Abstract
Thomsen-Friedenreich (TF) antigen, which plays an important role in the regulation of cancer cell proliferation, occurs in ∼90% of all human cancers and precancerous conditions. Although TF antigen has been known for almost 80 yr as a pancarcinoma antigen, the recognition mechanism between TF antigen and target protein has not been structurally characterized. A number of studies indicated that TF disaccharide is a potential ligand of the galactoside-binding galectins. In this work, we identified the TF antigen as a potential ligand of the antitumor galectin AAL (Agrocybe aegerita lectin) through glycan array analysis and reported the crystal structure of AAL complexed with the TF antigen. The structure provides a first look at the recognition mode between AAL and TF antigen, which is unique in a conservative (Glu-water-Arg-water) structural motif-based hydrogen bond network. Structure-based mutagenesis analysis further revealed the residues responsible for recognition specificity and binding affinity. Crystal structures of AAL complexed with two other TF-containing glycans showed that the unique TF recognition mode is kept intact, which may be commonly adopted in some cancer-related galectins. The finding provided the new target and approach for the antitumor drug design and relative strategy based on the AAL-TF recognition mode as a prototype model.—Feng, L., Sun, H., Zhang, Y., Li, D.-F., Wang, D.-C. Structural insights into the recognition mechanism between an antitumor galectin AAL and the Thomsen-Friedenreich antigen.
Keywords: glycan target for tumor on cell surface, complex structure between tumor-related protein and glycan, protein-glycan interaction mode
It is known that all human cancers are accompanied by changes of cellular glycosylation (1–3). Many of these changes usually lead to exposure of tumor-associated carbohydrate structure (4–6). Among others, the increased occurrence of Galβ1–3GalNAcα1-Ser/Thr is one of the most common glycosylation changes, which is usually referred to as Thomsen-Friedenreich (TF) antigen. The TF antigen is the core 1 structure of O-linked mucin type glycans. In cancer and precancerous conditions, TF antigen occurs in ∼90% of all human cancers (3). In many cases, the increased TF occurrence is in correlation with cancer progression and metastasis. It has been reported that cell surface glycoproteins with TF antigen play an important role in the regulation of cancer cell proliferation (6, 7). Therefore, the TF antigen has been used as a target in tumor diagnosis and therapy.
A number of studies indicated that TF disaccharide is a potential ligand of the endogenous galactoside-binding galectins, and their interactions may affect the growth of tumor cells (6, 8). Galectins are a widespread family of lectins characterized by a conserved carbohydrate recognition domain that specifically recognize β-galactose. To date, a vast array of the intracellular and extracellular functions of galectins has been identified. Among others, the cancer-associated galectins have been the focus of many studies (9). Therefore, interactions between galectin and TF antigen and its functional implications have gained much attention. It has been reported that galectin-1 and galectin-3 exert their cancer-related functions through specific recognition of the TF antigen expressed on the cell surface (6, 10). However, the binding interactions have not been structurally characterized; hence, the recognition mechanism remains to be clarified. So far, only the structures of two galectins, CGL2 and N-terminal domain of galectin-9, complexed with TF disaccharide have been determined (11, 12), but they did not show any interactions between the galectins and the GalNAc moiety unique for TF antigen. Therefore, it remains unknown whether and how galectins could specifically recognize TF antigen via the GalNAc moiety. Although TF antigen has been known for almost 80 yr as a pancarcinoma antigen, the structural mechanism of the recognition between TF antigen and target protein has not been elucidated based on 3-D structure. Consequently, the structural mechanism for galectin-carbohydrate recognition is based mainly on the structures of galectins complexed with lactose or N-acetyl lactose.
AAL (Agrocybe aegerita lectin) from the edible mushroom A. aegerita is an antitumor protein that exerts its tumor-suppressing function via apoptosis-inducing activity in cancer cells (13). We demonstrated that AAL belongs to a galectin family with a unique carbohydrate recognition domain that specifically recognizes β-galactose (14). Subsequently, the crystal structures of ligand-free AAL and its complex with lactose were determined (15). The structure-based mutagenesis analysis revealed that some critical structural elements, such as the dimerization and galactose binding site, are prerequisite for the functional performance of AAL. One interesting observation we made in previous studies was that the R85A mutation left AAL's lactose binding affinity intact but abolished its tumor cell apoptosis-inducing activity completely. One possibility under our ensuring investigations is that this mutation abolishes the specificity of AAL for its bioactivity-related ligand other than the lactose. Here we report the identification of TF antigen as a native ligand of AAL and the structures of AAL complexed with a TF antigen bearing the Thr residue essential for O-glycan and some TF derivatives. The structures reveal a unique recognition mode for AAL-TF interaction, which consists of a hydrogen bond network formed by a conserved structural motif (Glu66-water-Arg85-water). The structure-based mutagenesis analysis shows that residues Glu66 and Arg85 might contribute mainly to the recognition specificity and binding affinity, respectively. The sequence and structure comparisons indicate that the recognition mode observed in this study might be commonly adopted in some other galectins. The results shed light on the structural mechanism for AAL-TF recognition and elucidate the structural details of the AAL-TF binding mode, which should provide new targets and approaches for the antitumor drug design and relative strategy based on AAL-TF antigen recognition mode as a prototype model.
