Abstract
Selective autophagy involves the recognition and targeting of specific cargo, such as damaged organelles, misfolded proteins, or invading pathogens for lysosomal destruction1–4. Yeast genetic screens have identified proteins required for different forms of selective autophagy, including cytoplasm-to-vacuole targeting, pexophagy, and mitophagy, and mammalian genetic screens have identified proteins required for autophagy regulation5. However, there have been no systematic approaches to identify molecular determinants of selective autophagy in mammalian cells. To identify mammalian genes required for selective autophagy, we performed a high-content, image-based, genome-wide siRNA screen to detect genes required for the colocalization of Sindbis virus capsid protein with autophagolysosomes. We identified 141 candidate genes required for viral autophagy, which were enriched for cellular pathways related to mRNA processing, interferon signaling, vesicle trafficking, cytoskeletal motor function, and metabolism. Ninety-six of these genes were also required for Parkin-mediated mitophagy, indicating that common molecular determinants may be involved in autophagic targeting of viral nucleocapsids and autophagic targeting of damaged mitochondria. Murine embryonic fibroblasts lacking one of these gene products, the C2-domain containing protein, Smurf1, are deficient in the autophagosomal targeting of Sindbis and herpes simplex viruses and in the clearance of damaged mitochondria. Moreover, Smurf1-deficient mice display an accumulation of damaged mitochondria in heart, brain, and liver. Thus, our study identifies candidate determinants of selective autophagy, and defines Smurf1 as a newly recognized mediator of both viral autophagy and mitophagy.
To identify novel genes required for selective autophagy, we performed a genome-wide siRNA screen to detect changes in the colocalization of a red-labeled Sindbis virus (SIN) capsid protein with a green-labeled marker of autophagosomes, GFP-LC3 (Supplementary Fig. 1a) in SIN-infected HeLa/GFP-LC36 cells. Using correlative light and electron microscopy (EM), we confirmed that colocalized red and green puncta represented autophagic structures (primarily autolysosomes) containing numerous viral nucleocapsids (Fig. 1a). The predominance of viral nucleocapsids concentrated in these structures (relative to within the cytoplasm) is consistent with selective autophagic targeting of viral nucleocapsids (herein referred to as virophagy).
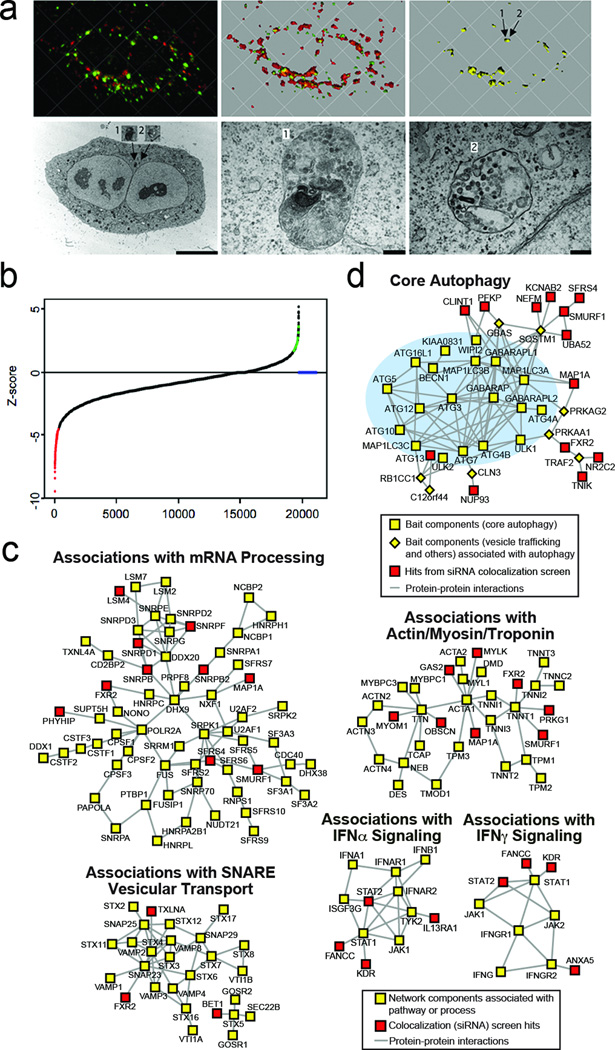
Figure 1. Genome-wide screen to identify cellular factors required for Sindbis virus capsid colocalization with autophagosomes.
