Abstract
Diapause is an adaptive response triggered by seasonal photoperiodicity to overcome unfavorable seasons. The photoperiodic clock is a system that controls seasonal physiological processes, but our knowledge about its physiological mechanisms and genetic architecture remains incomplete. The circadian clock is another system that controls daily rhythmic physiological phenomena. It has been argued that there is a connection between the two clocks. To examine the genetic connection between them, we analyzed the associations of five circadian clock genes (period, timeless, Clock, cycle and cryptochrome) with the occurrence of diapause in Drosophila triauraria, which shows a robust reproductive diapause with clear photoperiodicity. Non-diapause strains found in low latitudes were compared in genetic crosses with the diapause strain, in which the diapause trait is clearly dominant. Single nucleotide polymorphism and deletion analyses of the five circadian clock genes in backcross progeny revealed that allelic differences in timeless and cryptochrome between the strains were additively associated with the differences in the incidence of diapause. This suggests that there is a molecular link between certain circadian clock genes and the occurrence of diapause.
Introduction
Seasonal photoperiodic responses are observed in many organisms, including plants, fungi, birds, mammals, and arthropods [1]–[3]. Diapause is a photoperiodic response that often results in a delay in a specific developmental stage; it is broadly observed in insects and other arthropods and is considered to be an adaptation to unfavorable seasons or conditions.
Diapause often depends on day (or night) length and temperature and is observed over a wide range of taxa. However, the physiological processes and genetic architectures involved in diapause are still largely unknown [4]. In contrast, the circadian clock, which controls daily rhythmic physiological phenomena, including locomotor activity, eclosion, and oviposition [5]–[7], has been studied well at the molecular level [8]–[10]. Both types of biological clock involve time-dependent responses to light stimulation [11]. Since Bünning [12] first proposed a two-clock connection whereby the daily circadian clock forms the basis of the seasonal photoperiodic timer, there has been a long argument—over several decades—about whether insect photoperiodism follows this theory [10], [11]. Many studies, performed in various species, have resulted in several models for explaining photoperiodic function [13]–[19]. One of the most widely adopted strategies for testing the involvement of circadian clock oscillation in the photoperiodic clock is the Nanda-Hamner protocol [20]. Although this protocol often produces positive results [14], [21], [22], it has also failed under different conditions, or even with different strains within the same species [4], [19], [23]. An alternative, nonoscillatory, hourglass-like model has also been proposed [15], [19]. This model was successfully applied by Veerman and Vaz Nunes in a spider mite [18]. However, the species of spider mite used also demonstrated a strongly positive Nanda-Hamner effect, suggesting that the photoperiodic clock was based on circadian clock oscillation [24]. Similar inconsistent results from the two models have been obtained in other insects [25]–[27]. Thus, the extent—if any—of dependence of the photoperiodic clock on the circadian clock remains obscure.
To further define the involvement of circadian clocks with photoperiodic responses, we performed a genetic analysis of the two clocks to determine whether common genes were present in the two clock systems. We chose the fruit fly Drosophila triauraria, a close relative of Drosophila melanogaster, for our analysis [28]. Circadian clock mechanisms have been well studied in D. melanogaster, but photoperiodicity in this insect's reproductive diapause is difficult to detect [29]. In contrast, D. triauraria has a distinct reproductive diapause with a robust photoperiodic response. Ovarian development in D. triauraria occurs normally with long daylength at moderate temperatures (e.g. 15°C) but not under short daylength conditions even at the same temperature [30], [31]. Geographic variation in diapause intensity is known to occur, and non-diapause populations (i.e., those that do not enter diapause, even under short daylength conditions) have been described [30], [31].
Circadian clocks consist of feedback loops involving several genes that are well conserved across broad taxa, from fungi to mammals [32], [33]. In D. melanogaster, five major genes are known in the central components of the circadian clock: period (per), timeless (tim), Clock (Clk), cycle (cyc), and cryptochrome (cry) [10]. Two intracellular feedback loops function in the circadian clock oscillators encoded by four of these genes, namely per, tim, Clk, and cyc. These gene products function as PER/TIM and CLK/CYC heterodimers. The CLK/CYC heterodimer activates the transcription of per and tim, while simultaneously repressing the transcription of Clk. Likewise, PER/TIM heterodimers repress their own production. The CRY protein encoded by cry is photosensitive and mediates entrainment of the clock by light [34], [35]. At the onset of photophase, CRY stimulated by light binds to TIM and then promotes degradation of TIM by phosphorylation [35]. These five genes are therefore central components of the circadian clock and were the focus of our study.
Although D. triauraria has tested positive for the involvement of circadian clock oscillation in the photoperiodic clock by using the Nanda-Hamner protocol [36], clear genetic covariance has not been detected between circadian-based oscillation resonance and diapause incidence [37]. Alternatively, an hourglass timer has been considered, assuming that diapause-inducing substances accumulate gradually, late at night [36]–[38]. This assumption of an hourglass timer and the lack of genetic covariance between circadian-based oscillation resonance and diapause incidence do not necessarily deny the involvement of circadian clock genes in diapause. Indeed, the involvement of a circadian clock has been suggested as part of photoperiodic time measurement in this species [36], [39]. Furthermore, recent studies suggest non-clock functions of circadian clock genes in the diapause occurrence [40], [41].
Here, we describe distinct phenotypes between diapause and non-diapause strains of D. triauraria and analyze the involvement of circadian clock genes in the photoperiodic response. Our results clearly demonstrate a correlation between diapause and two genetic loci that include tim and cry, thus providing a crucial link to the occurrence of diapause in D. triauraria.
Methods
Flies
We used four strains of D. triauraria, ONM, OEB12, KMJ1, and KMJD2. All the strains were established from natural populations in Japan. ONM was obtained from Onuma (42.0°N, 140.4°E) in 1981 (this strain was kindly provided by SG Goto). Strain OEB12 was collected on Okinoerabujima Island (27.2°N, 128.3°E) in 2005. Strains KMJ1 and KMJD2 came from Kumejima Island (26.2°N, 126.5°E) in 2002 and 2003, respectively. Strains OEB12, KMJ1, and KMJD2 were provided from the Drosophila Stocks of Ehime University (http://kyotofly.kit.jp/cgi-bin/ehime/index.cgi). All strains were maintained in glass vials containing standard cornmeal–yeast–glucose–agar medium at 23°C and a daily cycle of 15 h light:9 h dark.