MATERIALS AND METHODS
Glycan array analysis
Purified AAL and mutant R85A with a His6 tag were used at 200 μg/ml to probe the plate glycan array, version 3.2, by Core H of the Consortium for Functional Glycomics (CFG; http://www.functionalglycomics.org/static/index.shtml). Bound galectins were detected using the specific anti-His6 monoclonal antibody and goat anti-rabbit IgG-Alexa Fluor 488 (Invitrogen, Carlsbad, CA, USA) in accordance with the consortium's standard protocol.
Crystallization and structure determination
The mutants R85A and E66A were introduced using QuickChange Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA, USA) and validated by DNA sequencing. The genes were inserted into the plasmid pET22b (Novagen, Darmstadt, Germany) and expressed in BL21(DE3; Novagen). The wild-type and mutant AAL were purified using an NTA Ni+ chelating column (Novagen) and then were loaded onto a Hiload Superdex75 16/60 column (GE Healthcare, Uppsala, Sweden) (16). The proteins were stored in 50 mM Tris buffer (pH 8.0) for crystallization or in PBS for other assays.
Crystallizations for all samples were performed at room temperature using the hanging-drop vapor-diffusion method. The protein solution, containing 10 mg/ml AAL and 10 mM TF antigen (CFG), was crystallized in 0.1 M Bis-Tris (pH 5.5), 25% PEG3350, 0.2 M lithium chloride, and 5% acetone. The protein solution was mixed in a 1:1 ratio with precipitate solution. Crystals generally appeared within 24 h and grew to their full sizes in 2 d. Similar procedures were used for the crystallization of the complexes AAL-TFN (Merck, Darmstadt, Germany), R85A-TFN, E66A-TFN, and AAL-GM1 (CFG).
The X-ray diffraction data were collected on a Rigaku R-Axis IV++ image plate (Rigaku, Tokyo, Japan) using Cu Kα radiation (λ=1.5418 Å) or at beam line 17A of Photo Factory (KEK, Tsukuba, Japan) at 85 K. The crystal used for data collection was dipped into cryoprotectants (paraffin oil) for ∼5 s with nylon cryoloops (Hampton Research, Aliso Viejo, CA. USA) before being flash-frozen at 85 K. All data sets were processed with the program MOSFLM (17) and merged by SCALA from the CCP4 program suite (18). The phase was solved by Phaser (19) using the wild-type AAL structure model. The ligands, including TF antigen, TFN, and GM1, were manually added using O (20); the structures were refined using program CNS (21). The quality of the models was assessed using PROCHECK (22). The figures were created using PyMOL (23). Carbohydrate-galectin interactions were analyzed using LIGPLOT (24).
Tumor cell apoptosis-inducing activity assay
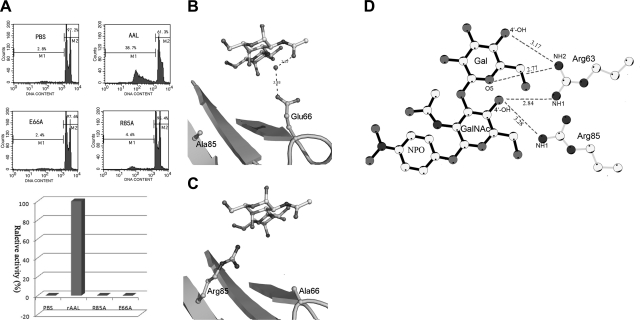
Tumor cell apoptosis-inducing activities of AAL and the mutants were assayed as described in our previous study (15). The HeLa cells (2×105) cultured with or without the AAL proteins, including E66A and R85A (300 μg/ml) at 37°C for 36 h, were harvested, washed with PBS, and fixed with 75% ethanol at 4°C for 2 h before the treatment of RNase A (0.25 mg/ml; Dingguo, Beijing, China) at 37°C for 1 h. After washing with PBS, the cells were stained with 50 μg/ml propidium iodide at room temperature for 10 min. Cell cycle analysis was performed with a BD FACSVantage SE flow cytometer (Becton Dickinson, Franklin Lakes, NJ, USA). The percentages of HeLa cells undergoing apoptosis (M2) in presence of different AAL samples were used to calculate the relative activities of mutants. The relative activities of PBS and AAL were normalized, and those of mutants were evaluated with the value of (M2_mutant − M2_PBS)/(M2_AAL − M2_PBS). All experiments were independently repeated 3 times, and the data were averaged.
Isothermal titration calorimetry (ITC)
ITC experiments were performed on an iTC200 (isothermal titration calorimeter, 200 μl cell; Microcal, Northampton, MA, USA) as described previously in detail (25). The proteins including wild-type AAL, mutants E66A and R85A, and the carbohydrates were dissolved in PBS. The titrations were performed at 25°C, with stirring at 1000 rpm. Titrations consisted of 39 injections of 1 μl and were separated by 250 s. The dissociation constant (Kd) was determined by Origin software provided by Microcal.