a, Correlative light and electron microscopy (EM) of HeLa-GFP-LC3 cell infected with SIN/mCherry.capsid virus. Top left, deconvolved image of red and green fluorescence channels; middle, 3-D surface reconstruction of red and green channels; right, yellow (red + green) colocalization channel. Arrows denote yellow puncta that correspond to “1” and “2” in EMs below. Bottom left, EM of identical cell; middle and right, high magnification images of insets “1” and “2”. Scale bars, left = 10 µm, middle and right = 200 nm. b, Ranked distribution of median z-scores for each siRNA pool in primary colocalization (virophagy) screen. Red, decreased colocalization; green, increased colocalization; blue, insufficient numbers of green or red puncta per cell or total number of cells per well for analysis. c, Maps of protein interactions in enriched network modules (see Supplementary Fig. 3b). d, Association of siRNA hits with autophagy network.
Screening of a human siGenome library containing 21,215 siRNA pools revealed that knockdown of 195 and 13 genes resulted in decreased or increased colocalization, respectively, (Fig. 1b, Supplementary Table 1, Supplementary Fig. 1b). Genes were re-screened with sets of 4 individual siRNAs (Supplementary Table 2; see column “J” of Supplementary Table 3 for siRNA sequences) to confirm our primary screen and rule out potential off-target effects of individual siRNAs; knockdown with two or more siRNAs resulted in decreased colocalization for 141 (72%) genes. (Fig. 2, Supplementary Fig. 1b and 2a). None of these 141 gene knockdowns decreased numbers of green puncta in uninfected cells (data not shown), indicating these genes function in virophagy but not in regulation of autophagy. There was no enrichment of siRNAs containing miRNA seed sequences among these genes (P=0.95) (Supplementary Tables 3 and 4), suggesting that bias due to miRNA-like off-target effects was unlikely. There was a low confirmation rate for siRNAs that increased colocalization (2 of 13 genes); therefore, we subsequently focused only on siRNAs that decreased colocalization.
Figure 2. Gene list for viral capsid/autophagosome colocalization (C) confirmation screen and secondary screens for survival of virus-infected cells (S) and Parkin-mediated mitophagy (M).
Shown are the number of individual siRNAs from a pool of 4 targeting each gene that scored positive in each screen. Red, genes with 2 or more positive siRNAs (confirmed hits); Green, genes with <2 positive siRNAs. See Supplementary Tables 2–8 for further details.
Bioinformatic analyses of the 141 confirmed hits required for SIN capsid/GFP-LC3 colocalization revealed enrichment for gene sets associated with biological processes and molecular functions including RNA splicing/processing, protein phosphorylation, transport, calcium-binding and the cytoskeleton (Supplementary Table 5, Supplementary Fig. 3a). Examination of our hits within a framework of functional cellular pathways revealed strongly enriched network modules associated with RNA processing, interferon (IFN)α and γ signaling, SNARE vesicular transport, cytoskeletal-associated components, and several metabolic pathways (Supplementary Fig. 3b, Fig. 1c). This is consistent with IFNγ’s function in selective microbial autophagy1 and the described role of the actin cytoskeleton in selective autophagy in yeast7 and mammalian8 cells. The enrichment of SNARE proteins suggests that in addition to a function in autophagosome formation and maturation9, 10, these proteins may be involved in the trafficking of selective cargo to the autophagosome. Twelve colocalization hits form primary interactions with core autophagy machinery and associated components11 (Fig. 1d). One colocalization screen hit, clathrin interaction 1 (CLINT) interacts with ATG8 components (GABARAPL1, MAP1LC3A, MAP1LC3B), which are crucial in the recognition of cargo during selective autophagy2. Another hit, ATG13, is a member of the core autophagy network, suggesting that it may have an as yet undefined function in selective autophagy, in addition to its role in the Atg1/ULK1 autophagy induction complex12. Five colocalization hits, SMURF1, NEFM, KCNAB2, SFRS4, and UBA52, interact with p62/SQSTM1, a known adaptor in diverse forms of selective autophagy2, including SIN capsid targeting to autophagosomes6.
Selective SIN autophagy (virophagy) promotes the survival of SIN-infected cells6. To determine if our identified candidate virophagy genes have a similar function, we screened our confirmation siRNA library for genes that decreased cell survival after SIN infection. Two or more siRNAs targeting 98 of the genes decreased cell survival after SIN infection (Fig. 2, Supplementary Tables 3 and 6, Supplementary Figs. 1b, 2a); colocalization and cell survival effects of individual siRNAs were significantly correlated (Supplementary Fig. 2b) (P=3.8E-08, Spearman correlation). This is consistent with a pro-survival function of autophagic targeting of SIN capsid in virally-infected cells6.
