Abstract
We tested the ability of 87 profilin point mutations to complement temperature-sensitive and null mutations of the single profilin gene of the fission yeast Schizosaccharomyces pombe. We compared the biochemical properties of 13 stable noncomplementing profilins with an equal number of complementing profilin mutants. A large quantitative database revealed the following: 1) in a profilin null background fission yeast grow normally with profilin mutations having >10% of wild-type affinity for actin or poly-l-proline, but lower affinity for either ligand is incompatible with life; 2) in the cdc3-124 profilin ts background, fission yeast function with profilin having only 2–5% wild-type affinity for actin or poly-l-proline; and 3) special mutations show that the ability of profilin to catalyze nucleotide exchange by actin is an essential function. Thus, poly-l-proline binding, actin binding, and actin nucleotide exchange are each independent requirements for profilin function in fission yeast.
INTRODUCTION
Profilins are small proteins that interact with at least three types of ligands: actin monomers (and the homologous Arp2 subunit of Arp2/3 complex); proteins with sequences adopting a type II poly-l-proline helix; and polyphosphoinositides (Machesky and Pollard, 1993; Schluter et al., 1997). Interaction with actin monomers suppresses growth at the pointed end of actin filaments but not the barbed end (Pollard and Cooper, 1984). This allows profilin–actin complexes to support the growth of uncapped barbed ends in cells. In addition, most profilins (amoeba [Mockrin and Korn, 1980; Nishida, 1985], vertebrate [Goldschmidt-Clermont et al., 1991; Perelroizen et al., 1996], yeast [Eads et al., 1998]) but not all profilins (plants [Perelroizen et al., 1996; Eads et al., 1998; Kovar et al., 2000]) stimulate the exchange of the adenine nucleotide bound to actin monomers. This nucleotide exchange activity is postulated to prevent ADP-actin monomers dissociated from filaments from being trapped in high-affinity complexes with ADF/cofilins (Blanchoin and Pollard, 1998; Didry et al., 1998). Potential protein ligands with proline-rich sequences include VASP (Reinhard et al., 1995), Enabled (Ahern-Djamali et al., 1999), Mena (Gertler et al., 1996; Lanier et al., 1999), N-WASP (Suetsugu et al., 1998), WAVE/Scar (Miki et al., 1998), verprolin/WIP (Ramesh et al., 1997), diaphanous/p140mDia (Watanabe et al., 1997), cappuccino (Manseau et al., 1996), Bni1p and Bnr1p (Evangelista et al., 1997; Imamura et al., 1997), cdc12 (Chang et al., 1997), drebrin and gephyrin (Mammoto et al., 1998), SMN (Giesemann et al., 1999), and aczonin (Wang et al., 1999). In the case of Mena, diaphanous, cappuccino, Bni1p, Bnr1p, and cdc12, genetic interactions support the biological significance of the interaction.
Null or conditional mutations established that loss of profilin function is lethal in mice (Lanier et al., 1999), flies (Cooley et al., 1992; Manseau et al., 1996), and fission yeast (Balasubramanian et al., 1994) and severely disabling in Dictyostelium (Haugwitz et al., 1994) and budding yeast (Haarer et al., 1990). The existence of more than one profilin gene complicates the situation in mice and Dictyostelium. The two yeast have single profilin genes.
Although profilin is just a small protein, its biological functions are sufficiently complex that it has not yet been possible to show definitively why it is essential for viability. To date 18 articles have described 66 designed mutations or combinations of mutations in human, bovine, budding yeast, Anacthamoeba, Dictyostelium, and plant profilins (supplementary material, Table 1). These results are consistent with the hypothesis that interactions of profilin with both poly-l-proline and with actin are important for biological function. However, with some exceptions, many of these experiments are incomplete in one way or another, especially in establishing the stability of mutant proteins and measuring quantitatively interactions with actin or poly-l-proline.
We report here the first comprehensive analysis of the interactions of profilin with actin and poly-l-proline with a large collection of mutants of fission yeast profilin. S. pombe is favorable for this analysis. An essentially complete genome sequence contains only one profilin gene, cdc3+. A null mutation of cdc3+ is lethal. The mutation E42K in the temperature-sensitive strain cdc3-124 disables profilin at 36°C but not at 25°C (Balasubramanian et al., 1994). At 25°C the cdc3-124 strain grows normally with morphology indistinguishable from wild-type. At 36°C cdc3-124 cells have defects in cytokinesis, resulting in elongated, dumbbell-shaped cells with up to eight nuclei and mislocalized actin filament patches. We made ∼90 point mutations, each designed from an atomic model of the protein to interfere with interactions with actin or poly-l-proline. We tested the ability of each mutant profilin to complement a profilin null mutation and a profilin temperature-sensitive mutation. We purified a selection of >30 of complementing and noncomplementing mutant proteins and characterized their stability and interactions with actin and poly-l-proline. This large database of quantitative information revealed that both actin binding and poly-l-proline binding are required independently for biological function. In addition, the data show that viability depends on both the affinities of profilin for each of these ligands and other functional attributes of the mutant protein. In particular, novel mutations reveal that ability of profilin to catalyze nucleotide exchange by actin is essential for viability of fission yeast and that at least one poly-l-proline ligand has different binding requirements than poly-l-proline.
MATERIALS AND METHODS
Strains, Media, and Chemicals
S. pombe strains used in this study were diploid profilin null strain VSP38 (Δcdc3::ura4+/cdc3+, ade6-216/ade6-210, leu1-32/leu1-32, ura4-D18/ura4-D18, h+/h−), haploid temperature-sensitive strain KYG491 (cdc3-124, ade6-216, ura4-D18, leu1-32, h+), and the matching wild-type haploid strain KGY247 (cdc3+, ade6-216, ura4-D18, leu1-32, h+). Dr. Kathy Gould of Vanderbilt University kindly provided these strains. EMM and ME media were purchased from Bio-101. εATP was from Molecular Probes (Eugene, OR). Poly-l-proline (MW 5000), phloxin-B, thiamine, amino acids, and nucleotides were from Sigma (St. Louis, MO). The poly-l-proline affinity column was prepared according to Kaiser et al. (1989).
Mutagenesis and Purification of S. pombe Profilin
Wild-type S. pombe profilin cDNA in expression vector pMW-172 was provided by Dr. Steven C. Almo of Albert Einstein College of Medicine. Selected residues of S. pombe profilin were mutated by reverse polymerase chain reaction (PCR) mutagenesis according to Ho et al. (1989), except that pfu polymerase (Stratagene, La Jolla, CA) was used to increase the fidelity and reduce adenine addition at the end of PCR products. Nucleotide sequences of all PCR products were confirmed by automated dideoxynucleotide sequencing. Plasmids of wild-type and mutant profilins were freshly transformed into competent Escherichia coli BL21(DE3) strain and cultured overnight without isopropyl-β-d-thiogalactopyranoside induction in LB media.
Purification of wild-type and mutant profilins was based on Kaiser et al. (1989). Briefly, a pellet of bacteria from 300 ml of LB media was suspended in 35 ml of Buffer L (25 mM Tris-Cl pH 7.5, 50 mM sucrose, 10 mM EDTA, 5 mM dithiotreitol [DTT], 1 mM phenylmethylsulfonyl fluoride) with 2 M urea, lysed by sonication, and centrifuged at 95,000 × g for 20 min. If the recombinant profilin was soluble the extract was applied to a 2.5 cm (diameter) × 4-cm (height) column of Whatman DEAE-cellulose, DE52, and washed with Buffer L. The flow through containing profilin was dialyzed against Buffer D (20 mM Tris-Cl pH 8.0, 20 mM KCl, 1 mM EDTA, 1 mM DTT). Recombinant profilins were further purified by affinity chromatography on poly-l-proline Sepharose. After adsorbing the sample, the column was washed with 3 M urea in Buffer D. Bound profilin was eluted with 8 M urea in Buffer D and refolded by dialysis against 1 mM EDTA, 1 mM DTT, 20 mM Tris-Cl, pH 8.0. Profilin mutants that did not bind poly-l-proline were purified by gel filtration on Superdex 2000. If the recombinant profilin was insoluble after extracting the bacteria, the pellet was dissolved in 8 M urea in Buffer D at 4°C. After removal of insoluble material by centrifugation at 95,000 × g for 20 min, the supernatant was adjusted to 4 M urea by addition of Buffer D and run through a DE52 anion exchange column in the same buffer. Profilin in the flow through was dialyzed against Buffer D and purified further as described above.
The concentration of S. pombe profilin was determined by absorbance at 280 nm with an extinction coefficient of 1.63 OD/mg/ml for wild-type and most mutant profilins. The extinction coefficient of P107W profilin was 2.22 OD/mg/ml. These extinction coefficients were measured in a Beckman XLI analytical ultracentrifuge by using absorbance and Raleigh interference optics with an assumed fringe displacement of 3.32 per milligram per milliliter.
Rabbit muscle Ca-ATP actin was prepared by the method of Spudich and Watt (1971) and purified by gel filtration on Sephacryl S-300 in Buffer A (0.2 mM ATP, 0.5 mM DTT, 0.1 mM CaCl2, 1 mM NaN3, 2 mM Tris-Cl pH 8.0).
Urea Denaturation
Profilins were added at a final concentration of 1.5 μM to 1 mM EDTA, 1 mM DTT, 20 mM Tris-Cl, pH 8.0, containing 0 to 8 M urea at room temperature. Intrinsic fluorescence was measured with a PTI spectrofluorimeter (Photon Technology Instruments, Santa Clara, CA) with excitation at 295 nm. The emission peak of profilin shifts from 330 nm (without urea) to 352 nm and increases in intensity (with 8 M urea) (Kaiser and Pollard, 1996). The difference between the emission intensities at 352 and 330 nm was plotted against concentration of urea. The urea concentration at the midpoint of the transition in intensity was used to measure stability.
Interaction of Profilin with Poly-l-Proline
Profilins were added at a final concentration of 1.5 μM to 1 mM EDTA, 1 mM DTT, 20 mM Tris-Cl, pH 8.0, containing 0 to 5 mM poly-l-proline at room temperature. Intrinsic fluorescence was measured with excitation at 295 nm and emission at 323 nm. The dissociation equilibrium constant (Kd) was calculated using Kaleidagraph software and Eq. 1 from the dependence of fluorescence change on the concentration of poly-l-proline.
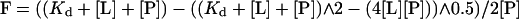 |
1 |
F is the relative fluorescence; [L] and [P] are the total concentrations of poly-l-proline and profilin.
Interaction of Profilin with Actin Monomers
Monomeric rabbit skeletal muscle actin at a concentration of 12 μM in 0.2 mM ATP, 0.5 mM DTT, 0.1 mM CaCl2, 1 mM NaN3, 2 mM Tris-Cl pH 8.0 was treated at 4°C with Dowex-1 (Bio-Rad AG1-x4 resin washed with water extensively and buffered with 1 M Tris-Cl, pH 7.5, and then buffered with 25 mM Tris-Cl pH 7.5 and 0.01% NaN3) to remove free ATP. εATP Mg-actin was prepared by adding 200 μM εATP, 200 μM EGTA, and 50 μM MgCl2 for 50 min at 4°C. Free εATP was removed with Dowex-1 and Mg-εATP actin was used within 3 h.
The time course of εATP dissociation from 1.2 μM Mg-εATP actin was measured by fluorescence with excitation at 360 nm and emission at 410 nm (Waechter and Engel, 1975; Nishida, 1985) after adding 500 μM ATP with 0–100 μM profilin. The rate constant (k−1) for the dissociation of εATP from actin was determined by fitting the time course with a single-exponential equation by using Kaleidagraph software. Profilin promotes dissociation of nucleotide from actin. The dependence of k−1 on profilin concentration gave the dissociation equilibrium constant (Kd) for actin and profilin according to Eq. 1, where F is k−1 and [L] and [P] represent the concentrations of profilin and actin.
Profilin mutant Y79R fails to promote nucleotide exchange, but inhibits nucleotide exchange by wild-type profilin. We used a competition assay to measure the affinity of Y79R profilin for actin in the presence of wild-type profilin. We measured the time course of dissociation of εATP from 1.2 μM εATP actin in the presence of 500 μM ATP, 1.5 μM wild-type profilin, and 0–100 μM Y79R. The apparent dissociation rate constant (kobs) from each time course was determined by fitting a single-exponential equation.
The total actin AT is either free Af, bound to wild-type profilin (Awt), or bound to mutant profilin (Amu).
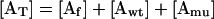 |
2 |
Three reactions contributed to the observed dissociation rate constant, kobs: dissociation from free actin (Af), actin bound to wild-type profilin (Awt), and actin bound to Y79R profilin (Amu).
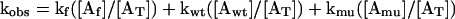 |
3 |
where kf, kwt, and kmu are rate constants for εATP dissociation from free actin, actin bound to wild-type profilin and actin bound to Y79R. All three of these rate constants were measured experimentally. Because kf = kmu, Eq. 3 is shortened to the following:
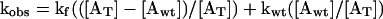 |
4 |
The dissociation equilibrium constant for actin and wild-type profilin is as follows:
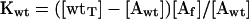 |
5 |
where wtT is total wild-type profilin. Under the conditions of this experiment essentially all of Y79R profilin is free, so the dissociation equilibrium constant for actin and Y79R profilin is as follows:
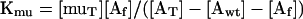 |
6 |
where muT is the total Y79R profilin.
We canceled Af and resolved [Awt] from Eqs. 5 and 6:
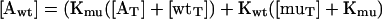 |
7 |
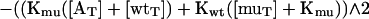 |
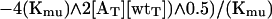 |
Eq. 7 was substituted into Eq. 4 and fit with Kaleidagraph software to give Kmu
For reasons that we do not understand 100 μM concentrations of wild-type or mutant profilin inhibit nucleotide dissociation from actin ∼10% compared with lower concentrations, so we corrected for this in the analysis of competition data.
Spontaneous actin polymerization was initiated by adding 20 μl of 10× KMEI (0.1 M imidazole, 0.5 M KCl, 10 mM MgCl2, 10 mM EGTA, pH 7.0) to 180 μl of 5 μM Mg-ATP muscle actin monomers, 0.1 μM pyrene-labeled actin monomers, and 0–50 μM profilin. The polymer concentration was monitored by pyrene fluorescence with excitation at 365 nm and emission at 407 nm (Kouyama and Mihashi, 1981).
