Abstract
Human embryonic (ESC) and induced pluripotent stem cells (iPSC) present exciting opportunities for studying development and in vitro disease modeling. However, reported variability in iPSC behavior has called their utility into question. We therefore constituted a test set of 16 iPSCs lines from 7 individuals of varying gender and health status, characterized them extensively for pluripotency, and evaluated their ability to terminally differentiate. Using standardized procedures in two independent laboratories, 13 of the iPSC lines gave rise to functional motor neurons with a range of efficiencies similar to ESCs. Although three iPSC lines were resistant to neural differentiation, early neuralization rescued their performance. Therefore, all lines in the test set passed a stringent test of differentiation capacity despite variations in expression of early pluripotency markers, transgenes and karyotype. This novel iPSC/ESC test set is a robust resource for those interested in the basic biology of stem cells and their applications.
Keywords: Induced pluripotent stem cells (iPS), embryonic stem cells (ES), motor neurons, directed differentiation, differentiation efficiency
Induced pluripotent stem cell (iPSC) lines present an opportunity to produce previously inaccessible cell-types needed for disease-related studies1. Importantly, iPSCs can be made from patients and their healthy relatives, allowing the genetic variants that either predisposed them to, or protected them from disease to be studied2–5. However, if patient-specific iPSCs are to become a standard resource, it is vital to understand how reliably they generate differentiated derivatives.
Among the concerns raised about iPSCs is that reprogramming may be incomplete, resulting in cell lines with variable gene expression or DNA methylation6–8. Indeed, it was reported that when human iPSC lines were differentiated toward a motor neuron identity, they uniformly failed to produce this neural subtype with the efficiency observed using ESCs9. Moreover, it has been suggested that differentiated progeny of iPSCs that harbor reprogramming proviruses have a problematic gene expression signature that can only be resolved by viral excision5. It is therefore critical to use a larger sample of cell lines systematically examined in parallel to determine whether cell line variability could potentially limit iPSC utility.
Critically, it remains to be addressed whether standard reprogramming, expansion and directed differentiation processes are robust enough to minimize noise caused by donor-to-donor variation, which could obscure disease-specific phenotypes. Finally, it has not been determined whether individual iPS cell lines behave similarly from laboratory to laboratory. To address these questions, we derived a set of cell lines that accounts for many sources of variation that might be encountered in the course of modeling development or disease. We then compared the ability of the lines to undergo directed differentiation as a stringent test of their pluripotency. We selected motor neurons as a model system because they are an example of the many differentiated human cell types that cannot be obtained by other means, and are specifically affected in ALS10.
After all cell lines were extensively characterized, we found that like ESCs, most iPSCs were capable of generating functional motor neurons under a standard differentiation protocol, while others required more efficient neuralization. The efficiency with which each individual line generated motor neurons was highly reproducible between two different laboratories, indicating that the collection can function robustly as a shared resource. Lastly, we have shown that this panel of iPSCs includes examples of interline, inter-donor and inter-sex variation in differentiation efficiency, which will be important parameters for future study. This test set has already served as the basis for a study of the epigenetic influences on stem cell differentiation potential by Bock et al., 201111. Now that these test set cell lines are available for distribution they should prove to be a valuable resource for many avenues of stem cell research.
Results
A Test Set of iPS cell lines
We assembled a test set of 16 human iPSC lines, including 2 previously reported2 and 14 novel lines (Table 1). This set included lines from seven individuals of both sexes whose age ranged from 29 to 82 years. All iPSCs were derived by retroviral transduction of skin fibroblasts. Most lines were produced using only three factors (OCT4, KLF4, SOX2) but as a control, two lines were derived using cMYC2. To allow evaluation of the effects of individual genetic background, we included independent lines derived from the same subjects (2 lines from each of 2 donors, 3 lines from each of 2 donors and 4 lines from 1 donor). Finally, we included lines from both healthy controls (n=10) and patients with ALS (n=6). To determine whether reprogramming systematically influences stem cell properties, we compared the performance of these iPSC lines to 6 ESC lines (Table 1).
Table 1. Human stem cell lines used for comparative study.
16 human iPSC lines were used for comparison with each other and with 6 ESC lines. iPSC lines include 14 newly generated 3-factor lines from 2 ALS patients and 5 control patients, and 2 previously published 4-factor lines from 1 ALS patient. This cohort of human stem cell lines allows comparisons to be made between ESCs and iPSCs, between 3- factor and 4-factor iPSC lines, male vs. female lines, between lines derived from the same donor and those derived from another donor, and between cells derived from ALS patients and control donors.