To investigate whether the identified candidate virophagy genes also function in other forms of selective autophagy, we performed a secondary screen for autophagy of damaged mitochondria (mitophagy). We used HeLa cells that express an mCherry fusion of Parkin, a cytosolic E3 ubiquitin ligase that translocates to depolarized mitochondria to induce mitophagy after treatment with uncoupling agents (such as CCCP: carbonyl cyanide m-chlorphenylhydrazaone) 13. Of the 141 confirmed colocalization hits, 2 or more siRNAs targeting 96 (68%) genes decreased mitophagy (Fig. 2, Supplementary Tables 3 and 7, Supplementary Figs. 1b, 2a, 2b). Host factors involved in involved in viral autophagy and mitophagy overlapped significantly (P=0.019, Spearman correlation). The minority of genes that only scored positive in either the virophagy confirmation or the mitophagy secondary screen may have a role in targeting some, but not other cargoes, for selective autophagy; however, the lack of overlap may also reflect different sensitivities of the two screens. Mitophagy hits consisted of several mitochondria-associated components14 (NME2, MDH1, NTHL1, PDK1, COX8A, MRPS2, MRPS10, NDUFB9 and BLOC1S) and interactors of mitochondria-associated components (Supplementary Fig. 4).
We focused further on one gene, SMURF1 (Smad-Ubiquitin Regulatory Factor 1), encoding a HECT-domain ubiquitin ligase that targets several cytoplasmic proteins for degradation15. SMURF1 was a confirmed hit in all three confirmation or secondary screening assays (see Supplementary Fig. 5 for representative raw data from colocalization confirmation screen); is present in two of the enriched networks (mRNA processing and actin cytoskeleton) (Fig. 1c); and is a predicted interacting partner of the autophagy adaptor, p62/SQSTM12, 4 (Fig. 1d).
We confirmed that SMURF1 is not required for general autophagy, but is a bona fide mediator of selective autophagy, including virophagy and mitophagy. siRNA knockdown of SMURF1 in HeLa cells, unlike knockdown of the essential autophagy protein, ATG7, did not alter general starvation-induced autophagy (Supplementary Fig. 6a). Furthermore, Smurf1−/− murine embryonic fibroblasts (MEFs) displayed normal levels of starvation-induced LC3-II conversion, p62 degradation, and ultrastructural evidence of autophagosome and autolysosome accumulation (Fig. 3a, b).
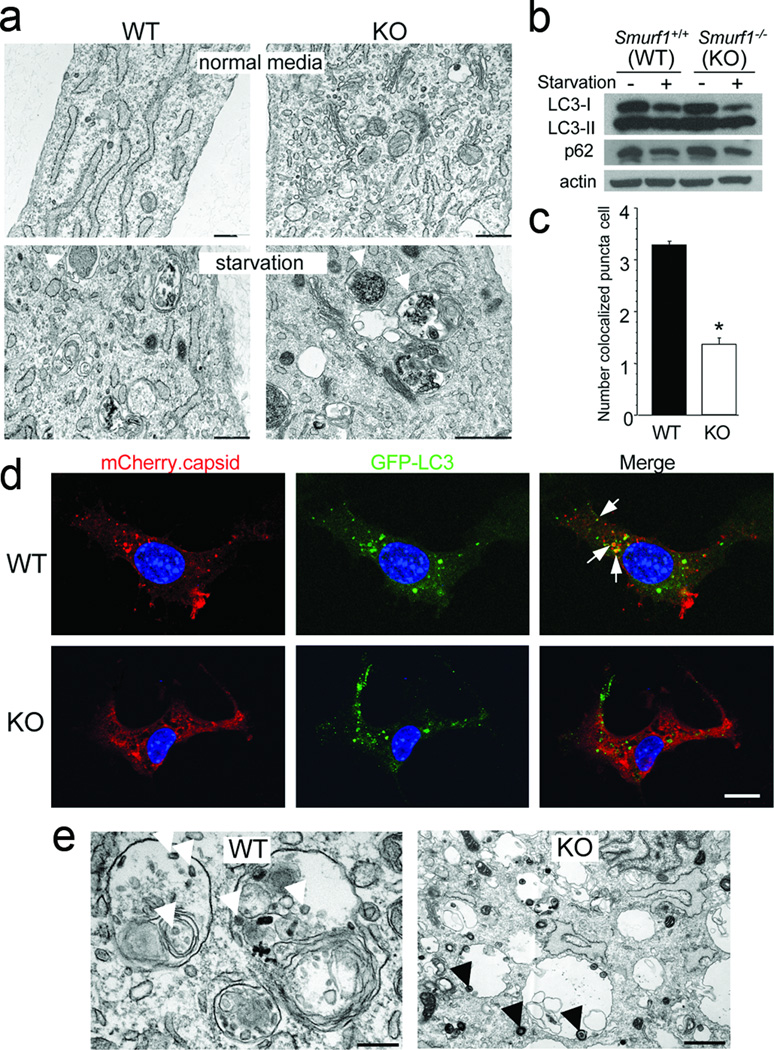
Figure 3. Smurf1 functions in virophagy but not in starvation-induced autophagy.