Genetic Complementation of Profilin-Null Strain and Temperature-sensitive Strain
Wild-type and mutant profilins cDNAs in pREP 81 plasmid under the control of the weakest thiamine-repressible nmt1 promoter were transformed into profilin null or ts strains by electroporation. Transformed cells were grown on plates with selective media at 32°C for the null strain and 25°C for the ts strain. Complementation of heterozygous null strains was assessed by random sporulation assays at 32°C.
Wild-type and ts profilin strains with complementing plasmids were grown in liquid media at 25°C. While still in exponential phase cells were diluted to OD595 of 0.25 and then grown for 12 h at 36°C while maintaining the exponential phase by diluting the culture. Cells were mounted on glass slides and DIC micrographs were taken with a digital camera. The length and width of 100–200 cells were measured using Metamorph (Universal Imaging, West Chester, PA) software.
Fluorescence Microscopy
Wild type, ts strain, and ts strains with plasmids carrying wild-type or mutant profilins were grown at 25°C and then shifted to 36°C for 8 h. Cells were collected at both temperatures and fixed in 70% ethanol. Cells were rehydrated in H2O and stained with 10 μg/ml 4,6-diamidino-2-phenylindole (DAPI) and 50 μg/ml Calcofluor in H2O at room temperature for 10 min. Images of cells stained with Calcofluor and DAPI were taken sequentially (Balasubramanian et al., 1994) with a digital camera.
Immunoblot Analyses
Rabbit was immunized with purified recombinant S. pombe profilin and antibodies purified from serum by adsorption to and elution from an immunoblot with immobilized profilin (Pollard, 1984)
Cells growing exponentially in 20 ml of liquid medium at 25°C or for 4 h at 36°C were spun down and resuspended in 200 μl of lysis buffer (25% sucrose, 20 mM imidazole pH 7.0, 5 mM EDTA, 5 mM DTT, 1 mM phenylmethylsulfonyl fluoride and 1 tablet of protease inhibitor cocktail [Roche Molecular Biochemicals, Indianapolis, IN] per 25 ml). Cells were lysed at 4°C with glass beads in a Fastprep agitator (Bio-101) for 4 × 18 s at a speed of 5.5. We added 75 μl more lysis buffer and lysates were collected by poking a hole at the bottom of tube followed by centrifugation at 3000 rpm for 1 min. Lysates were centrifuged for 20 min at 4°C in an Eppendorf centrifuge. Supernatants were assayed for protein by Bradford assay (Bio-Rad, Richmond, CA) and prepared for gel electrophoresis by boiling 5 min in SDS sample buffer. Samples containing roughly 25 μg of protein were run on 16% polyacrylamide gels in SDS. Standards consisted of 0.3–50 ng of purified S. pombe profilin mixed with 25 μg of lysate of the cdc3 ts strain, which contains no detectable profilin under our lysis conditions. Proteins were transferred to polyvinylidene difluoride membranes (Millipore, Bedford, MA) overnight at 23 V in buffer consisting of 10 mM 3-(cyclohexylamino)propanesulfonic acid pH 11.0 and 10% methanol. The membrane was blocked with Tris-buffered saline/Tween 20 (TBST) (10 mM Tris-Cl pH 7.5, 150 mM NaCl, 0.1% Tween 20) containing 5% dry milk for 4 h and incubated with affinity-purified anti-profilin antibody (1:100 dilution) in TBST with 1% dry milk for 4 h. Membranes were washed three times in TBST over 20 min. Membranes were treated with a 1:10,000 dilution of goat anti-rabbit antibody conjugated with horseradish peroxidase (Zymed Laboratories, South San Francisco, CA) in TBST with 1% dry milk for 1 h and bound antibodies were visualized with enhanced chemiluminescence reagents (Amersham Pharmacia Biotech, Piscataway, NJ). Films were scanned and the profilin content of each sample calculated by comparison with the internal standard curve.
RESULTS
Design of Point Mutations to Affect Specific Ligand Interactions
Our aim was to find point mutations of profilin that are stably folded but eliminate interactions with one and only one class of ligand, actin (Arp2/3 complex), poly-l-proline, or phosphatidylinositol biphosphate (PIP2), so that we could test which interactions are essential for viability. Our strategy was to replace amino acids known to be involved with each interaction. Structural studies of profilins from different species have defined the binding sites for actin (Schutt et al., 1993) and poly-l-proline (Archer et al., 1993; Mahoney et al., 1997, 1999). The PIP2 binding site is less well defined (Sohn et al., 1995; Chaudhary et al., 1998). We used atomic structures to align the sequences of bovine (Schutt et al., 1993), Saccharomyces cerevisiae (Eads et al., 1998), and Acanthamoeba profilins (Vinson et al., 1993; Fedorov et al., 1994). The sequence of S. pombe profilin was aligned with these sequences without ambiguity. This alignment was used to identify residues of S. pombe profilin involved with each interaction (Figure 1). We used different strategies to eliminate the binding of each ligand.
Figure 1.
Mutations designed to cover all residues involved with binding various ligands, e.g., the binding with actin (a), poly-l-proline (p), and PIP2 (+), marked on top of the wild-type S. pombe profilin sequence. Some residues may participate in both actin and PIP2 binding. Each point mutant is represented by the mutated amino acid that aligns with the wild-type sequence. E42K (*) is the mutation of profilin in cdc3-124 ts strain. C89S (*) is a control mutant for structure stability. Mutants that failed to complement both null (32°C) and ts (36°C) strains are bold and underlined; mutants that complemented the ts at 36°C but not the null strain at 32°C are bold only. Four of these noncomplementing mutants complement null strain at 25°C (i).
Actin Binding.
About 20 amino acids from profilin contribute to a large interface of ∼2250 Â2 with actin in the complex of bovine profilin with β-actin (Schutt et al., 1993). To create steric interference between the proteins, we first replaced many residues, one at a time with amino acids with bulky side chains such as Trp, Tyr, or Phe (Figure 1). Second, four conserved profilin residues with positive charges (K67, R72, K81, K84) pair with Glu residues on actin. This suggested that ionic interactions between these residues might stabilize the binding. We substituted up to six different residues for these four basic residues. We also introduced single Glu residues at seven places on the interface of profilin with actin to alter its electrostatic potential. In bovine profilin, two longer loops flank both ends of strand β6. Some residues in these loops interact with actin but have no counterpart in S. pombe profilin.
Poly-l-proline Binding.
Residues involved with poly-l-proline binding are the most conserved across all species. The crystal structure of poly-l-proline bound to profilin (Mahoney et al., 97) shows two types of interactions: hydrophobic interactions involving the pyrrolidine ring of poly-l-proline with aromatic side chains of conserved amino acids on profilin; and hydrogen bonds between backbone carbonyl oxygens of poly-l-proline and side chains on profilin. We substituted a variety of amino acids for conserved residues in the poly-l-proline binding site in an attempt to disrupt both types of interactions.
PIP2 Binding.
The binding site for PIP2 is not well defined, but is likely to involve positively charged amino acids (Fedorov et al., 1994). S. pombe profilin has six Lys, four Arg, and one His. We mutated each to Glu, Ala, Leu, and Trp (or Tyr). Four of these residues are also part of actin binding interface (Figure 1).
Complementation of S. pombe Strains with Temperature-sensitive or Null Mutations of Profilin
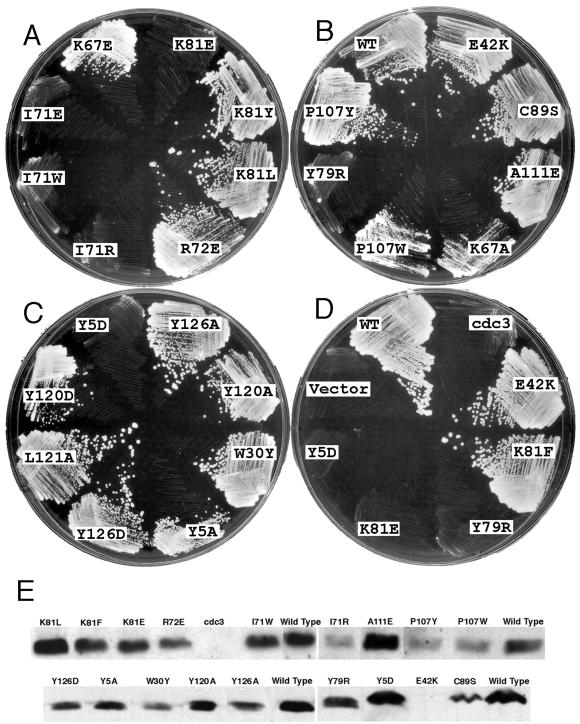
We transformed a haploid strain carrying a profilin-ts allele (cdc3-124) and a diploid strain containing one profilin-null allele with plasmids carrying each of 85 profilin mutants under the regulation of the weakest thiamine-suppressible nmt1 promoter in plasmid Rep81. This test included 48 mutants designed to interfere with actin binding, nine mutants targeted to poly-l-proline binding, and 46 mutants targeted to PIP2 binding (18 of these overlapped with mutants for actin binding). We tested two additional point mutants, E42K (the mutation found in the cdc3-124 ts allele) and C89S (a mildly unstable core mutation). Neither of these residues participates directly in any known binding interaction. We tested for complementation of the ts strain at 36°C (Figure 2, A–D, and Table 1) and did random sporulation assays at 32 and 25°C to examine whether mutants complemented the null allele (Table 1). A few mutants were also tested for complementation of the ts strain by using the strongest nmt1 promoter in plasmid Rep3 (Table 1).
Figure 2.
(A–D) Complementation of cdc3-124 ts strain at 36°C by expression of mutant profilins from the weakest nmt-1 promoter in pRep81 plasmid in the absence of thiamine. (A) Actin binding mutants. (B) Actin binding mutants, wild-type profilin (WT), mutant profilin (E42K) for cdc3-124 strain, and structure stability control mutant C89S. (C) Poly-l-proline binding mutants. (D) Mutants with profound loss of poly-l-proline binding Y5D, actin binding K81E, and nucleotide exchange activity Y79R, mutant with increased nucleotide exchange activity K81F, ts mutant (E42K), wild-type (WT) profilin, vector control as well as cdc3-124 strain. (E) Immunoblots to measure the expression levels of mutant profilins in cdc3-124 ts strain shifted to 36°C for 4 h. Approximately 25 μg of protein was loaded in each lane. Table 1 lists the quantitation of these results. No profilin was detected in the cdc3-124 strain without transformation or when transformed with mutant E42K.
Table 1.
Summary of results
| Mutants | Complementation
|
Mutant
proteins
|
Mutant in cdc3 ts strain
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Rep81
|
Rep3
|
Urea (M) | PLP
|
Nucleotide
exchange
|
Polymerization
|
Expression level 36°C 4 h (μg/10 mg protein) | Average length/ width 36°C 12 h (nm) | Increased cell density 36°C 12 h (fold) | ||||
| Null
|
Ts
|
Ts
|
||||||||||
| 25°C | 32°C | 36°C | 36°C | Affinity fraction of wild typea | Affinity fraction of wild typea | Acceleration fraction of wild typea | fraction of wild type affinitya | |||||
| Controls | ||||||||||||
| Wild type | Yes | Yes | Yes | Yes | 4.5 | (55 μM) | (0.21 μM)b | (2.3-fold) | 1.0 | 0.67 | 12.4/3.9 | 9.0 |
| C89S | Yes | Yes | Yes | NT | 3.2 | 1.1 | 1.0b | 1.0 | NT | 0.53 | 13.0/3.9 | 8.9 |
| E42K | Yes | No | Yes | Yes | 2.2 | NT | NT | NT | NT | N/D | 18.6/4.7 | 6.7 |
| PLP binding mutants | ||||||||||||
| L121A | NT | Yes | Yes | NT | NT | 0.11 | NT | NT | NT | 0.36 | 14.7/4.5 | 9.2 |
| W30Y | NT | Yes | Yes | NT | 4.5 | 0.32 | NT | NT | NT | 0.28 | 14.7/4.6 | 8.1 |
| Y5A | Yes | Yes | Yesc | NT | 4.0 | 0.26 | 0.7b | 0.77 | NT | 0.53 | 21.3/5.4 | 6.0 |
| Y5D | No | No | No | No | 3.3 | 0.011 | 0.68b | 1.3 | 0.5 | 0.60 | 25.5/5.9 | 4.5 |
| Y120A | NT | No | Yes | NT | 4.1 | 0.037 | 0.15b | 1.1 | NT | 0.44 | 20.6/5.5 | 6.0 |
| Y120D | No | No | Yes | NT | 3.9 | 0.031 | 1.4b | 0.92 | NT | 0.66 | 22.9/5.6 | 5.4 |
| L121R | No | No | No | NT | 1.6 | NT | NT | NT | NT | NT | NT | NT |
| Y126D | No | No | Yesc | NT | 4.1 | 0.031 | 0.31b | 1.2 | NT | 0.41 | 17.3/4.7 | 4.7 |
| Actin binding mutants | ||||||||||||
| K67A | No | No | Yesc | NT | 4.4 | 0.91 | <0.01 | 0 | <0.025 | NT | 15.1/4.7 | 7.5 |
| K67E | No | No | Yesc | Yesc | 4.2 | 1.0 | <0.01 | 0 | <0.025 | NT | 22.3/5.9 | 5.1 |
| K67L | No | No | Yesc | Yes | P/F | NT | NT | NT | NT | NT | NT | NT |
| K67W | No | No | No | NT | NT | NT | NT | NT | NT | NT | NT | NT |
| I69W | No | No | Yesc | NT | P/F | NT | NT | NT | NT | NT | NT | NT |
| I71E | No | No | No | No | 4.0 | 1.1 | <0.01 | 0 | <0.025 | 0.63 | 26.7/6.4 | 4.9 |
| I71R | No | No | No | No | 3.8 | 1.0 | <0.01 | 0 | <0.025 | 0.28 | 25.1/6.3 | 4.5 |
| I71W | No | No | No | NT | 4.5 | 1.1 | <0.01 | 0 | <0.025 | 1.0 | 24.3/6.3 | 4.5 |
| R72E | No | No | Yesc | Yes | 3.6 | 0.59 | NT | NT | NT | 0.49 | 21.8/5.5 | 4.4 |
| S77W | Yes | No | Yes | NT | P/F | NT | NT | NT | NT | NT | NT | NT |
| Y79R | No | No | No | No | 3.3 | 0.42 | 0.053d | 0 | 0.13 | 0.62 | 27.2/6.6 | 5.0 |
| K81A | NT | Yes | Yes | NT | 4.4 | 1.0 | 0.12b | 1.5 | NT | NT | 13.4/4.2 | 8.8 |
| K81E | No | No | No | No | 4.5 | 1.0 | <0.01 | 0 | <0.025 | 0.67 | 25.0/5.8 | 4.5 |
| K81F | NT | Yes | Yes | NT | 4.5 | 0.77 | 0.027b | 4.4 | 0.2 | 0.79 | 13.0/4.4 | 8.3 |
| K81L | NT | Yes | Yes | NT | 4.0 | 0.9 | 0.25b | 6.3 | NT | 1.0 | 13.2/4.4 | 8.9 |
| K81Y | NT | Yes | Yes | NT | 4.5 | 1.0 | 0.018b | 3.7 | NT | 0.35 | 13.2/4.6 | 8.6 |
| K84W | Yes | No | No | NT | P/F | NT | NT | NT | NT | NT | NT | NT |
| G86W | Yes | No | No | NT | NT | NT | NT | NT | NT | NT | NT | NT |
| P107W | No | No | Yesc | NT | 4.2 | 1.1 | 0.04b | 3.3 | 0.5 | 0.39 | 19.9/6.2 | 4.4 |
| P107Y | No | No | Yes | NT | 3.6 | 1.1 | 0.055b | 0.54 | NT | 0.38 | 20.2/5.6 | 8.3 |
| A111E | No | No | Yes | NT | 4.0 | 0.91 | 0.023b | 0.92 | NT | 1.2 | 15.9/4.8 | 6.9 |
| Column | 1 | 2 | 3 | 4 | 5 | 6a | 7a | 8a | 9a | 10 | 11 | 12 |
P/F, Partially folded; N/D, not detectable; NT, not tested.
