmons et al., 2008), making their activities potentially susceptible to fluctuation of α-KG and/or 2-HG. This study is directed toward understanding how 2-HG functions as an oncometabolite and determining the functional relationship between α-KG reduction and 2-HG elevation.
RESULTS
2-HG Inhibits the Activity of α-KG-Dependent Histone Demethylases In Vitro
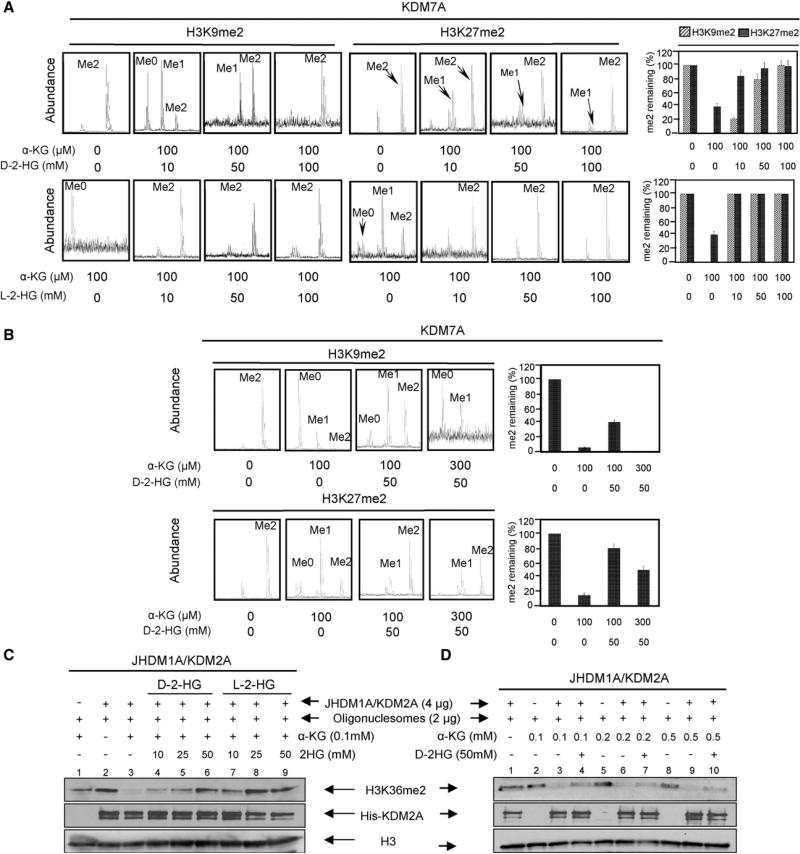
To test the hypothesis that changes in concentrations of α-KG and/or 2-HG may affect the activities of these dioxygenases, we first examined in vitro effect of 2-HG on CeKDM7A, a Caenorhabditis elegans dual specificity histone demethylase that recognizes both dimethylated H3K9 and H3K27, using synthetic methylated H3K9 and H3K27 peptides as substrates. Mass spectrometric analysis demonstrated the removal of one or two methyl groups from both peptides by CeKDM7A in an α-KG-dependent manner (Figure 1A). Addition of 50 mM and 100 mM of D-2-HG resulted in partial and nearly complete inhibition of CeKDM7A, respectively (Figure 1A). The same result was obtained using D-2-HG synthesized from two distinct routes (see Figures S1A and S1B available online), excluding the possibility that the observed inhibition was due to contamination in D-2-HG. We also examined the effect of L-2-HG and found it was more potent than D-2-HG in inhibiting CeKDM7A (Figure 1A).
Figure 1. 2-HG Is a Competitive Inhibitor of α-KG for Histone Demethylases.
(A) 2-HG inhibits Caenorhabditis elegans KDM7A demethylase activity. CeKDM7A activities toward H3K9me2 and H3K27me2 peptides were assayed in the presence of increasing concentrations of either D-2-HG or L-2-HG as indicated. The demethylated products were analyzed by mass spectrometry (left) and mean activity values of duplicated assays, represented by percentage of remaining methylated peptides (right), are shown. Error bars represent ± standard deviation (SD) for triplicate experiments.
(B) α-KG rescues 2-HG inhibition of CeKDM7A demethylase activity. Error bars represent ± SD for triplicate experiments.
(C) 2-HG inhibits human JHDM1A/KDM2A demethylase activity. Purified recombinant JHDM1A demethylase activity was assayed in the presence of various concentrations of D-2-HG and L-2-HG as indicated.
(D) α-KG reverses the inhibitory effect of D-2-HG on JHDM1A. JHDM1A activity was assayed in the presence of 50 mM D-2-HG and various concentrations of α-KG.
See also Figure S1.
To further examine the mode of interaction between α-KG and D-2-HG, we incubated CeKDM7A with a fixed concentration (50 mM) of D-2-HG and increasing amount of α-KG. A partial inhibition of KDM7A toward both H3K9me2 and H3K27me2 peptides was observed in the presence of 50 mM D-2-HG and 100 μM α-KG. Addition of 300 μM α-KG was capable of reversing the inhibition of CeKDM7A by 50 mM D-2-HG (Figure 1B), indicating that D-2-HG is a weak competitive inhibitor against α-KG toward the CeKDM7A demethylase. The lower binding affinity of 2-HG than α-KG is likely due to the hydroxyl moiety being a weaker ligand of the catalytic Fe (II) center than the keto group in α-KG.
We next determined the effect of 2-HG on human histone H3K36 demethylase JHDM1A/KDM2A using nucleosomes as a substrate. Consistent with the results from CeKDM7A, we found that both enantiomers of 2-HG inhibited KDM2A with D-2-HG being less potent than L-2-HG (Figure 1C; Figure S1C). Moreover, increasing α-KG concentrations counteracted D-2-HG inhibition on KDM2A (Figure 1D; Figure S1D).
To confirm the potency of both D- and L-2-HG in competing with α-KG, we determined the inhibition constants (Ki) for D-2-HG, L-2-HG, and N-oxalylglycine (N-OG), an α-KG analog commonly used as a competitive inhibitor of dioxygenases (Cunliffe et al., 1992; Epstein et al., 2001) toward KDM5B/JARID1B/PLU-1, a H3K4 specific demethylase whose alterations have been found in both prostate (Xiang et al., 2007) and breast cancer (Lu et al., 1999; Yamane et al., 2007). These experiments revealed that L-2-HG (Ki = 0.628 ± 0.036 mM) has a similar potency as N-OG (Ki = 0.653 ± 0.128 mM) and is ~17-fold more potent than D-2-HG (Ki = 10.87 ± 1.85 mM) in inhibiting KDM5B/JARID1B/PLU-1 (Figure S1E).
Together, these results demonstrate that both 2-HG enantiomers act as weak antagonists of α-KG to inhibit α-KG-dependent histone demethylases with D-2-HG being significantly less potent than L-2-HG.
2-HG Occupies the Same Space as α-KG Does in the Active Site of CeKDM7A
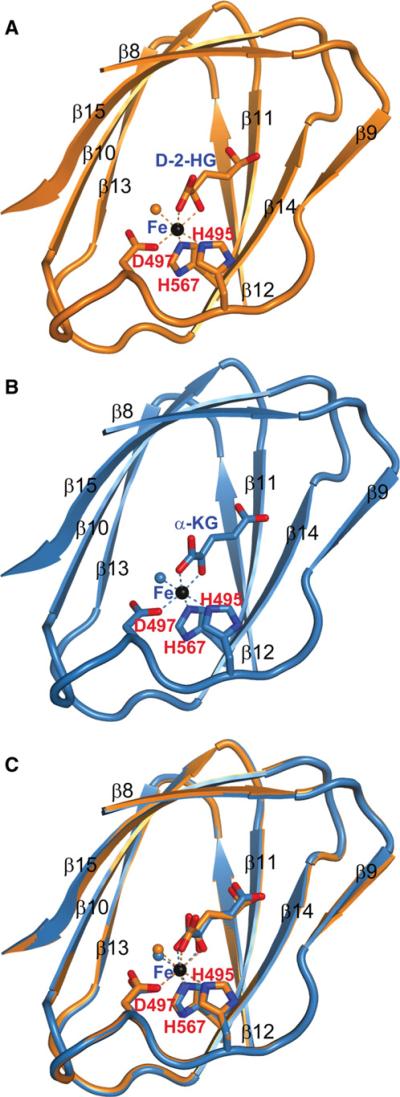
To gain mechanistic insights of 2-HG inhibition, we determined the structure of CeKDM7A bound with D-2-HG at 2.1 Å (Figure 2; Table S1 and Figure S2). Like other JmjC-domain-containing histone demethylase (Chen et al., 2006; Han et al., 2007), the JmjC domain of the catalytic core of CeKDM7A also forms a jelly roll motif with the Fe(II) coordinated by side chains of three highly conserved residues (D497, H495, and H567) within the JmjC domain (Figure 2A; Figure S2A). Notably, D-2-HG binds to the catalytic core in close proximity of Fe (II). We also solved the structure of CeKDM7A bound with α-KG at 2.25 Å (Figure 2B). Comparison of these two structures reveals that D-2-HG adopts a nearly identical orientation as α-KG with one notable difference: whereas the Fe (II) is coordinated by two oxygen atoms in the keto carboxyl end of α-KG, it is coordinated by one oxygen atom and a hydroxyl group in D-2-HG (Figure 2C; Figure S2B). These results provide a structural basis supporting D-2-HG as a competitor of α-KG.