a, EM analysis of WT and Smurf1−/− MEFs cultured in normal media or EBSS (starvation) for 4 hrs. Arrowheads, representative autophagosomes; arrows, representative autolysosomes. Scale bars, 500 nm. b, Western blot analyses of LC3-I/II and p62 levels in MEFs of indicated genotype. c, Quantitation of colocalization of SIN mCherry.capsid and GFP-LC3 in indicated MEFs 16 hrs after SIN-mCherry.capsid/GFP-LC3 infection. Data shown represent mean ± s.e.m. number of colocalized red and green puncta per cell for 50 cells per well in triplicate samples. *, P<0.001 vs. WT, t-test. d, Representative confocal micrographs of images used for quantitation in (c). Arrows, colocalized red and green puncta. Scale bar, 15 mm. e, Representative EMs of indicated MEFs infected with HSV-1 (strain 17termA). White arrows, partially degraded viral nucleocapsids inside autolysosomes; black arrows, intact viral nucleocapsids inside viral vesicles. Scale bar, 0.5 µm. For a–e, similar results were obtained in 3–5 independent experiments.
However, a significant decrease in mCherry.capsid/GFP-LC3 colocalization was observed in SIN-infected Smurf1−/− MEFs (Fig. 3c, d). Similar to p626, Smurf1 and SIN capsid protein co-immunoprecipitate in SIN-infected MEFs and HeLa cells (Supplementary Fig. 7a, b). Smurf1 is not required for the interaction between p62 and SIN capsid (Supplementary Fig. 7c). The interaction between Smurf1 and SIN capsid may be relevant for targeting SIN capsid for autophagosomal degradation, as levels of SIN capsid were increased in Smurf1−/− MEFs and SMURF1 siRNA-treated HeLa cells. (Supplementary Fig. 7d–f). Increased SIN capsid levels in Smurf1-deficient cells cannot be explained by increased capsid production since viral growth was similar in Smurf1−/− and WT MEFs (Supplementary Fig. 7g, h), or by changes in proteasomal degradation since SIN capsid levels were not altered by treatment with the proteasome inhibitor MG132 (Supplementary Fig. 7d), and SIN capsid ubiquitination was not detected (data not shown). SIN-infected Smurf1−/− MEFs had accelerated cell death (despite similar viral titers) as compared to WT controls (Fig. 3g). Thus, Smurf1 interacts with SIN capsid; Smurf1 is required for SIN capsid targeting to autophagosomes and degradation through a proteasome-independent pathway; and Smurf1-dependent degradation of SIN capsid promotes cell survival.
To determine whether Smurf1 is required for the autophagic targeting of other viruses, we performed EM of WT and Smurf1−/− MEFs infected with a mutant strain of herpes simplex virus type 1 harboring a deletion of ICP34.5, a potent inhibitor of viral autophagy16–18 (Fig. 3e). As reported17, the majority of cytoplasmic HSV-1 virions in WT MEFs were inside autolysosomal structures and appeared partially degraded. In contrast, in Smurf1−/− MEFs, the majority of cytoplasmic HSV-1 virions were inside single-membraned vesicles involved in HSV-1 cytoplasmic egress and displayed intact structure. This lack of autophagic targeting of HSV-1 in Smurf1−/− MEFs was not due to a general defect in autophagy, since HSV-1 infection induced autophagy similarly in Smurf1−/− and WT MEFs (Supplementary Fig. 6b). Thus, Smurf1 is required for the autophagic targeting of both a positive-strand RNA (Sindbis) and a double-stranded DNA (herpes simplex) virus.
Next, we examined the role of SMURF1 in mitophagy. In HeLa cells, all four SMURF1 siRNAs decreased SMURF1 protein expression expression (Supplementary Fig. 8a) and inhibited Parkin-mediated CCCP-induced mitophagy as effectively as an siRNA targeting p62, a mediator of mitophagy in some previous reports19, 20, and siRNA targeted against the essential autophagy protein, Atg7 (Supplementary Fig. 8b, c). The magnitude of each individual siRNA’s effect on SMURF1 protein expression knockdown correlated with the magnitude of inhibition of Parkin-mediated autophagy. Therefore, in the mitophagy confirmation screen, two of the SMURF1 siRNAs were likely false negatives; indeed, the number of Parkin-expressing cells in wells treated with these siRNAs was low (data not shown), precluding meaningful statistical analyses. A similar finding was true in the viral colocalization screen.