Assay for ovarian diapause
We conducted a photoperiodic response assay under two light-cycle conditions, 15 h light:9 h dark (long daylength, LD) and 10 h light:14 h dark (short daylength, SD). These conditions were chosen because the critical daylength of ONM (the diapause strain) was 13 h [37].
Flies were allowed to lay eggs for 1 day at 23°C under the LD light cycle, and then the food vials with laid eggs were moved to either LD or SD conditions at 15°C. Virgin females were collected within 8 h after eclosion and placed into new food vials under the same conditions. Sixteen days after eclosion, the flies were dissected in PBS and ovary development was examined. Females containing only oocytes that did not develop beyond stage 7 were defined as being in diapause [42], [43]. This assay was conducted on all four original strains described above and on hybrid F1 females from crosses between ONM (the diapause strain) and the other three strains (non-diapause strains). We also examined the progeny from reciprocal backcrosses of strains ONM and OEB12.
DNA extraction
Genomic DNA of the two strains ONM and OEB12 was extracted individually with a DNeasy 96 Tissue kit (QIAGEN, Valencia, CA) in accordance with the protocol for animal tissues in the QIAGEN manual. DNA was eluted in 200 µl of buffer AE, of which 1 µl was used for amplification by PCR.
Strain-specific molecular markers of circadian rhythm genes
To identify strain-specific molecular markers for the five circadian rhythm genes (per, tim, Clk, cyc, and cry), we amplified several regions within each of the genes by using PCR using ExTaq polymerase (TaKaRa, Shiga, Japan). Primers for the PCR and sequencing for genotyping are shown in Table 1. Amplified PCR products were purified with an ExoSAP-IT PCR-cleanup kit (USB, Cleveland, OH) and then sequenced with an Applied Biosystems (Foster City, CA) 3130xl Genetic Analyzer. Sequencing reactions were conducted with a BigDye Terminator v1.1/v3.1 Cycle Sequencing Kit (Applied Biosystems). Purification with ExoSAP-IT and cycle sequencing reactions with the BigDye Terminator were performed according to the manufacturer's instructions.
Table 1. Primers used for PCR and sequencing.
| Gene | Primer sequences | Annealing temp. for PCR | |
| per | F | 5′-TTCTGCTGCGTCATCTCCATGC-3′ | 51°C |
| R | 5′-GGAACTCTTACTGTCATAGTAGTC-3′ | ||
| S | 5′-GACTTTGTGCACATCAAGGA-3′ | ||
| tim | F | 5′-TGATACCVYTGCTGGAGAATGCC-3′ | 63°C |
| R | 5′-TNGTGTCDATGTGCTCCATRTCC-3′ | ||
| S | 5′-GAACCAGGAGTCCATCTCCA-3′ | ||
| Clk | F | 5′-TGGTCAARTTYGTKGGYTACTTTC-3′ | 55°C |
| R | 5′-SGCATAGCTGACMACYTTRTGRGTG-3′ | ||
| S | 5′-GAAATGAGCATYATYGATCCSAC-3′ | ||
| cyc | F | 5′-CAAAAYCACAGYGAGATCGAGAARCG-3′ | 53°C |
| R | 5′-ATRTAGTCAATYTCGYTYGTCC-3′ | ||
| S | 5′-TGAGCTCTCCTCCATGATCC-3′ | ||
| cry | F | 5′-AGCGAATGTGATCTGGTTCC-3′ | 50°C |
| R | 5′-ATGCTCAGGCAGATCTCGTT-3′ | ||
| S | 5′-ACACAGGCTCGCAACTGGAC-3′ |
F, forward primer; R, reverse primer; S, sequence primer.
Generation of backcross progeny, and phenotyping and genotyping
We used two strains, the diapause strain ONM and the non-diapause strain OEB12, to generate backcross progeny (BC). First, to obtain F1 females, we performed two reciprocal crosses, ONM females ×OEB12 males, and OEB12 females × ONM males. We refer to the F1 flies from the former cross as F1(ONM×OEB12) and those from the latter cross as F1(OEB12×ONM). Then, BC females were produced by backcrossing both of the reciprocal F1 females to the OEB12 males. All of these crosses were performed under LD conditions at 23°C, and the mated F1 females were allowed to lay eggs under the same conditions for 1 day in a food vial. The food vials with laid eggs were then moved to two different conditions: LD and SD at 15°C. Ovarian development was inspected for diapause on day 16 post-eclosion, as described above. Dissected females were kept at −20°C for DNA extraction and genotyping. Genotyping was conducted for the five genes by examining strain-specific molecular markers with sequencing after PCR amplification. We used the following conditions for PCR to genotype the genes: 1.2 µl 10× ExTaq buffer, 0.96 dNTPs, 3.0 µl of 2-µM forward primer, 3.0 µl of 2-µM reverse primer, 0.15 µl ExTaq polymerase, 1.0 µl DNA template, and 2.69 µl distilled water for a final reaction volume of 12 µl. Reaction conditions were 94°C for 2 min plus 40 cycles of 94°C for 30 s, 50 to 63°C annealing for 30 s (51°C for per, 63°C for tim, 55°C for Clk, 53°C for cyc, 50°C for cry), 72°C for 1 min 30 s, plus a final 72°C extension for 7 min.
Eclosion rhythm
Circadian rhythms were measured by using the pattern of adult eclosion under 12 h light:12 h dark at 23°C. In our preliminary experiments, we found only white flies (indicating that they had just eclosed) at the onset of the photoperiod. We therefore counted eclosed flies every hour during the 12 h of photophase. The white flies found at the onset of photophase were counted as those eclosed during the 1 h before the onset of photophase. The degree of rhythmicity in eclosion was measured by using Winfree's R-values [44]. The R-values were calculated on the basis of summation of the diel distributions of eclosion. First, the 8-h period of the day containing the highest number of eclosions was identified. The R-value was then calculated by dividing the number of eclosions within the remaining 16 h (i.e., outside the 8-h period) by the number within the highest 8 h and multiplying by 100. R-values of less than 30 were considered to be “highly rhythmic,” whereas those over 90 were “arrhythmic” [45].
Statistics
We applied the generalized linear model assuming a binomial distribution of diapause occurrence (diapause or not) to analyze the effects of allelic differences in the clock genes on diapause incidence. Statistical calculations for the generalized linear model were performed by using the “glm” function in the “stats” package of the R statistical software package version 2.8.0 ([46], http://www.r-project.org/). The best-fit model was determined by using stepwise model selection with the Akaike information criterion (AIC) contained within the “MASS” package in R, choosing the AIC with the least score as the best fit. In the model selection, we included not only the five genes as main factors but also all possible interactions among the five genes.