Figure 2. 2-HG and α-KG Bind to the Same Site in Histone Demethylases.
(A) The structure of D-2-HG bound to CeKDM7A JmjC domain. D-2-HG and CeKDM7A are shown in stick and cartoon representation, respectively. Secondary structural elements of CeKDM7A are indicated. Fe (II) is colored in black, Fe (II) coordination is represented by dotted lines and water molecule is shown as orange ball.
(B) The structure of α-KG bound to CeKDM7A JmjC domain, illustrated as in (A).
(C) Superimposition of structures shown in (A) and (B). See also Figure S2.
2-HG Inhibits the Activity of Multiple Histone Demethylases In Vivo
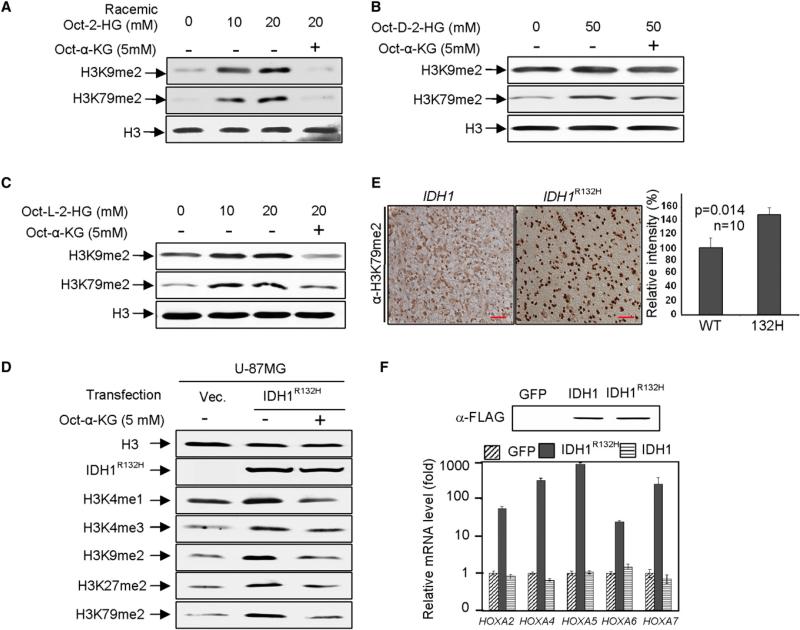
Inhibition of histone demethylases by 2-HG in vitro and binding of 2-HG and α-KG to the same site in the catalytic center of CeKDM7A led us to determine the effect of 2-HG on genome wide histone methylation in vivo. To this end, we synthesized cell-permeable α-KG and racemic octyl-2-HG and verified their structures by NMR (Figures S3A–S3D). Addition of 10 mM octyl-2-HG to the cultured U-87MG cells resulted in a significant accumulation of intracellular 2-HG as determined by GC-MS assay (Figure S3E) and increase of dimethylation on H3K9 and H3K79 by 5- and 10-fold, respectively (Figure 3A; Figure S3F). Addition of cell-permeable octyl-α-KG reversed the increase of both H3K9 and H3K79 dimethylation, providing in vivo evidence supporting the competitive interaction between 2-HG and α-KG. We also synthesized enantiomer specific cell-permeable 2-HG and compared their inhibitory potency. Consistent with in vitro assay, treatment of U-87MG cells with either cell-permeable D- or L-2-HG increased dimethylation on both H3K9 and H3K79 with octyl-D-2-HG being less potent than octyl-L-2-HG (Figures 3B and 3C; Figures S3G and S3H).
Figure 3. Reduced Activity of IDH1 and Elevated 2-HG Increase Genome-Wide Histone Methylations and Alter Gene Expression.
(A–C) Cell-permeable octyl-2-HG increases histone methylation. H3K9me2 and H3K79me2 levels of U-87MG cells treated with racemic octyl-2-HG (A), octyl-D-2-HG (B), and octyl- L-2-HG (C) were analyzed by western blotting.
(D) Histone methylation increased by IDH1R132H overexpression can be rescued by addition of cell-permeable octyl-α-KG. Specified histone methylation levels of U-87MG cells expressing IDH1R132H were analyzed by western blotting.
(E) Elevated H3K79 dimethylation in IDH1R132H gliomas. IDH1 wild-type or heterozygous for R132H glioma samples were subjected to IHC analysis for H3K79me2 methylation. Scale bars represent 50 μm. Shown are representative IHC results (left) and mean values of IHC quantification (right). Error bars represent ± SD for triplicate experiments. Complete results of all 20 samples are presented in Figure S3K.
(F) Reduction of IDH1 activity activates HOXA genes. HOXA mRNA levels were analyzed by qRT-PCR in U-87MG cells after forced expression of wild-type or R132H mutant IDH1. Error bars represent ± SD for triplicate experiments. See also Figure S3.
R132H Mutation of IDH1 Alters Histone Methylation in Human Glioma Cells and Tumor Samples
We next ectopically expressed IDH1R132H in U-87MG cells and determined the levels of multiple histone methylation markers. Comparing with cells expressing empty vector, the ectopic expression of wild-type increased α-KG by 20% in U-87MG cells, ectopic expression of IDH1R132H mutant resulted in a near 60% reduction of α-KG by 60% and <20-fold increase in D-2-HG (Figure S3I). A visible increase in H3K4 monomethylation, H3K27 dimethylation, H3K4 trimethylation, H3K9 dimethylation, and H3K79 dimethylation was observed (Figure 3D; Figure S3J). Addition of cell-permeable octyl-α-KG restored histone demethylation. Together, these results indicate that in addition to CeKDM7A and KDM2A, 2-HG and mutant IDH1 inhibit wide range of histone demethylases, including those involved in the demethylation of H3K4, H3K9, H3K27, and H3K79, and both inhibitions by 2-HG and IDH1 mutant can be reversed by the addition of cell-permeable α-KG.
These results led us to determine whether IDH1 mutation could affect histone methylation in primary tumors. We analyzed H3K79 dimethylation in a panel of 20 human glioma samples, 10 containing wild-type IDH1 and 10 bearing mutated IDH1 (R132H) (see Table S2). H3K79 dimethylation levels were found to be significantly elevated in glioma samples that harbor IDH1 mutation compared to tumor samples that are similar grade but have wild-type IDH1 (Figure 3E; Figure S3K). To further substantiate this result, we determined the expression of several HOXA genes whose increased expression is associated with increased H3K79 dimethylation in MLL-rearranged mouse leukemia and human AML patients (Krivtsov et al., 2008). qRT-PCR analysis demonstrated that the expression of these HOXA genes was increased in cells with forced expression of the IDH1R132H (Figure 3F). Collectively, these results demonstrate that either expression of mutant IDH1 or increase of 2-HG results in an inhibition of histone demethylases in vivo.
Reduction of IDH Results in an Inhibition of Histone Demethylases
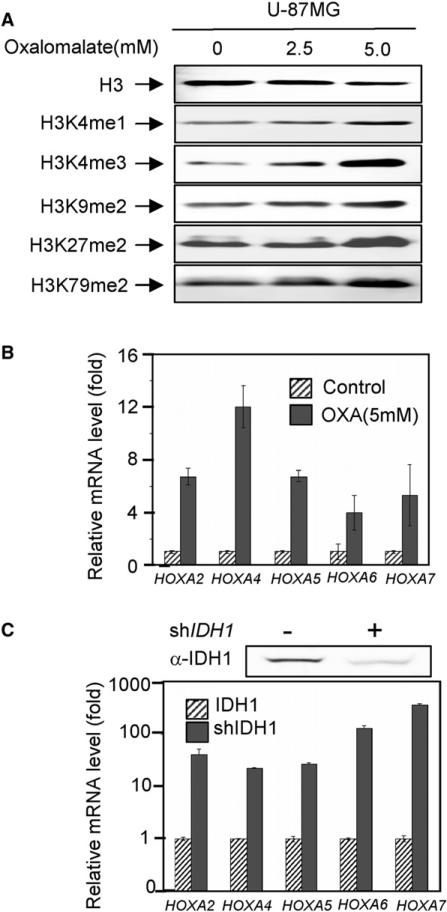
Given the previous observations that mutations in IDH1 or IDH2 cause both α-KG reduction and 2-HG accumulation and the current finding that 2-HG acts as an antagonist of α-KG in vitro, we sought to determine whether reducing the activity of IDH1 and IDH2 could cause similar increase in histone methylation. To this end, we treated cells with oxalomalate, a competitive inhibitor of IDH1 and IDH2 that would decrease both cytoplasmic and mitochondrial α-KG. We found that this treatment led to a dose-dependent increase of trimethylation of H3K4, dimethylation at H3K9, H3K27, and H3K79, and a modest increase in H3K4 mono-methylation (Figure 4A; Figure S4). The differences between different histone demethylases in their responses to oxalomalate treatment probably reflect their different affinities toward α-KG. To further support the above observation, we also determined the expression of the same panel of HOXA genes (see Figure 3F) and found that expression of these HOXA genes was increased in cells treated with oxalomalate (Figure 4B) as well as in cells depleted for IDH1 by shRNA knockdown (Figure 4C). Similar conclusion was also obtained with two additional α-KG-dependent dioxygenases (see below). As both oxalomalate treatment and IDH1 knockdown reduced α-KG without 2-HG accumulation, these results indicate that inhibition of IDH1 could cause similar effect as 2-HG treatment, providing additional evidence supporting a competitive mode between α-KG and 2-HG.