We further examined the role of Smurf1 in mitophagy by assessing mitochondrial clearance in CCCP-treated Smurf1−/− MEFs. Unlike in HeLa cells, Parkin overexpression did not promote mitophagy in MEFs of either genotype (data not shown). However, 25–30% of MEFs treated with 10 µm CCCP showed changes in mitochondrial morphology. In WT MEFs with damaged mitochondria (swollen or fragmented appearance), partial mitochondrial clearance occurred with compaction of the remaining mitochondria around the nucleus (Fig. 4a). In contrast, in Smurf1−/− cells with damaged mitochondria, virtually no mitochondrial clearance occurred and there was diffuse accumulation of fragmented mitochondria throughout the cytoplasm (Fig. 4a, arrows). This phenotypic difference was confirmed using two independent methods of quantitation, including assessment of the total percentage of CCCP-treated cells that displayed diffuse accumulation of abnormal mitochondria (Fig. 4b) and the measurement of fractional mitochondrial surface area per cell (Fig. 4c).
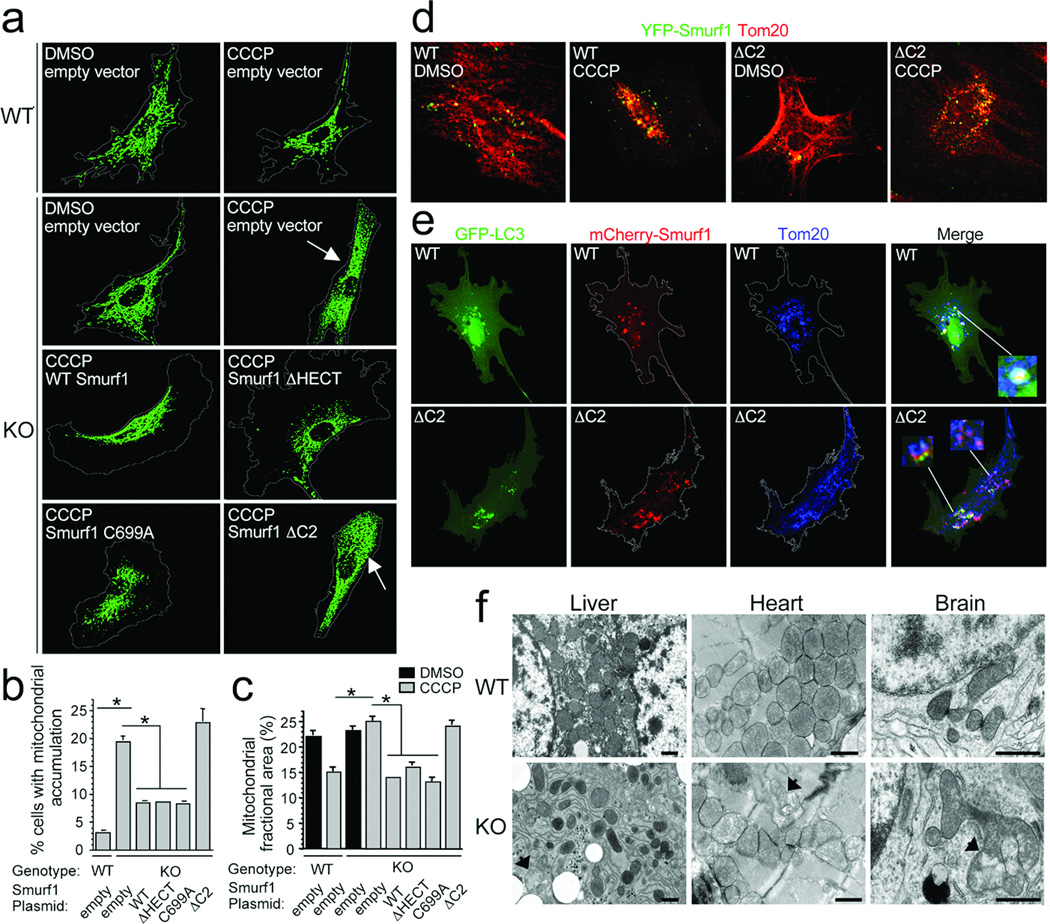
Figure 4. Smurf1 functions in mitophagy.