The numerical values for the mutants in column 6–9 are the fractional affinities or activities compared with wild type (e.g. “0.037” indicates that the mutant retains 3.7% of wild type activity).
Affinity of wild type (Kd=0.21 μM) and fractional activity of mutants determined by nucleotide exchange.
Complements but grows slower; colonies are small.
By inhibition of wild type promoted nucleotide exchange.
Although profilin is essential for viability of S. pombe (Balasubramanian et al., 1994) and all of our point mutations were designed to interfere with ligand binding, a remarkable 64 of 87 mutants complemented both the null and ts mutations in the absence of thiamine. Eight mutants failed both complementation tests: Y5D, K67W, I71E, I71R, I71W, Y79R, K81E, and L121R. Eleven more mutants failed to complement the null allele, but managed to complement the ts allele (K67A, K67E, K67L, I69W, R72E, P107Y, P107W, A111E, Y120A, Y120D, Y126D). Another two mutants complemented the null allele at 25°C but not at 32°C (K84W and G86W). E42K and S77W complemented the ts strain and null strain at 25°C but not 32°C. The substitutions in all 23 noncomplementing mutants are located on a surface where profilin interacts with either actin or poly-l-proline except E42K, which is on the “back” side of the protein. Expression of ts profilin E42K from the Rep81 promoter in the absence of thiamine rescued the E42K ts mutant strain (cdc3-124) at the restrictive temperature of 36°C, suggesting that the concentration of active profilin in the cdc3-124 ts strain is too low for growth at the restrictive temperature. Mutants S77W, K84W, and G86W rescued the null at 25° but not at 32°C, indicating these profilin alleles are also temperature sensitive.
At the restrictive temperature used for complementation tests, 36°C, most mutant profilins were expressed from plasmids in the cdc3-124 background at approximately the same level as genomic wild-type profilin, judging from immunoblot assays (Figure 2E and Table 1). No profilin was detected in the cdc3-124 ts strain under our lysis conditions unless transformed with a profilin plasmid. Wild-type strain KGY247 has 0.68 μg of profilin per 10 mg of total protein. cdc3-124 transformed with wild-type profilin has 0.67 μg of profilin per 10 mg of total protein. All mutant profilins were expressed at a similar level (0.28–1.2 μg/10 mg protein), except for the cdc3-124 mutant E42K, which was undetectable.
Structural Stability of Mutants
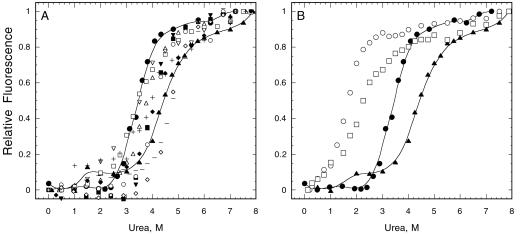
We used urea denaturation to test the stability of noncomplementing mutant proteins (Figure 3, A and B, and Table 1) to rule out folding defects as a trivial explanation for the failure of mutants to complement either ts or null alleles. For controls in this and the following assays we examined up to six complementing mutants of the same residue. Wild-type S. pombe profilin undergoes a shift in intrinsic fluorescence over a narrow range of urea concentrations as the protein makes a transition from folded to unfolded. Making the usual two state (folded/unfolded) assumption (Tanford, 1968; Santoro and Bolen, 1988), the concentration of urea at the midpoint of this transition reflects the equilibrium constant for folding. A difference from wild type represents a change in stability. The transition urea concentration for both recombinant and native wild-type S. pombe profilin is 4.5 M urea. This is higher than S. cerevisiae and Acanthamoeba profilins (3.4–3.5 M). Substituting tryptophan for another residue changed the intrinsic fluorescence of the folded protein, but did not interfere with the urea denaturation experiments.
Figure 3.
Urea denaturation of purified profilins monitored by intrinsic fluorescence. (A) Mutants that are equal to or more stable than C89S (●) were considered as stable, including poly-l-proline binding mutants Y5A (▵), Y5D (□), Y120D (▪), Y120A (▾), Y126D (○) and actin binding mutants Y79R (▿), K81E (⋄), K81Y (−), P107Y (+), A111E (♦), and wild-type profilin (▴). (B) Unstable mutants E42K(□) and L121R (○) denature at lower urea concentrations than C89S (●) and wild-type profilin (▴).
Mutant C89S served as a control for the influence of stability on physiological function. Although much less stable than wild-type profilin, with a transition concentration of 3.2 M urea, C89S rescued both the profilin ts and null alleles as well as wild-type profilin (Figure 2B and Table 1). Cys 89 is buried on strand β7, outside the binding surface for actin, far from the poly-l-proline binding surface and is unlikely to contribute to PIP2 binding. C89S bound poly-l-proline with a Kd value of 50 μM (Figure 4A) and actin with a Kd value of 0.21 μM (Figure 5A) (not significantly different from wild-type profilin). This argues that an abnormal phenotype for any mutant with a transition concentration >3.2 M urea is not due simply to unstable structure. Folding instability seems to account for the temperature sensitivity of cdc3-124 where the substitution E42K is far from any known ligand binding site. Its transition concentration was 2.2 M urea (Figure 3B). Recombinant E42K protein was extremely difficult to purify from bacteria and aggregated at 4°C without urea.
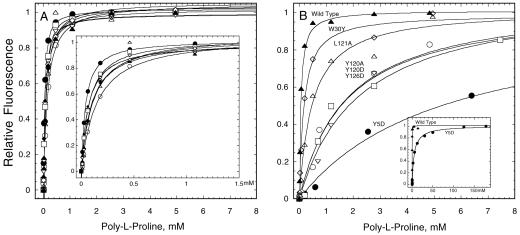
Figure 4.
Binding of profilins to poly-l-proline measured by intrinsic fluorescence. The dependence of the fluorescence changes on profilin concentration is fit with binding isotherms to measure the equilibrium constant. (A) Wild-type (▴) and actin binding mutants Y79R (○), K81E (♦), K81Y (▿), P107W (▵), P107Y (⋄), A111E (□), and minimally stable mutant C89S (●). Inset: Curve fits at low concentrations of poly-l-proline. (B) Wild-type profilin (▴) and poly-l-proline binding mutants W30Y (⋄), L121A (▵), Y120A (□), Y120D (○), Y126D (▿), and Y5D (●). Inset: Curve fit for wild-type and Y5D over a much larger range of poly-l-proline (150 mM).
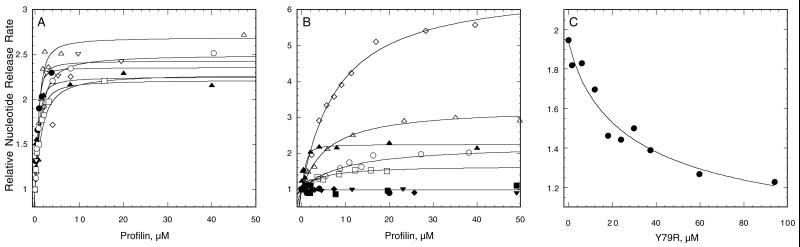
Figure 5.
Nucleotide release assays using 1.2 μM εATP muscle actin and a range of profilin concentrations. The dependence of the rates of nucleotide release relative to actin alone on the concentration of profilin are fit with binding isotherms to measure the equilibrium constant. (A) Wild-type (▴), C89S (●) and poly-l-proline mutants Y5A (⋄), Y5D (▵), Y120A (▿), Y120D (□), and Y126D (○). (B) Actin binding mutants I71R (▪), I71W (▾), Y79R (●), and K81E (♦) have no detectable effect on the rate; P107W (▵), P107Y (□), and A111E (○) required higher concentrations than wild-type to promote exchange but maximum rates similar to wild-type (▴); K81F (⋄) requires higher concentrations than wild-type but increases nucleotide release more than wild-type. (C) Effect of a range of concentrations of Y79R on nucleotide release catalyzed by 1.5 μM wild-type profilin.
We limited our biochemical characterization to stable mutants with transition midpoints higher than the C89S control at 3.2 M urea. This eliminated E42K and L121R (Figure 3B). We also set aside K67L, I69W, K84W, and S77W, because they lacked a distinct urea denaturation transition and appeared to be partially unfolded without urea (our unpublished results). We eliminated K67W, designed as an actin binding mutant, because it failed to bind the poly-l-proline affinity column. We did not test G86W, but assumed that its temperature-sensitive phenotype is due to structural instability. We set aside K67A and K67E, because they aggregated in the actin polymerization buffer.
This screen yielded 13 structurally stable mutants that failed to complement the profilin ts and/or profilin null strains, consisting of four mutants designed to compromise poly-l-proline binding (Y5D, Y120A, Y120D, Y126D) and nine designed to compromise actin binding (I71E, I71R, I71W, R72E, Y79R, K81E, P107W, P107Y, A111E). For controls, we included structurally stable mutants that complemented both null and ts strains: three designed to compromise poly-l-proline binding (Y5A, W30Y, L121A) and four potential actin binding mutants (K81A, K81F, K81L, K81Y).
Poly-l-Proline Binding by Wild-Type and Mutant S. pombe Profilins
We tested each purified profilin for poly-l-proline binding by using the enhancement of intrinsic fluorescence as the assay (Figure 4, A and B, and Table 1). The dissociation equilibrium constant (Kd) of recombinant wild-type S. pombe profilin was 55 μM proline residues. Native wild-type profiin has a Kd value of 46 μM. For a decamer, the minimal length of poly-l-proline for high-affinity binding (Perelroizen et al., 1994; Petrella et al., 1996), this corresponds to a Kd value of 5.5 and 4.6 μM, respectively. This affinity of S. pombe profilin for poly-l-proline is slightly higher than Acanthamoeba profilin (twofold) and fivefold higher than recombinant S. cerevisiae profilin (Petrella et al., 1996; Eads et al., 1998). All stable mutants in the actin binding site bound poly-l-proline normally (Figure 4A and Table 1).
The stable mutants that targeted the poly-l-proline binding site had a range of lower affinities for poly-l-proline (Figure 4B and Table 1), but all bound actin normally (Figure 5A and Table 1). Y5D, the only poly-l-proline mutant that failed to complement both null and ts allele, had the greatest loss of affinity for poly-l-proline, 94-fold. Mutants Y120A, Y120D, and Y126D had moderately lower affinity for poly-l-proline (between 27- and 32-fold; Figure 4B and Table 1) and complemented the profilin ts strain but not the profilin null strain (Figure 2C). Mutations Y5A, W30Y, and L121A reduced the affinity for poly-l-proline <10-fold (Figure 4B and Table 1). S. pombe tolerated this small reduction in poly-l-proline binding, because all of these mutants complemented both the ts and null strains (Figure 2C and Table 1). Thus, moderate loss of affinity for poly-l-proline compromised biological function and a 100-fold loss of affinity was incompatible with life.
Characterization of Actin Binding by S. pombe Wild-Type and Mutant Profilins
Nucleotide Release Assay.
We used the ability of profilin to promote nucleotide release from monomeric actin to measure the affinity of profilin for actin (Figure 5, A–C). In the low-salt conditions of our assay, Mg-εATP-rabbit muscle G-actin exchanged its bound εATP with an excess of unlabeled ATP with first order kinetics. The kobs of 0.0065 s−1 is the dissociation rate constant for εATP (k−1), the rate-limiting step. A saturating concentration of wild-type S. pombe profilin increased the rate of exchange by 2.3-fold, up to 0.015 s−1, similar to S. cerevisiae profilin (Eads et al., 1998). The dissociation equilibrium constant (Kd) for profilin binding actin was determined from the dependence of k−1 on profilin concentration (Figure 5, A and B). By this criterion wild-type S. pombe profilin bound rabbit muscle ATP-actin monomers with a Kd value of 0.21 μM (Table 1). S. pombe profilin may bind its own actin somewhat better, because a similar profilin from another lower eukaryote, Acanthamoeba, binds its own ATP-actin more strongly (0.1 μM) than muscle actin (0.48 μM) (Vinson et al., 1998).