| Cell Type |
Donor Fibroblast |
Cell Line | ALS Diagnosis |
Reprogramming Factors |
Sex | Donor Age |
Reference |
|---|---|---|---|---|---|---|---|
| iPS | 11 | 11a | healthy control | OCT4/SOX2/KLF4 | M | 36 | this report |
| iPS | 11 | 11b | healthy control | OCT4/SOX2/KLF4 | M | 36 | this report |
| iPS | 11 | 11c | healthy control | OCT4/SOX2/KLF4 | M | 36 | this report |
| iPS | 15 | 15b | healthy control | OCT4/SOX2/KLF4 | F | 48 | this report |
| iPS | 17 | 17a | healthy control | OCT4/SOX2/KLF4 | F | 71 | this report |
| iPS | 17 | 17b | healthy control | OCT4/SOX2/KLF4 | F | 71 | this report |
| iPS | 18 | 18a | healthy control | OCT4/SOX2/KLF4 | F | 48 | this report |
| iPS | 18 | 18b | healthy control | OCT4/SOX2/KLF4 | F | 48 | this report |
| iPS | 18 | 18c | healthy control | OCT4/SOX2/KLF4 | F | 48 | this report |
| iPS | 20 | 20b | healthy control | OCT4/SOX2/KLF4 | M | 55 | this report |
| iPS | 27 | 27b | SOD1G85S | OCT4/SOX2/KLF4 | F | 29 | this report |
| iPS | 27 | 27e | SOD1G85S | OCT4/SOX2/KLF4 | F | 29 | this report |
| iPS | 29 | 29A | SOD1L144F | OCT4/SOX2/KLF4/cMYC | F | 82 | Dimos et al., 20082 |
| iPS | 29 | 29B | SOD1L144F | OCT4/SOX2/KLF4/cMYC | F | 82 | Dimos et al., 20082 |
| iPS | 29 | 29d | SOD1L144F | OCT4/SOX2/KLF4 | F | 82 | this report |
| iPS | 29 | 29e | SOD1L144F | OCT4/SOX2/KLF4 | F | 82 | this report |
| ES | - | HuES 3 | - | - | M | - | Cowan et al., 200423 |
| ES | - | HuES 6 | - | - | F | - | Cowan et al., 200423 |
| ES | - | HuES 9 | - | - | F | - | Cowan et al., 200423 |
| ES | - | HuES 13 | - | - | M | - | Cowan et al., 200423 |
| ES | - | HuES 3 HB9:GFP |
- | - | M | - | Di Giorgio et al., 200812 |
| ES | - | R1 | - | - | M | - | James et al., 200624 |
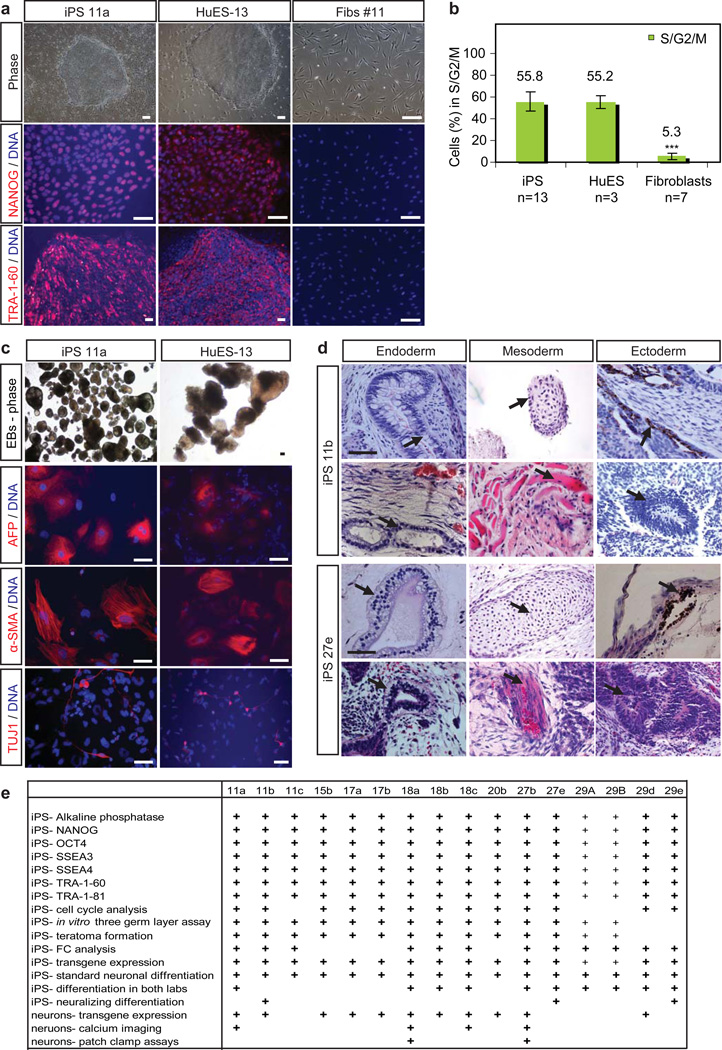
To verify the newly-derived iPSC lines were indeed pluripotent stem cells, we assessed expression of the pluripotency markers alkaline phosphatase, NANOG, OCT4, SSEA3, SSEA4, TRA-1-60 and TRA-1-81. In addition to exhibiting extracellular staining patterns for both SSEA and TRA-1 proteins, all colonies showed distinct nuclear staining when assayed for NANOG and OCT4 immunoreactivity (Fig. 1a, Supplementary Fig. 1a). All iPSC lines generated compact colonies with a morphology (Fig. 1a, Supplementary Fig. 1a) and cell cycle profile comparable to those of ESC controls (Fig. 1b, Supplementary Fig. 1b). We next determined that the test set could differentiate into all three embryonic germ layers as detected by expression of neuron-specific tubulin (TUJ1), smooth muscle actin (αSMA), and endodermal α-fetoprotein (AFP) after 16 days of differentiation in vitro (Fig. 1c, Supplementary Fig. 1c). Lastly, we tested the ability of several lines to generate teratomas in immune-compromised mice. Following injection, each line (14/14 tested) formed teratomas containing complex tissue structures characteristic of all three embryonic germ layers (Fig. 1d, Supplementary Fig. 1d). Based on histological criteria these structures included neuronal fibers, hair follicles, melanocytes, keratin pearls, muscle cells, cartilage, glands and goblet cells. Thus our test set resource contains lines that have been extensively characterized and meet the most stringent criteria for pluripotency (Figure 1e).
Figure 1. Characterization of pluripotency in the test-set of iPSC lines.