Results
Diapause incidence
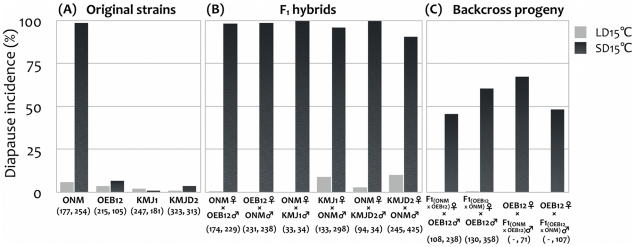
We defined diapause as occurring in those 16-day-old females that contained only oocytes that did not develop beyond stage 7 (i.e., no yolk accumulation in the egg chambers). Using this criterion, no, or very little, diapause was observed in the four strains, or in the hybrid F1 and BC females, when they were reared under LD 15°C conditions (Fig. 1). However, striking differences were observed among females reared under SD 15°C conditions (Figs. 1 and 2): the high-latitude strain ONM displayed a very substantial diapause incidence (98.8%), whereas the low-latitude strains OEB12, KMJ1, and KMJD2 exhibited very low diapause incidences (6.7%, 1.1%, and 3.8%, respectively) (Fig. 1A). All females that were F1 hybrids between the diapause strain ONM and the non-diapause strains displayed high diapause incidences under SD 15°C conditions (90.8% to 100%, Fig. 1B); these incidences were similar to those observed in the diapause strain ONM. These results strongly indicate that the diapause strain had a dominant genetic effect on the phenotypes of heterozygous females.
Figure 1. Diapause incidences in the original strains (A), F1 hybrids (B), and BC progeny (C) of Drosophila triauraria.
LD, long daylength conditions (15 h light:9 h dark); SD, short daylength conditions (10 h light:14 h dark). Under both conditions the temperature was 15°C from egg to adult. Diapause incidences under LD are shown as gray bars, and those under SD are shown as black bars. Crosses from which F1 and BC females were obtained are shown under each bar. F1(ONM×OEB12) and F1(OEB12×ONM) indicate F1 flies obtained from ONM♀×OEB12♂ and OEB12♀×ONM♂, respectively. Numbers of dissected females are shown in parentheses: within the parentheses the values at left are those under LD conditions and those at right are under SD conditions.
Figure 2. Pairs of ovaries dissected from 16-day-old female Drosophila triauraria reared under short daylength conditions at 15°C.
Top: non-diapause strain (OEB12). Bottom: diapause strain (ONM).
Circadian clock genes of D. triauraria and strain-specific molecular markers
We used the PCR primers shown in Table 1 to amplify genomic regions of the circadian clock genes per, tim, Clk, cyc, and cry. The PCR products were sequenced and BLAST searches confirmed their high degrees of similarity to the corresponding genes in D. melanogaster, as follows: per 86.8% (738/850 bp), tim 85.7% (275/321 bp), Clk 89.3% (134/150 bp), cyc 87.6% (686/783 bp), and cry 81.4% (544/668 bp). These regions are all exonic regions in D. melanogaster.
We identified at least one molecular marker—either a single nucleotide polymorphism (SNP) or insertion/deletion within each gene—that was useful for distinguishing allelic differences between the diapause strain ONM and the non-diapause strain OEB12 (Table 2): two strain-specific SNPs in per, an ONM-specific 12-base deletion and one strain-specific SNP in tim, one strain-specific SNP in Clk, three strain-specific SNPs in cyc, and one strain-specific SNP and an OEB12–specific 12-base deletion in cry. For each gene, we confirmed the strain specificity of these markers in more than 30 flies of each strain. We also confirmed the heterozygosity in the locations of the sequences from F1 flies between the strains. Furthermore, we found no contradictions in the genotyping results in 329 BC flies. These strain-specific markers were found in either coding or noncoding regions of sequences corresponding to those of D. melanogaster in the alignment results, as shown in Table 2 (coding regions are shown as boldface characters). These strain-specific markers were used for genotypic analysis of diapause in BC females.
Table 2. Strain-specific SNPs/deletions in the five clock genes in Drosophila triauraria.
| per | ONM | GAAGAGCACCTTCTGCGTGATGT(180bp)GCAGCTACAAGGGTAAGTGG* * |
| OEB12 | GAAGAGCACGTTCTGCGTGATGT(180bp)GCAGCTACAAGGGTGAGTGG* * | |
| tim | ONM | TTTATTTA------------AGTATTAT(45bp)GAGGATATATCTAATC************ * |
| TTTATTTATAAATTATCTTATTTATTAT(45bp)GAGGATATCTCCAATC************ * | ||
| OEB12 | TTTATTTATAAATTATCTTATYWATTAT(45bp)GAGGATATATCYAATC<1/emph>************ * | |
| TTTATTTATAAATTATCTTATYWATTAT(45bp)GAGGATATTTCYAATC************ * | ||
| Clk | ONM | TATATGCCGTTCGAGGTGCTAGGCACCTCTGGTTATGATTACTATCACTT* |
| OEB12 | TATATGCCGTTYGAGGTGCTGGGCACYTCYGGYTAYGATTACTATCACTT* | |
| cyc | ONM | GAGCTTAAGATG(32bp)CGTGGTAGGTTGT(146bp)GTGCCCTAGGGA* * * |
| OEB12 | GAGCTCAAGATG(32bp)CGTGGTGGGTTGT(146bp)GTGCCCCAGGGA* * * | |
| cry | ONM | AGAGGTAACAGGTTCAGTTACGGYTGTATTGCTTTAGTTTTAACTTTWTT************ * |
| OEB12 | AGAGGTAACAGGTTCAGTTACGGCTGTACTGCTTTAGTTTTAACTTTATT************ * | |
| AGAGGTAAC------------GGTTGTATTGTTTTAGTTTTAACTTTATT************ * |
Asterisks indicate SNPs/deletions used as strain-specific markers. Boldface characters show coding regions equivalent to the D. melanogaster sequences used for comparison. Two types of sequences were found in ONM and OEB12 in the case of tim, and in OEB12 in the case of cry.