Protein Data Bank (PDB) accession number
Structure factors and coordinates of the AAL-TF antigen, AAL-TFN, AAL-GM1, and the mutant E66A-TFN, R85A-TFN have been deposited in the PDB with the accession codes 3AFK, 3M3C, 3M3Q, 3M3E, and 3M3O, respectively.
RESULTS AND DISCUSSION
TF disaccharide is the bioactivity-related ligand for AAL
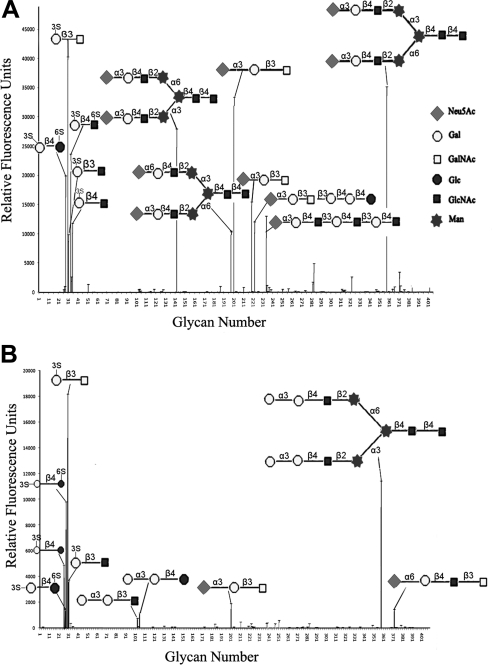
Our previous study has shown that mutant R85A of AAL maintains the lactose-binding activity but loses its tumor cell apoptosis-inducing activity (15). This finding suggests that lactose seems not a bioactivity-related ligand for AAL. To investigate which oligosaccharide is the possible bioactive ligand involved in AAL-inducing tumor cell apoptosis, glycan array analyses of AAL and mutant R85A were performed first. The entire list of glycans tested and the results of the binding assays can be found online (http://www.functionalglycomics.org/glycomics/HServlet?operation=view&sideMenu=no&psId=primscreen_2300#, 2302#). The results show that glycans with high affinity to AAL contain either TF disaccharide (Galβ1–3GalNAc), lactose (Galβ1–4Glc, Lac), or N-acetyl lactose (Galβ1–4GlcNAc, LacNAc) (Fig. 1A). AAL has the highest affinity to the sulfated TF disaccharide fixed to a spacer arm Sp8 (-CH2CH2CH2NH2), [3OSO3] Galβ1–3GalNAcα-Sp8 in the array. This result indicates that the fixed sulfated TF disaccharide is a suitable ligand for AAL.
Figure 1.
Sugar-binding specificities of AAL and mutant R85A. A) Binding of AAL to carbohydrates, from the CFG glycan array. Error bars = sd. Sulfate group of the sugar is shown as 3S. B) Glycan array result of mutant R85A.
It is well known that the array analysis is generally not exact enough for precise investigation. Therefore, to detect the interaction between AAL/R85A and TF antigen with greater accuracy, ITC was used to measure the exact binding affinity as Kd of the wild AAL and mutant R85A with TF antigen. The results show that the affinity of mutant R85A to TF is significantly decreased to 15%, compared with that of AAL (see Table 1). In correlation, the tumor cell apoptosis-inducing activity of mutant R85A is dramatically lost in the bioassay (see Fig. 5A). Together, these observations indicate that TF antigen should be one of the bioactivity-related ligands for AAL.
Table 1.
Dissociation constants (Kd) of AAL and mutants, determined by ITC at 298 K
| Protein |
Kd (μM) |
||
|---|---|---|---|
| Lactose | TF antigen | TF p-nitrophenyl | |
| AAL | 165.29 ± 12.27 | 251.89 ± 17.38 | 86.21 ± 3.17 |
| AAL-E66A | 171.53 ± 3.50 | 89.29 ± 2.68 | 37.45 ± 0.74 |
| AAL-R85A | 116.28 ± 5.71 | 1655.63 ± 47.70 | 250.63 ± 16.52 |
Values are derived from the simultaneous fit of ≥2 independent titrations to a 1-set-of-sites model.
Figure 5.
Bioactivity and 3-D structure analyses of mutants E66A and R85A. A) Relative tumor cell apoptosis-inducing activities of E66A and R85A, assayed by flow cytometry using the PI staining method. Top panel: representative bivariate plots of the FACS analysis. Bottom panel: relative activities of E66A and R85A compared with AAL. B, C) Recognition sites relative to the GalNAc moiety observed in structures of R85A-TFN (B) and E66A-TFN (C). They both lose the (Arg85-water-Glu66-water) motif-based hydrogen bond network. D) More direct interactions between protein and ligand in the E66A-TFN complex. Residues and TFN ligand are shown as ball-and-stick model. Distance of hydrogen bonds is labeled on dashed lines.