Testing the effects of profilin mutations on actin nucleotide exchange revealed an unanticipated degree of complexity (Table 1). No profilin mutation enhanced affinity for actin or inhibited the rate of nucleotide exchange below that of actin alone, but various mutations reduced affinity for actin in combination with nucleotide exchange activity ranging from 0 to 6 times that of wild-type profilin.
Profilins with mutations that complement neither null nor ts profilin mutations (I71E, I79R, I71W, Y79R, K81E) had no detectable effect on nucleotide exchange (Figure 5B and Table 1), even at a concentration of 100 μM (our unpublished results). They might have a Kd value for actin >250 μM, or might bind actin without affecting nucleotide exchange. To detect the latter situation, we used a competition assay to test whether 30 μM mutant profilin inhibited nucleotide exchange by 1 μM wild-type profilin. I71E, I71R, I71W, and K81E do not interfere with nucleotide exchange catalyzed by wild-type profilin, but Y79R inhibited nucleotide exchange down to the level of actin alone. The concentration dependence of this inhibition by Y79R gave a Kd value of 4.0 μM for ATP-actin (Figure 5C). Thus, the affinity of Y79R for actin is 19-fold lower than wild type, but it has no effect on nucleotide release when bound to actin.
Mutant profilins that failed to complement the null mutation, but allowed the ts mutant to grow (P107W, P107Y, A111E), had modestly reduced affinities for actin (25–44-fold) and when bound to actin all catalyzed nucleotide exchange normally (Figure 5B and Table 1). R72E belongs to this group of mutants, but we did not test its actin binding.
A selection of mutant profilins that complemented both the null and ts mutants (K81A, K81F, K81L, K81Y) had normal to moderately reduced affinity (4–56-fold) for actin, and several of these low-affinity profilins had remarkably enhanced ability to catalyze nucleotide exchange, up to 6.3-fold better than wild-type profilin (Figure 5B and Table 1). Enhanced nucleotide exchange appeared to compensate completely for weak binding.
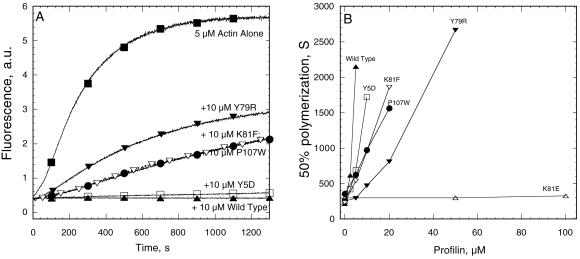
Effect on Actin Polymerization.
Profilin inhibits spontaneous actin polymerization by preventing actin dimer and trimer formation and pointed end elongation (Pollard and Cooper, 1984). Although this assay does not measure affinity directly owing to complicated effects of profilin on multiple steps in the reaction, we could compare the activity of mutant profilins with the concentration dependence of inhibition by wild-type profilin (Figure 6, A and B, and Table 1). Our stability benchmark C89S and the severe poly-l-proline binding mutant Y5D inhibited spontaneous actin polymerization within a factor of 2 of wild-type profilin. Mutants that failed to bind actin monomers in the nucleotide exchange assay (I71E, I71R, I71W, K81E) also failed to inhibit spontaneous actin polymerization (Figure 6B and Table 1). Y79R inhibited spontaneous actin polymerization even though it did not affect nucleotide exchange (Figure 6, A and B). K81F and P107W both inhibited spontaneous actin polymerization better than they bound to actin in the nucleotide exchange assay, but neither bound actin well by this criterion.
Figure 6.
Effect of profilins on spontaneous polymerization of 5 μM Mg-actin with 0.1 μM pyrene-labeled actin measured by pyrene fluorescence. (A) Time course of polymerization of actin alone (▪) and with wild-type profilin (▴), poly-l-proline binding mutant Y5D (□), and actin binding mutants Y79R (▾), K81F (▿), and P107W (●). (B) Dependence of the time required for half maximal polymerization on the concentrations of various profilin mutants. Symbols are same as in A with the addition of K81E (▵) that does not inhibit actin polymerization at even 100 μM.
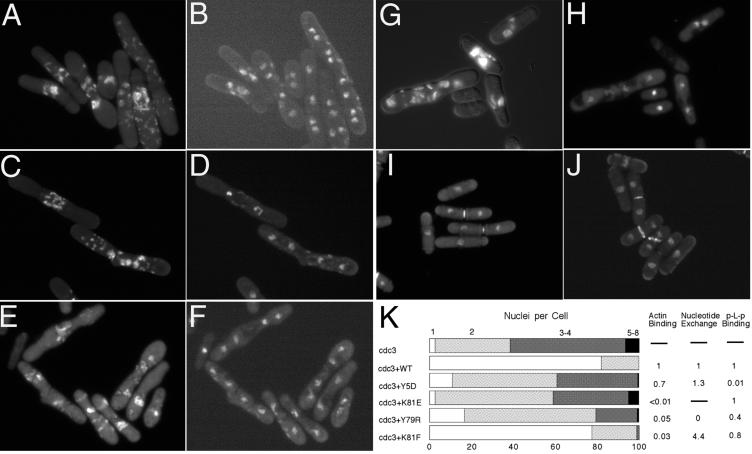
Phenotype of cdc3-124 Cells Expressing Wild-Type and Mutant Profilins
We studied the morphology of log phase cdc3-124 cells depending upon wild-type and mutant profilins for growth at the restrictive temperature by DIC microscopy (to observe shape and measure size) and fluorescence microscopy after staining with DAPI (to count the number of nuclei) and Calcofluor (to assess septum formation) (Figure 7, A–K, and Table 1). At 25°C, cdc3-124 cells were indistinguishable from wild-type cells in terms of their shape, size, number of nuclei, and septum staining. Most cells had one nucleus (76%); the remaining cells had two nuclei. Compact septa were present in the middle of 18% of cells. Expression of wild-type and mutant profilins in cdc3-124 strain at 25°C did not alter morphology.
Figure 7.
Morphology of cdc3-124 ts strain expressing various profilins from pRep 81 plasmid under the control of the weakest nmt1 promoter after incubation for 8 h at 36°C in the absence of thiamine. Cells were fixed and stained with DAPI and Calcofluor. Calcofluor staining was recorded first (A, C, E, G). After bleaching of the Calcofluor, DAPI staining was recorded (B, D, F, H). (A and B) cdc3-124 without transformation. (C and D) Y5D. (E and F) K81E. (G and H) Y79R. Calcofluor and DAPI were recorded in the same time for wild-type profilin (I) and K81F (J). (K) Quantitation of nucleus number in comparison with biochemical data.
At 36°C, cdc3-124 cells were defective in cytokinesis (Balasubramanian et al., 1994). After 8 h at 36°C elongated or dumbbell-shaped cells accumulated two (36%), four (54%), or even more (6.8%) nuclei (Figure 7K). Calcofluor staining was stronger than normal and most cells had abnormal septa, usually concentrated between the nuclei but also dispersed elsewhere (Figure 7, A and B). Lengths ranged from 12 to 57 μm. Episomal expression of wild-type profilin completely suppressed the cdc3-124 phenotype at 36°C. Cell size and the number of nuclei were indistinguishable from cdc3-124 at 25°C and wild-type cells at 25°C or 36°C (Figure 7, I and K, and Table 1).
Poly-l-Proline Binding Mutants.
Y5D profilin with only 1% of wild-type affinity for poly-l-proline failed to complement either null or ts cells. cdc3-124 cells expressing Y5D at 36°C had a terminal phenotype similar to cdc3-124 cells (Figure 7, C, D, and K).
Profilin mutants retaining 3–32% of wild-type affinity for poly-l-proline were generally effective in vivo in proportion to their affinity for poly-l-proline. Profilin mutants with only 3–4% of wild-type affinity for poly-l-proline (Y120A, Y120D, Y126D) did not rescue profilin null strains but allowed cdc3-124 cells to grow at 36°C in spite of defects in cytokinesis. On average these cells were ∼50% longer than wild-type cells (Table 1) and 54–57% had two nuclei and 22–25% cells had more than two nuclei. Calcofluor staining showed defects similar to cdc3-124 at the restrictive temperature. Profilin mutants W30Y and L121A retaining 32 and 11% of wild-type affinity for poly-l-proline complemented the null mutation and completely corrected the cytokinesis defects of cdc3-124 cells at 36°C.
Mutant Y5A is an exception. This mutation reduced affinity for poly-l-proline by only 3.8-fold, similar to mutant W30Y with full biological activity. Nevertheless, cdc3-124 cells with Y5A grew slowly at 36°C with a clear cytokinesis defect: cells were 50% longer than wild-type cells and 71% had two or more nuclei. The Y5A mutation may compromise binding to a key biological ligand more than binding to poly-l-proline.
Actin Binding Mutants.
Profilin mutants with actin binding reduced <1% of wild-type rescued neither null nor profilin ts cells. For example, cdc3-124 cells expressing profilin mutant K81E with <1% wild-type affinity did not grow at 36°C and had the same morphological defects as cdc3-124 cells (Figure 7, E, F, and K).
Profilin mutants with 2–10% wild-type affinity for actin in the nucleotide exchange assay failed to rescue profilin null cells and varied in their ability to rescue profilin ts cells. Expression of profilin mutants P107W, P107Y, and A111E allowed the ts strain to grow at 36°C but the cells were moderately enlarged (Table 1) with defects in Calcofluor staining similar to cdc3-124 at 36°C.
The behavior of two mutant profilins in these rescue assays argues for the importance of profilin in actin nucleotide exchange. Mutant Y79R, which retained 5% of affinity for actin in the competitive nucleotide exchange assay and 13% affinity in the polymerization assay but no nucleotide exchange activity, failed to complement null cells. cdc3-124 cells expressing Y79R did not grow at 36°C, although fewer cells had multiple nuclei than without this mutant profilin (Figure 7, G, H, and K). On the other hand, K81F retained 3% of wild-type actin binding by nucleotide exchange and 20% by the polymerization assay, but had the novel ability to stimulate nucleotide exchange 4 times better than wild-type profilin. K81F not only complemented the null mutation but also allowed the ts strain to grow at 36°C with nearly as few morphological defects as wild-type profilin (Figure 7J). At the restrictive temperature 77.5% of these cells had one nucleus, 21.4 had two nuclei, and only 1% had more than two nuclei (Figure 7K). The only defects were a slight increase septal materials or irregular septum shape or septum mislocalization in about one-third of the cells with stained septa. Apparently, these minor defects do not hinder cell division.
DISCUSSION
By creating and characterizing a large collection of point mutations, we now have a clearer picture of the biochemical requirements for profilin function in fission yeast. We paid particular attention to two factors that might complicate interpretation of complementation experiments: protein stability and expression levels. We concentrated on those noncomplementing mutant profilins that are stable and expressed at levels close to endogenous profilin in the wild-type strain.
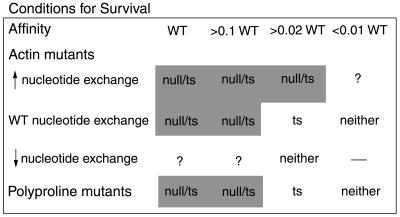
We were fortunate that the cdc3-124 mutation has a dramatic phenotype at the restrictive temperature, yet episomal expression of wild-type profilin completely suppresses these abnormalities. Interpretation of the functional tests in vivo depended on quantitative analysis of the biochemical properties. Comparison of quantitative biochemical assays with cellular structure and function established how affinity and catalytic activity are related to viability (Figure 8). Exploring a spectrum of biochemical properties required a survey of many different mutants, because it was impossible to predict these properties from the structures. Previous studies of mutant profilins relied largely on qualitative biochemical characterization. Few articles reported binding constants for profilin mutants tested in vivo (supplemental materials, Table 1).
Figure 8.
Dependence of complementation of null and ts mutants on the affinity of profilin mutants for actin and poly-l-proline. Reduced affinities within 10-fold of wild-type profilin (>0.1 WT) are tolerated both in profilin null and ts background; loss of affinities between 10- and 50-fold (>0.02 WT) are able to rescue the ts allele but not the null; >100-fold loss of affinities (<0.01 WT) are incompatible with life. Altered nucleotide exchange activity of actin binding mutants also determined their complementing ability. “?” indicates no mutants to test in these categories in the group with affinity >0.02 WT.
For practical reasons, we used readily available ligands (poly-l-proline and rabbit muscle actin) to characterize our mutant profilins rather than S. pombe actin and the potential poly-l-proline ligands Cdc12p (Chang et al., 1997), verprolin, and Wsp1p. Although using these yeast ligands would have been desirable, none has been purified in quantities sufficient for testing numerous mutants. We feel that the use of rabbit muscle actin is justified by the fact that the profilin binding sites are almost identical on vertebrate and S. pombe actins. Among 22 residues that interact directly with profilin in the crystal structure (Schutt et al., 1993) only residue 371 differs; histidine in muscle actin and tyrosine in S. pombe actin. This must account for the small differences of affinity of, for example, Acanthamoeba profilin for amoeba and muscle actin (Vinson et al., 1998) and the threefold difference in the ability of S. cerevisiae profilin to promote nucleotide release from rabbit muscle and yeast actins (Eads et al., 1998). Nevertheless, the loss of affinities of mutant yeast profilins for poly-l-proline and muscle actin parallel closely the severity of their phenotypes. On the other hand, profilin residues that interact with actin are very variable. Thus, differences in the ability of profilins to promote nucleotide exchange by actin are attributable to profilin alone.
Structural Stability
Protein instability is a potential trivial explanation for the failure of any point mutation in a complementation test. The cdc3-124 mutation E42K illustrates how an apparently harmless amino acid substitution on the surface of the protein well away from any known ligand binding site disables the protein by making it severely unstable. Deletions of residues from the C terminus (Kaiser and Pollard, 1996) also compromise the stability of profilins.