Genotyping with the five circadian clock genes
Dominance of the diapause character in strain ONM was already known from the diapause incidence in F1 females (Fig. 1A, B). Therefore, we examined the effect of the circadian genes by backcrossing F1 females with OEB12 males (i.e., of the non-diapause strain) and genotyping the BC females. If a gene affected the diapause phenotype, then its frequency of occurrence in females homozygous for the OEB12 alleles would be expected to be low, whereas that in females heterozygous for ONM/OEB12 alleles would be expected to be high.
We genotyped 329 BC females in total: 116 BC females from the backcross using F1(ONM×OEB12) females, and 213 BC females from the backcross of F1(OEB12×ONM) females. The frequencies of occurrence of the circadian clock genes in females homozygous for the OEB12 allele were not significantly different from those in females heterozygous for the ONM and OEB12 alleles, except in the case of cyc (χ2 = 7.30, P = 0.007) (Table 3). The relative positions of the genes, as expected from the recombination values (Table 4), were similar to those in D. melanogaster, which are per on X, tim on 2L, Clk and cyc on 3L, and cry on 3R (FlyBase, http://flybase.org/).
Table 3. Segregation ratio for each gene.
| Frequency | ||||
| Gene | Homo | Hetero | χ2 | P |
| per | 154 | 175 | 1.340 | 0.247 |
| tim | 150 | 179 | 2.556 | 0.110 |
| Clk | 165 | 164 | 0.003 | 0.956 |
| cyc | 140 | 189 | 7.298 | 0.007 |
| cry | 182 | 147 | 3.723 | 0.054 |
“Frequency” shows the numbers of flies homozygous and heterozygous for each gene. A χ2-test was conducted under the expectation of a 1:1 segregation ratio between homozygous and heterozygous flies.
Table 4. Recombination values between genes.
| Recombination values (%) | ||||
| tim | Clk | cyc | cry | |
| per | 58.97 | 50.15 | 48.63 | 51.06 |
| tim | 55.62 | 54.10 | 55.32 | |
| Clk | 23.40 | 31.91 | ||
| cyc | 40.12 | |||
Association between circadian clock genes and diapause incidence
In our statistical analysis we compared the AICs of all possible combinations of factors (i.e., the five genes) to find the best-fit model. The model that included “diapause = tim+cyc+cry” had the smallest AIC (390.2) (Table 5). In this model, tim and cry were significant (P<<0.0001), but cyc was not significant (P = 0.079). The AICs of models including and excluding cyc were similar: the AIC for the model “diapause = tim+cry” was 391.3. Other models that included other interactions and main factors did not give AICs smaller than 390.2; that is, none of the interactions or the two main factors per and Clk had any effect on diapause incidence.
Table 5. The final model selected as the generalized linear model.
| Source | Estimate | SE | z value | P (>|z|) |
| (Intercept) | −1.27 | 0.27 | −4.67 | <<0.0001 |
| tim | 1.44 | 0.26 | 5.47 | <<0.0001 |
| cyc | 0.45 | 0.26 | 1.76 | 0.079 |
| cry | 1.48 | 0.27 | 5.48 | <<0.0001 |
glm (formula = Diapause ∼ tim+cyc+cry, family = binomial). Null deviance: 446.17 on 328 degrees of freedom. Residual deviance: 382.20 on 325 degrees of freedom. AIC: 390.2.
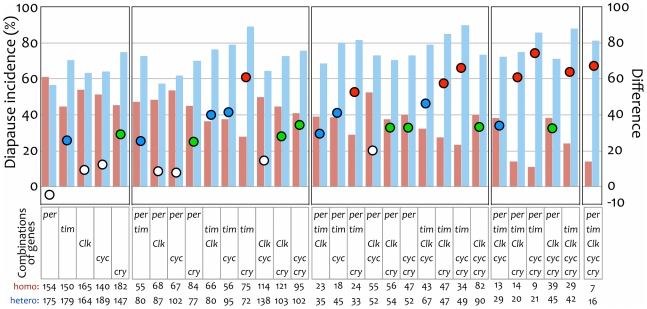
We summarized the diapause incidences in flies of each genotype in Figure 3. Here, the rates of homozygous and heterozygous flies for each gene set are shown as red and blue bar graphs, respectively. Differences between homozygous and heterozygous flies are plotted as circles; the difference values were calculated as “diapause incidence in heterozygous females – diapause incidence in homozygous females” for each gene set. Stronger genetic effects on the phenotype should result in greater differences between flies that carry homozygous alleles vs. those that carry heterozygous alleles. As found above, tim and cry were highly associated with diapause occurrence.
Figure 3. Diapause incidences for each genotype of BC progeny of Drosophila triauraria under short daylength conditions, and their differences between heterozygous and homozygous females.
Red and blue bars show diapause incidences (%) of flies homozygous and heterozygous for the genes shown as “Combinations of genes” in the middle of the figure, respectively. Numbers of dissected flies are shown at the bottom of the figure for homozygous and heterozygous, respectively. Plotted circles show differences in diapause incidences between heterozygous and homozygous flies of each gene set. The differences indicate the strength of the genetic effects of each gene set on the phenotype: a gene set with a strong genetic effect will show a higher difference. Blue, differences involving tim; green, differences involving cry; red, differences involving both tim and cry; white, differences involving neither tim nor cry. Scale for diapause incidence is shown on the left side, and that for difference is on the right side.
Eclosion rhythm
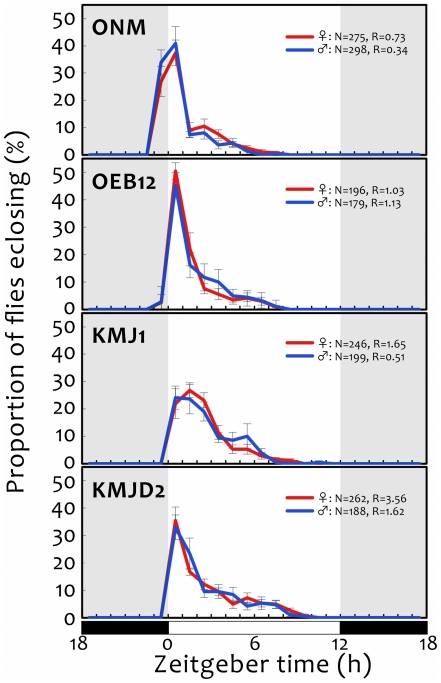
We determined the eclosion rhythms of 1843 flies (275 females and 298 males from ONM, 196 females and 179 males from OEB12, 246 females and 199 males from KMJ1, and 262 females and 188 males from KMJD2) (Fig. 4). The Winfree's R-values were 0.73 for females and 0.34 for males from ONM; 1.03 for females and 1.13 for males from OEB12; 1.65 for females and 0.51 for males from KMJ1; and 3.56 for females and 1.62 for males from KMJD2. These results show that the eclosion patterns were strongly rhythmic. Furthermore, the phases of the rhythmic patterns clearly differed between diapause and non-diapause strains. About 30% of flies of the diapause strain ONM had eclosed within 1 h before the onset of photophase, and about 40% had eclosed within 1 h after that. In contrast, all three non-diapause strains had barely eclosed before the onset of photophase (2.6% of females and 2.8% of males from OEB12 and 0% of both sexes from KMJ1 and KMJD2).