Figure 4. INHIBITION of IDH1 Reduces Histone Demethylase Activities In Vivo.
(A) Inhibition of IDH1 activity increases histone methylation. Specified histone methylation levels in U-87MG cells treated with increasing concentrations of oxalomalate were analyzed by western blotting.
(B and C) Reduction of IDH1 activity activates HOXA genes. HOXA mRNA levels were analyzed by qRT-PCR in U-87MG cells after knocking down IDH1 (C), and treatment of oxalomalate (B). Error bars represent ± SD for triplicate experiments. See also Figure S4.
α-KG-dependent prolyl hydroxylases and collagen prolyl-4-hydroxylases are inhibited by reduced IDH1 activity or R132H
In addition to histone demethylases, mammalian cells express a large number of dioxygenases that also utilize α-KG as a key substrate (Iyer et al., 2009; Loenarz and Schofield, 2008). To determine how broadly IDH1 mutations affect α-KG-dependent dioxygenases, we investigated the effect of reduced IDH1 activity and increased 2-HG on two additional α-KG-dependent dioxygenases; prolyl hydroxylases (PHDs) and collagen prolyl-4-hydroxylase (C-P4H).
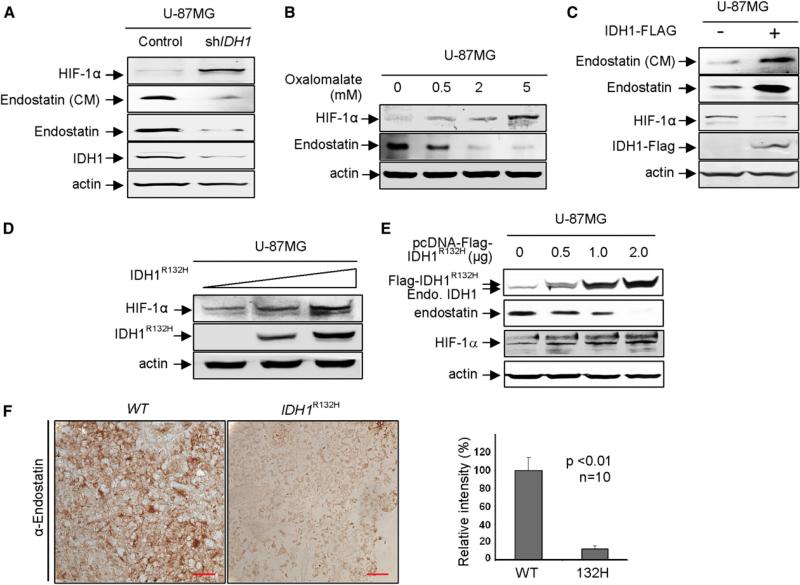
We first determined the effect of reduced function of IDH1 on the levels of HIF-1α and endostatin. HIF-1α is a transcription factor whose function is linked to metabolism, angiogenesis and tumorigenesis, and whose protein level is downregulated under normoxic conditions by PHD-mediated hydroxylation and subsequent hydroxylation-targeted ubiquitination (Bruick and McKnight, 2001; Epstein et al., 2001; Ivan et al., 2001; Jaakkola et al., 2001). Endostatin is a 20-kDa secretory peptide implicated in inhibiting angiogenesis and tumor growth and its production is catalyzed by C-P4H (Krane, 2008; O'Reilly et al., 1997). We found that knocking down IDH1 by shRNA increased HIF-1α and decreased endostatin (Figure 5A). Likewise, inhibition of endogenous IDH1 by oxalomalate treatment similarly increased HIF-1α and decreased endostatin (Figure 5B). Conversely, ectopic expression of wild-type IDH1 decreased HIF-1α and increased endostatin (Figure 5C). In addition to PHDs, the stability and steady-state levels of HIF-1α protein are also regulated by FIH-1 (factor inhibiting HIF-1α) (Lando et al., 2002; Mahon et al., 2001), an asparaginyl hydrogenase whose activity is dependent on α-KG and could therefore also be affected by reduced function of IDH1. Unlike knocking down PHD2 that caused an evident increase of HIF-1α, knocking down FIH-1 in 293T cells did not appreciably affect the steady-state level of HIF-1α (Figure S5A). This result indicates that, at least in 293T cells, FIH-1 does not appear to play a major role in HIF-1α degradation, a notion that is consistent with a recent study in Fih-1 deletion mice showing that FIH-1 plays little or no discernible role in altering HIF stability and function (Zhang et al., 2010). We therefore conclude that the increase of HIF-1α in cells with reduced function of IDH1 is largely caused by the impairment of PHDs.
Figure 5. IDH1 Function Supports the Activity of α-KG-Dependent Dioxygenases In Vivo.
(A–E) The effects of reducing or increasing IDH1 function on two α-KG-dependent dioxygenases, PHDs and C-P4H, were examined in U-87MG cells after knockdown IDH1 (A), treatment of oxalomalate, a competitive inhibitor of IDH1 (B), overexpression of wild-type IDH1 (C), and tumor-derived IDH1R132H mutant (D and E). The protein levels of HIF-1α, endostatin, and ectopically and endogenously expressed IDH1 were determined by western blotting.
(F) Decreased endostatin in IDH1R132H gliomas. IDH1 wild-type or heterozygous for R132H glioma samples were subjected to IHC analysis for endostatin. Shown are representative IHC results (left) and mean values of IHC quantification (right). Scale bars represent 50 μm. Error bars represent ± SD for triplicate experiments. Complete results of all 20 samples are presented in Figure S5C. See also Figure S5.
To determine how tumor-derived IDH1 mutants affect these two dioxygenases, we ectopically expressed IDH1R132H in U-87MG cells and determined the steady-state levels of both endostatin and HIF-1α proteins. We observed a dose-dependent increase of HIF-1α and decrease of endostatin with increasing expression of IDH1R132H (Figures 5D and 5E). Ectopic expression of IDH1R132H did not significantly affect the levels of collagen XVIII mRNA (Figure S5B), supporting the notion that decreased endostatin protein on IDH1R132H expression is due to inhibition of C-P4H. An evident endostatin decrease and HIF-1α increase were seen in cells in which IDH1R132H is expressed at a level similar to that of endogenous wild-type IDH1 (lane 2, Figure 5E), excluding the possibility that observed changes in the activity of both PHDs and C-P4H dioxygenases are caused by the grossly overexpression of IDH1 mutant.
We also performed immunohistochemical staining of endostatin in the same panel of 10 IDH1 wild-type and 10 IDH1 mutated gliomas (Table S2). We found that tumors with the R132H mutation expressed significantly less endostatin than those containing wild-type IDH1 (Figure 5F; Figure S5C). Together, these results demonstrate that the activities of α-KG-dependent PHDs and C-P4H are supported by the function of IDH1 and impaired by tumor-derived mutant IDH1.
2-HG Treatment Inhibits PHD and C-P4H
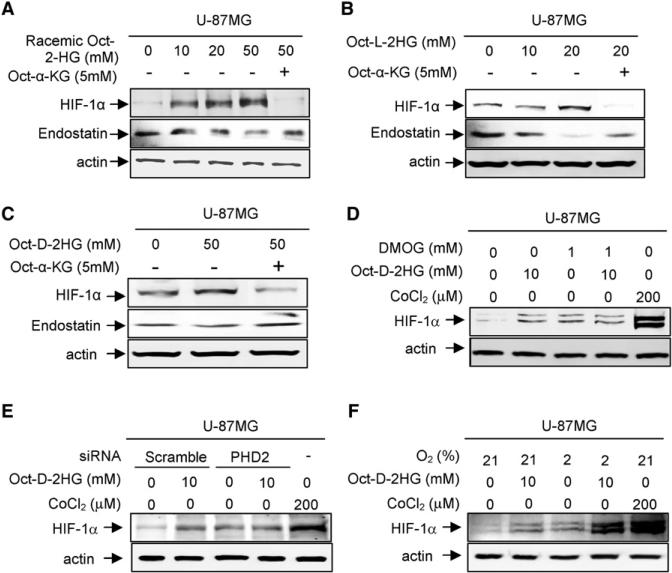
We next examined more directly the effect of 2-HG on these two dioxygenases in cells. Treatment of U-87MG cells with either cell-permeable racemic mixture or enantiomer-specific 2-HG increased HIF-1α and decreased endostatin (Figures 6A–6C). Similar increase of HIF-1α was also observed in 293T cells treated with octyl-D-2-HG (Figure S6A). Consistent with the inhibition of histone demethylases, octyl-D-2-HG exhibited less potent inhibition on both dioxygenases than octyl-L-2-HG and addition of cell-permeable octyl-α-KG substantially suppressed the effect of 2-HG.
Figure 6. 2-HG Inhibits the Activity of α-KG-Dependent Dioxygenases In Vivo.
(A–C) Cell-permeable 2-HG increases HIF-1α and decreases endostatin. U-87MG cells were treated with racemic octyl-2-HG (A), octyl-L-2-HG (B), and octyl-D-2-HG (C). The steady state levels of endostatin and HIF-1α proteins were determined by direct western blotting.
(D) DMOG treatment abolishes 2-HG effect on HIF-1α induction.
(E) PHD2 knock down abolishes 2-HG effect on HIF-1α induction.
(F) 2-HG treatment further induces HIF-1α in hypoxic U-87MG cells. See also Figure S6.