We measured the stability of mutated profilins by urea denaturation, a test similar to thermal denaturation. At the midpoint of these titrations half of the protein is unfolded. Few previous studies of profilin mutants included this key information (supplementary materials, Table 1). Because mutant C89S denatured at 3.2 M urea but was indistinguishable from wild-type profilin in ligand binding and ability to complement the profilin null mutation, we conclude that any mutant denaturing at a urea concentration >3.2 M is stable and that failure to function in vivo is attributable to loss of one or more biochemical activities. Of the profilin mutants with amino acid substitutions on the surface that failed to function in complementation tests, one-third were unstable by this criterion. Wild-type S. pombe profilin is remarkably stable (4.5 M) compared with S. cerevisiae profilin (3.4 M) and Acanthamoeba profilins (3.5 M). S. pombe profilin mutant K67A is also much more stable (4.4 M) than the corresponding S. cerevisiae mutant K66A (2.7 M) (Wolven et al., 2000). It is not known whether other organisms tolerate unstable mutant profilins better than S. pombe.
Strategy for the Poly-l-Proline Site.
The poly-l-proline binding site on profilin includes the phylogenetically most conserved amino acids, notably, W2, Y5, W30, Y120, and Y126. These aromatic residues contribute to binding poly-l-proline through hydrophobic interactions and donating hydrogen bonds to backbone carbonyls (Petrella et al., 1996; Mahoney et al., 1997). We avoided tryptophan-2, because in our experience with Acanthamoeba profilin, mutation of this tryptophan usually compromised stability (Vinson and Pollard, unpublished observations). Similarly, human profilin mutant W3F eluted from a poly-l-proline affinity column with 3 M urea, which might indicate a loss of stability (Ostrander et al., 1999). Human profilin W3N lost not only poly-l-proline binding but also actin binding; it too may have been poorly folded, although this was not tested (Bjorkegren-Sjogren et al., 1997). We explored other residues in the poly-l-proline binding site, namely, Y5, Y120, L121, and Y126, because substitutions were tolerated and all are crucial for poly-l-proline binding.
Strategy for the Actin Site.
Profilin tolerated many mutations of the actin binding surface without a loss of structural stability. However, mutants with tryptophan substitutions on the surface were frequently unstable (K67W, I69W, S77W, K84W).
Conditions for Survival
Ability of profilin to provide fission yeast with biological function depends on its affinity for both actin and poly-l-proline and ability to catalyze nucleotide exchange on actin (Figure 8 and Table 1). Profilin mutations reducing affinity for poly-l-proline or actin >100-fold were unable to complement either profilin null or ts strains, so both poly-l-proline binding and actin binding are essential for the organism. Mutant profilins with affinities for either ligand reduced 20–50-fold complement the ts strain but not the null. Residual function of E42K at the restrictive temperature appears to allow these partially disabled mutant proteins to support viability. The ability of E42K expression from the nmt1 promoter to complement the ts strain verifies residual function of E42K profilin at the restrictive temperature in spite of no protein detected on immunoblots. Profilins with near wild-type affinity complemented both null and ts strains.
Because the phenotype of cdc3-124 cells expressing profilin mutants with drastic losses in affinity for either poly-l-proline (Y5D) or actin (K81E) or actin nucleotide exchange (Y79R) were indistinguishable, all three activities are likely to be required simultaneously for normal function. Similarly, mutants of S. cerevisiae profilin designed to interfere with poly-l-proline binding (W29A, W2A, Y119A) have the same phenotype as profilins with mutations in the actin binding site (Wolven et al., 2000). These budding yeast profilin mutants may benefit from more detailed biochemical analysis, because it is impossible to predict the properties of these substitutions.
Poly-l-Proline Ligand Binding
Poly-l-proline binding is essential for profilin function in fission yeast. With one interesting exception, the defects in poly-l-proline binding paralleled the loss of activity in complementation assays. Residue Y5 is noteworthy, because S. pombe mutant Y5A with only a 3-fold loss of affinity for poly-l-proline barely complemented the ts strain. We speculate that Y5 is important for interaction with a type II polyproline helix containing residues other than proline, perhaps for binding cdc12 during cytokinesis (Chang et al., 1997). This residue was mutated twice previously. Human profilin with the Y6F substitution eluted from a poly-l-proline affinity column with 3.5 M urea (7.5 M urea for wild type), so it either lost stability or affinity for poly-l-proline (Sohn et al., 1995). No other properties were reported for this mutant. Maize profilin with the same Y6F substitution bound actin normally and bound poly-l-proline with affinity twofold higher than wild type (Gibbon et al., 1998). This mutant profilin was more active than wild-type profilin in disrupting the actin cytoskeleton of stamen hair cells.
Actin Binding and Nucleotide Exchange Promoted by Profilin
Although viability generally correlated with affinity for actin, simple loss of affinity for actin cannot explain the phenotypes of all of the actin site mutants. Mutant profilins Y79R, A111E, P107W, and K81F all had actin affinities in the range of 2–5% of wild type but varied in biological function. Their ability to complement profilin null and ts strains paralleled their nucleotide exchange activity. K81F (also K81Y, Table 1) had better nucleotide exchange activity than wild-type profilin and complemented both null and ts strains. A111E and P107W had nucleotide exchange activity similar to wild-type profilin and complemented the ts but not the null strain. Y79R lacking nucleotide exchange could not even complement the ts strain, like mutants without detectable actin binding. These data strongly indicate that nucleotide release is an essential function of profilin in S. pombe.
The biological relevance of profilin promoting nucleotide exchange by actin monomers has been debated since its discovery (Mockrin and Korn, 1980). Profilins vary widely in ability to catalyze nucleotide exchange: human profilin increases nucleotide exchange 40- to 1000-fold relative to actin alone (Goldschmidt-Clermont et al., 1992; Perelroizen et al., 1996; Selden et al., 1999); Acanthamoeba profilin increases nucleotide exchange 8- to 17-fold (Mockrin and Korn, 1980; Vinson et al., 1998); and S. cerevisiae profilin increases nucleotide exchange only threefold (Eads et al., 1998). In each case, the rate of exchange depends not only on the profilin but also on the type of actin, bound nucleotide (ADP or ATP), bound divalent cation (Mg- or Ca-), and buffer conditions.
Plant profilins raise serious questions about the importance of nucleotide exchange activity. Profilins from Arabidopsis, birch, and maize lack nucleotide exchange activity in assays with muscle and plant actins (Perelroizen et al., 1996; Eads et al., 1998; Kovar et al., 2000), although they bind actin (Giehl et al., 1994; Kovar et al., 2000) and promote assembly of actin sequestered by thymosin-β4 (Perelroizen et al., 1996; Ballweber et al., 1998). The relevance of the latter activity is not clear given the absence of thymosin from the Arabidopsis genome. Arabidopsis profilin complemented both the profilin null strain of S. cerevisiae and the cdc3-124 ts strain of S. pombe (Christensen et al., 1996). Expression of maize profilin in Dictyostelium lacking both endogenous profilins suppressed the aberrant cell shape, and increased actin filament staining and developmental defects (Karakesisoglou et al., 1996).
On the other hand, studies of actin filament dynamics have revived the interest in a role for the nucleotide exchange activity of profilin. In vitro profilin overcomes the inhibition of ADP exchange by ADF/cofilins and recycles ATP-actin monomers back to actin filaments (Blanchoin and Pollard, 1998, 1999; Didry et al., 1998; Wolven et al., 2000). When actin turnover is driven by continuous sonication, profilin increases the polymer concentration by accelerating the rate-limiting exchange of ATP for ADP bound to actin monomers (Selden et al., 1999). In S. cerevisiae an actin mutation with a high rate of intrinsic nucleotide exchange overcomes a profilin mutation that reduces its affinity for actin both in in vitro filament turnover assays and in vivo (Wolven et al., 2000). This supports the importance of actin nucleotide exchange under physiological conditions. Our analysis both in vivo and in vitro of fission yeast profilin mutants that only differ in their nucleotide exchange activity provides the strongest evidence so far of the crucial requirement of nucleotide exchange activity for viability, at least in fission yeast.
Additional work is required to reconcile these differences among species. We suggest that the following be considered. 1) S. pombe may rely on the nucleotide exchange activity of profilin more than S. cerevisiae and Dictyostelium, because S. pombe absolutely requires profilin, whereas S. cerevisiae and Dictyostelium are viable (although very sick) without profilin. 2) Other molecules may supplement the nucleotide exchange activity of profilin in S. cerevisiae and Dictyostelium. This is unlikely in S. pombe because null strains are not viable unless provided with a profilin having nucleotide exchange activity. 3) The crucial variable among these species may be the intrinsic rate of nucleotide exchange by actin alone. Maize pollen actin has a higher intrinsic exchange rate than other actins (Kovar et al., 2000) and S. cerevisiae actin with a high exchange rate suppresses defects in profilin (Wolven et al., 2000). Nothing is yet known about the nucleotide exchange rate of S. pombe actin and all of these actins still need to be compared rigorously under identical conditions. 4) The concentrations and activities of other actin binding proteins, particularly ADF/cofilins, need to be considered.
The residue corresponding to fission yeast profilin Y79 is potentially important for nucleotide exchange. Mutant Y79R lacks nucleotide exchange activity and all plant profilins known to lack nucleotide exchange activity (Arabidopsis, birch, and maize) have R at this position (Perelroizen et al., 1996; Eads et al., 1998; Kovar et al., 2000). In addition to the 10 profilins from these three species, we checked 26 other plant species with 39 profilin sequences. Out of a total of 49 plant profilins 47 have R at this position. Moreover, only two profilins from species other than plants (Drosophila and shrimp) use R at this position. The nucleotide exchange activity of these two profilins is not known. On other hand, all vertebrate profilins (13 sequences from human, cow, mouse, rat, chicken, frog, and fish) have Asp at this position and human profilin has the best actin nucleotide exchange activity. Eleven profilin sequences from yeast, ciliate, Dictyostelium, Acanthamoeba, and Physarum use Tyr at this position. Profilins of S. pombe, S. cerevisiae, and Acanthamoeba all have moderate nucleotide exchange activity. Twelve profilin sequences from sea urchin, nematode, sponge, Entamoeba, and trypanosome have a nonpolar residue Leu, Ile, Met, or Phe at this position; their ability to catalyze nucleotide exchange is unknown. Fission yeast profilin K81 also appears to influence nucleotide exchange activity. Substitutions K81F, K81Y, and K81L all increased the nucleotide exchange activity dramatically. Interestingly, either K or R is used at this position among 87 other profilins sequences from 69 species. In a more detailed study of the mechanism of nucleotide exchange, we intend to test a wider range of amino acid substitutions for Y79.
PIP2 Binding
Our work to date does not address the physiological function of profilin binding to PIP2. This question is more complicated experimentally than investigating actin and poly-l-proline binding owing to the fact that the PIP2 binding site is likely to overlap the actin binding site. It is believed that PIP2 binding involves positively charged amino acid(s) on the protein surface interacting with negatively charged phosphate groups of the lipid head group. Among the 11 positively charged residues of S. pombe profilin, only three (K67, R72, K81) are conserved among yeast, amoeba, and vertebrate profilins. These three amino acids all contribute to actin binding. Mutations in these three basic residues gave rise to noncomplementing proteins. Glu substitution for K or R (Table 1, K67E, R72E, K81E) gave the weakest suppression of the cytokinesis defect of cdc3-124. Similarly, alanine substitutions for S. cerevisiae profilin K66, R71, and R80 compromised the ability of profilin to complement profilin null cells (Wolven et al., 2000). To learn whether altered PIP2 binding might explain the K81E phenotype, we tested its binding to PIP2 micelles by gel filtration (Machesky et al., 1990). This mutant profilin bound micelles the same as wild-type profilin (our unpublished results). This is evidence that the complete failure of biological function of mutant K81E is due to its loss of actin binding, not PIP2 binding. Residue R88 of human profilin (corresponding to S. pombe K81) also appears to be more important for actin binding than PIP2 binding. Profilin R88L lacked ability to inhibit actin polymerization or stimulate nucleotide exchange, with only a fourfold loss of PIP2 binding (Sohn et al., 1995).
On the other hand, S. pombe profilin with substitutions of Glu for seven of the other eight (nonconserved) basic residues complemented both the null and ts strains. H100E was a minor exception, because a very small fraction of cells depending on this profilin had a cytokinesis defect. Substitutions of these positively charged amino acids in other profilins (human K25Q and K53I; Dictyostelium K114E; and S. cerevisiae R75A, R75G, and R75E) either maintained wild-type PIP2 binding or complemented profilin null cells (Haarer et al., 1993; Sohn et al., 1995; Lee et al., 2000).
Other Comparisons with Previous Studies
Eighteen previous reports described 66 mutations of various profilins, 45 of which were point mutations (supplemental materials, Table 1). Twenty-four of these point mutations were tested for biological function in homologous or heterologous systems; 14 of these 24 mutants are characterized with at least one biochemical experiment. In general our data agree with the biological tests of previously studied mutants.
Of the reported substitutions in S. cerevisiae profilin six are identical to ours (Y119A = Y120A, R75A = R76A, R75E = R76E, K66A = K67A, R71A = R72A, R71E = R72E) and one is similar (R80A ∼ K81A). Although the tests differed, five pairs of these mutants appeared to have similar phenotypes. For example, budding yeast Y119A was functional in normal medium but not in 2.5% formamide at 36°C, whereas fission yeast Y120A partially complemented the profilin ts strain but not the null strain. One exception is budding yeast R80A that was conditionally lethal in 2.5% formamide at 36°C, whereas fission yeast K81A complemented the null like wild-type profilin. The other exception is budding yeast R71E with wild-type function, whereas fission yeast R72E only partially complemented the profilin ts strain and failed to complement the null strain.
Human profilin R74E (comparable to S. pombe R72) had only 2% of wild-type affinity for actin (Korenbaum et al., 1998). Human profilin H113S (comparable to S. pombe Y120) failed to bind poly-l-proline beads (Bjorkegren-Sjogren et al., 1997). Neither was as effective as wild-type profilin in stimulating Cdc42-induced actin polymerization in extracts of neutrophils (Yang et al., 2000). S. pombe R72E and Y120A/D failed to complement the profilin null strain, but did complement the ts strain.
Substitution of different residues at homologous positions often give similar results in biological tests in diverse organisms. S. cerevisiae mutants R80G and R75G functioned normally like our closest mutants R81A and R76A. Substitution F59A in bovine profilin (comparable to S. pombe F57) reduced actin binding 14-fold but even low-level expression still partially corrected the defects of Dictyostelium lacking both profilin isoforms. S. pombe profilin with a different substitution in this residue (F57D) partially rescued the ts strain. Two mutations of the second conservative Trp (human W31F and S. cerevisiae W29A) were not characterized for stability (Ostrander et al., 1999; Wolven et al., 2000). S. pombe profilin W30Y is stable with only a threefold loss of affinity for poly-l-proline and functions normally in vivo. Maize profilin Y6F (the same position as S. pombe Y5) was discussed above.