Crystal structure of AAL-complexed TF antigen reveals the structural mechanism for AAL-TF antigen recognition
As the core 1 structure O-linked mucin type glycan, an intact TF antigen consists of 3 moieties: an N-acetyl galactose linking with a galactose and a Ser/Thr residue (Galβ1–3GalNAcα1-Ser/Thr). The moiety of GalNAc is unique for TF antigen. In this work, we refer to the TF antigen without the Ser/Thr residue as the TF disaccharide. In this study we take the glycan (Galβ1–3GalNAcα1-Thr) as a TF antigen complexed with AAL in the structural determination, which should show the AAL-TF recognition in O-glycosylation state. The structure of AAL-TF antigen was solved at 1.95-Å resolution using the molecular replacement method, with the AAL structure previously determined (15) as the search model. The statistics of data collection and structure refinement are summarized in Table 2.
Table 2.
Summary of structure determination and refinement for AAL complexed with glycan ligands
| Data set | AAL-TF antigen | AAL-TFN | E66A-TFN | R85A-TFN | AAL-GM1 |
|---|---|---|---|---|---|
| Data collection | |||||
| Space group | P1 | P212121 | P21 | P3221 | P21 |
| Cell dimensions | |||||
| a, b, c (Å) | 40.85, 43.44, 56.06 | 60.59, 62.42, 96.07 | 56.81, 53.24, 103.4 | 42.56, 42.56, 125.3 | 50.86, 66.71, 56.21 |
| α, β, γ (deg) | 102.08, 98.15, 118.06 | 90, 90, 90 | 90, 95.08. 90 | 90, 90, 120 | 90, 93.18, 90 |
| Resolution range (Å) | 36.37–1.95 | 43.5–2.01 | 47.84–2.10 | 41.77–2.09 | 33.5–2.18 |
| Solvent content (%) | 38.5 | 47.4 | 38.1 | 28.71 | 50.55 |
| Subunits/asymmetric unit | 2 | 2 | 4 | 1 | 2 |
| Unique reflections | 21402 | 24896 | 34472 | 8441 | 19377 |
| Multiplicitya | 2.7 (2.7) | 7.2 (7.3) | 2.9 (2.9) | 5.9 (5.8) | 3.6 (3.0) |
| Completeness (%)a | 92.0 (76.1) | 100 (100) | 95 (92) | 100 (100) | 95 (98.7) |
| Mean I/σ (I)a | 39 (22.9) | 14.4 (6.1) | 6.3 (2.8) | 19.2 (5.3) | 9.2 (7.1) |
| Rmerg (%)a | 2.1 (3.7) | 8.7 (14.1) | 10 (34.8) | 4.7 (29.3) | 12 (23) |
| Refinement | |||||
| B_overall, by Patterson (Å2) | 14.5 | 27 | 18.9 | 31.4 | 28.0 |
| R factor | 0.187 | 0.188 | 0.205 | 0.218 | 0.206 |
| R_free | 0.209 | 0.221 | 0.257 | 0.256 | 0.238 |
| RMS bonds (Å) | 0.0049 | 0.0053 | 0.0059 | 0.006 | 0.0064 |
| RMS angles (deg) | 1.38 | 1.43 | 1.41 | 1.43 | 1.40 |
| Ramachandran distribution | |||||
| Most favored (%) | 89.6 | 96.8 | 94.8 | 94.3 | 95.5 |
| Allowed (%) | 9.8 | 3.2 | 4.8 | 4.5 | 4.5 |
| Disallowed (%) | 0.7 | 0 | 0.5 | 1.3 | 0 |
Values in parentheses are for last shell.
Overall structure
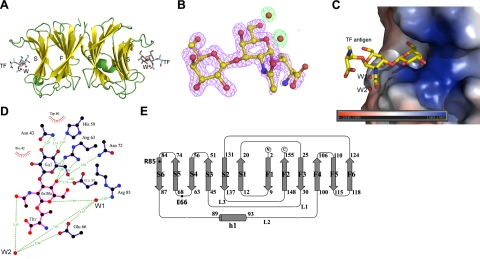
The general fold of AAL in the complex shows a high level of similarity to ligand-free AAL with Cα RMSD of 0.44 Å. One AAL dimer and 2 TF antigen molecules are in the asymmetric unit, with each TF antigen binding an AAL protomer (Fig. 2A). The subunit structure adopts a typical prototype galectin fold in which two 6-stranded antiparallel β sheets (F1-F6 and S1-S6) jointly form a β-sandwich structure. The S1–S6 strands create a concave surface on which the TF antigen is bound, while the F1–F6 strands provide the interacting surface for the dimerization. All the residues involved in TF antigen binding are located on S3–S6 sheet, except Glu66, which is on the loop connecting S4 and S5 (Fig. 2A, E). Electron density maps show that the TF antigen in the complex is well ordered and that the carbohydrate ring in the TF is in the chair conformation (Fig. 2B).
Figure 2.
Overall structure of AAL-TF antigen complex. A) Dimer structure of AAL-TF antigen complex. TF antigen and water molecules bound to the CRD concave of AAL are shown in a ball-and-stick model. B) Fo-Fc omit electron density map around TF antigen and 2 conservative water molecules nearby is calculated without the ligand and contoured at 3σ. C) Carbohydrate recognition site showing a positively charged cavity bound to a TF antigen with 2 water molecules. D) Interactions between TF antigen and AAL. Residues Pro42, Asn43, His59, Arg63, Asn72, Trp80, and Glu83 are involved in galactose moiety recognition (blue), while Glu66 and Arg85 with 2 water molecules form a hydrogen bond network in recognition with the GalNAc moiety (magenta). Arg63 and Glu83 are attended to both galactose and GalNAc moieties. E) Topological diagram of AAL secondary structure.