Consistent with the hypothesis that the effect of 2-HG in stabilizing HIF-1α protein in vivo is achieved by antagonizing the binding of α-KG to PHD2, treatment of cells with a cell-permeable NOG, dimethyloxalylglycine (DMOG), or knocking down PHD2 both resulted in an accumulation of HIF-1α protein and activation of several HIF-1α target genes (Figures 6D and 6E; Figures S6B–S6E), but treatment of cells with the combination of cell-permeable octyl-D-2-HG and either DMOG or siPHD2 did not cause a further increase of HIF-1α. Treatment of cells with CoCl2, a mimetic of hypoxia and a chemical inducer of HIF-1α, accumulated HIF-1α to a level significantly higher than either DMOG or siPHD2 treatment, excluding the possibility that the lack of further HIF-1α accumulation by the octyl D-2-HG in DMOG- or siPHD2-treated cells is because that HIF-1α has already accumulated to the maximal level by DMOG or siPHD2 treatment. Further supporting the notion that 2-HG induces HIF-1α through an inhibition of PHD2 by competing off α-KG, addition of octyl-2-HG to hypoxic cells in which the activity of PHD is reduced by the decrease cosubstrate oxygen, but not impaired α-KG binding, resulted in further HIF-1α accumulation (Figure 6F).
Ectopic Expression of Tumor-Derived IDH1 and IDH2 Mutants Reduces TET-Catalyzed 5hmC Production
Very recently, a class of α-KG-dependent dioxygenase, the TET family of proteins, was discovered that catalyze the conversion of 5-methylcytosine (5mC) to 5-hydroxylmethycytosine (5hmC) (Iyer et al., 2009; Tahiliani et al., 2009). Mammalian cells express three TET genes, TET1, TET2, and TET3. Although the founding member, TET1, was initially identified as the gene fused with mixed-lineage leukemia (MLL) in chromosomal ten-eleven-translocation in rare cases of AML and suspected to be an onco-gene (Lorsbach et al., 2003; Ono et al., 2002), it was recently discovered that TET2 is mutationally inactivated in ~15% of myeloid cancers, including 22% of AML (Delhommeau et al., 2009; Langemeijer et al., 2009). Given the dependence of TET catalytic activity on α-KG, we set forth to determine whether IDH1 and IDH2 mutations and D-2-HG would affect TET activities and DNA cytosine hydroxymethylation and methylation.
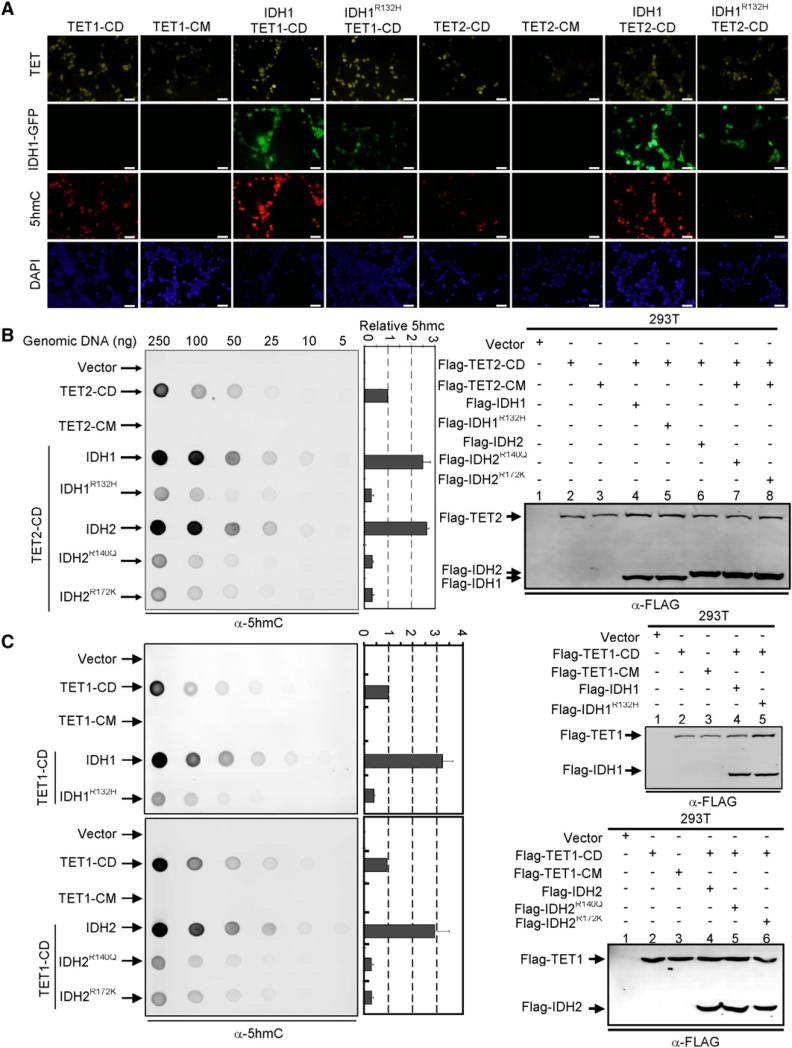
The level of 5hmC in most cells is very low, but is substantially increased in cells transiently transfected with plasmids expressing the wild-type catalytic domain of TET (TET1-CD and TET2-CD) protein that can be readily detected by immunofluorescence using an antibody specifically recognizing 5hmC (Ito et al., 2010; Tahiliani et al., 2009) (Figure 7A). Immunofluorescence with the anti-5hmC antibody revealed that coexpression of wild-type IDH1 with TET1-CD or TET2-CD caused a significant increase of 5hmC signal (Figure 7A), suggesting that the concentration of α-KG is a rate-limiting factor of TET2 catalyzed hydroxylation of 5-methylcytosine in TET1-overexpressing cells. Notably, cotransfection of TET1-CD or TET2-CD with IDH1R132H reduced the 5hmC signal to a barely detectable low level. Essentially the same result was also obtained for IDH2. Both TET1- and TET2-catalyzed 5mC-to-5hmC conversions were substantially increased by the coexpression with wild-type IDH2, but nearly completely inhibited by the coexpression of either IDH2R140Q or IDH2R172K mutants (Figures S7A and S7B). Together, these results demonstrate an inhibitory effect of mutant IDH1 and IDH2 toward the hydroxylase activity of the TET family proteins.
Figure 7. Tumor-Derived IDH1 and IDH2 Mutants Inhibit the 5hmC Production by TET1 and TET2.
(A) HEK293 cells were transiently transfected with plasmids expressing indicated proteins. Thirty-six to forty hours after the transfection, cells were fixed and stained with antibodies specific to Flag to determine the expression of TET protein, to 5hmC to determine the levels of 5hmC, and to DAPI to view the cell nuclei or visualized for green fluorescence to determine the expression of IDH1 proteins. Scale bars represent 50 μm. Additional results on the inhibition of TET1 and TET2 function by IDH2 mutants are presented in Figures S7A and S7B.
(B and C) HEK293 cells were transiently transfected as described in (A). Thirty-six to forty hours after the transfection, genomic DNAs were isolated from the transfected cells, spotted on nitrocellulose membranes and immunoblotted with an antibody specific to 5hmC. Quantification of 5hmC was calculated from three independent assays. The expression of individual proteins was determined by immunoblotting as shown in the right. One representative quantification of 5hmC level determined from the assays using 50 ng genomic DNA is included and the rest of the quantifications are presented in Figures S7C–S7E. Error bars represent ± SD for triplicate experiments. See also Figure S7.
To confirm this result, we isolated genomic DNA from HEK293T cells transiently transfected with TET1 or TET2 individually or in combination with either wild-type or mutant IDH1 and IDH2, and determined 5hmC levels by dot-blot that allowed for more quantitative measurement than the immunofluorescence. These experiments demonstrate that ectopic expression of the wild-type, but not the mutant of TET1 or TET2, resulted in high levels of 5hmC in the cells comparing with cells transfected with control vector (Figures 7B and 7C). Coexpression with wild-type IDH1 or IDH2 caused a significant increase of 5hmC. For example, in the assays using 50 ng genomic DNA, TET2-catalyzed 5hmC production was increased by 149% and 166% by the coexpression of wild-type IDH1 or IDH2, respectively. In contrast, coexpression of TET2-CD with three tumor-derived mutants all caused a substantial decrease of TET2-mediated 5hmC production, resulting in a 70% reduction of 5hmC by the coexpression of IDH1R132H, 66% reduction by both IDH2R140Q and IDH2R172K (Figure 7B; Figure S7C). Virtually the same result was also obtained for TET1-catalyzed 5hmC production that was increased by 222% and 203% by the coexpression of wild-type IDH1 or IDH2, respectively, but reduced by 60%, 69%, and 68% by the coexpression of IDH1R132H, IDH2R140Q, and IDH2R172K, respectively (Figure 7C; Figures S7D and S7E).