Others have tested the biological functions of point mutations of nine residues that we did not test in S. pombe. Three are nonconserved amino acids (human H119E, Dictyostelium K114E, and S. cerevisiae H81A [Suetsugu et al., 1998; Lee et al., 2000; Wolven et al., 2000]). Three (S. cerevisiae S1A, Q3A, D7A; Wolven et al., 2000) are outside the binding sites for known ligands. Three more mutants not tested in S. pombe (human W3F, W3N, and S. cerevisiae W2A) are known or likely to be structurally unstable.
Table S1.
Supplementary Table 1. Summaries of all previously described profilin mutants
| Mutant | Stability | Actin
binding
|
Plp
binding
|
PIP2 | In vivo assay | Reference | Comparable data in S. pombe | ||
|---|---|---|---|---|---|---|---|---|---|
| Quantitative | Non-quantitative | Quantitative | Non-quantitative | ||||||
| Bovine F59A | N/T | Kd: 24 μMa (wt: 2.3 μM) Ca2+-actin | Lostb | N/T | No changec | N/T | Normal localization in mouse SW3T3d; partial rescue of Dictyostelium profilin null with expression only 10% of endogenous leveld | Schluter et al., 1998 | F57D complements null and ts, but only partially suppresses cytokinesis phenotype in ts at 36°C |
| Bovine V60E | N/T | Kd: 12 μMa (wt: 2.3 μM) Ca2+-actin | Lostb | N/T | No changec | N/T | N/T | Schluter et al., 1998 | G58Y, G58W suppress null and ts like wild type |
| Bovine G120F | N/T | Kd: 42 μMa (wt: 2.3 μM) Ca2+-actin | Lostb | N/T | No changec | N/T | N/T | Schluter et al., 1998 | P107Y, P107W complement ts, but not null; strong phenotype in ts; actin binding reduced to 6% and 4% |
| Bovine K125A | N/T | Kd: 5 μMa (wt: 2.3 μM) Ca2+-actin | Reducedb | N/T | No changec | N/T | N/T | Schluter et al., 1998 | K112A, K112E, K112L, K112W suppress null and ts like wild type |
| Human W3Fe | N/T | N/T | No
changef; co-IP with antiprofilin serum |
N/T | Binds plp column | No change | Episomal expression suppresses lethal budding yeast profilin null; overexpression attenuates abnormal morphologyd | Ostrander et al., 1999 | N/T |
| Human W31Fe | N/T | N/T | No changef; co-IP with anti-profilin serum | Kd: 1244 μM (wt: 204 μM) | Binds plp column | No change | Episomal expression suppresses lethal budding yeast profilin null; overexpression attenuates abnormal morphologyd | Ostrander et al., 1999 | W30Y suppresses null and ts like wild type; 33% plp binding, stable as WT |
| Human W3F W31Fe | N/T | N/T | N/T | N/T | Does not bind plp column | N/T | Episomal expression does not suppress lethal budding yeast profilin nulld | Ostrander et al., 1999 | N/T |
| Human Y6F | N/T | N/T | N/T | N/T | Eluted with 3.5 M urea | Wild type levelg | N/T | Sohn et al., 1995 | Y5D, Y5A see Table 1 |
| Human D8A | N/T | N/T | N/T | N/T | Eluted with 7.5 M urea | Kd: 0.025 μM (wt: 0.21 μM) | N/T | Sohn et al., 1995 | N/T |
| Human L10R | Aggregated | N/T | N/T | Sohn et al., 1995 | N/T | ||||
| Human K25Q | N/T | N/T | N/T | N/T | Eluted with 3.5 M urea | Wild type levelg | N/T | Sohn et al., 1995 | R24A, E, L, Y suppress null and ts like wild type |
| Human K53I | N/T | N/T | N/T | N/T | Eluted with 7.5 M urea | Wild type levelg | N/T | Sohn et al., 1995 | N/T |
| Human R74L | N/T | N/T | N/T | N/T | Eluted with 7.5 M urea | Wild type levelg | N/T | Sohn et al., 1995 | R72E, see Table 1; R72A suppresses ts incompletely; R72Y, R72W suppress null and ts like wild type |
| Human R88L | Wild typeh | Nucleotide exchange need 25 times higher concentration to reach wild type level | Much reduced inhibition of polymerization | N/T | Eluted with 7.5 M urea | Kd: 0.60 μM (wt: 0.21 μM) | N/T | Sohn et al., 1995 | K81L increases nucleotide exchange activity 6.3-fold; actin binding 25%; suppresses null and ts like wild type; K81E lost actin binding completely, but normal PIP2 binding; K81A, F, Y see Table 1 |
| Human R88L K90E | N/T | N/T | N/T | N/T | Eluted with 7.5 M urea | N/T | N/T | Sohn et al., 1995 | N/T |
| Human H119D | N/T | N/T | N/T | N/T | Eluted with 7.5 M urea | Wild type levelg | N/T | Sohn et al., 1995 | L106E, W, Y suppress null and ts like wild type |
| Human G121D | N/T | N/T | N/T | N/T | Eluted with 7.5 M urea | Wild type levelg | N/T | Sohn et al., 1995 | G108 E, Y, W suppress null and ts like wild type |
| Human K125Q | N/T | N/T | N/T | N/T | Eluted with 7.5 M urea | N/T | N/T | Sohn et al., 1995 | K112 A, E, Y, W suppress null and ts like wild type |
| Human W3N | N/T | Kd: 0.93 μM (wt: 0.28 μM) | Reduced inhibition of polymerizationi | N/T | Does not bind plp beads | PIP2 binds 2.93-fold more W3N than wild type | N/T | Bjorkergren-Sjogren et al., 1997; Bjorkegren et al., 1993 | N/T |
| Human H133S | N/T | Kd: 0.32 μM (wt: 0.28 μM) | Slightly reduced inhibition of polymerizationi | N/T | Does not bind plp beads | Wild type levelj | Less effective on profilin-enhanced Cdc42-induced actin nucleation in high-speed supernatant of lysed rabbit neutrophils | Bjorkegren-Sjogren et al., 1997; Bjorkegren et al., 1993; Yang et al., 2000 | Y120A, Y120D see Table 1 |
| Human G14A | N/T | N/T | N/T | N/T | Does not bind plp beads | N/T | N/T | Bjorkegren et al., 1993 | N/T |
| Human H119E (PI,PII) | N/T | N/T | GST-H119E beads failed to bind actin from bovine tissue lysate | N/T | GST-H119E beads binds pLp containing ligands from bovine tissue lysate; (GST-wt binds N-WASP, wt PI: 60 nMk wt PII: 400 nMk) | N/T | PI H119E inhibits EGF stimulated microspike formation when WASP is overexpressed (COS cells); PI H119E inhibits neurite extension in neuroblastoma cells; PI H119E blocks actin filament clustering induced by WAVE in COS cells; PI H119E suppresses cdc42-induced microspike and Rac-induced membrane ruffles in swiss3T3 cells | Suetsugu et al., 1998; Miki et al., 1998; Suesugu et al., 1999 | L106E, R, W suppress null and ts like wild type |
| Human R55D | N/T | Kd: 0.84 μM (wt: 0.34 μM) nucleotide exchange: 0.12 S−1 (wt 0.08 S−1, actin alone 0.0021 S−1) | N/T | N/T | N/T | N/T | Korenbaum et al., 1998 | N/T | |
| Human ΔP96 ΔT97 | N/T | Kd: 1.1 μM (wt: 0.34 μM) nucleotide exchange: 0.13 S−1 (wt: 0.08 S−1, actin alone: 0.0021 S−1) | N/T | Binds plp column like wild type | Wild type level by filter assay | Do not change the F-actin organization like wt in SW3T3 of porcine aortic endothelial cells | Korenbaum et al., 1998; Hajkova et al., 1997 | N/T | |
| Human K69N | N/T | Kd: 0.56 μM (wt: 0.34 μM) | N/T | N/T | N/T | N/T | Korenbaum et al., 1998 | K67A, E, L, W. See Table 1 | |
| Human R74E | N/T | Kd: 16 μM (wt: 0.34 μM); no inhibition of actin polymerization; nucleotide exchange: N/T | N/T | N/T | N/T | Less effective on profilin-enhanced Cdc42-induced actin nucleation in high-speed supernatant of lysed rabbit neutrophils | Korenbaum et al., 1998; Yang et al., 2000 | R72E, see Table 1; R72A suppresses ts incompletely; R72Y, R72W suppress null and ts like wild type | |
| Human K90E | N/T | Kd: N/T no inhibition of actin polymerization; nucleotide exchange: 0.07 S−1 (wt: 0.08 S−1) |
N/T | N/T | N/T | N/T | Korenbaum et al., 1998 | N/T | |
| Human K125N | N/T | Kd: 0.18 μM (wt: 0.34 μM); nucleotide exchange: 0.05 S−1 (wt: 0.08 S−1) | N/T | N/T | N/T | N/T | Korenbaum et al., 1998 | K112 A, E, Y, W suppress null and ts like wild type | |
| Budding yeast Δ3C-terminal | N/T | N/T | Binds DNaseI beads | N/T | Reduced binding to plp beads | N/T | Low copy suppresses ΔPFY; failed to show a two-hybrid interaction with Bni1p | Haarer et al., 1993; Evangelista et al., 1997 | N/T |
| Budding yeast Δ4C-terminal | N/T | N/T | Binds DNaseI beads | N/T | Much reduced binding to plp beads | N/T | Low copy suppresses ΔPFY | Haarer et al., 1993 | N/T |
| Budding yeast Δ6C-terminal | N/T | N/T | Reduced binding to DNaseI beads | N/T | No binding to plp beads | N/T | Low copy partially suppress ΔPFY; high copy suppresses ΔPFY | Haarer et al., 1993 | N/T |
| Budding yeast Δ7,Δ8C-terminal | N/T | N/T | Greatly reduced binding to DNaseI beads | N/T | No binding to plp beads | N/T | Low copy does not suppress ΔPFY; high copy partially suppresses ΔPFY | Haarer et al., 1993 | N/T |
| Budding yeast R81G | N/T | N/T | Binds DNaseI beads like wild type; no inhibition of actin polymerization at low concentrationl | N/T | Binds plp beads, eluted with 6 M urea | Wild type level | Low copy suppresses ΔPFY | Haarer et al., 1993 | K81E, L, A,
F, Y, W see Table 1 Key residue for actin binding |
| Budding yeast R72E | N/T | N/T | Binds DNaseI beads like wild type; no inhibition of actin polymerization at low concentrationl | N/T | Bind plp beads, eluted with 3 M urea | Lost (microfiltration) | Low copy suppresses ΔPFY; does not suppress ΔCAP (WT suppresses ΔCAP) | Haarer et al., 1993; Vojtek et al., 1991 | R72E, see Table 1; R72A suppresses ts incompletely; R72Y, R72W suppress null and ts like wild type |
| Budding yeast R76G | N/T | N/T | Binds DNaseI beads like wild type; reduced inhibition of actin polymerization at low concentrationl | N/T | Bind plp beads, eluted with 3 M urea | Normal | Low copy suppresses
ΔPFY; suppresses ΔCAP |
Haarer et al., 1993; Vojtek et al., 1991 | R76A, E, L, Y suppress null and ts like wild type |
| Budding yeast R76G R81K | N/T | N/T | Binds DNaseI beads like wild type; no inhibition of actin polymerization at low concentrationl | N/T | Bind plp beads, eluted with 3 M urea | Normal | Low copy suppresses
ΔPFY; do not suppress ΔCAP |
Haarer et al., 1993; Vojtek et al., 1991 | N/T |
| Budding yeast R76E | N/T | N/T | Binds DNaseI beads like wild type; reduced inhibition of actin polymerization at low concentrationl | N/T | Bind plp beads, eluted with 3 M urea | Normal | Low copy
suppresses ΔPFY; suppresses ΔCAP |
Haarer et al., 1993; Vojtek et al., 1991 | R76A, E, L, Y suppress null and ts like wild type |
| Budding yeast R76G R81G | N/T | N/T | Binds DNaseI beads | N/T | N/T | N/T | Low copy suppresses ΔPFY; do not suppress ΔCAP | Haarer et al., 1993; Vojtek et al., 1991 | N/T |
| Budding yeast R72G R81K | N/T | N/T | Binds DNaseI beads | N/T | N/T | N/T | Low copy suppresses ΔPFY; do not suppress ΔCAP |
Haarer et al., 1993; Vojtek et al., 1991 | N/T |
| Budding yeast R72G R76G R81K | N/T | N/T | Binds DNaseI beads | N/T | N/T | N/T | Low copy suppresses ΔPFY | Haarer et al., 1993 | N/T |
| Budding yeast R76G R81K H82D | N/T | N/T | Binds DNaseI beads | N/T | N/T | N/T | Low copy suppresses ΔPFY | Haarer et al., 1993 | N/T |
| Budding yeast R75A | N/T | N/T | N/T | N/T | N/T | N/T | Integrate into ΔPFY, phenotype on YPD: wt; phenotype on YPD+ formamide: wt | Wolven et al., 2000 | N/T |
| Budding yeast R90A K92A | N/T | N/T | N/T | N/T | N/T | N/T | Integrate into ΔPFY, phenotype on YPD: wt; phenotype on YPD+ formamide: wt | Wolven et al., 2000 | N/T |
| Budding yeast R23A R75A | N/T | N/T | N/T | N/T | N/T | N/T | Integrate into ΔPFY, phenotype on YPD: wt; phenotype on YPD+ formamide: wt | Wolven et al., 2000 | N/T |
| Budding yeast R23A R92A | N/T | N/T | N/T | N/T | N/T | N/T | Integrate into ΔPFY, phenotype on YPD: wt; phenotype on YPD+ formamide: wt | Wolven et al., 2000 | N/T |
| Budding yeast K66A | 2.6 M urea (3.1 M for wt) | Lost binding to PFY-plp beadsm; lost inhibition of actin polymerization; no effect on nucleotide exchange | N/T | Same as wild type | N/T | Integrate into ΔPFY, phenotype on YPD; conditional 25–34°C; phenotype on YPD + formamide: lethal at 37°C; phenotype suppressed by act1-157 (higher nucleotide exchange rate); synthetic lethal with a few cofilin mutant strains | Wolven et al., 2000 | K67A, E, L, W. See Table 1 | |
| Budding yeast R71A | N/T | N/T | N/T | N/T | N/T | N/T | Integrate into ΔPFY, phenotype on YPD: wt; phenotype on YPD + formamide: lethal at 37°C | Wolven et al., 2000 | R72E, see Table 1; R72A suppresses ts incompletely; R72Y, R72W suppress null and ts like wild type |