(a) iPSC colonies were morphologically identical to hESC colonies, and express pluripotency markers NANOG and TRA-1-60, unlike the patient fibroblasts from which they were derived. Scale bars are 200 µm. (b) iPSC lines showed cell cycle profiles similar to that of ESCs and different to their parental fibroblasts. The percentage of cells undergoing different stages of the cell cycle was determined by propidium iodide staining and FC. The percentage of cells in S,G2 and M phase was found for each cell lines assayed, and then averaged for each category (c) Like hESCs, iPSC lines generated cell types of all three embryonic germ layers (endoderm- AFP, mesoderm- α-SMA, ectoderm- TUJ1) in vitro, as embryoid bodies (EBs), scale =100µm, and (d) when injected into mouse kidney capsules and allowed to form teratomas in vivo, scale=50µm. Representative images of H&E-stained sections are shown for lines 11b and 27e. Glands and goblet cells (endoderm), cartilage and muscle (mesoderm), pigmented neural epithelium and neural rosettes (ectoderm) are shown in the top and bottom panels respectively for both lines. (e) Summary chart depicting assays by which iPSC lines in the test set were characterized. Pluripotency assays (small +) for 29A and B were previously published by Dimos et al., 2008.
A majority of iPSC lines generate electrically active motor neurons using standard procedures
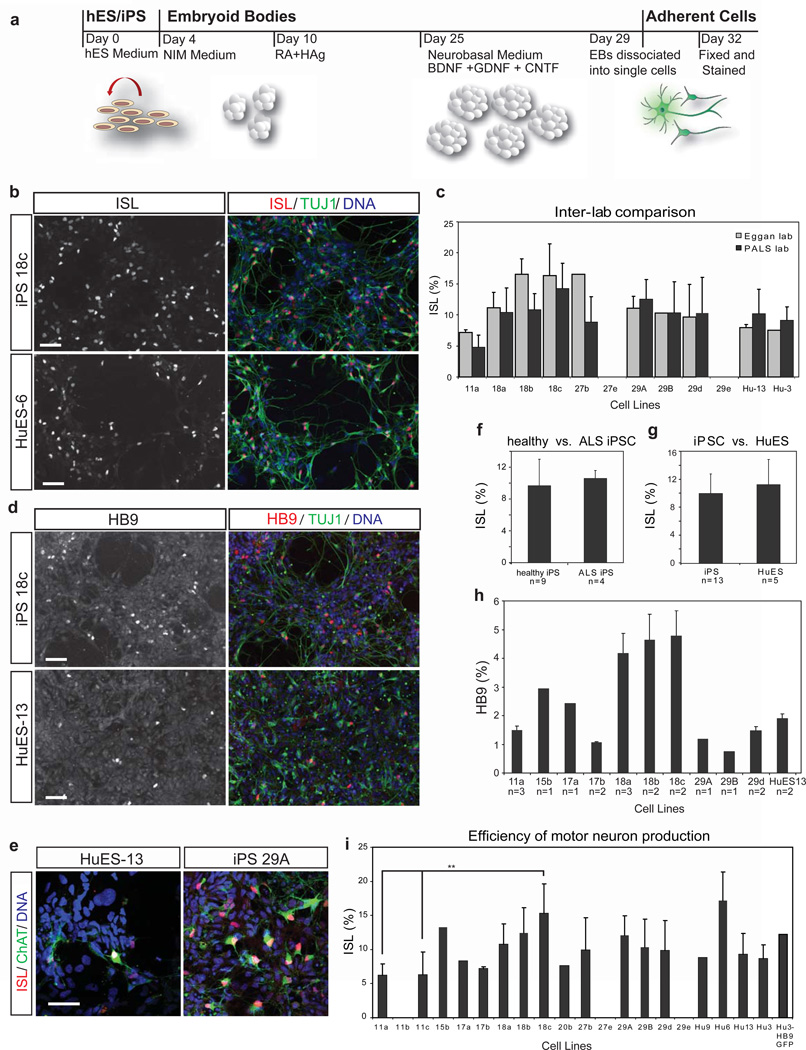
As a rigorous quantitative test of the potential for iPSCs to undergo terminal differentiation, we determined the efficiency with which each line could generate spinal motor neurons. Using standard procedures involving retinoic acid and induction of the sonic hedgehog pathway12–13 (Fig. 2a), the majority (19/22) of the iPSC and ESC lines generated TUJ1+ cells with a neuronal morphology that also expressed the motor neuron marker ISLET 1/2 (ISL) (Fig. 2b). To confirm the reliability of the lines as a resource for the community, we transferred the entire test set from the laboratory in which they were derived to a geographically distinct facility, which then repeated the directed differentiation experiments. Consistent with reproducible properties for each cell line, the same lines were found to generate ISL+ neurons (Fig. 2c). Indeed, the differentiation efficiency for each line (ranging from 4–15% ISL+/total nuclei) showed no difference between the two laboratories (Fig. 2c; ANOVA f=1.132, p=0.301, Supplementary Table 1). Thus, the test set will also serve as a robust resource for research in other centers.
Figure 2. iPSCs show similar capacity for directed motor neuron differentiation to ESCs.