Figure 4. Time-series proportions of Drosophila triauraria flies eclosing under 12-h light:12-h dark cycles at 23°C.
Eclosing flies were counted every hour over several days. Counted numbers of flies were pooled and the proportions calculated for each data point. Error bars represent ± SEM. R numbers are Winfree's R values.
Discussion
Using diapause and non-diapause strains, we studied the association between five circadian clock genes and the diapause occurrence, that is photoperiodic response, in D. triauraria. Because all four strains that we used had clear eclosion rhythms, we considered that their circadian clocks were functioning normally. Clear differences were found in diapause incidence under SD conditions between the strain from a high-latitude location (ONM, the diapause strain) and the strains from low-latitude locations (OEB12, KMJ1, and KMJD2, the non-diapause strains). Our genotyping analysis revealed that molecular markers within the two genes tim and cry were associated with this difference in diapause incidence, specifically under SD conditions (not under LD conditions); the three genes (per, Clk, and cyc) had no significant effects.
The involvement of circadian clock genes in diapause induction has been discussed for a long time [4], [11], [19], [47], [48]. Recently, some circadian clock genes have been suggested to be associated with diapause in insects [40], [49]–[56]. Studies using putative mutant strains [49]–[51] or the natural variations in diapause incidence that are often observed with latitudinal clines [40], [52] have shown that tim is the gene most likely associated with diapause occurrence. The different isoforms encoded by tim alleles associated with diapause occurrence in D. melanogaster differ in their interactions with the CRY protein, and their circadian photoresponsiveness are different [57]; this seem to be suggestive, even though their findings do not explain the differences in diapause incidence. Here, we found that the occurrence of diapause was associated with the presence of molecular markers in tim and cry. The proteins encoded by these two genes are the first components in the photoresponse of the circadian clock: CRY is a circadian photoreceptor that triggers TIM degradation in response to light [35]. Involvement of CRY in diapause occurrence has also been suggested in the flesh fly Sarcophaga similis [58].
per has also been examined in some other insect species, and both positive [53]–[55], [59] and negative [60], [61] results for involvement in diapause have been reported. As these cases, the involvement of per was likely partial or depended on the species. Our study clearly showed no association of the per locus with diapause in D. triauraria, even though at least one locus on the X chromosome is expected to be responsible for diapause in this species [38]. We in fact detected a difference in diapause incidence that seemed to indicate an X-chromosome effect. This difference was observed between two BC females obtained from backcrosses using OEB12 females and hybrid F1 males (χ2 = 5.5152, df = 1, P = 0.0189; Fig. 1C). In the crosses no recombination was expected, because we had used hybrid male flies. The BC females therefore had different X chromosome pairs: the BC females from the cross with F1(OEB12×ONM) males had two X chromosomes from OEB12, and the BC females from the cross with F1(ONM×OEB12) males had a pair of X chromosomes from ONM and OEB12.
There had been little study of the involvement of Clk and cyc in diapause until recently [53], when control of the occurrence of diapause by cyc was reported in the bean bug Riptortus pedestris by using RNAi [54], [55]. However, we found no association of Clk and cyc with the diapause occurrence, even though a marginal effect was detected for cyc (P = 0.079, Table 5).
To our knowledge, no one has performed a genetic linkage analysis between the genes responsible for diapause and multiple circadian clock genes in one species at the same time. As with the circadian clock, the photoperiodic clock mechanism can be made up of the interactions of several genes. Taking into consideration these interactions among genes, we can expect genetic linkage analysis with multiple genes to supply useful information. We performed an examination with the five genes simultaneously and then analyzed the interactions between them. We detected no clear interactions in our statistical analysis, even between tim and cry; this lack of interaction is indicated by the parallel nature of the blue and red lines in Figure 5. Instead, an additive effect of tim and cry was observed (Fig. 3): the red circles (involvement of both of tim and cry) showed large differences in diapause incidence between heterozygous and homozygous flies; the blue (tim) and green (cry) showed intermediate differences; and the white (neither tim nor cry) had distinctly weak differences. This additivity is also indicated in Figure 5 as separated parallel blue and red lines. These findings suggest that tim and cry have independent effects on the occurrence of diapause, unlike their action in the circadian clock. The occurrence of diapause might not be based on circadian clock function; alternatively, even if it is based on clock function, natural variations in the photoperiodic response of diapause might depend simply on variations within one or two genes. It seems reasonable for some of the circadian clock genes to be involved in the occurrence of diapause but for the circadian clock function itself not to be involved, as suggested in previous studies [40], [41]. However, the involvement of cry suggests that the photoresponse function is common to both clocks.
Figure 5. Diapause incidences in allelic combinations of two of the five genes of BC females of Drosophila triauraria.
“homo,” homozygous for alleles from the non-diapause strain; “hetero,” heterozygous for those from the diapause and non-diapause strains. In the cases of per, Clk, and cyc (rows 1, 3, and 4), the blue and red lines almost overlap in each graph, indicating that there was no allelic effect on diapause incidence. In the cases of tim and cry (rows 2 and 5), the blue lines are always higher than the red lines in each graph, indicating that there is an effect of allelic differences on diapause incidence; furthermore, the blue and red lines are completely parallel in the combination of tim and cry, indicating that there is no interaction effect between them. These effects were confirmed by the statistical analysis.