| Budding yeast R80A | N/T | N/T | N/T | N/T | N/T | N/T | Integrate into ΔPFY, phenotype on YPD: wt; phenotype on YPD + formamide: lethal at 37°C | Wolven et al., 2000 | K81A, E, L,
A, Y, F see Table 1 |
| Budding yeast R66A R80A | N/T | N/T | N/T | N/T | N/T | N/T | Integrate into ΔPFY, phenotype on YPD: wt; phenotype on YPD + formamide: N/T | Wolven et al., 2000 | N/T |
| Budding yeast H81A | N/T | N/T | N/T | N/T | N/T | N/T | Integrate into ΔPFY, phenotype on YPD: wt; phenotype on YPD + formamide: wt | Wolven et al., 2000 | L82W, L82Y suppress null and ts like wild type |
| Budding yeast D73A D74A | N/T | N/T | N/T | N/T | N/T | N/T | Integrate into ΔPFY, phenotype on YPD: wt; phenotype on YPD + formamide: wt | Wolven et al., 2000 | N/T |
| Budding yeast D7A | N/T | N/T | N/T | N/T | N/T | N/T | Integrate into ΔPFY, phenotype on YPD: wt; phenotype on YPD + formamide: wt | Wolven et al., 2000 | N/T |
| Budding yeast Q3A | N/T | N/T | N/T | N/T | N/T | N/T | Integrate into ΔPFY, phenotype on YPD: wt; phenotype on YPD + formamide: wt | Wolven et al., 2000 | N/T |
| Budding yeast S1A | N/T | N/T | N/T | N/T | N/T | N/T | Integrate into ΔPFY, phenotype on YPD: wt; phenotype on YPD + formamide: wt | Wolven et al., 2000 | N/T |
| Budding yeast W2A | N/T | N/T | N/T | N/T | N/T | N/T | Integrate into
ΔPFY, phenotype on YPD: conditional
(25–34°C) |
Wolven et al., 2000 | N/T |
| Budding yeast W29A | N/T | N/T | N/T | N/T | N/T | N/T | Integrate into
ΔPFY, phenotype on YPD: lethal |
Wolven et al., 2000 | W30Y suppresses null and ts like wild type; 33% plp binding, stable as WT |
| Budding yeast Y119A Y125A | N/T | N/T | N/T | N/T | N/T | N/T | Integrate into
ΔPFY, phenotype on YPD: conditional (25–34°C); phenotype on YPD + formamide: lethal at 37°C |
Wolven et al., 2000 | N/T |
| Budding yeast Y119A | N/T | N/T | N/T | N/T | N/T | N/T | Integrate into
ΔPFY; phenotype on YPD: wt; phenotype on YPD + formamide: lethal at 37°C |
Wolven et al., 2000 | Y120A, Y120D see Table 1 |
| S. pombe E42K | N/T | N/T | N/T | N/T | N/T | N/T | Ts phenotype; strong cytokinesis defects | Balasubramanan et al., 1994 | See Table 1 |
| DictyPII W3N | N/T | N/T | Slightly reducedn | N/T | Lost plp bindingn | Wild type leveln | Only high expression can rescue the null phenotype | Lee et al., 2000 | N/T |
| DictyPII K114E | N/T | N/T | Profoundly reducedn | N/T | Normaln | Wild type leveln | Low expression can rescue the null phenotype (reduction of F-actin and complete development) | Lee et al., 2000 | A116E suppresses null and ts nearly like wild type |
| Acantha moeba PII 122PLV | 1.5 M urea (wt: 3.5 M) | Yes | N/T | Kd: 13,200 μM (wt: 100 μM) | N/T | N/T | Kaiser et al., 1996 | N/T | |
| Acantha moeba PII 122PSSL D | 1.5 M urea (wt: 3.5 M) | Yes | N/T | Kd: 14,700 μM (wt: 100 μM) | N/T | N/T | Kaiser et al., 1996 | N/T | |
| Acantha moeba PII 117PLV | 0.9 M urea (wt: 3.5 M) | Yes | N/T | No binding | N/T | N/T | Kaiser et al., 1996 | N/T | |
| Acantha moeba PII S76C | 3.5 M urea (wt: 3.5 M) | Yes | N/T | Kd: 100 μM (wt: 100 μM) | N/T | N/T | Kaiser et al., 1996 | N/T | |
| Acantha moeba PII S92C | 3.5 M urea (wt: 3.5 M) | Yes | N/T | Kd: 100 μM (wt: 100 μM) | N/T | N/T | Kaiser et al., 1996 | N/T | |
| Maize ZmPro1 Y6F | N/T | Kd: 0.09 μM (wt: 1.2 μM) | Kd: 130 μM (wt: 275 μM) | N/T | Nuclear displacement: 4.5 min with 100 μM Y6F (wt: 6.6 min with 100 μM) | Gibbon et al., 1999 | Y5A, Y5D see Table 1 | ||
Table S2.
Supplementary Table 2. List of corresponding profilin mutations in S. pombe and other species based on sequence alignment and their positions in atomic structures
| Residues in S. pombe | S. pombe | S. cerevisiae | Dictyostelium | Maize | Human | Cow |
|---|---|---|---|---|---|---|
| Y5 | A*, D* | Y6F | T6F | |||
| R24 | A, E, Y, L | K25Q | ||||
| W30 | Y | W29A | W31F | |||
| F57 | D*, Y, W | F59A | ||||
| G58 | Y, W | V60E | ||||
| K67 | E*, A*, L*, W* | K66A* | K67N | |||
| R72 | E*, W, Y, A | R71E*, A* | R74L, E* | |||
| R76 | A, E, L, Y | R75G, E*, A* | ||||
| K81 | W, A*, E*, L*, Y, F* | R80G, A* | R88L | |||
| L82 | W, Y | H81A | ||||
| L106 | E, R, Y | H119D, E* | ||||
| P107 | E, W*, Y* | G120F | ||||
| G108 | E, W, Y | G121D | ||||
| K112 | E, L, W, A | K125Q, N | K125A* | |||
| A116 | E, Y | K114E | ||||
| Y120 | D*, A* | Y119A* | H133S | |||
| S1 | S1A | |||||
| W2 | W2A* | W3N* | W3N*, F* | |||
| Q3 | Q3A | |||||
| D7 | D7A | D8A | ||||
| L10 | L10R | |||||
| G14 | G14A | |||||
| Q51 | K53I | |||||
| P53 | R55D | |||||
| No | K90E | |||||
| Same | 7 | 1 | 2 | 1 | ||
| New in S. pombe | K15, R18, E42, I69*, I71*, S77*, Y79*, K84*, G86*, C88, K93, H100, A111*, L121*, Y126* |
N/T, not tested;
, informative.
Assayed by steady-state critical concentration for polymerization; Ca2+-actin was used in this assay, on the assumption that profilin acts only as a sequestering protein (Perelroizen et al., 1996).
Spontaneous polymerization of 8 μM Ca2+-actin with 12 μM profilin, using 10% pyrene-labeled actin (0.8 μM). Pyrene-labeled actin has a low affinity for profilin (Malm, 1984).
Profilin was eluted from a plp column by urea gradient. This assay cannot distinguish a loss affinity for plp from a loss of stability.
Cross species.
Mutant may not be stable because it was eluted by 3 M urea from plp column.
Antibody was not purified. It is not a direct proof of binding, because many other proteins coprecipitated.
Assayed by the inhibition of PLC-γ activity.
Assayed by protease sensitivity.
12 μM β/γ actin was polymerized in MgCl2 or KCl with 12 μM profilin.
Assayed by gel filtration using profilin and PIP2 micelles with various molar ratios.
Kd is not accurate with immobilized GST fusion protein.
This assay used 2.85 μM profilin, +2 μM actin. Higher profilin concentration needed to be tested.
Profilin was immobilized on plp beads. Actin was brought down with an increasing profilin concentration. Note: repetitively washing beads will allow profilin-actin dissociation, so reduced binding between profilin and actin might not be detectable.
Results are from abstract.
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health Research Grant GM-26338 to T.D.P. and by the Lucille P. Markey Charitable Trust through the Graduate Program in Cellular and Molecular Medicine at Johns Hopkins Medical School. We are grateful to Susan Forsburg, Magdalena Bezanilla, Wei-Lih Lee, Laurent Blanchoin, Don Kaiser, Harry Higgs, Jean-Baptiste Marchand, and Kurt Amann of the Salk Institute for their help with the design and execution of these experiments. We also thank Dr. Kathy Gould of Vanderbilt University for providing yeast strains, and Dr. Steven Almo of the Albert Einstein College of Medicine for providing a wild-type profilin cDNA.
REFERENCES
- Ahern-Djamali SM, Bachmann C, Hua P, Reddy SK, Kastenmeier AS, Walter U, Hoffmann FM. Identification of profilin and src homology 3 domains as binding partners for Drosophila Enabled. Proc Natl Acad Sci USA. 1999;96:4977–4982. doi: 10.1073/pnas.96.9.4977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archer SJ, Vinson VK, Pollard TD, Torchia DA. Secondary structure and topology of Acanthamoeba profilin-I as determined by heteronuclear magnetic resonance spectroscopy. Biochemistry. 1993;32:6680–6687. doi: 10.1021/bi00077a022. [DOI] [PubMed] [Google Scholar]
- Balasubramanian MK, Hirani BR, Burke JD, Gould KL. The Schizosaccharomyces pombe cdc3+ gene encodes a profilin essential for cytokinesis. J Cell Biol. 1994;125:1289–1301. doi: 10.1083/jcb.125.6.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballweber E, Giehl K, Hannappel E, Huff T, Jockusch BM, Mannherz HG. Plant profilin induces actin polymerization from actin:β-thymosin complexes and competes directly with β-thymosins and with negative co-operativity with DNase I for binding to actin. FEBS Lett. 1998;425:251–255. doi: 10.1016/s0014-5793(98)00240-3. [DOI] [PubMed] [Google Scholar]
- Bjorkegren-Sjogren C, Korenbaum E, Nordberg P, Lindberg U, Karlsson R. Isolation and characterization of two mutants of human profilin I that do not bind poly(L-proline) FEBS Lett. 1997;418:258–264. doi: 10.1016/s0014-5793(97)01376-8. [DOI] [PubMed] [Google Scholar]
- Blanchoin L, Pollard TD. Interaction of actin monomers with Acanthamoeba actophorin (ADF/cofilin) and profilin. J Biol Chem. 1998;273:25106–25111. doi: 10.1074/jbc.273.39.25106. [DOI] [PubMed] [Google Scholar]
- Blanchoin L, Pollard TD. Mechanism of interaction of Acanthamoeba actophorin (ADF/cofilin) with actin filaments. J Biol Chem. 1999;274:15538–15546. doi: 10.1074/jbc.274.22.15538. [DOI] [PubMed] [Google Scholar]
- Chang F, Drubin D, Nurse P. Cdc12p, a protein required for cytokinesis in fission yeast, is a component of the cell division ring and interacts with profilin. J Cell Biol. 1997;137:169–182. doi: 10.1083/jcb.137.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhary A, Chen J, Gu QM, Witke W, Kwiatkowski DJ, Prestwich GD. Probing the phosphoinositide 4.5-bisphosphate binding site of human profilin I. Chem Biol. 1998;5:273–281. doi: 10.1016/s1074-5521(98)90620-2. [DOI] [PubMed] [Google Scholar]
- Christensen HEM, Ramachandran S, Tan CT, Surana U, Dong CH, Chua NH. Arabidopsis profilins are functionally similar to yeast profilins: identification of a vascular bundle-specific profilin and a pollen-specific profilin. Plant J. 1996;10:269–279. doi: 10.1046/j.1365-313x.1996.10020269.x. [DOI] [PubMed] [Google Scholar]
- Cooley L, Verheyen E, Ayers K. Chickadee encodes a profilin required for intercellular cytoplasmic transport during Drosophila oogenesis. Cell. 1992;69:173–184. doi: 10.1016/0092-8674(92)90128-y. [DOI] [PubMed] [Google Scholar]
- Didry D, Carlier MF, Pantaloni D. Synergy between actin depolymerizing factor/cofilin and profilin in increasing actin filament turnover. J Biol Chem. 1998;273:25602–25611. doi: 10.1074/jbc.273.40.25602. [DOI] [PubMed] [Google Scholar]
- Eads JC, Mahoney NM, Vorobiev S, Bresnick AR, Wen KK, Rubenstein PA, Haarer BK, Almo SC. Structure determination and characterization of Saccharomyces cerevisiae profilin. Biochemistry. 1998;37:11171–11181. doi: 10.1021/bi9720033. [DOI] [PubMed] [Google Scholar]
- Evangelista M, Blundell K, Longtine M, Chow C, Adames N, Pringle J, Peter M, Boone C. Bni1p, a yeast formin linking cdc42p and the actin cytoskeleton during polarized morphogenesis. Science. 1997;276:118–122. doi: 10.1126/science.276.5309.118. [DOI] [PubMed] [Google Scholar]
- Fedorov AA, Magnus KA, Graupe MH, Lattman EE, Pollard TD, Almo SC. X-ray structures of isoforms of the actin-binding protein profilin that differ in their affinity for phosphatidylinositol phosphates. Proc Natl Acad Sci USA. 1994;91:8636–8640. doi: 10.1073/pnas.91.18.8636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gertler FB, Niebuhr K, Geinhard M, Wehland J, Soriano P. Mena, a relative of VASP and Drosophila Enabled, is implicated in the control of microfilament dynamics. Cell. 1996;87:227–239. doi: 10.1016/s0092-8674(00)81341-0. [DOI] [PubMed] [Google Scholar]
- Gibbon B, Zonia LE, Kovar DR, Hussey PJ, Staiger CJ. Pollen profilin function depends on interaction with proline-rich motifs. Plant Cell. 1998;10:981–993. doi: 10.1105/tpc.10.6.981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giehl K, Valenta R, Rothkegel M, Ronsiek M, Mannherz HG, Jockusch BM. Interaction of plant profilin with mammalian actin. Eur J Biochem. 1994;226:681–689. doi: 10.1111/j.1432-1033.1994.tb20096.x. [DOI] [PubMed] [Google Scholar]