(a) Protocol for directed differentiation of human stem cell lines into motor neurons. Cells were differentiated as EBs from day 0–29 in media formulations containing morphogens, including retinoic acid (RA), a small molecule agonist of the sonic hedgehog pathway (HAg), and neurotrophic factors BDNF, GDNF, and CNTF. EBs were dissociated and single cells plated for adherent culture on day 29. On day 32 cultures were analyzed. (b) Representative immunostaining results for iPSC (18c) and ESC (HuES-6) cultures show many ISL+ TUJ1+ motor neurons (scale = 50 µm). (c) The percentage of all nuclei which were ISL+ was quantified from differentiations performed independently in the Eggan and PALS labs. Data sets from lines differentiated in both labs are compared here, are highly similar, and have reproducible, characteristic %ISL efficiencies. 29e and 27e did not differentiate efficiently in either lab. (d) Efficiency of motor neuron differentiation was also measured by an alternative marker of motor neuron identity, HB9 (scale = 50 µm). (e) Many ISL+ motor neurons were also ChAT+, indicating proper maturation toward a cholinergic transmitter phenotype. (f) iPSC lines from control and ALS patients differentiated into ISL+ motor neurons with similar efficiencies, (g) as did ESCs and iPSCs. (h) The percentages of HB9+ nuclei were compared for a subset of iPSC lines and HuES-13. While comparisons again suggest donor- or line-specific differences, iPSC lines were overall equally capable of generating HB9+ motor neurons as HuES-13 (mean ± SD). (i) % ISL+ data from both labs was pooled for each iPSC and ESC line, and comparisons between lines showed generally similar performance, with significant differences between iPSC line 18c and iPSC lines 11a and 11c (p<0.05).
To confirm that the ISL+ neurons were motor neurons, we quantified the expression of other markers indicative of this neural sub-type. We found that the iPSC lines producing the most ISL+ neurons also produced the largest number of cells expressing the motor neuron specific transcription factor HB914 (Fig. 2d). On average, iPSC lines produced as many HB9+ neurons as did the established hESC line HuES-13 (Fig. 2h). In addition, in a tested subset of lines ISL+ neurons were immunopositive for ChAT, the enzyme required for acetylcholine synthesis (Fig. 1e). These differentiated cultures, which also contained a variety of motor neuron progenitors, expressed the neural marker NCAM (Supplementary Fig. 2a), and expressed significant levels of mRNA encoding the markers HB9, ChAT, OLIG2, and CHT1 (Supplementary Fig. 2b). Finally, unlike PAX6+ progenitors in these cultures, ISL+ neurons were never observed to be actively cycling as measured by Ki67 immunostaining (Supplementary Fig. 2c). Although there were quantitative differences in motor neuron generation among the lines (Fig. 2i, Supplementary Table 2), they did not reflect overall differences between iPSC and ESC lines (Fig. 2f; Supplementary Table 1) or between healthy control and ALS iPSC lines (Fig. 2g; Supplementary Table 1). Moreover, four-factor lines did not underperform relative to three-factor lines (Supplementary Table 1). Therefore, most iPSC lines in the resource can generate spinal motor neurons with a reproducible efficiency that is equivalent to that of gold standard hESC lines.
In order to demonstrate that the motor neurons we produced were functional, we compared the electrophysiological properties of motor neurons from four iPSC lines with motor neurons from two ESC lines. We first monitored intracellular Ca2+ dynamics using the Ca2+-sensitive dyes Fura Red AM (Fig. 3 a–b) and Fluo-4 AM. This was performed both in the absence of exogenous stimulation (Fig. 3c, h, j) to monitor spontaneous activity, and following application of either kainate to activate ionotropic glutamate receptors, or KCl to depolarize the membrane and open voltage-gated Ca2+ channels (Figure 3d–g, i, k). Spontaneous calcium transients were visible in the cell bodies and processes of multiple cells from each line, even without treatment (Fig. 3c, h, j; Supplementary Fig. 3a, Supplementary Video 1). Upon exposure to kainate, increases in Ca2+ levels were observed in 78% of cells with neuronal morphology (n=132 cells). Many kainate-responsive cells also exhibited Ca2+ transients upon exposure to KCl (Fig. 3i, k). Immunostaining confirmed that many of the cells that responded to kainate and KCl in these mixed cultures were ISL+ motor neurons (Fig. 3b, i; Supplementary Fig. 3b).
Figure 3. ESC and iPSC derived neurons are physiologically active.
(a) Image of iPSC 11a-derived neurons filled with Fura Red AM and Fluo-4 AM dyes. The Fura Red channel is shown. The field illustrated is that imaged in panels b–g. Activity of labeled cells is represented in panels h and i. scale =100 µM. (b) ISL immunostaining of 11a field in a–g showing ISL+ neurons (star) and ISL- neurons (arrow). (c) Spontaneous electrical activity in cultured iPSC-derived neurons visualized by a ‘subtracted image’ that shows the difference in pixel intensities between two images acquired 1.7s apart in the Fluo 4 channel. Higher grey values represent increased pixel intensity. (d–g) Identically exposed pseudocolored averages of ten Fluo 4 AM images taken: (d) during the control period before addition of KA, (e) following treatment with 100 µM KA, (f) after washing following KA administration, and (g) following treatment with 50 mM KCl. Warmer colors represent increased fluorescence intensity. (h) Plot of Fluo 4/Fura Red intensity ratio in the somata of the two cells indicated by the star and arrow in a–c showing spontaneous activity. (i) Fluo 4/Fura Red intensity ratio of cells in a–c during sequential administration of KA and KCl indicated by bars above graph. (j) Examples of Fluo 4/Fura Red ratios from cell bodies of single spontaneously active cells in cultures of ESC R1-derived neurons, and iPSC 11a, 18a, 18c, and 27b-derived neurons as well as one example of a non-responsive, non-active cell in an R1 culture (NR). (k) Response of cells in (j) to KA and KCl. Sample voltage-clamp traces from (l) ESC and (m) iPSC 18a-derived neurons. (n) Blow-up of an iPSC 27b-derived neuron recording reveals typical sodium currents (left), which are blocked by 500 nM TTX (right). (o) Current-clamp recordings of single action potentials in ESC and iPSC 27b-derived neurons as well as multiple action potentials in an iPSC 18a-derived neuron.