2-HG Inhibits the Activity of TET 5-Methylcytosine Hydroxylases
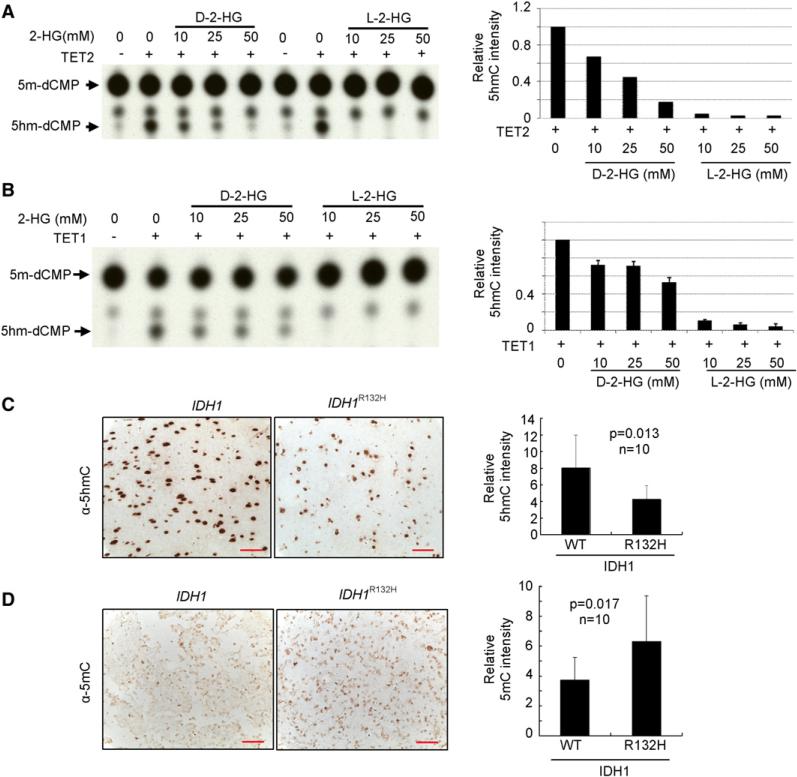
We next tested whether 2-HG may function as an inhibitor of α-KG-dependent TET hydroxylases. We carried out in vitro enzymatic assay to test this possibility using purified Flag-tagged mouse TET catalytic domains as well as their corresponding catalytic mutants following previous published procedure (Ito et al., 2010). Omission of α-KG completely abolished the activity of TET in catalyzing the conversion of 5mC to 5hmC, confirming the dependence of TET activity on α-KG (Figures 8A and 8B). In the presence of 0.1 mM of α-KG, addition of 10 mM D-2-HG resulted in a partial (33%) inhibition of TET2 and addition of 50 mM D-2-HG resulted in more inhibition (83%) of TET2 (Figure 8A). D-2-HG exhibited a less pronounced inhibitory effect toward TET1, reducing the 5hmC production by 28% and 47%, respectively, when 10 and 50 mM D-2-HG were added to the reaction (Figure 8B). These results indicate that D-2-HG is a weak inhibitor of TET hydroxylases. We also examined the effect of L-2-HG and found it was more potent than D-2-HG in inhibiting both TET2 and TET1 with 10 mM L-2-HG capable of inhibiting most of TET1 and TET2 activity in the presence of 0.1 mM α-KG.
Figure 8. Glioma Harboring Mutant IDH1 Have Decreased 5hmC.
(A and B) Recombinant catalytic domains and corresponding catalytic mutants of murine TET2 (A) or TET1 (B) protein was produced and purified from insect Sf9 cells, and incubated with double-stranded DNA oligonucleotides containing a fully methylated MspI site in the presence of Fe (II) and α-KG (0.1 mM). Recovered oligonucleotides were digested with MspI, end labeled with T4 DNA kinase, digested with DNaseI and phosphodiesterase, and analyzed by TLC. Error bars represent ± SD for triplicate experiments.
(C) IDH1 wild-type or heterozygous for R132H glioma samples were subjected to IHC analysis for 5hmC. Shown are representative IHC results (left) and mean values of IHC quantification (right). Scale bars represent 50 μm. Error bars represent ± SD for triplicate experiments. Complete results of all 20 samples are presented in Figure S8A.
(D) IDH1 wild-type or heterozygous for R132H glioma samples were subjected to IHC analysis for 5mC. Scale bars represent 50 μm. Shown are representative IHC results (left) and mean values of IHC quantification (right). Error bars represent ± SD for triplicate experiments. Complete results of all 20 samples are presented in Figure S8B.
In normal mouse brain, 5hmC constitutes a surprisingly high level of total nucleotides in many different cell types, ranging from 0.2% in granule cells to 0.6% in Purkinje cells (Kriaucionis and Heintz, 2009; Munzel et al., 2010). It is currently not clear the scope and level of 5hmC in primary tumors. We analyzed 5hmC by immunohistochemistry (IHC) in the same panel of 20 human glioma samples (Table S2). Notably, 5hmC was readily detectable by IHC in all glioma samples we have examined regardless of their IDH1 status. Glioma samples harboring a mutant IDH1, however, accumulate significantly lower 5hmC than those containing wild-type IDH1. The average relative intensity of 5hmC was 8.04 ± 3.97 in glioma with wild-type IDH1 and reduced to and 4.27 ± 1.62 (p = 0.013) in IDH1-mutated gliomas (Figure 8C; Figure S8A). This result provides in vivo evidence in human tumor supporting the conclusion that IDH1 mutations reduce the levels of 5hmC.
Promoter DNA methylation profiling analysis has recently revealed that a subset of glioblastoma, proneural subgroup previously identified by gene expression profiling and exhibiting features of increased PDGRF gene expression and IDH1 mutation (Verhaak et al., 2010), displays hypermethylation at a large number of loci (Noushmehr et al., 2010), suggesting a potential link between IDH1 mutation and increased DNA methylation. Given that TET-catalyzed 5hmC production from 5mC, we therefore determined 5mC by immunohistochemistry in the same panel of 20 human glioma samples. In contrast to 5hmC levels, gliomas with IDH1 mutations accumulate significantly higher 5mC than those containing wild-type IDH1. The average relative intensity of 5mC was 3.75 ± 1.49 in glioma with wild-type and increased to 6.33 ± 3.02 (p = 0.017) in gliomas harboring a mutant IDH1 (Figure 8D; Figure S8B). This result provides in vivo evidence in human tumor that IDH1 mutations reduce the levels of 5hmC with an associated increase of 5mC.
DISCUSSION
D-2-HG Is a Weak Antagonist of α-KG
In this study, we provide evidence that D-2-HG is an antagonist of α-KG and inhibits multiple α-KG-dependent dioxygenases. Notably, both enantiomers of 2-HG, especially D-2-HG that accumulated in IDH1 and IDH2 mutated tumors, are weak inhibitors in competing with α-KG. In the presence of 0.1 mM α-KG, 10 mM D-2-HG exhibits a clear, but only a partial inhibitory effect toward KDM7A histone demethylase and TET methylcytosine hydroxylases. In other words, as much as 100-fold molar excess of D-2-HG over α-KG is needed to cause a significant inhibitory effect toward α-KG-dependent dioxygenases. This weak activity may be explained by the fact that the hydroxyl moiety in D-2-HG is a weaker ligand of the catalytic Fe (II) center than the keto group in α-KG. We argue that the requirement for such a high concentration of D-2-HG to inhibit this class of enzymes, although seemingly supraphysiological, is pathophysiologically relevant to 2-HG-mediated tumorigenesis. IDH1-mutated gliomas accumulated D-2-HG to a very high level, between 5–35 μmol/g with an average of 15.48 μmol/g (Dang et al., 2009). The concentrations of α-KG in the same cohort of IDH1-mutated glioma are between 0.016 to 0.085 μmol/g with an average of 0.0415 μmol/g indicating that the ratio of D-2-HG and α-KG would reach an average of 373-fold. Our findings therefore support the notion that although D-2-HG may not play a significant role in the regulation of α-KG-dependent dioxygenases in normal cells because of its low level, it could play an important role under pathological conditions in tumor cells expressing mutated IDH1 or IDH2. The weak activity of D-2-HG also helps to explain why tumor cells need to accumulate and can tolerate such a high level of D-2-HG.
Does Reduced α-KG Sensitize α-KG-Dependent Enzymes to the Inhibition by 2-HG?
Joining the loss-of-function mutations targeting fumarate hydratase (FH) and different subunits of succinate dehydrogenase (SDH) (Baysal et al., 2000; Tomlinson et al., 2002), the discovery of IDH1 and IDH2 mutations in human cancers further highlights the direct link between metabolic dysregulation and tumorigenesis. Mutations targeting FH and SDH lead to similar increase in the PHD substrate, HIF-1α (Isaacs et al., 2005; Pollard et al., 2005; Selak et al., 2005). Furthermore, succinate, the substrate of SDH that is accumulated in cells with reduced or inactivation of SDH, has been shown to directly inhibit multiple α-KG-dependent enzymes, including histone demethylases (Smith et al., 2007). Thus, a common feature of the mutations in these three metabolic enzymes is the reduced activity of α-KG-dependent dioxygenases, either indirectly by the accumulation of competitive inhibitors, namely fumarate, succinate, and 2-HG, or directly through the reduction of IDH1 and IDH2 catalytic activity in α-KG production. It will be interesting to determine whether histone and DNA methylations are similarly altered in SDH and FH mutated tumor cells.