a, Representative mitochondrial morphology in Smurf1+/+ (WT) and Smurf1+/+ (KO) MEFs transfected with indicated construct and treated with DMSO or 10 µM CCCP for 24 hrs. b, Quantitation of percentage of total cells with a diffuse accumulation of abnormal fragmented mitochondria and lack of mitochondrial clearance. Results shown represent combined data from 3–5 experiments per condition with triplicate wells (of at least 100 cells per well) analyzed for each condition per experiment. Shown are mean ± s.e.m. for average values from each experiment. Similar results were observed in each independent experiment. *, P< 0.001, Student t-test. c, Measurement of mitochondrial fractional area (% of total cellular area) in MEFs treated as in (a). Results shown represent mean ± s.e.m. for 50 cells per condition. d, Representative confocal micrographs of KO MEFs transfected with YFP-Smurf1 wild-type (WT) or YFP-Smurf1ΔC2 (ΔC2) and treated for 4 hrs with DMSO or CCCP. e, Representative confocal micrographs of KO MEFs transfected with GFP-LC3 and mCherry-Smurf1 wild-type (WT) or mCherry-Smurf1ΔC2 (ΔC2) and treated for 4 hrs with CCCP. Inset, upper right, formation of completed autophagosome around a damaged mitochondrion associated with WT Smurf1; insets, lower right, incomplete autophagosomes or absence of LC3 signal around mitochondria associated with Smurf1ΔC2. See also Supplementary Fig. 10–11 for enlarged images. f, Representative EMs of indicated tissues from 10 month-old WT (Smurf1+/+) or KO (Smurf1−/−) mice. Arrows, representative abnormal mitochondria. Scale bars, 1 µm. Similar abnormalities were observed throughout entire EM tissue section for each mouse and in tissue samples from 3 different mice for each genotype.
To evaluate the mechanism of action of Smurf1, we compared the effects of WT and mutant Smurf1 expression plasmids on rescue of selective autophagy in Smurf1−/− MEFs (Fig. 4a–c). We focused on mitophagy rather than SIN capsid virophagy due to the resistance of MEFs to SIN infection after plasmid transfection. The defect in mitophagy in Smurf1−/− MEFs was partially rescued by WT Smurf1 transfection. Smurf1ΔHECT21, lacking the HECT domain that catalyzes ubiquitin ligation onto target proteins, or Smurf1 C699A21, a catalytically inactive point mutant, rescued the mitochondrial clearance defect as efficiently as WT Smurf1. Thus, in addition to its known role in targeting proteins for proteasomal degradation via ubiquitination, Smurf1 has a ubiquitin ligase activity-independent function in mediating the selective degradation of damaged mitochondria.
In contrast, a Smurf1 mutant lacking the C2 domain, Smurf1ΔC2, was completely defective in mitophagy rescue in CCCP-treated Smurf1−/− MEFs (Fig. 4a–c), despite similar levels of expression as transfected WT Smurf1 (Supplementary Fig. 9a). The C2 domain of Smurf1 was not required for Smurf1 co-immunoprecipitation with p62 (Supplementary Fig. 9b), suggesting that Smurf1 does not function in selective autophagy by recruiting p62. C2 domains (including those of protein kinase C and Smurf1) bind membrane phospholipids and function in protein targeting to the plasma membrane and/or membrane subcellular compartments22, 23. This raised the possibility that Smurf1 might function in the targeting of selective autophagy cargo through interaction with the nascent autophagosome membrane.
To investigate this possibility, we examined the subcellular localization of WT Smurf1 and Smurf1ΔC2 with damaged mitochondria and autophagosomes (Fig. 4d). In Smurf1−/− MEFs transfected with WT YFP-Smurf1, CCCP treatment induced the colocalization of YFP-Smurf1 with damaged mitochondria. In Smurf1−/− MEFs transfected with YFP-Smurf1ΔC2, increased numbers of fragmented and swollen mitochondria were observed in basal conditions and these increased further upon CCCP treatment. These abnormal mitochondria colocalized with YFP-Smurf1ΔC2 whereas normal reticular-appearing mitochondria rarely colocalized with YFP-Smurf1ΔC2. YFP-Smurf1 C699A, displayed the same subcellular staining pattern as WT YFP Smurf1 (data not shown). Thus, Smurf1 colocalizes with damaged mitochondria in a C2 domain-independent manner.