To further demonstrate that iPSC-derived neurons express the repertoire of voltage-gated ion channels characteristic of active neurons, we made electrophysiological recordings using whole-cell patch clamping. All cells with a neuronal morphology derived from HuES3 Hb9:GFP (n=9 cells), iPSC line 18a (n=10 cells), and iPSC line 27b (n=10 cells) showed fast voltage-activated inward currents followed by slow outward currents, consistent with voltage-activated sodium and potassium currents, respectively (Figure 3l, m). Inward currents (n=5/5) were blocked by tetrodotoxin, an inhibitor of voltage-gated sodium channels (Fig. 3n). In addition, depolarizing stimuli in current-clamp mode elicited single action potentials in both ESC (n=2) and iPSC-derived neurons (n=2), as well as repetitive firing in a neuron derived from iPSC line 18a (Fig. 3o). Therefore, we conclude that both ESC and iPSC-derived neurons generated from the cell lines in the resource are similarly functional at a physiological level.
Contribution of other variables to differentiation
Although all cell lines were capable of generating motor neurons, we systematically examined some parameters that have been implicated in differentiation efficiency and so would be of interest to potential users of this resource. The majority of iPSC lines (9 out of 15; Supplementary Fig. 6) exhibited genomic stability at both early (p13) and late (p42) passages. The other six lines (29d, 27b, 29e, 11a, 11b, 15b) acquired disparate abnormalities of varying severity at later passages. However, lines that became karyotypically abnormal did not produce motor neurons with a significantly different efficiency from normal lines (p=0.932, Supplementary Table 1). Since it is not known how these chromosomal changes may affect behavior of any given cell type, these lines should be used with caution in studies making phenotypic comparisons.
We, and others, have reported that reprogramming transgenes can continue to be expressed in patient-specific iPSC lines, but whether they interfere with differentiation has not been fully studied2,5. We therefore used qRT-PCR to quantify relative levels of transcription of the reprogramming transgenes from both their endogenous loci and from the integrated retrovirus (Fig. 4a). In most cases, levels of viral transcription were either undetectable or very low when compared to levels of transcripts from the endogenous loci; indeed, viral SOX2 was never detected (Supplementary Fig. 7a). However, a subset of iPSC lines (11b, 11c, 15b, 18b, 18c, 27b, 27e and 29e) continued to express varying levels of viral KLF4 both in the undifferentiated state and following differentiation to motor neurons (Fig. 4a, Supplementary Fig. 7b–d). Moreover, viral OCT4 transcripts were present in three of the iPSC lines (15b, 18c, and 27b) both before and after differentiation. Strikingly, there was no correlation between the total level of aggregated transgene expression in iPSCs and the efficiency of motor neuron differentiation as judged by %ISL+ neurons (R2=0.1687). In addition, we performed immunofluorescence staining for OCT4 and the motor neuron marker ISL (Fig. 4b, Supplementary Fig. 7e). Remarkably, a line such as 15b that showed persistent transgene expression generated motor neurons with high abundance, even though many of the motor neurons expressed nuclear OCT4. Therefore, although examples of lines that display karyotypic variation and persistent transgene expression are available in the test set, they had no detectable effect on rates of motor neuron differentiation.
Figure 4. Persistent transgene expression does not inhibit differentiation.
(a) Quantitative RT-PCR was used to measure relative levels of transcript from endogenous genes ‘e’ and viral transgenes ‘v’ of the reprogramming factors OCT4 and KLF4 in undifferentiated iPSCs and ESCs, and in day 32 neuron cultures. Transgene expression or silencing in the undifferentiated cells is maintained after differentiation. Relative levels in undifferentiated HuES-3 were set as 1. (b) Day 32 motor neuron cultures were co-stained for ISL and OCT4. HuES-3 and iPSC 17a-derived cultures, which do not express viral OCT4, did not stain for OCT4. However, iPSC 15b-derived cultures, which do express viral OCT4, contained many OCT4+ ISL+ motor neurons and OCT4+ISL+ cells. Arrow = OCT4+ ISL+, arrow head = OCT4+ ISL−, chevron = OCT4− ISL+.
Finally, we looked at the contribution of age, sex, and donor genotype to the outcome of differentiation in our test set. There was no correlation between donor age and the percentage of ISL+ neurons generated (R2= 0.0084). However, there was a significant difference in differentiation efficiency found between male and female lines (ANOVA p=0.048, Supplementary Table 3). These sex-specific differences could result from variable processes such as X chromosome inactivation. Lastly, we compared the ability of independent lines from several of the donors to differentiate into motor neurons. Southern blot analysis (Supplementary Fig. 8a–c) confirmed that lines from donors 11, 18, and 29 arose from distinct reprogramming events. Subsequently, inspection of the differentiation data (Fig. 2c, d and i) showed that all three iPSC lines from donor 18 produced many motor neurons, whereas the lines from donor 11 performed less well. The difference between line 18c, the best of the three from that donor, and the two lines from donor 11 that generated motor neurons was indeed significant (p<0.05; Fig. 1d), and when comparisons between averaged differentiation efficiencies of each donor were made, a significant difference was found (ANOVA p= 0.006, Supplementary Table 4, Supplementary Figure 8d). These results further demonstrate that the cell lines included in the test set may provide an opportunity for other researchers to investigate the effects that gender and other donor-specific phenomenon, have in directed differentiation.