We previously showed that mutant IDH1 retains its ability to associate with wild-type subunit and forms a catalytic inactive heterodimer, leading to the dominant inhibition of wild-type IDH1 (Zhao et al., 2009). This model is supported by the recent crystal structural analysis of wild-type:mutant IDH1 heterodimer showing that the R132H mutation hinders the conformational changes from the initial ICT-binding state to the pretransition state (Yang et al., 2010). In other words, the IDH1 enzyme activity in IDH1-mutated tumor cells is reduced to ~25% of normal cells. Production of D-2-HG by the mutant IDH1 and IDH2 homodimers raises a question of relative contribution of α-KG reduction and D-2-HG accumulation to tumorigenesis. We showed here that treatment of cells with either 2-HG enantiomer or inhibition of IDH1 and/or IDH2 activity alone in the absence of D-2-HG accumulation causes similar effects of reducing the activity of multiple α-KG-dependent dioxygenases. It is possible that IDH1 or IDH2 mutations alone do not reduce cellular level of α-KG sufficiently low to have a significant tumorigenic consequence, but nonetheless sensitize α-KG-dependent dioxygenases to the inhibitory effect by the large amounts of 2-HG accumulated in the cell. Hence, α-KG reduction and D-2-HG accumulation cooperatively contribute to tumorigenesis. The findings that a cell-permeable α-KG derivative can reverse the effects of 2-HG inhibition and that D-2-HG is a weak competitive inhibitor suggest that drugs mimicking α-KG merit exploration as a therapy for tumors that harbor an IDH1 or IDH2 mutation, either alone or in combination with inhibitors targeting mutant IDH1 and IDH2 to reduce the 2-HG production.
IDH1 and IDH2 Mutations Alter Histone and DNA Methylation
Notable among the α-KG-dependent dioxygenases whose activities are reduced by IDH1 and IDH2 mutations are the families of histone demethylases and TET 5-methylcytosine hydroxylases, leading to genome-wide increase and decrease of histone methylation and 5-hydroxymethylcytosine, respectively. We demonstrate that both histone demethylases and TET hydroxylases are inhibited by D-2-HG. The exact function of hydroxylation of 5-methylcytosine, whether representing a distinct transcriptional regulation or an intermediate of DNA demethylation, is currently unclear but has been speculated to play a role in epigenetic control. In the case of histone demethylases, multiple histone modification markers are affected. Recently, it was reported that expression of IDH1R132H suppresses TET2 activity and the mutations of IDH1 and IDH2 genes occur in a mutual exclusive manner with that of TET2 gene in AML (Figueroa et al., 2010). Our finding that expression of mutants IDH1 or IDH2 and D-2-HG inhibit the activity of TET2 in catalyzing 5mc-to-5hmC conversion not only support, but also provide biochemical basis for the mutual exclusivity of IDH1/2 and TET2 gene mutations. Together with the previous finding that IDH1-mutated gliomas display hypermethylation at a large number of loci (Noushmehr et al., 2010), these findings suggest that instead of altering the expression of a single or a few specific genes, mutations in IDH1 and IDH2 may change the expression of potentially large number of genes. Given that both mutations of IDH1 and TET2 have been reported to occur at a very early stage during glioma and leukemia development (Delhommeau et al., 2009; Langemeijer et al., 2009; Watanabe et al., 2009), alteration of histone and DNA methylations resulting from IDH1 and IDH2 mutations may contribute to tumorigenesis through altering epigenetic control and potentially the fates of stem or progenitor cells.
EXPERIMENTAL PROCEDURES
Cell Culture, Transfection, Western Blotting, and Chemical Treatments
Procedures for cell culture, transfection, and western blotting are described in the Supplemental Experimental Procedures. Treatments of cells with cell-permeable α-KG or 2-HG were carried out by adding the octyl-α-KG ester or octyl-2-HG ester to the culture medium to a final concentration 4–6 hr before harvesting as indicated. Dimethyloxalylglycine (DMOG; Frontier Scientific) treatments were carried out by adding DMOG to the culture medium to a final concentration of 1 mM, 6 hr before harvesting. When both DMOG and octyl-2-HG were used for cell treatment, DMOG (1 mM final concentration) was added to the medium 2 hr before the addition of octyl-2-HG (10 mM final concentration), and cells were harvested 4 hr after octyl-2-HG was added. CoCl2 treatments were carried out by adding CoCl2 to the medium to a final concentration of 200 μM, 6 hr before harvesting.
Crystallography Study
Purification and crystallization of His-tagged CeKDM7A were described in Supplemental Experimental Procedures. Data sets were collected on beamline BL17U at Shanghai Synchrotron Radiation Facility (SSRF). All data were processed using the program HKL2000. The crystals of CeKDM7A in complex with D-2-HG contain two molecules in one asymmetric unit. The structures of CeKDM7A with D-2-HG or α-KG were determined by molecular replacement method using CeKDM7A structure and the models were manually built with COOT. All refinements were performed using the refinement module phenix.refine of PHENIX package. The model quality was checked with the PROCHECK program, which showed good stereochemistry according to the Ramachandran plot for all structures. The structure superimposition was performed with COOT. All structure figures were generated by PyMol.
Enzymatic Assays
To assay human JHDM1A/KDM2A demethylase activity toward H3K36me2, His tagged JHDM1A was first obtained by transforming pET28a-JHDM1A into Escherichia coli BL21 and protein expression was induced by addition of 1 mM IPTG at 30° C when cell density reaches 0.5 OD600 units. Cells were lysed by sonication and Ni-NTA agarose was used to purify His-JHDM1A fusion proteins. Histone demethylase assay was carried out by incubating 2 μg oligonucleosomes, 4 μg purified His-JHDM1A, and/or 10–50 mM L- or D-2-HG in histone demethylation buffer [50 mM HEPES (pH 8.0), 625 μM Fe(NH4)2(SO4)2, 0.1–0.5 mM α-KG, 2 mM ascorbate] at 37° C for 2–3 hr and the reactions were stopped by the addition of SDS loading buffer and subsequently analyzed by western blotting using anti-H3K36me2 antibody. To measure CeKDM7A demethylase activity toward H3K9me2 and H3K27me2, two synthetic dimethylated peptides H3K9me2 [ARTKQTARK (me2)STGGKA] and H3K27me2 [QLATKAARK (me2)SAPAS] were used as substrates. Demethylase assays were carried out in the presence of 10 μg enzyme, 1 μg peptide in 20 μl buffer 20 mM Tris-HCl (pH 7.5), 150 mM NaCl, 50 μM (NH4)2Fe(SO4)2, 100 μM α-KG, 2 mM Vc, 10 mM PMSF for 3 hr. The demethylation reaction mixture was desalted by passing through a C18 ZipTip (Millipore). To examine the inhibitory effect of 2-HG, various concentrations of 2-HG were incubated with KDM7A briefly before adding other reaction mixtures. The samples were analyzed by a MALDI-TOF/TOF mass spectrometer.
Three different assays were performed for TET-catalyzed 5mC-to-5hmC conversion. For in vivo assay using immunofluorescence, plasmids expressing Flag tagged TET proteins were either singularly transfected or cotransfected with plasmid expressing GFP-IDH fusion protein to HEK293T cell. Thirty-six to forty hours after transfection, cells were fixed with 4% paraformaldehyde in PBS for 15 min and then washed with cold PBS. Cells were permeabilized with 0.4% Triton X-100 in PBS for 15 min. For 5mC and 5hmC staining, DNA was denatured with 2 N HCl for 30 min. and then neutralized with 100 mM Tris-HCl (pH 8.5) for 10 min. After washing three times with PBS, samples were blocked for 1 hr with 5% goat serum, 1% BSA 0.05% Tween20 in PBS. The primary antibodies were added and incubated at 4° C overnight. After washing three times with PBS, cells were incubated with secondary antibodies and DAPI for 30 min, followed by triple wash with PBS. Images were recorded using Olympus immunofluorescence microscope DP71 and Olympus software. Antibodies to FLAG (Sigma-Aldrich, diluted at 1:1000), 5-hydroxymethylcytosine (Active Motif, Carlsbad, CA, dilution at 1:4000), 5-methylcytosine (clone 162 33 D3, Calbiochem, San Diego, CA, diluted at 1:500 1000) were purchased commercially.
For dot-blot assays, we followed the procedures described previously (Ito et al., 2010). Briefly, genomic DNA was spotted on nitrocellulose membranes. The membrane was baked at 80° C and then blocked with 5% skimmed milk in TBST for 1 hr, followed by the incubation with the anti-5hmC antibody overnight at 4° C and HRP-conjugated anti-rabbit IgG secondary antibody for 1 hr at room temperature. After washing three times with TBST, the membrane was treated with ECL and scanned by a Typhoon scanner. The quantification of dot-blot was done by Image-Quanta software (GE Healthcare).
In vitro TET-catalyzed 5mC-to-5hmC conversion was assayed as described previously (Ito et al., 2010) and described in detail in the Supplemental Experimental Procedures. Briefly, 5 μg purified proteins were incubated with 0.5 μg double-stranded oligonucleotide substrates in 50 mM HEPES (pH 8), 75 μM Fe(NH4)2(SO4)2, 2 mM ascorbate, and 0.1 mM α-KG with or without a various amount of 2-HG for 3 hr at 37° C. Oligonucleotide substrates were purified and then digested with MspI. 5′-end of the digested DNA was treated with calf alkaline phosphatase and labeled with [γ-32P]ATP and T4 polynucleotide kinase. Labeled fragments were ethanol-precipitated and digested with 10 μg of DNase I and 10 μg Phosphodiesterase I in the presence of 15 mM MgCl2, 2 mM CaCl2 at 37° C. One microliter digestion product was spotted on a PEI-cellulose TLC plate (Merck) and separated in an isobutyric acid/water/ammonium hydroxide (66:20:2) running buffer.