AAL-TF recognition
The TF antigen is bound to the surface concave formed by the S3–S6 sheet with insertion of the β-galactose ring into a wedge-like cavity (Fig. 2C). This spatial arrangement restrains the overall freedom of the galactoside and, in particular of the 4′-hydroxyl group to the axial epimer. Residues Pro42 and Trp80 form hydrophobic interactions with TF antigen, while Asn43, His59, Arg63, Asn72, and Glu83 directly interact with the Gal moiety of TF antigen via hydrogen bonds (Fig. 2D). The recognition mode of AAL with the Gal moiety of TF antigen is basically identical to that observed in the AAL-lactose complex (15).
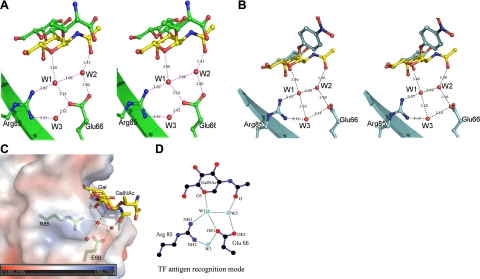
For GalNAc moiety of the TF antigen, which is corresponding to the glucose moiety in lactose, a unique recognition mode via a hydrogen bond network mediated by 2 water molecules, W1 and W2, is observed (Figs. 2D and 3A). In this network, the residue Arg85 interacts with atom O5 of the galactoside ring in GalNAc of the TF antigen through water molecule W1, while residue Glu66 mainly contributes to recognition with the N-acetyl group through water molecule W2 (Fig. 3A). Interestingly, these two paths are closely connected each other via hydrogen bonds between residues Arg85 and Glu66 and water molecules to form a unique recognition motif for the TF antigen (Fig. 3A).
Figure 3.
Recognition mode between AAL and TF antigen unique in a (Arg85-water-Glu66-water) motif-based hydrogen bond network. A, B) Stereo view of AAL-TF antigen (A) and AAL-TFN (B) complexes. Glycan ligands are shown as sticks; unique moiety GalNAc is highlighted in yellow. C) Special interactions between AAL and GalNAc moiety in the AAL-TF recognition site. D) Consensus recognition mode between galectin and TF antigen, unique in a water molecule-mediated hydrogen bond network.
In fact, lectins using water molecules to recognize the carbohydrate is frequently observed. For example, the peanut lectin employs the water bridges for generating carbohydrate specificity (26). Diego et al. (27) also reported that water molecules on the surface of the carbohydrate recognition domain of galectins were related to the galectins' affinity for carbohydrate ligand recognition. It seems that lectins take water molecules into the recognition for carbohydrate as an effective strategy suitable for the flexibility and variability of glycans.
Conformational requirement for GalNAc moiety ensuring structural element in recognition
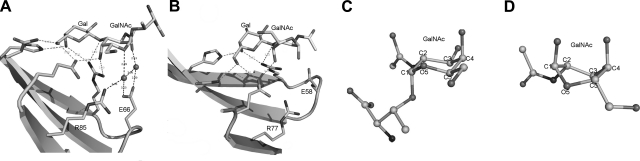
In addition to AAL, the structures of two galectins, galectin-9 and CGL2, have also been reported as TF disaccharide-complexed forms (11, 12), but the ligands in these structures do not have the Thr/Ser residues conjugated to their GalNAc moiety. Compared with AAL-TF antigen (Fig. 4A), none of the structures show any interactions between the galectins and the GalNAc moiety while exhibiting the same interacting mode for the Gal moiety (Fig. 4B). Hence, these two complex structures have not provided the structural information on the recognition between the galectins and GalNAc moiety of TF antigen. Structure comparisons of AAL-TF with the complexes of galectin-9 and CGL2 show that the GalNAc carbohydrate rings exist as the chair conformation in AAL-TF but the twisted-boat conformation in galectin-9 and CGL2. In the boat conformation, the atom O5 and acetyl group of GalNAc moiety move rather far from the counterpart residues of the galectins, with distances of 7.9 Å (CGL2) and 5.46 Å (galectin-9) to Arg85 and 6.01 Å (CGL) and 9.21Å (galectin-9) to Glu66, respectively. The detailed analysis reveals that the residue moiety linked to the GalNAc with α linkage should significantly stabilize the chair conformation of the GalNAc. These observations indicate that the chair conformation of GalNAc of TF antigen is required for the suitable recognition of galectins, and the attachment of the residue moiety to the GalNAc in α linkage is necessary for ensuring a recognizable conformation of the TF antigen.
Figure 4.