Suboptimal lines are rescued by active neuralization
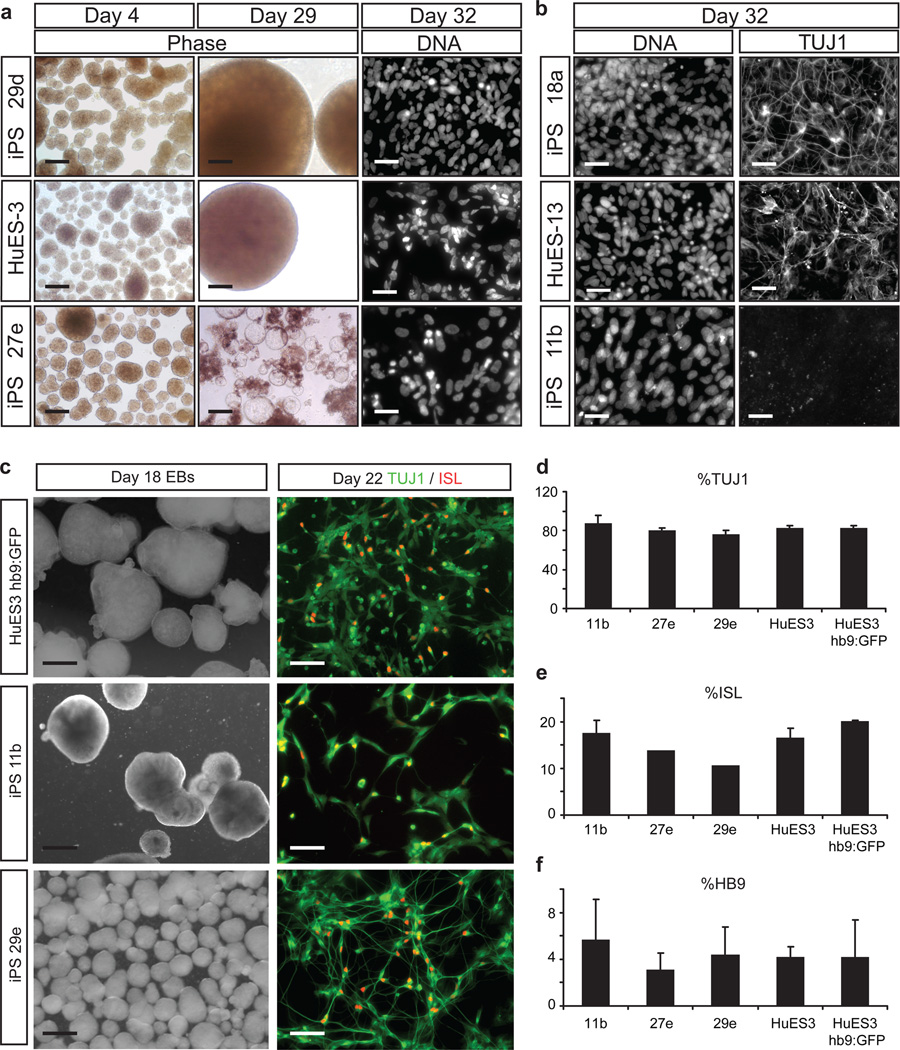
While the majority of our cell lines reproducibly generated motor neurons, there were three lines (11b, 27e, and 29e) that uniformly failed to do so in both laboratories. Although all three formed embryoid bodies (EB) (Fig. 5a, n=3–7 independent experiments per line), the EBs from two lines -27e and 29e- became cystic and disaggregated (Fig. 5a). This defect was reflected in a significant decrease in total yield of differentiated cells (Supplementary Fig. 5a, Supplementary Table 5) and by the failure of these two lines to generate TUJ1+ neurons (Supplementary Fig. 4b, Supplementary Table 6). A third line -11b- formed EBs with normal morphology but produced fewer than 10% TUJ1+ neurons (Fig. 5b). This was significantly lower than 18a (Holm-Sidak, T=5.037, p=0.002), and was in contrast to lines differentiating appropriately, which produced in excess of 25% TUJ1+ cells (Supplementary Fig. 4b). An indication that early blockade of differentiation might occur in some lines was obtained when we used expression of the antigens SSEA3 and TRA-1-60 to quantify the relative proportion of pluripotent cells within each iPSC line by flow cytometry (Supplementary Fig. 5a–e). One of the recalcitrant iPSC lines (27e) showed higher median fluorescence intensity of TRA-1-60 staining than all others (Supplementary Fig. 5d, Supplementary Table 7), suggesting it might be less prone to spontaneous differentiation.
Figure 5. Sub-optimal iPSC lines can be rescued using SMAD inhibition.
(a) During EB differentiation, iPSC lines 27e and 29e showed abnormal EB morphology and survival compared to lines that behaved normally (HuES-3 and 29d shown), phase scale = 500 µm, DNA scale = 129 µm. (b) Although EBs from iPSC line 11b had typical morphology, day 32 cultures showed decreased neuronal TUJ1 staining compared to all other normal lines (HuES-13 and 18a shown), scale = 129 µm. (c) Representative phase and immunostaining images for previously defective iPSC lines 29e, 11b, and control ESC lines HuES-3 and HuES-3-hb9:GFP. Phase image scales are 500 µm, immunostaining image scales are 100 µm. Quantification of immunostaining in differentiated cultures derived from the three previously problematic iPSC lines (11b, 27e, 29e) and ESC controls (d) percentage of TUJ1+ cells; (e) percentage of ISL+ cells; (f) percentage of HB9+ cells. Mean ± SEM.