The dominance of the diapause character in the diapause strain is apparent from the results in F1 females (Fig. 1B). The finding of a diapause incidence of about 50% in BC females suggests that the occurrence of diapause is controlled by a single locus. However, this is unlikely, because our genotyping analysis using molecular markers indicated that there were additive effects of multiple loci (Fig. 3), or, at least, two loci located in the regions including tim and cry and another locus on the X chromosome (but not per). The difference in diapause incidences between BC females homozygous for all five genes and those heterozygous for all five was 67.0% (see a red circle on the far-right column in Fig. 3). It was 60.9% between BC females homozygous for both tim and cry and heterozygous for both (the alleles of the other three genes were not fixed) (see a red circle on the 12th column from the left in Fig. 3), indicating that the incidence of diapause was strongly influenced by the two regions including tim and cry. However, note that 28.0% of BC females entered the diapause state even if they had tim and cry alleles from the non-diapause strain (see a red bar on the 12th column from the left in Fig. 3), indicating that there are other genes or regions that control diapause, besides the two regions including the two markers. This is consistent with the findings of a previous study that the differences in photoperiodic response in D. triauraria are due to genes at three or four loci, at least one of which is on the X chromosome [38]. Currently, however, we cannot exclude the possibility that the strong effects we detected are caused by genes located near tim and cry, not by tim and cry themselves.
Recently, two independent QTL mapping studies revealed that single genes control natural variations in the incidence of diapause in D. melanogaster: one is Dp110, which encodes insulin-regulated phosphatidylinositol 3-kinase [62], and the other is couch potato (cpo), which encodes an RNA-binding protein [63]. In the process leading to photoperiodic diapause, at least two components must be considered: one by which photoperiodic information is received and time is calculated, and the other, endogenous, process by which clock information is transmitted to responsive organs, such as the ovaries in reproductive diapause in Drosophila [55], [64]. The two abovementioned genes are considered responsible for the latter endogenous process, but not for the former one [29], [62]–[69]. It is not known whether tim and cry are associated with the photoperiodic timer or the endogenous process. We are interested in whether allelic differences in Dp110 and cpo are associated with differences in the incidence of diapause between D. triauraria strains. Both genes are located close to cry in D. melanogaster (cpo 90D1-E1, cry 91F11, Dp110 92F3).
Our results have provided us with clues as to what needs to be investigated next. We are currently establishing recombinant inbred lines between diapause and non-diapause strains of D. triauraria. Use of these recombinant inbred lines will enable us to perform fine mapping and to study the functions of tim and cry in the photoperiodic response, as well as the interactions between these two genes.
Acknowledgments
We thank Dr Shin G Goto, Dr Masahito T Kimura, and Ehime University for providing the strains, as well as Dr Hiroshi Matsubayashi and Dr Takashi Ohsako of the Drosophila Genetic Resource Center, Kyoto, for their helpful suggestions on experimental techniques. We also thank Dr Timothy Karr for kindly providing valuable comments on the manuscript.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported in part by a grant from the National Bio-Resource Project from the Ministry of Education, Science, Sports and Culture of Japan. No additional external funding received for this study. The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Suzuki L, Johnson CH. Algae know the time of day: circadian and photoperiodic programs. J Phycol. 2001;37:933–942. [Google Scholar]
- 2.Schults TF, Kay SA. Circadian clocks in daily and seasonal control of development. Science. 2003;301:326–328. doi: 10.1126/science.1085935. [DOI] [PubMed] [Google Scholar]
- 3.Bradshaw WE, Holzapfel CM. Evolution of animal photoperiodism. Annu Rev Ecol Evol Syst. 2007;38:1–25. [Google Scholar]
- 4.Saunders DS, Lewis RD, Warman GR. Photoperiodic induction of diapause: opening the black box. Physiol Entomol. 2004;29:1–15. [Google Scholar]
- 5.Merrow M, Spoelstra K, Roenneberg T. The circadian cycle: daily rhythms from behaviour to genes. EMBO Rep. 2005;6:930–935. doi: 10.1038/sj.embor.7400541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sheeba V, Chandrashekaran MK, Joshi A, Sharma VK. A case for multiple controlling different circadian rhythms in Drosophila melanogaster. J Insect Physiol. 2001;47:1217–1225. doi: 10.1016/s0022-1910(01)00107-x. [DOI] [PubMed] [Google Scholar]
- 7.Panda S, Hogenesch JB, Kay SA. Circadian rhythms from flies to human. Nature. 2002;417:329–335. doi: 10.1038/417329a. [DOI] [PubMed] [Google Scholar]
- 8.Wager-Smith K, Kay SA. Circadian rhythm genetics: from flies to mice to humans. Nat Genet. 2000;26:23–27. doi: 10.1038/79134. [DOI] [PubMed] [Google Scholar]
- 9.Helfrich-Förster C. Neurobiology of the fruit fly's circadian clock. Genes Brain Behav. 2005;4:65–76. doi: 10.1111/j.1601-183X.2004.00092.x. [DOI] [PubMed] [Google Scholar]
- 10.Hardin PE. The circadian timekeeping system of Drosophila. Curr Biol. 2005;15:R714–22. doi: 10.1016/j.cub.2005.08.019. [DOI] [PubMed] [Google Scholar]
- 11.Danks HV. How similar are daily and seasonal biological clocks? J Insect Physiol. 2005;51:609–619. doi: 10.1016/j.jinsphys.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 12.Bünning E. Die endogene tagesrhythmik als grundlage der photoperiodischen reaktion. Ber Dtsch Bot Ges. 1936;54:590–607. [Google Scholar]