- Giesemann T, Rathke-Hartlieb S, Rothkegel M, Bartsch JW, Buchmeier S, Jockusch BM, Jockusch H. A role for polyproline motifs in the spinal muscular atrophy protein SMN. J Biol Chem. 1999;274:37908–37914. doi: 10.1074/jbc.274.53.37908. [DOI] [PubMed] [Google Scholar]
- Goldschmidt-Clermont PJ, Furman MI, Wachsstock D, Safer D, Nachmias VT, Pollard TD. The control of actin nucleotide exchange by thymosinβ4 and profilin. A potential regulatory mechanism for actin polymerization in cells. Mol Biol Cell. 1992;3:1015–1024. doi: 10.1091/mbc.3.9.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldschmidt-Clermont PJ, Machesty LM, Doberstein SK, Pollard TD. Mechanism of the interaction of human platelet profilin with actin. J Cell Biol. 1991;113:1081–1089. doi: 10.1083/jcb.113.5.1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haarer BK, Lillie SH, Adams AEM, Magdolen V, Bandlow W, Brown SS. Purification of profilin from Saccharomyces cerevisiae and analysis of profilin-deficient cells. J Cell Biol. 1990;110:105–114. doi: 10.1083/jcb.110.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haarer BK, Petzold AS, Brown SS. Mutational analysis of yeast profilin. Mol Cell Biol. 1993;13:7684–7673. doi: 10.1128/mcb.13.12.7864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haugwitz M, Noegel AA, Karakesisoglou J, Schleicher M. Dictyostelium amoebae that lack G-actin-sequestering profilins show defects in F-actin content, cytokinesis, and development. Cell. 1994;79:303–314. doi: 10.1016/0092-8674(94)90199-6. [DOI] [PubMed] [Google Scholar]
- Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- Imamura H, Tanaka K, Hihara T, Umidawa M, Kamei T, Takahashi K, Sasaki T, Takai Y. Bni1p and Bnr1p: downstream targets of the Rho family small G-proteins which interact with profilin and regulate actin cytoskeleton in Saccharomyces cerevisiae. EMBO J. 1997;16:2745–2755. doi: 10.1093/emboj/16.10.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser DA, Goldschmidt-Clermont PJ, Levine BA, Pollard TD. Characterization of renatured profilin purified by urea elution from poly-l-proline agarose columns. Cell Motil Cytosleleton. 1989;14:251–262. doi: 10.1002/cm.970140211. [DOI] [PubMed] [Google Scholar]
- Kaiser DA, Pollard TD. Characterization of actin and poly-l-proline binding sites of Acanthamoeba profilin with monoclonal antibodies and by mutagenesis. J Mol Biol. 1996;255:89–107. doi: 10.1006/jmbi.1996.0070. [DOI] [PubMed] [Google Scholar]
- Karakesisoglou I, Schleicher M, Gibbon BC, Staiger CJ. Plant profilins rescue the aberrant phenotype of profilin-deficient Dictyostelium cells. Cell Motil Cytoskeleton. 1996;34:36–47. doi: 10.1002/(SICI)1097-0169(1996)34:1<36::AID-CM4>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- Korenbaum E, Nordberg P, Bjorkegren-Sjogren C, Schutt CE, Lindberg U, Karlsson R. The role of profilin in actin polymerization and nucleotide exchange. Biochemistry. 1998;37:9274–9283. doi: 10.1021/bi9803675. [DOI] [PubMed] [Google Scholar]
- Kouyama T, Mihashi K. Fluorimetry study of N-(1-pyrenyl) iodoacetamide-labeled F-actin. Local structural change of actin protomer both on polymerization and on binding of heavy meromyosin. Eur J Biochem. 1981;114:33–38. [PubMed] [Google Scholar]
- Kovar DR, Drobak BK, Staiger CJ. Maize profilin isoforms are functionally distinct. Plant Cell. 2000;12:583–598. doi: 10.1105/tpc.12.4.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanier LM, Gates MA, WItke W, Menzies AS, Wehman AM, Macklis JD, Kwiatkowski D, Soriano P, Gertler FB. Mena is required for neurulation and commissure formation. Neuron. 1999;22:313–325. doi: 10.1016/s0896-6273(00)81092-2. [DOI] [PubMed] [Google Scholar]
- Lee SS, Karakesisoglou I, Noegel AA, Rieger D, Schleicher M. Dissection of functional domains by expression of point-mutated profilins in Dictyostelium mutants. Eur J Cell Biol. 2000;79:92–103. doi: 10.1078/S0171-9335(04)70011-4. [DOI] [PubMed] [Google Scholar]
- Machesky LM, Goldschmidt-Clermont PJ, Pollard TD. The affinities of human platelet and Acanthamoeba profilin isoforms for polyphophoinositides account for their relative abilities to inhibit phospholipase C. Cell Regul. 1990;1:937–950. doi: 10.1091/mbc.1.12.937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machesky LM, Pollard TD. Profilin as a potential mediator of membrane cytoskeletal communication. Trends Cell Biol. 1993;3:381–385. doi: 10.1016/0962-8924(93)90087-h. [DOI] [PubMed] [Google Scholar]
- Mahoney NM, Janmey PA, Almo SC. Structure of the profilin-poly-l-proline complex involved in morphogenesis and cytoskeletal regulation. Nat Struct Biol. 1997;4:953–960. doi: 10.1038/nsb1197-953. [DOI] [PubMed] [Google Scholar]
- Mahoney NM, Rozwarski DA, Fedorov E, Fedorov AA, Almo SC. Profilin binds proline-rich ligands in two distinct amide backbone orientations. Nat Struct Biol. 1999;6:666–671. doi: 10.1038/10722. [DOI] [PubMed] [Google Scholar]
- Mammoto A, Sasaki T, Asakura T, Hotta I, Imamura H, Takahashi K, Matsuura Y, Shirao T, Takai Y. Interactions of drebrin and gephyrin with profilin. Biochem Biophys Res Commun. 1998;243:86–89. doi: 10.1006/bbrc.1997.8068. [DOI] [PubMed] [Google Scholar]
- Manseau L, Calley J, Phan H. Profilin is required for posterior patterning of the Drosophila oocyte. Development. 1996;122:2109–2116. doi: 10.1242/dev.122.7.2109. [DOI] [PubMed] [Google Scholar]
- Miki H, Suetsugu S, Takenawa T. WAVE, a novel WASP-family protein involved in actin reorganization induced by Rac. EMBO J. 1998;17:6932–6941. doi: 10.1093/emboj/17.23.6932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mockrin SC, Korn RD. Acanthamoeba profilin interacts with G-actin to increase the rate of exchange of actin-bound adenosine 5′-triphosphate. Biochemistry. 1980;19:5359–5362. doi: 10.1021/bi00564a033. [DOI] [PubMed] [Google Scholar]
- Nishida E. Opposite effects of cofilin and profilin from porcine brain on rate of exchange of actin-bound adenosine 5′-triphosphate. Biochemistry. 1985;24:1160–1164. doi: 10.1021/bi00326a015. [DOI] [PubMed] [Google Scholar]
- Ostrander DB, Ernst EG, Lavoie TB, Gorman JA. Polyproline binding is an essential function of human profilin in yeast. Eur J Biochem. 1999;262:26–35. doi: 10.1046/j.1432-1327.1999.00354.x. [DOI] [PubMed] [Google Scholar]
- Perelroizen I, Didry D, Christensen H, Chua NH, Carlier MF. Role of nucleotide exchange and hydrolysis in the function of profilin in actin assembly. J Biol Chem. 1996;271:12302–12309. doi: 10.1074/jbc.271.21.12302. [DOI] [PubMed] [Google Scholar]
- Perelroizen I, Marchand JB, Blanchoin L, Didry D, Carlier MF. Interaction of profilin with G-actin and poly(L-proline) Biochemistry. 1994;33:8472–8478. doi: 10.1021/bi00194a011. [DOI] [PubMed] [Google Scholar]
- Petrella EC, Machesky LM, Kaiser DA, Pollard TD. Structural requirements and thermodynamics of the interaction of proline peptides with profilin. Biochemistry. 1996;35:16535–16543. doi: 10.1021/bi961498d. [DOI] [PubMed] [Google Scholar]
- Pollard TD. Purification of a high molecular-weight actin filament gelation protein from Acanthamoeba that shares antigenic determinants with vertebrate spectrins. J Cell Biol. 1984;99:1970–1980. doi: 10.1083/jcb.99.6.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard TD, Cooper JA. Quantitative analysis of the effect of Acanthamoeba profilin on actin filament nucleation and elongation. Biochemistry. 1984;23:6631–6641. doi: 10.1021/bi00321a054. [DOI] [PubMed] [Google Scholar]
- Ramesh N, Anton IM, Hartwig JH, Geha RS. WIP, a protein associated with Wiskott-Aldrich syndrome protein, induces actin polymerization and redistribution in lymphoid cells. Proc Natl Acad Sci USA. 1997;94:14671–14676. doi: 10.1073/pnas.94.26.14671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhard M, Giehl K, Abel K, Haffner C, Jarchau T, Hoppe V, Jockusch BM, Walter U. The proline-rich focal adhesion and microfilament protein VASP is a ligand for profilins. EMBO J. 1995;14:1583–1589. doi: 10.1002/j.1460-2075.1995.tb07146.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoro MM, Bolen DW. Unfolding free energy changes determined by the linear extrapolation method. 1. Unfolding of phenylmethanesulfonyl α-chymotrypsin using different denaturants. Biochemistry. 1988;27:8063–8068. doi: 10.1021/bi00421a014. [DOI] [PubMed] [Google Scholar]
- Schluter K, Jockusch BM, Rothkegel M. Profilins as regulators of actin dynamics. Biochim Biophys Acta. 1997;1359:97–109. doi: 10.1016/s0167-4889(97)00100-6. [DOI] [PubMed] [Google Scholar]
- Schutt C, Myslik JC, Rozychi MD, Goonesekere NCW, Lindberg U. The structure of crystalline profilin-β-actin. Nature. 1993;365:810–816. doi: 10.1038/365810a0. [DOI] [PubMed] [Google Scholar]
- Selden LA, Kinosian HJ, Estes JE, Gershman LC. Impact of profilin on actin-bound nucleotide exchange and actin polymerization dynamics. Biochemistry. 1999;38:2769–2778. doi: 10.1021/bi981543c. [DOI] [PubMed] [Google Scholar]
- Sohn RH, Chen J, Koblan KS, Bray PF, Goldschmidt-Clermont PJ. Localization of a binding site for phosphatidylinositol 4,5-bisphosphate on human profilin. J Biol Chem. 1995;270:21114–21120. doi: 10.1074/jbc.270.36.21114. [DOI] [PubMed] [Google Scholar]
- Spudich JA, Watt S. The regulation of rabbit skeletal muscle contraction. I. Biochemical studies of the interaction of the tropomyosin-troponin complex with actin and the proteolytic fragments of myosin. J Biol Chem. 1971;246:4866–4871. [PubMed] [Google Scholar]
- Suetsugu S, Miki H, Takenawa T. The essential role of profilin in the assembly of actin for microspike formation. EMBO J. 1998;17:6516–6526. doi: 10.1093/emboj/17.22.6516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanford C. Protein denaturation. Adv Protein Chem. 1968;23:121–275. doi: 10.1016/s0065-3233(08)60401-5. [DOI] [PubMed] [Google Scholar]
- Vinson VK, Archer SJ, Lattman EE, Pollard TD, Torchia DA. Three dimensional structure of Acanthamoeba profilin-I. J Cell Biol. 1993;122:1277–1283. doi: 10.1083/jcb.122.6.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinson VK, De La Cruz EM, Higgs HN, Pollard TD. Interactions of Acanthamoeba profilin with actin and nucleotides bound to actin. Biochemistry. 1998;37:10871–10880. doi: 10.1021/bi980093l. [DOI] [PubMed] [Google Scholar]
- Waechter F, Engel J. The kinetics of the exchange of G-actin-bound 1,N6-ethenoadenosine 5′-triphosphate with ATP as followed by fluorescence. Eur J Biochem. 1975;57:453–459. doi: 10.1111/j.1432-1033.1975.tb02320.x. [DOI] [PubMed] [Google Scholar]
- Wang X, Kibschull M, Laue MM, Lichte B, Petrasch-Parwez E, Kilimann MW. Aczonin, a 550-kD putative scaffolding protein of presynaptic active zones, shares homology regions with Rim and Bassoon and binds profilin. J Cell Biol. 1999;147:151–162. doi: 10.1083/jcb.147.1.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe N, Madaule P, Reid T, Ishizaki T, Watanabe G, Kakizuka A, Saito Y, Nakao K, Jockusch B, Narumiya S. p140mDia, a mammalian homolog of Drosophila diaphanous, is a target protein for Rho small GTPase and is a ligand for profilin. EMBO J. 1997;16:3044–3056. doi: 10.1093/emboj/16.11.3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolven AK, Belmont LD, Mahoney NM, Almo SC, Drubin DG. In vivo importance of actin nucleotide exchange catalyzed by profilin. J Cell Biol. 2000;150:895–903. doi: 10.1083/jcb.150.4.895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Huang M, DeBiasio J, Pring M, Joyce M, Miki H, Takenawa T, Zigmond SH. Profilin enhances Cdc42-induced nucleation of actin polymerization. J Cell Biol. 2000;150:1001–1012. doi: 10.1083/jcb.150.5.1001. [DOI] [PMC free article] [PubMed] [Google Scholar]