We next determined whether the C2 domain of Smurf1 was required for the colocalization of damaged mitochondria with autophagosomes (Fig. 4e, Supplementary Fig. 10–11). In cells expressing WT mCherry-Smurf1, mitochondria were mostly compacted around the nucleus, and numerous autophagosomes were observed surrounding structures that labeled positive for both mCherry-Smurf1 and the mitochondrial marker, Tom20. In constrast, in cells expressing mCherry-Smurf1ΔC2, mCherry-Smurf1ΔC2- and Tom20-postive mitochondria were rarely found inside autophagosomes. In many regions, GFP-LC3-positive linear or cup-shaped structures were observed near mCherry-Smurf1ΔC2-positive mitochondria, but complete autophagosomes surrounding these mitochondria could not be detected. Thus, the C2 domain of Smurf1 is not required for its targeting to damaged mitochondria, but is required for damaged mitochondria to be normally engulfed by autophagosomes. It is not yet known whether this requirement reflects a direct role for the C2 domain in binding to autophagosomal membrane phospholipids or is a more indirect consequence of other, as-yet-undescribed, effects of the C2 domain in mitophagy.
To investigate whether Smurf1 may function in selective autophagy in vivo, we performed EM analyses of cerebellum, liver, and hearts of 10 month-old WT and Smurf1−/− mice21. In all three organs, Smurf1−/− mice exhibited an accumulation of abnormal mitochondria that were either swollen, fragmented, and/or contained abnormal cristae (Fig. 4f). This phenotype is consistent with a defect in mitophagy and mitochondrial quality control; however, we cannot rule out unknown triggers of mitochondrial damage in these animals. In the livers of Smurf1−/− mice, mitochondria were spatially disorganized and surrounded by networks of dilated endoplasmic reticulum (ER), perhaps reflecting a defect in mitochondrial targeting by isolation membranes (which are believed to originate from the ER1) and/or a defect in selective autophagy of the ER. There was a marked accumulation of lipid droplets in the livers of Smurf1−/− mice (Supplementary Fig. 12a), which may be consistent with selective degradation of lipid droplets by autophagy (lipophagy) in hepatocytes24. Furthermore, the granule cell layer of the cerebellum and cardiomyocytes of Smurf1−/− mice had increased numbers of p62/SQSTM1 aggregates (Supplementary Fig. 12b). Unlike findings in brains and hearts of mice lacking core autophagy genes25, p62 aggregate accumulation in these tissues was not associated with ubiquitin accumulation. This is consistent with a role for Smurf1 in selective autophagy but not in the form of basal autophagy that is involved in protein quality control25.
Together, our data in Smurf1−/− MEFs and in Smurf1−/− mice suggest a crucial function for Smurf1 in selective autophagy, including in the autophagic targeting of genetically distinct viruses, in the autophagic targeting of mitochondria, and, more speculatively, in the potential autophagic targeting of other cellular targets such as hepatic lipid droplets and ER. The mechanism by which Smurf1 functions in selective autophagy is independent of its E3 ubiquitin ligase activity, but rather involves its C2 membrane-targeting domain. We propose that the C2 domain of Smurf1 may participate in the delivery of selective autophagic substrates to the nascent autophagosome. Thus, Smurf1 has parallel functions in two distinct cellular degradation pathways, targeting specific proteins for degradation by the ubiquitin-proteasomal pathway15 (via its E3 ubiquitin ligase activity) and targeting selective cargo for degradation by the autophagy pathway (via its C2 domain).
Our findings in Smurf1−/− MEFs and mice illustrate that our high-content imaged based genome-wide screen successfully reveals novel candidate determinants of selective autophagy. More broadly, the identification of a set of 96 genes that may dually function in viral autophagy and mitophagy (but not in basal autophagy) suggests the existence of a common molecular network for targeting diverse unwanted cytoplasmic cargo to the lysosome. This network identification provides a basis for a more global understanding of the mechanisms involved in selective autophagy.
METHODS SUMMARY
High-content image-based genome wide siRNA screen
A genome-wide siRNA library (Dharmacon) containing 21,125 SMART pools was used for reverse transfection of HeLa/GFP-LC3 cells, followed by infection with SIN/mCherry.capsid virus, high content imaging using a Pathway855 automated microscope (BD Biosciences), quantitative image analysis, statistical analysis, and bioinformatic analysis as described in Supplementary Information. Primary hits were evaluated in three confirmation/secondary screens using the 4 individual siRNAs from each pool, including a screen for viral capsid/autophagsosome colocalization, cell survival during SIN infection, and Parkin-induced mitophagy.
Functional analyses of Smurf1
See Supplementary Information.
Full Methods and any associated references are available in the Supplementary Information.