Since the three recalcitrant lines were nevertheless able to initially form EBs, we asked whether they could be coaxed into the motor neuron differentiation pathway by pushing them toward a more neural fate at the beginning of the differentiation process. We combined our EB protocol with dual SMAD inhibition, similar to that reported by Chambers et al., 2009, for the first 9 days using SB431542 and the structural analog of dorsomorphin, LDN19318915–16. Comparing the three underperforming iPSC lines (11b, 27e, 29e) to that of two ESC lines, we found that the previously defective lines were all neuralized in this optimized protocol and gave rise to the same high abundance of TUJ1+ cells as did the ESC lines (>75%; Fig. 5c, d). Critically, the three iPSC lines that previously could not generate neurons now robustly produced ISL+ (Fig. 5c and e) and HB9+ (Fig. 5f) motor neurons by day 21 at levels indistinguishable from those of either the control ESCs (Fig. 5d–e) and from the other 13 iPSC lines (Fig. 2h, i). Thus, although 3 lines in our human stem cell resource underperformed using a basal differentiation protocol, they could be rescued through a neuralizing protocol to efficiently generate spinal motor neurons.
Discussion
To evaluate iPSCs as a research tool, we generated a large panel of cell lines from multiple donors, then examined aspects of their pluripotency and ability to generate terminally-differentiated motor neurons. The results of our comparisons confirm the remarkable value of iPSC lines for in vitro studies, and demonstrate that they can perform as well as standard ESC lines. When procedures were standardized, this observation held true for experiments performed in two geographically distinct laboratories. The analyses provided here serve as a quality control for this stem cell resource, while also providing sufficient data on specific aspects of variability to allow investigators to select lines that will be of particular relevance to their own research.
Our study is not the first to compare human iPSC and ESC lines, but it is the most extensive comparison of their ability to generate a specific terminally-differentiated cell thus far. Most studies have used panels of four or fewer iPSC lines17–22, limiting the possibilities for understanding variability between cell lines or drawing general conclusions about functional similarities between iPSCs and ESCs. Similar to a comparable study9, we find that variation exists between the differentiation efficiencies of individual iPSC lines. However, contrary to the conclusions of that study, which were that the differentiation capacity of iPSCs needs be improved in order to match that of ESCs, we found that iPSC lines could be made to differentiate on average as well as ESC lines. Whether this is due to differences in protocols for reprogramming and motor neuron differentiation between the two studies, or whether it reflects the larger sample analyzed here, remains to be determined.
While all cell lines in our test set were capable of generating motor neurons, the standard protocol for motor neuron production did reveal significant quantitative differences in the propensity of the lines for terminal differentiation. These differences were highly reproducible, suggesting that they represent intrinsic characteristics of the lines. Our initial hypothesis was that the poorly performing lines would be identified by anomalies in standard tests for stem cell quality. However, all cell lines tested expressed pluripotency markers, could form the three germ layers in vitro, and in teratomas. Moreover, although variations in karyotype and transgene expression were observed, they were not accurate predictors of differentiation capacity. Fortunately, a solution for identifying such predictors has now been proposed by Bock et al.11, who used our test set to search for epigenetic and transcriptional differences that correlate with differentiation potential. They used the lines we describe here, to develop a scorecard for stem cell quality that predicted our motor neuron differentiation results (Fig. 2i) with remarkable precision.
We anticipate that one of the major uses of the cell lines provided through this resource will be to model ALS. Importantly, our data demonstrate that several conditions that are necessary for reliable disease modeling are met. First, since ALS is not a developmental disease, our finding that iPSCs carrying an ALS-triggering mutation differentiated similarly to those from healthy controls is as expected. Second, although lines from different healthy donors taken together showed donor-related variation in differentiation efficiency, the pairwise comparisons did not reach significance. This increases the chances that phenotypic differences we may eventually observe between ALS cases and controls are related to disease. Nevertheless, since we found real line-to-line differences, it will be essential to confirm that any phenotypes are ALS-related by silencing the mutant SOD1 disease gene. Lastly, the remarkable concordance between the results from two different laboratories reported here suggests that, once ALS-related phenotypic differences are discovered, they will prove sufficiently reproducible to serve as a general paradigm for the field.
Supplementary Material
Acknowledgements
We thank H. Mitsumoto, J. Montes, P. Kaufmann, J. Andrews for collecting skin biopsies, K. Koszka, A. Sproul, A. Hon, A. Garcia-Diaz for technical assistance, M. Park, A. Meissner, C. Bock for manuscript assistance, as well as S. Brenner-Morton and T. Jessell for providing Islet antibodies. This work was funded by Project A.L.S., NYSTEM and NIH GO grant 1RC2 NS069395-01. G.L.B. is an HSCI/NIH Trainee. E.K. is an EMBO Postdoctoral Fellow. B.J.W. is supported by NIH Training Grant 5T32GM007592. C.J.W. supported by grants from NINDS & NICHD. K.E. is an HHMI early career scientist.
Footnotes
Author Contributions
G.F.C., M.W.A, and D.H.O. maintained human fibroblasts. C.T.R. and J.T.D. reprogrammed all iPSC lines. G.L.B. and E.K. expanded all iPSC lines. G.L.B. and E.K. led and contributed equally to all other experiments and analyses in the Eggan Lab. G.F.C., M.W.A. and D.H.O. led and contributed equally to all other experiments and analyses in the Project ALS Lab. D.J.K. performed FC analysis. A.B.M. and D.J.W. designed and performed Ca2+ imaging. B.J.W. performed recordings. M.Y. assisted with teratomas L.D. assisted with quantitative analysis. S.M. assisted with stem cell culture. G.L.B., E.K. and K.E., G.C., M.W.A, C.H. and H.W., conceived the experiments and wrote the manuscript.