Recognition site in AAL-TF antigen (A) and CGL2-TF disaccharide (B) complexes, comparatively showing that the distinct interactions between the galectin and the GalNAc moiety occur in the AAL-TF antigen, which corresponds to a chair conformation of GalNAc (C), while missing in the CGL2-TF disaccharide, which otherwise corresponds to a twisted-boat conformation of GalNAc (D). Structural data for CGL2-TF disaccharide is from PDB 1ULG.
Roles of residues Glu66 and Arg85 in TF antigen recognition identified by mutagenesis analysis
To reveal the specific roles of the critical residues Glu66 and Arg85 in interactions between AAL and TF antigen, the mutations of E66A and R85A are constructed and analyzed extensively. First, the affinities of the mutants are investigated by ITC. For all the titrations, the experimental data could be fitted to a 1-set-of-sites model, indicating that the two sites in the lectin dimer are independent and equivalent. The Kd values of binding to TF antigen and lactose derived from the thermodynamic analysis for mutant E66A and R85A parallel to the wild AAL as a control are listed in Table 1. The results show that the binding affinities of mutants to TF antigen are dramatically increased about 2-fold for E66A while significantly reduced to 15% for mutant R85A, compared with that of AAL. In contrast, the mutants E66A and R85A have binding affinities toward lactose similar to that of AAL.
The tumor cell apoptosis-inducing activities of the mutants were assayed with a BD FACSVantage SE flow cytometer. The percentages of HeLa cells undergoing apoptosis in the presence of different samples were used to calculate the relative activities of mutants. The relative activities of PBS and AAL were normalized. All experiments for AAL, E66A, R85A, and PBS were repeated independently 3 times, and the data were averaged. The results show that both mutant proteins E66A and R85A totally lose their tumor cell apoptosis-inducing activity (Fig. 5A).
To understand the structural basis of above observations further, three crystal structures of AAL and its mutants E66A and R85A complexed with TF p-nitrophenyl (TFN; Galβ1–3GalNAcα1-p-nitrophenyl), an analog of TF antigen, were determined in parallel at 2.10-, 2.09-, and 2.01-Å resolution, respectively; their statistical analyses are summarized in Table 2. The general structure of AAL-TFN is very similar to the structure of AAL-TF antigen, with a Cα RMSD of 0.421 Å for the proteins and 0.429 Å for TF disaccharide without the attached groups (Thr and p-nitrophenyl), and the recognition mode observed in the AAL-TF antigen structure remains intact in the structure (Fig. 3B), which shows a reasonable model for the mutant analysis. However, in both structures of E66A-TFN and R85A-TFN, the unique hydrogen bond network constituted by Glu66, Arg85, and two conservative water molecules in the AAL-TF structure is entirely destroyed (Fig. 5B, C), which should be the structural reason for abolishing the bioactivity of both mutants. Nevertheless, in the E66A-TFN complex structure, accompanying the disruption of interactions in the hydrogen bond network, residues Arg85 and Arg63 are reoriented to form some more direct contacts, including R85NH1-GalNAcO4, R63NH1-GalNAcO4, R632NH-GalO4 and R632NH-GalO5 (Fig. 5D), which should strengthen the binding ability of the mutant and lead to evident increase of the binding affinity of mutant E66A, although its bioactivity is lost completely. For mutant R85A, the complex structure shows that the hydrogen bond network is destroyed, and only the interaction between Glu66 and the N-acetyl group of the GalNAc moiety is maintained (Fig. 5B), which corresponds to a significantly reduced affinity to 15% for TFN binding. The above data demonstrate that residue Glu66 is mainly involved in the specificity in the recognition of AAL with TF antigen via interaction with the acetyl group of the GalNAc moiety, while residue Arg85 is involved in the determination of the binding affinity of AAL to TF antigen, mainly through interactions with the carbohydrate ring of GalNAc. These two residues are essential for the suitable recognition mode of AAL with TF antigen.
The AAL-TF antigen recognition mode may be a common paradigm in galectins
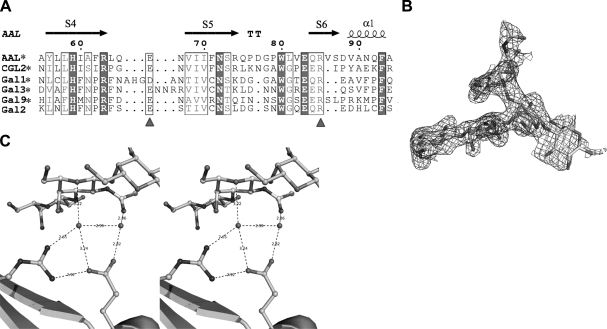
To date it is known that some galectins, including galectin 1, 3, 9 and CGL2 can also bind with TF antigen (6, 10, 11, 12). The structure-based sequence alignment of AAL with these galectins shows that the residues Arg85 and Glu66 of AAL, which are essential for the recognition with TF antigen, are very conservative in all these galectins (Fig. 6A). It implies that these galectins may also adopt the recognition mode unique in a (Glu66-water-Arg85-water) structural motif-based hydrogen bond network as a paradigm for recognizing TF-antigen.
Figure 6.