- 13.Vaz Nunes M. A double circadian oscillator model for quantitative photoperiodic time measurement in insects and mites. J Theor Biol. 1998;194:299–311. doi: 10.1006/jtbi.1998.0767. [DOI] [PubMed] [Google Scholar]
- 14.Vaz Nunes M, Saunders DS. Photoperiodic time measurement in insects: a review of clock models. J Biol Rhythms. 1999;14:84–104. doi: 10.1177/074873049901400202. [DOI] [PubMed] [Google Scholar]
- 15.Lees AD. Some effects of temperature on the hour glass photoperiodic clock in the aphid Megoura viciae. J Insect Physiol. 1986;32:79–89. [Google Scholar]
- 16.Vaz Nunes M, Veerman A. Photoperiodic time measurement in the spider mite Tetranychus urticae: a novel concept. J Insect Physiol. 1982;28:1041–1053. [Google Scholar]
- 17.Vaz Nunes M, Veerman A. ‘Bistability’ experiments and the photoperiodic clock in the spider mite Tetranychus urticae. Entomol Exp Appl. 1997;84:195–197. [Google Scholar]
- 18.Veerman A, Vaz Nunes M. Analysis of the operation of the photoperiodic counter provides evidence for hourglass time measurement in the spider mite Tetranychus urticae. J Comp Physiol A. 1987;160:421–430. [Google Scholar]
- 19.Veerman A. Photoperiodic time measurement in insects and mites: a critical evaluation of the oscillator-clock hypothesis. J Insect Physiol. 2001;47:1097–1109. doi: 10.1016/s0022-1910(01)00106-8. [DOI] [PubMed] [Google Scholar]
- 20.Nanda KK, Hamner KC. Studies on the nature of the endogenous rhythm affecting photoperiodic responses of Biloxi soybean. Bot Gaz. 1958;120:14–25. [Google Scholar]
- 21.Takeda M, Skopik SD. Photoperiodic time measurement and related physiological mechanisms in insects and mites. Annu Rev Entomol. 1997;42:323–349. doi: 10.1146/annurev.ento.42.1.323. [DOI] [PubMed] [Google Scholar]
- 22.Tauber E, Kyriacou BP. Insect photoperiodism and circadian clocks: models and mechanisms. J Biol Rhythms. 2001;16:381–390. doi: 10.1177/074873001129002088. [DOI] [PubMed] [Google Scholar]
- 23.Danks HV. Studying insect photoperiodism and rhythmicity: components, approaches and lessons. Eur J Entomol. 2003;100:209–221. [Google Scholar]
- 24.Vaz Nunes M, Veerman A. A ‘dusk’ oscillator affects photoperiodic induction of diapause in the spider mite, Tetranychus urticae. J Insect Physiol. 1986;32:1029–1034. [Google Scholar]
- 25.Saunders DS, Lewis RD. The photoperiodic clock and counter mechanism in two species of flies: evidence for damped circadian oscillators in time measurement. J Comp Physiol A. 1988;163:365–371. [Google Scholar]
- 26.Veerman A, Beekman M, Veenendaal RL. Photoperiodic induction of diapause in the large white butterfly, Pieris brassicae: evidence for hourglass time measurement. J Insect Physiol. 1988;34:1063–1069. [Google Scholar]
- 27.Kimura Y, Masaki S. Hourglass and oscillator expression of photoperiodic diapause response in the cabbage moth Mamestra brassicae. Physiol Entomol. 1993;18:240–246. [Google Scholar]
- 28.Bock IR, Wheeler MR. The Drosophila melanogaster species group. Univ Texas Publs Stud Genet. 1972;7:7213. [Google Scholar]
- 29.Tatar M, Chien SA, Priest NK. Negligible senescence during reproductive dormancy in Drosophila melanogaster. Am Nat. 2001;158:248–258. doi: 10.1086/321320. [DOI] [PubMed] [Google Scholar]
- 30.Kimura MT. Geographic variation and genetic aspects of reproductive diapause in Drosophila triauraria and Drosophila quadraria. Physiol Entomol. 1983;8:181–186. [Google Scholar]
- 31.Kimura MT. Geographic variation of reproductive diapause in the Drosophila auraria complex (Diptera: Drosophilidae). Physiol Entomol. 1984;9:425–431. [Google Scholar]
- 32.Dunlap JC. Molecular bases for circadian clocks. Cell. 1999;96:271–290. doi: 10.1016/s0092-8674(00)80566-8. [DOI] [PubMed] [Google Scholar]
- 33.Harmer SL, Panda S, Kay SA. Molecular bases of circadian rhythms. Annu Rev Cell Dev Biol. 2001;17:215–253. doi: 10.1146/annurev.cellbio.17.1.215. [DOI] [PubMed] [Google Scholar]
- 34.Emery P, So WV, Kaneko M, Hall JC, Rosbash M. CRY, a Drosophila clock and light-regulated cryptochrome, is a major contributor to circadian rhythm resetting and photosensitivity. Cell. 1998;95:669–679. doi: 10.1016/s0092-8674(00)81637-2. [DOI] [PubMed] [Google Scholar]
- 35.Ceriani MF, Darlington TK, Staknis D, Más P, Petti AA, et al. Light-dependent sequestration of TIMELESS by CRYPTOCHROME. Science. 1999;285:553–556. doi: 10.1126/science.285.5427.553. [DOI] [PubMed] [Google Scholar]
- 36.Yoshida T, Kimura MT. The photoperiodic clock of Drosophila triauraria: involvement of a circadian oscillatory system. J Insect Physiol. 1993;39:223–228. [Google Scholar]
- 37.Yoshida T, Kimura MT. The photoperiodic clock of Drosophila triauraria: involvement of two processes in the night-length measurement system. J Insect Physiol. 1993;39:101–106. [Google Scholar]
- 38.Kimura MT, Yoshida T. A genetic analysis of photoperiodic reproductive diapause in Drosophila triauraria. Physiol Entomol. 1995;20:253–256. [Google Scholar]
- 39.Yoshida T, Kimura MT. Relation of the circadian system to the photoperiodic clock in Drosophila triauraria (Diptera: Drosophilidae): an approach from analysis of geographic variation. Appl Entomol Zool. 1994;29:499–505. [Google Scholar]
- 40.Tauber E, Zordan M, Sandrelli F, Pegoraro M, Osterwalder N, et al. Natural selection favors a newly derived timeless allele in Drosophila melanogaster. Science. 2007;316:1895–1898. doi: 10.1126/science.1138412. [DOI] [PubMed] [Google Scholar]
- 41.Stehlík J, Závodská R, Shimada K, Šauman I, Koštál V. Photoperiodic induction of diapause requires regulated transcription of timeless in the larval brain of Chymomyza costata. J Biol Rhythms. 2008;23:129–139. doi: 10.1177/0748730407313364. [DOI] [PubMed] [Google Scholar]
- 42.King RC. New York: Academic Press; 1970. Ovarian development in Drosophila melanogaster. [Google Scholar]
- 43.Kimura MT. Quantitative response to photoperiod during reproductive diapause in the Drosophila auraria species-complex. J Insect Physiol. 1990;36:147–152. [Google Scholar]
- 44.Winfree AT. An integrated view of resetting of a circadian clock. J Theor Biol. 1970;28:327–374. doi: 10.1016/0022-5193(70)90075-5. [DOI] [PubMed] [Google Scholar]