References
- 1.Kiskinis E, Eggan K. Progress toward the clinical application of patient-specific pluripotent stem cells. J Clin Invest. 2010;120:51–59. doi: 10.1172/JCI40553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dimos JT, et al. Induced pluripotent stem cells generated from patients with ALS can be differentiated into motor neurons. Science. 2008;321:1218–1221. doi: 10.1126/science.1158799. [DOI] [PubMed] [Google Scholar]
- 3.Park IH, et al. Disease-specific induced pluripotent stem cells. Cell. 2008;134:877–886. doi: 10.1016/j.cell.2008.07.041. d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee G, et al. Modelling pathogenesis and treatment of familial dysautonomia using patient-specific iPSCs. Nature. 2009;461:402–406. doi: 10.1038/nature08320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Soldner F, et al. Parkinson's disease patient-derived induced pluripotent stem cells free of viral reprogramming factors. Cell. 2009;136:964–977. doi: 10.1016/j.cell.2009.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chin MH, et al. Induced pluripotent stem cells and embryonic stem cells are distinguished by gene expression signatures. Cell Stem Cell. 2009;5:111–123. doi: 10.1016/j.stem.2009.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Doi A, et al. Differential methylation of tissue- and cancer-specific CpG island shores distinguishes human induced pluripotent stem cells, embryonic stem cells and fibroblasts. Nat Genet. 2009;41:1350–1353. doi: 10.1038/ng.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stadtfeld M, et al. Aberrant silencing of imprinted genes on chromosome 12qF1 in mouse induced pluripotent stem cells. Nature. 2010;465:175–181. doi: 10.1038/nature09017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hu BY, et al. Neural differentiation of human induced pluripotent stem cells follows developmental principles but with variable potency. Proc Natl Acad Sci USA. 2010;107:4335–4340. doi: 10.1073/pnas.0910012107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kanning KC, Kaplan A, Henderson CE. Motor Neuron Diversity in Development and Disease. Annu Rev Neurosci. 2010 doi: 10.1146/annurev.neuro.051508.135722. [DOI] [PubMed] [Google Scholar]
- 11.Bock C, Kiskinis E, Verstappen G. Reference maps of human ES and iPS cell variation enable high-throughput characterization of pluripotent cell lines. Cell. 2011 doi: 10.1016/j.cell.2010.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Di Giorgio FP, Boulting GL, Bobrowicz S, Eggan KC. Human embryonic stem cell-derived motor neurons are sensitive to the toxic effect of glial cells carrying an ALS-causing mutation. Cell Stem Cell. 2008;3:637–648. doi: 10.1016/j.stem.2008.09.017. [DOI] [PubMed] [Google Scholar]
- 13.Wichterle H, Lieberam I, Porter JA, Jessell TM. Directed differentiation of embryonic stem cells into motor neurons. Cell. 2002;110:385–397. doi: 10.1016/s0092-8674(02)00835-8. [DOI] [PubMed] [Google Scholar]
- 14.Arber S, et al. Requirement for the homeobox gene Hb9 in the consolidation of motor neuron identity. Neuron. 1999;23:659–674. doi: 10.1016/s0896-6273(01)80026-x. [DOI] [PubMed] [Google Scholar]
- 15.Chambers SM, et al. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat Biotechnol. 2009;27:275–280. doi: 10.1038/nbt.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou J, et al. High-efficiency induction of neural conversion in human ESCs and human induced pluripotent stem cells with a single chemical inhibitor of transforming growth factor beta superfamily receptors. Stem Cells. 2010;28:1741–1750. doi: 10.1002/stem.504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Taura D, et al. Adipogenic differentiation of human induced pluripotent stem cells: comparison with that of human embryonic stem cells. FEBS Lett. 2009;583:1029–1033. doi: 10.1016/j.febslet.2009.02.031. [DOI] [PubMed] [Google Scholar]
- 18.Tokumoto Y, Ogawa S, Nagamune T, Miyake J. Comparison of efficiency of terminal differentiation of oligodendrocytes from induced pluripotent stem cells versus embryonic stem cells in vitro. J Biosci Bioeng. 2010;109:622–628. doi: 10.1016/j.jbiosc.2009.11.013. [DOI] [PubMed] [Google Scholar]
- 19.Xi J, et al. Comparison of contractile behavior of native murine ventricular tissue and cardiomyocytes derived from embryonic or induced pluripotent stem cells. FASEB J. 2010 doi: 10.1096/fj.09-145177. [DOI] [PubMed] [Google Scholar]
- 20.Armstrong L, et al. Human induced pluripotent stem cell lines show stress defense mechanisms and mitochondrial regulation similar to those of human embryonic stem cells. Stem Cells. 2010;28:661–673. doi: 10.1002/stem.307. [DOI] [PubMed] [Google Scholar]
- 21.Ghosh Z, et al. Persistent donor cell gene expression among human induced pluripotent stem cells contributes to differences with human embryonic stem cells. PLoS One. 2010;5 doi: 10.1371/journal.pone.0008975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grigoriadis AE, et al. Directed differentiation of hematopoietic precursors and functional osteoclasts from human ES and iPS cells. Blood. 2010;115:2769–2776. doi: 10.1182/blood-2009-07-234690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cowan CA, et al. Derivation of embryonic stem-cell lines from human blastocysts. N Engl J Med. 2004;350:1353–1356. doi: 10.1056/NEJMsr040330. [DOI] [PubMed] [Google Scholar]
- 24.James D, Noggle SA, Swigut T, Brivanlou AH. Contribution of human embryonic stem cells to mouse blastocysts. Dev Biol. 2006;295:90–102. doi: 10.1016/j.ydbio.2006.03.026. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.