Conservation of the (Arg85-water-Glu66-water) motif-based recognition mode. A) Sequence alignment of CRD for AAL, CGL2, Gal 1, 2, 3, and 9. Triangles indicate critical residues (Glu66 and Arg85 in AAL), which are invariant in these galectins. Asterisks indicate galectins identified as being able to bind with TF antigen. Sequence data of CGL2 and Gal 1, 2, 3, and 9 are from the U.S. National Center for Biotechnology Information (NCBI) protein database. B) Fo-Fc omit electron density map for the TF-containing pentasaccharide observed in AAL-GM1 complex structure. C) Recognition sites between AAL and TF GalNAc moiety of GM1, showing that the distinct recognition mode is kept intact as in the AAL-TF antigen complex structure (see Fig. 3D).
In addition, in the glycan array analysis, we found that AAL can also bind with a TF-containing glycan, ganglosides GM1 (Galβ1–3GalNAcβ1–4[Neuα2–3]Galβ1–4Glc). GM1 is a pentasaccharide containing a TF disaccharide moiety. The structure of AAL-GM1 complex was then determined at 2.18-Å resolution (Table 2). Electron density maps show that GM1 in the complex is well ordered (Fig. 6B). The structure shows that, though there is an extended carbohydrate chain linked to the TF disaccharide, the conservative (Arg85-water-Glu66-water) hydrogen bond network in interaction with GalNAc of TF disaccharide remains intact (Fig. 6C). The observations from the structure of TF-containing derivative and the sequence alignment of TF-binding galectins cooperatively show that the recognition mode for TF antigen, as observed in AAL, may be commonly adopted by TF-binding galectins.
The results presented in this article identified the antitumor-related ligand and found the ligand-AAL recognition model with detailed structural elements, including active sites, critical residues for the bioactivity, and the structural mechanism for the induction of bioactivity, which should provide a new target and approach for antitumor drug design and relative strategy, based on AAL-TF antigen recognition mode as a prototype model.
Acknowledgments
This work was supported by the “973” and “863” Research Programs of China (2006CB806502, 2006CB910901, 2006AA02E135) and the Ministry of Health, China (2009ZX09103-676). The glycan array analysis was conducted by the Protein-Carbohydrate Interaction Core, and the TF antigen and GM1 pentasaccharide were provided by the Carbohydrate Synthesis/Protein Expression Core of the Consortium for Functional Glycomics (CFG), funded by the National Institute of General Medical Sciences grant GM62116. The authors thank David F. Smith and Jamie Heimburg-Molinaro (Consortium for Functional Glycomics) for screening the glycan array, and Yi Han, Yanxia Jia, and Chunchun Liu (Institute of Biophysics, Chinese Academy of Sciences) for their help with in-house X-ray data collection, ITC assay, and flow cytometric assay, respectively. The authors are also grateful to the Photo Factory (08G007; KEK, Tsukuba, Japan), and the Beijing and Shanghai Synchrotron Radiation Facility for providing access to the synchrotron radiation facilities for the X-ray diffraction experiment. The authors also thank Dr. Yonglin Hu for carefully checking and editing the manuscript.
REFERENCES
- 1. Baldus S. E., Engelmann K., Hanisch F.-G. (2004) MUC1 and the MUCs: a family of human mucins with impact in cancer biology. Crit. Rev. Clin. Lab. Sci. 41, 189–231 [DOI] [PubMed] [Google Scholar]
- 2. Hanisch F. G. B. S. (1997) The Thomsen-Friedenreich (TF) antigen: a critical review on the structural, biosynthetic and histochemical aspects of a pancarcinoma-associated antigen. Histol. Histopathol. 12, 263–281 [PubMed] [Google Scholar]
- 3. Yu L.-G. (2007) The oncofetal Thomsen–Friedenreich carbohydrate antigen in cancer progression. Glycoconj. J. 24, 411–420 [DOI] [PubMed] [Google Scholar]
- 4. Springer G. F. (1997) Immunoreactive T and Tn epitopes in cancer diagnosis, prognosis, and immunotherapy. J. Mol. Med. 75, 594–602 [DOI] [PubMed] [Google Scholar]
- 5. Slovin S.F., Musselli G. R., Fernandez C., Diani M., Verbel D., Danishefsky S., Livingston P., Scher H. I. (2005) Thomsen-Friedenreich (TF) antigen as a target for prostate cancer vaccine: clinical trial results with TF cluster (c) -KLH plus QS21 conjugate vaccine in patients with biochemically relapsed prostate cancer Cancer Immunol. Immunother. 54, 694–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Udo Jeschke U. K., Wiest I., Schulze S., Kuhn C., Friese K., Walzel H. (2006) Binding of galectin-1 (gal-1) to the Thomsen–Friedenreich (TF) antigen on trophoblast cells and inhibition of proliferation of trophoblast tumor cells in vitro by gal-1 or an anti-TF antibody. Histochem. Cell Biol. 126, 437–444 [DOI] [PubMed] [Google Scholar]