- 45.Saunders DS. Amsterdam: Elsevier; 2002. Insect Clocks, third ed. [Google Scholar]
- 46.Ihaka R, Gentleman R. R: a language for data analysis and graphics. J Comput Graph Stat. 1996;5:299–314. [Google Scholar]
- 47.Saunders DS. Erwin Bünning and Tony Lees, two giants of chronobiology, and the problem of time measurement in insect photoperiodism. J Insect Physiol. 2005;51:599–608. doi: 10.1016/j.jinsphys.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 48.Bradshaw WE, Holzapfel CM. What season is it anyway? Circadian tracking vs. photoperiodic anticipation in insects. J Biol Rhythms. 2010;25:155–165. doi: 10.1177/0748730410365656. [DOI] [PubMed] [Google Scholar]
- 49.Kostál V, Shimada K. Malfunction of circadian clock in the non-photoperiodic-diapause mutants of the Drosophilid fly, Chymomyza costata. J Insect Physiol. 2001;47:1269–1274. doi: 10.1016/s0022-1910(01)00113-5. [DOI] [PubMed] [Google Scholar]
- 50.Pavelka J, Shimada K, Kostál V. TIMELESS: a link between fly's circadian and photoperiodic clocks? Eur J Entomol. 2003;100:255–265. [Google Scholar]
- 51.Goto SG, Han B, Denlinger DL. A nondiapausing variant of the flesh fly, Sarcophaga bullata, that shows arrhythmic adult eclosion and elevated expression of two circadian clock genes, period and timeless. J Insect Physiol. 2006;52:1213–1218. doi: 10.1016/j.jinsphys.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 52.Mathias D, Jacky L, Bradshaw WE, Holzapfel CM. Geographic and developmental variation in expression of the circadian rhythm gene, timeless, in the pitcher-plant mosquito, Wyeomyia smithii. J Insect Physiol. 2005;51:661–667. doi: 10.1016/j.jinsphys.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 53.Syrová Z, Dolezel D, Saumann I, Hodkova M. Photoperiodic regulation of diapause in linden bugs: are period and Clock genes involved? Cell Mol Life Sci. 2003;60:2510–2515. doi: 10.1007/s00018-003-3227-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ikeno T, Tanaka S, Numata H, Goto SG. Photoperiodic diapause under the control of circadian clock genes in an insect. BMC Biol. 2010;8:116. doi: 10.1186/1741-7007-8-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bradshaw WE, Holzapfel CM. Circadian clock genes, ovarian development and diapause. BMC Biol. 2010;8:115. doi: 10.1186/1741-7007-8-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kostál V. Insect photoperiodic calendar and circadian clock: Independence, cooperation, or unity? J Insect Physiol. 2011;57:538–556. doi: 10.1016/j.jinsphys.2010.10.006. [DOI] [PubMed] [Google Scholar]
- 57.Sandrelli F, Tauber E, Pegoraro M, Mazzotta G, Cisotto P, et al. A molecular basis for natural selection at the timeless locus in Drosophila melanogaster. Science. 2007;316:1898–900. doi: 10.1126/science.1138426. [DOI] [PubMed] [Google Scholar]
- 58.Goto SG, Numata H. Possible involvement of distinct photoreceptors in the photoperiodic induction of diapause in the flesh fly Sarcophaga similis. J Insect Physiol. 2009;55:401–407. doi: 10.1016/j.jinsphys.2008.11.008. [DOI] [PubMed] [Google Scholar]
- 59.Han B, Denlinger DL. Length variation in a specific region of the period gene correlates with differences in pupal diapause incidence in the flesh fly, Sarcophaga bullata. J Insect Physiol. 2009;55:415–418. doi: 10.1016/j.jinsphys.2009.01.005. [DOI] [PubMed] [Google Scholar]
- 60.Saunders DS, Henrich VC, Gilbert LI. Induction of diapause in Drosophila melanogaster: photoperiodic regulation and the impact of arrhythmic clock mutations on time measurement. Proc Natl Acad Sci U S A. 1989;86:3748–3752. doi: 10.1073/pnas.86.10.3748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Saunders DS. The circadian basis of ovarian diapause regulation in Drosophila melanogaster: is the period gene causally involved in photoperiodic time measurement? J Biol Rhythms. 1990;5:315–331. doi: 10.1177/074873049000500404. [DOI] [PubMed] [Google Scholar]
- 62.Williams KD, Busto M, Suster ML, So AK, Ben-Shahar Y, et al. Natural variation in Drosophila melanogaster diapause due to the insulin-regulated PI3-kinase. Proc Natl Acad Sci U S A. 2006;103:15911–15915. doi: 10.1073/pnas.0604592103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schmidt PS, Zhu CT, Das J, Batavia M, Yang L, et al. An amino acid polymorphism in the couch potato gene forms the basis for climatic adaptation in Drosophila melanogaster. Proc Natl Acad Sci U S A. 2008;105:16207–16211. doi: 10.1073/pnas.0805485105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schiesari L, Kyriacou CP, Costa R. The hormonal and circadian basis for insect photoperiodic timing. FEBS Letters. 2011;585:1450–1460. doi: 10.1016/j.febslet.2011.02.026. [DOI] [PubMed] [Google Scholar]
- 65.Bellen HJ, Kooyer S, D'Evelyn D, Pearlman J. The Drosophila Couch potato protein is expressed in nuclei of peripheral neuronal precursors and shows homology to RNA-binding proteins. Genes Dev. 1992;6:2125–2136. doi: 10.1101/gad.6.11.2125. [DOI] [PubMed] [Google Scholar]
- 66.Clancy DJ, Gems D, Harshman LG, Oldham S, Stocker H, et al. Extension of life-span by loss of CHICO, a Drosophila insulin receptor substrate protein. Science. 2001;292:104–106. doi: 10.1126/science.1057991. [DOI] [PubMed] [Google Scholar]
- 67.Britton JS, Lockwood WK, Li L, Cohen SM, Edgar BA. Drosophila's insulin/PI3-kinase pathway coordinates cellular metabolism with nutritional conditions. Dev Cell. 2002;2:239–249. doi: 10.1016/s1534-5807(02)00117-x. [DOI] [PubMed] [Google Scholar]
- 68.Hwangbo DS, Gershan B, Tu MP, Palmer M, Tatar M. Drosophila dFOXO controls lifespan and regulates insulin signalling in brain and fat body. Nature. 2004;429:562–566. doi: 10.1038/nature02549. [DOI] [PubMed] [Google Scholar]
- 69.Wittwer F, Jaquenoud M, Brogiolo W, Zarske M, Wustemann P, et al. Susi, a negative regulator of Drosophila PI3-kinase. Dev Cell. 2005;8:817–827. doi: 10.1016/j.devcel.2005.04.002. [DOI] [PubMed] [Google Scholar]