Clinic Samples, Immunohistochemistry, and Histopathological Analyses
All glioma samples were acquired from Affiliated Huashan Hospital of Fudan University. A physician or nurse practitioner obtained informed consent from the patients. The procedures related to human subjects were approved by Ethic Committee of the Institutes of Biomedical Sciences (IBS), Fudan University. Primers used for amplifying and sequencing IDH1 are described in Supplemental Experimental Procedures. Tissue sections from glioma samples were deparaffinized twice by xylene and then hydrated. Hydrogen peroxide (0.6%) was used to eliminate endogenous peroxidase activity. The sections were blocked with goat serum in TBS for 30 min. Sections were then incubated with either anti-endostatin antibody (Abcam) or anti-H3K79me2 antibody (Abcam) at 1:500 dilution overnight at 4° C. Secondary antibody was then applied and incubated at 37° C for 1 hr. Sections were developed with DAB kit and stopped with water. To quantify the positive area of staining in samples, five fields from each sample were randomly selected and microscopically examined by a pathologist and a neurobiologist in a double-blind manner. Images were captured using a charge-coupled device camera and analyzed using Motic Images Advanced software (version 3.2, Motic China Group Co. Ltd). Cells showing either cytoplasmic or nuclear signals (brown) were counted as positive. The average positive area was calculated by dividing the positively stained areas over total area.
Supplementary Material
Significance.
IDH1 and IDH2 genes are mutated in >75% of low grade gliomas and secondary glioblastoma multiforme (GBM) and in ~20% of acute myeloid leukemia (AML). Two distinct alterations are caused by the tumor-derived mutations in IDH1 or IDH2: loss of its normal catalytic activity in the production of α-ketoglutarate (α-KG) and gain of the catalytic activity to produce 2-hydroxygulatrate (2-HG). We report here that 2-HG is a competitive inhibitor of multiple α-KG-dependent dioxygenases, including histone demethylases, prolyl hydroxylases, collagen prolyl-4-hydroxylase, and the TET family of 5-methylcytosine hydroxylases, and results in genome-wide change of histone and DNA methylation.
ACKNOWLEDGMENTS
We thank members of Fudan MCB laboratory for their valuable inputs and support throughout this study, Nara Lee for helping for assaying human histone demethylase activity, staff members of Beamline BL17U at Shanghai Synchrotron Radiation Facility (SSRF) for data collection, and Sara Jackson for the critical reading of the manuscript. This work is supported by the 985 Program from the Chinese Ministry of Education; state key development programs of basic research of China (2009CB918401, 2009CB918600), Chinese National Science Foundation grants (30971485/C0706, 30600112, 30871255, 11079016), National Basic Research Program of China (2011CB965300, 2011CB910600), Shanghai key basic research projects, China (09JC1402300), and grants from NIH (K.L.G. and Y.X.).
Footnotes
ACCESSION NUMBERS
The atomic coordinates of the complex have been deposited in the Protein Data Bank with accession number 3PUQ (CeKDM7A in complex with α-KG) and 3PUR (CeKDM7A in complex with D-2-HG).
SUPPLEMENTAL INFORMATION
Supplemental Information includes Supplemental Experimental Procedures, eight figures, and two tables and can be found with this article online at doi:10.1016/j.ccr.2010.12.014.
REFERENCES
- Aghili M, Zahedi F, Rafiee E. Hydroxyglutaric aciduria and malignant brain tumor: a case report and literature review. J. Neurooncol. 2009;91:233–236. doi: 10.1007/s11060-008-9706-2. [DOI] [PubMed] [Google Scholar]
- Baysal BE, Ferrell RE, Willett-Brozick JE, Lawrence EC, Myssiorek D, Bosch A, van der Mey A, Taschner PE, Rubinstein WS, Myers EN, et al. Mutations in SDHD, a mitochondrial complex II gene, in hereditary paraganglioma. Science. 2000;287:848–851. doi: 10.1126/science.287.5454.848. [DOI] [PubMed] [Google Scholar]
- Bruick RK, McKnight SL. A conserved family of prolyl-4-hydroxylases that modify HIF. Science. 2001;294:1337–1340. doi: 10.1126/science.1066373. [DOI] [PubMed] [Google Scholar]
- Chen Z, Zang J, Whetstine J, Hong X, Davrazou F, Kutateladze TG, Simpson M, Mao Q, Pan CH, Dai S, et al. Structural insights into histone demethylation by JMJD2 family members. Cell. 2006;125:691–702. doi: 10.1016/j.cell.2006.04.024. [DOI] [PubMed] [Google Scholar]
- Clifton IJ, McDonough MA, Ehrismann D, Kershaw NJ, Granatino N, Schofield CJ. Structural studies on 2-oxoglutarate oxygenases and related double-stranded beta-helix fold proteins. J. Inorg. Biochem. 2006;100:644–669. doi: 10.1016/j.jinorgbio.2006.01.024. [DOI] [PubMed] [Google Scholar]
- Couture JF, Collazo E, Ortiz-Tello PA, Brunzelle JS, Trievel RC. Specificity and mechanism of JMJD2A, a trimethyllysine-specific histone demethylase. Nat. Struct. Mol. Biol. 2007;14:689–695. doi: 10.1038/nsmb1273. [DOI] [PubMed] [Google Scholar]
- Cunliffe CJ, Franklin TJ, Hales NJ, Hill GB. Novel inhibitors of prolyl 4-hydroxylase. 3. Inhibition by the substrate analogue N-oxaloglycine and its derivatives. J. Med. Chem. 1992;35:2652–2658. doi: 10.1021/jm00092a016. [DOI] [PubMed] [Google Scholar]
- Dang L, White DW, Gross S, Bennett BD, Bittinger MA, Driggers EM, Fantin VR, Jang HG, Jin S, Keenan MC, et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009;462:739–744. doi: 10.1038/nature08617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delhommeau F, Dupont S, Della Valle V, James C, Trannoy S, Masse A, Kosmider O, Le Couedic JP, Robert F, Alberdi A, et al. Mutation in TET2 in myeloid cancers. N. Engl. J. Med. 2009;360:2289–2301. doi: 10.1056/NEJMoa0810069. [DOI] [PubMed] [Google Scholar]
- Ducray F, El Hallani S, Idbaih A. Diagnostic and prognostic markers in gliomas. Curr. Opin. Oncol. 2009;21:537–542. doi: 10.1097/CCO.0b013e32833065a7. [DOI] [PubMed] [Google Scholar]
- Epstein AC, Gleadle JM, McNeill LA, Hewitson KS, O'Rourke J, Mole DR, Mukherji M, Metzen E, Wilson MI, Dhanda A, et al. C. elegans EGL-9 and mammalian homologs define a family of dioxygenases that regulate HIF by prolyl hydroxylation. Cell. 2001;107:43–54. doi: 10.1016/s0092-8674(01)00507-4. [DOI] [PubMed] [Google Scholar]
- Figueroa ME, Abdel-Wahab O, Lu C, Ward PS, Patel J, Shih A, Li Y, Bhagwat N, Vasanthakumar A, Fernandez HF, et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell. 2010;18:553–567. doi: 10.1016/j.ccr.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross S, Cairns RA, Minden MD, Driggers EM, Bittinger MA, Jang HG, Sasaki M, Jin S, Schenkein DP, Su SM, et al. Cancer-associated metabolite 2-hydroxyglutarate accumulates in acute myelogenous leukemia with isocitrate dehydrogenase 1 and 2 mutations. J. Exp. Med. 2010;207:339–344. doi: 10.1084/jem.20092506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Z, Liu P, Gu L, Zhang Y, Li H, Chen S, Chai J. Structural basis for histone demethylation by JHDM1. Frontier Science. 2007;1:52–61. [Google Scholar]
- Isaacs JS, Jung YJ, Mole DR, Lee S, Torres-Cabala C, Chung YL, Merino M, Trepel J, Zbar B, Toro J, et al. HIF overexpression correlates with Biallelic loss of fumarate hydratase in renal cancer: novel role of fumarate in regulation of HIF stability. Cancer Cell. 2005;8:143–153. doi: 10.1016/j.ccr.2005.06.017. [DOI] [PubMed] [Google Scholar]
- Ito S, D'Alessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010;466:1129–1133. doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivan M, Kondo K, Yang H, Kim W, Valiando J, Ohh M, Salic A, Asara JM, Lane WS, Kaelin WG., Jr. HIFalpha targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science. 2001;292:464–468. doi: 10.1126/science.1059817. [DOI] [PubMed] [Google Scholar]
- Iyer LM, Tahiliani M, Rao A, Aravind L. Prediction of novel families of enzymes involved in oxidative and other complex modifications of bases in nucleic acids. Cell Cycle. 2009;8:1698–1710. doi: 10.4161/cc.8.11.8580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaakkola P, Mole DR, Tian YM, Wilson MI, Gielbert J, Gaskell SJ, Kriegsheim A, Hebestreit HF, Mukherji M, Schofield CJ, et al. Targeting of HIF-alpha to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science. 2001;292:468–472. doi: 10.1126/science.1059796. [DOI] [PubMed] [Google Scholar]
- Krane SM. The importance of proline residues in the structure, stability and susceptibility to proteolytic degradation of collagens. Amino Acids. 2008;35:703–710. doi: 10.1007/s00726-008-0073-2. [DOI] [PubMed] [Google Scholar]
- Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324:929–930. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krivtsov AV, Feng Z, Lemieux ME, Faber J, Vempati S, Sinha AU, Xia X, Jesneck J, Bracken AP, Silverman LB, et al. H3K79 methylation profiles define murine and human MLL-AF4 leukemias. Cancer Cell. 2008;14:355–368. doi: 10.1016/j.ccr.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lando D, Peet DJ, Gorman JJ, Whelan DA, Whitelaw ML, Bruick RK. FIH-1 is an asparaginyl hydroxylase enzyme that regulates the transcriptional activity of hypoxia-inducible factor. Genes Dev. 2002;16:1466–1471. doi: 10.1101/gad.991402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langemeijer SM, Kuiper RP, Berends M, Knops R, Aslanyan MG, Massop M, Stevens-Linders E, van Hoogen P, van Kessel AG, Raymakers RA, et al. Acquired mutations in TET2 are common in myelodysplastic syndromes. Nat. Genet. 2009;41:838–842. doi: 10.1038/ng.391. [DOI] [PubMed] [Google Scholar]
- Loenarz C, Schofield CJ. Expanding chemical biology of 2-oxoglutarate oxygenases. Nat. Chem. Biol. 2008;4:152–156. doi: 10.1038/nchembio0308-152. [DOI] [PubMed] [Google Scholar]
- Lorsbach RB, Moore J, Mathew S, Raimondi SC, Mukatira ST, Downing JR. TET1, a member of a novel protein family, is fused to MLL in acute myeloid leukemia containing the t(10;11)(q22;q23). Leukemia. 2003;17:637–641. doi: 10.1038/sj.leu.2402834. [DOI] [PubMed] [Google Scholar]
- Lu PJ, Sundquist K, Baeckstrom D, Poulsom R, Hanby A, Meier-Ewert S, Jones T, Mitchell M, Pitha-Rowe P, Freemont P, Taylor-Papadimitriou J. A novel gene (PLU-1) containing highly conserved putative DNA/chromatin binding motifs is specifically up-regulated in breast cancer. J. Biol. Chem. 1999;274:15633–15645. doi: 10.1074/jbc.274.22.15633. [DOI] [PubMed] [Google Scholar]
- Mahon PC, Hirota K, Semenza GL. FIH-1: a novel protein that interacts with HIF-1alpha and VHL to mediate repression of HIF-1 transcriptional activity. Genes Dev. 2001;15:2675–2686. doi: 10.1101/gad.924501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardis ER, Ding L, Dooling DJ, Larson DE, McLellan MD, Chen K, Koboldt DC, Fulton RS, Delehaunty KD, McGrath SD, et al. Recurring mutations found by sequencing an acute myeloid leukemia genome. N. Engl. J. Med. 2009;361:1058–1066. doi: 10.1056/NEJMoa0903840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munzel M, Globisch D, Bruckl T, Wagner M, Welzmiller V, Michalakis S, Muller M, Biel M, Carell T. Quantification of the sixth DNA base hydroxymethylcytosine in the brain. Angew. Chem. Int. Ed. Engl. 2010;49:5375–5377. doi: 10.1002/anie.201002033. [DOI] [PubMed] [Google Scholar]
- Noushmehr H, Weisenberger DJ, Diefes K, Phillips HS, Pujara K, Berman BP, Pan F, Pelloski CE, Sulman EP, Bhat KP, et al. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell. 2010;17:510–522. doi: 10.1016/j.ccr.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Reilly MS, Boehm T, Shing Y, Fukai N, Vasios G, Lane WS, Flynn E, Birkhead JR, Olsen BR, Folkman J. Endostatin: an endogenous inhibitor of angiogenesis and tumor growth. Cell. 1997;88:277–285. doi: 10.1016/s0092-8674(00)81848-6. [DOI] [PubMed] [Google Scholar]
- Ono R, Taki T, Taketani T, Taniwaki M, Kobayashi H, Hayashi Y. LCX, leukemia-associated protein with a CXXC domain, is fused to MLL in acute myeloid leukemia with trilineage dysplasia having t(10;11) (q22;q23). Cancer Res. 2002;62:4075–4080. [PubMed] [Google Scholar]
- Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Siu IM, Gallia GL, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard PJ, Briere JJ, Alam NA, Barwell J, Barclay E, Wortham NC, Hunt T, Mitchell M, Olpin S, Moat SJ, et al. Accumulation of Krebs cycle intermediates and over-expression of HIF1alpha in tumours which result from germline FH and SDH mutations. Hum. Mol. Genet. 2005;14:2231–2239. doi: 10.1093/hmg/ddi227. [DOI] [PubMed] [Google Scholar]
- Rzem R, Veiga-da-Cunha M, Noel G, Goffette S, Nassogne MC, Tabarki B, Scholler C, Marquardt T, Vikkula M, Van Schaftingen E. A gene encoding a putative FAD-dependent L-2-hydroxyglutarate dehydrogenase is mutated in L-2-hydroxyglutaric aciduria. Proc. Natl. Acad. Sci. USA. 2004;101:16849–16854. doi: 10.1073/pnas.0404840101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selak MA, Armour SM, MacKenzie ED, Boulahbel H, Watson DG, Mansfield KD, Pan Y, Simon MC, Thompson CB, Gottlieb E. Succinate links TCA cycle dysfunction to oncogenesis by inhibiting HIF-alpha prolyl hydroxylase. Cancer Cell. 2005;7:77–85. doi: 10.1016/j.ccr.2004.11.022. [DOI] [PubMed] [Google Scholar]
- Simmons JM, Muller TA, Hausinger RP. Fe(II)/alpha-ketoglutarate hydroxylases involved in nucleobase, nucleoside, nucleotide, and chromatin metabolism. Dalton Trans. 2008:5132–5142. doi: 10.1039/b803512a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith EH, Janknecht R, Maher LJ., 3rd Succinate inhibition of alpha-ketoglutarate-dependent enzymes in a yeast model of paraganglioma. Hum. Mol. Genet. 2007;16:3136–3148. doi: 10.1093/hmg/ddm275. [DOI] [PubMed] [Google Scholar]
- Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, Rao A. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlinson IP, Alam NA, Rowan AJ, Barclay E, Jaeger EE, Kelsell D, Leigh I, Gorman P, Lamlum H, Rahman S, et al. Germline mutations in FH predispose to dominantly inherited uterine fibroids, skin leiomyomata and papillary renal cell cancer. Nat. Genet. 2002;30:406–410. doi: 10.1038/ng849. [DOI] [PubMed] [Google Scholar]
- Topcu M, Jobard F, Halliez S, Coskun T, Yalcinkayal C, Gerceker FO, Wanders RJ, Prud'homme JF, Lathrop M, Ozguc M, Fischer J. L-2-Hydroxyglutaric aciduria: identification of a mutant gene C14orf160, localized on chromosome 14q22.1. Hum. Mol. Genet. 2004;13:2803–2811. doi: 10.1093/hmg/ddh300. [DOI] [PubMed] [Google Scholar]
- Tsukada Y, Fang J, Erdjument-Bromage H, Warren ME, Borchers CH, Tempst P, Zhang Y. Histone demethylation by a family of JmjC domain-containing proteins. Nature. 2006;439:811–816. doi: 10.1038/nature04433. [DOI] [PubMed] [Google Scholar]
- Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, Miller CR, Ding L, Golub T, Mesirov JP, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward PS, Patel J, Wise DR, Abdel-Wahab O, Bennett BD, Coller HA, Cross JR, Fantin VR, Hedvat CV, Perl AE, et al. The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting alpha-ketoglutarate to 2-hydroxyglutarate. Cancer Cell. 2010;17:225–234. doi: 10.1016/j.ccr.2010.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Nobusawa S, Kleihues P, Ohgaki H. IDH1 mutations are early events in the development of astrocytomas and oligodendrogliomas. Am. J. Pathol. 2009;174:1149–1153. doi: 10.2353/ajpath.2009.080958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Y, Zhu Z, Han G, Ye X, Xu B, Peng Z, Ma Y, Yu Y, Lin H, Chen AP, Chen CD. JARID1B is a histone H3 lysine 4 demethylase up-regulated in prostate cancer. Proc. Natl. Acad. Sci. USA. 2007;104:19226–19231. doi: 10.1073/pnas.0700735104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamane K, Tateishi K, Klose RJ, Fang J, Fabrizio LA, Erdjument-Bromage H, Taylor-Papadimitriou J, Tempst P, Zhang Y. PLU-1 is an H3K4 demethylase involved in transcriptional repression and breast cancer cell proliferation. Mol. Cell. 2007;25:801–812. doi: 10.1016/j.molcel.2007.03.001. [DOI] [PubMed] [Google Scholar]
- Yan H, Parsons DW, Jin G, McLendon R, Rasheed BA, Yuan W, Kos I, Batinic-Haberle I, Jones S, Riggins GJ, et al. IDH1 and IDH2 mutations in gliomas. N. Engl. J. Med. 2009;360:765–773. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang B, Zhong C, Peng Y, Lai Z, Ding J. Molecular mechanisms of “off-on switch” of activities of human IDH1 by tumor-associated mutation R132H. Cell Res. 2010;20:1188–1200. doi: 10.1038/cr.2010.145. [DOI] [PubMed] [Google Scholar]
- Zhang N, Fu Z, Linke S, Chicher J, Gorman JJ, Visk D, Haddad GG, Poellinger L, Peet DJ, Powell F, Johnson RS. The asparaginyl hydroxylase factor inhibiting HIF-1alpha is an essential regulator of metabolism. Cell Metab. 2010;11:364–378. doi: 10.1016/j.cmet.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S, Lin Y, Xu W, Jiang W, Zha Z, Wang P, Yu W, Li Z, Gong L, Peng Y, et al. Glioma-derived mutations in IDH1 dominantly inhibit IDH1 catalytic activity and induce HIF-1alpha. Science. 2009;324:261–265. doi: 10.1126/science.1170944. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.