- 7. Lu-Gang Yu B. J., Fernig D. G., Milton J. D., Smith J. A., Gerasimenko O. V., Jones M., Rhodes J. M. (1998) Stimulation of proliferation in human colon cancer cells by human monoclonal antibodies against the TF antigen (galactose 1–3 N-acetyl-galactosamine). Int. J. Cancer 73, 424–431 [DOI] [PubMed] [Google Scholar]
- 8. Leffler H., Barondes S. H. (1986) Specificity of binding of three soluble rat lung lectins to substituted and unsubstituted mammalian beta-galactosides. J. Biol. Chem. 261, 10119–10126 [PubMed] [Google Scholar]
- 9. Liu F.-T., Rabinovich G. A. (2005) Galectins as modulators of tumour progression. Nat. Rev. Cancer 5, 29–41 [DOI] [PubMed] [Google Scholar]
- 10. Yu L.-G., Andrews N., Zhao Q., McKean D., Williams J. F., Connor L. J., Gerasimenko O. V., Hilkens J., Hirabayashi J., Kasai K., Rhodes J. M. (2007) Galectin-3 interaction with Thomsen-Friedenreich disaccharide on cancer-associated MUC1 causes increased cancer cell endothelial adhesion. J. Biol. Chem. 282, 773–781 [DOI] [PubMed] [Google Scholar]
- 11. Nagae M., Nishi N., Murata T., Usui T., Nakamura T., Wakatsuki S., Kato R. (2006) Crystal structure of the galectin-9 N-terminal carbohydrate recognition domain from Mus musculus reveals the basic mechanism of carbohydrate recognition. J. Biol. Chem. 281, 35884–35893 [DOI] [PubMed] [Google Scholar]
- 12. Walser P. J., Haebel P. W., Kunzler M., Sargent D., Kues U., Aebi M., Ban N. (2004) Structure and functional analysis of the fungal galectin CGL2. Structure 12, 689–702 [DOI] [PubMed] [Google Scholar]
- 13. Zhao C., Sun H., Tong X., Qi Y. (2003) An antitumour lectin from the edible mushroom Agrocybe aegerita. Biochem. J. 374, 321–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yang N., Tong X., Xiang Y., Zhang Y., Liang Y., Sun H., Wang D. C. (2005) Molecular character of the recombinant antitumor lectin from the edible mushroom Agrocybe aegerita. J. Biochem. 138, 145–150 [DOI] [PubMed] [Google Scholar]
- 15. Yang N., Li D.-F., Feng L., Xiang Y., Liu W., Sun H., Wang D.-C. (2009) Structural basis for the tumor cell apoptosis-inducing activity of an antitumor lectin from the edible mushroom Agrocybe aegerita. J. Mol. Biol. 387, 694–705 [DOI] [PubMed] [Google Scholar]
- 16. Yang N., Tong X., Xiang Y., Zhang Y., Sun H., Wang D. C. (2005) Crystallization and preliminary crystallographic studies of the recombinant antitumour lectin from the edible mushroom Agrocybe aegerita. Biochim. Biophys. Acta 1751, 209–212 [DOI] [PubMed] [Google Scholar]
- 17. Rossmann M. G., van Beek C. G. (1999) Data processing. Acta Crystallogr. D Biol. Crystallogr. 55, 1631–1640 [DOI] [PubMed] [Google Scholar]
- 18. Collaborative Computational Project (1994) Collaborative computational project number 4: providing programs for protein crystallography. Acta Crystallogr. D. Biol. Crystallogr. 50, 760–763 15299374 [Google Scholar]
- 19. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jones T. A., Zou J. Y., Cowan S. W., Kjeldgaard M. (1991) Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47, 110–119 [DOI] [PubMed] [Google Scholar]
- 21. Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., Warren G. L. (1998) Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54, 905–921 [DOI] [PubMed] [Google Scholar]
- 22. Laskowski R. A., MacArthur M. W., Moss D. S., Thornton J. M. (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26, 283–291 [Google Scholar]
- 23. DeLano W. L. (2005) Pymol, v. 0.98 DeLano Scientific, San Francisco, CA, USA [Google Scholar]
- 24. Wallace A. C., Laskowski R. A., Thornton J. M. (1995) LIGPLOT: A program to generate schematic diagrams of protein-ligand interactions. Prot. Eng. 8, 127–134 [DOI] [PubMed] [Google Scholar]
- 25. Wiseman T., Williston S., Brandts J. F., Lin L.-N. (1989) Rapid measurement of binding constants and heats of binding using a new titration calorimeter. Anal. Biochem. 179, 131–137 [DOI] [PubMed] [Google Scholar]
- 26. Natchiar K., Srinivas O., Mitra N., Surolia A., Jayaraman N., Vijayan M. (2006) Structural studies on peanut lectin complexed with disaccharides involving different linkages: further insights into the structure and interactions of the lectin. Acta Crystallogr. D Biol. Crystallogr. 62, 1413–1422 [DOI] [PubMed] [Google Scholar]
- 27. Gauto D. F., Di Lella S., Guardia C. M. A., Estrin D. O. A., Marti M. A. (2009) Carbohydrate-binding proteins: dissecting ligand structures through solvent environment occupancy. J. Phys. Chem. B 113, 8717–8724 [DOI] [PubMed] [Google Scholar]