Supplementary Material
Acknowledgements
We thank Mridula Vishwanath, Shuguang Wei and Bruce Posner for assistance with high-throughput siRNA screening; William Sun for information technology support; Kurt Scudder for assistance with image analysis algorithms; Angela Diehl for expert medical illustration; Victor Stollar, Margaret McDonald, Richard Kuhn and Richard Youle for helpful discussions and providing reagents; Abhijit Bugde for assistance in the UTSW Live Cell Imaging Facility; and Laurie Mueller and Tom Januszewski for assistance with electron microscopy. This work was supported by NIH grants AI109617 (B.L.), UL1 RR024982 (G.X., Y.X.), AI062773 (R.J.X.), DK83756 (R.J.X.), DK086502 (R.J.X.) and DK043351 (R.J.X. and A.N.); NSF grant DMS-0907562 (G.X.); and the Center for Cancer Research, National Cancer Institute Intramural Research Program (Y.E.Z.).
Footnotes
Author Contributions
A.O., R.S., M.N., M.R., J.W., Y.Z., K.L.-P., and B.L. designed the experiments. A.O., R.S., Z.Z., and Q.S. performed the experiments. G.X., A.N., C.F., R.X., and Y.X. performed statistical and bioinformatic analyses. A.O., R.S., and B.L. wrote the manuscript.
Author Information
Reprints and permissions information is available at www.nature.com/reprints. The authors declare no competing financial interests. Readers are welcome to comment on the online version of this article at www.nature.com/nature.
REFERENCES
- 1.Levine B, Mizushima N, Virgin HW. Autophagy in immunity and inflammation. Nature. 2011;469:323–335. doi: 10.1038/nature09782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Noda NN, Ohsumi Y, Inagaki F. Atg8-family interacting motif crucial for selective autophagy. FEBS Lett. 2010;584:1379–1385. doi: 10.1016/j.febslet.2010.01.018. [DOI] [PubMed] [Google Scholar]
- 3.Wild P, et al. Phosphorylation of the autophagy receptor optineurin restricts Salmonella growth. Science. 2011;333:228–233. doi: 10.1126/science.1205405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Komatsu M, Ichimura Y. Selective autophagy regulates various cellular functions. Genes Cells. 2010;15:923–933. doi: 10.1111/j.1365-2443.2010.01433.x. [DOI] [PubMed] [Google Scholar]
- 5.Lipinski MM, et al. A genome-wide siRNA screen reveals multiple mTORC1 independent signaling pathways regulating autophagy under normal nutritional conditions. Dev Cell. 2010;18:1041–1052. doi: 10.1016/j.devcel.2010.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Orvedahl AO, et al. Autophagy protects against Sindbis virus infection of the central nervous system. Cell Host & Microbe. 2010;7:115–127. doi: 10.1016/j.chom.2010.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Monastyrska I, Rieter E, Klionsky DJ, Reggiori F. Multiple roles of the cytoskeleton in autophagy. Biol Rev Camb Philos Soc. 2009;84:431–448. doi: 10.1111/j.1469-185X.2009.00082.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee JY, et al. HDAC6 controls autophagosome maturation essential for ubiquitin-selective quality-control autophagy. Embo J. 2010;29:969–980. doi: 10.1038/emboj.2009.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Longatti A, Tooze SA. Vesicular trafficking and autophagosome formation. Cell Death Differ. 2009;16:956–965. doi: 10.1038/cdd.2009.39. [DOI] [PubMed] [Google Scholar]
- 10.Nair U, et al. SNARE proteins are required for macroautophagy. Cell. 2011;146:290–302. doi: 10.1016/j.cell.2011.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Behrends C, Sowa ME, Gygi SP, Harper JW. Network organization of the human autophagy system. Nature. 2010;466:68–76. doi: 10.1038/nature09204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mizushima N. The role of the Atg1/ULK1 complex in autophagy regulation. Curr Opin Cell Biol. 2010;22:132–139. doi: 10.1016/j.ceb.2009.12.004. [DOI] [PubMed] [Google Scholar]
- 13.Narendra D, Tanaka A, Suen DF, Youle RJ. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J Cell Biol. 2008;183:795–803. doi: 10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pagliarini DJ, et al. A mitochondrial protein compendium elucidates complex I disease biology. Cell. 2008;134:112–123. doi: 10.1016/j.cell.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xing L, Zhang M, Chen D. Smurf control in bone cells. J Cell Biochem. 2010;110:554–563. doi: 10.1002/jcb.22586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Talloczy Z, et al. Regulation of starvation- and virus-induced autophagy by the eIF2alpha kinase signaling pathway. Proc Natl Acad Sci U S A. 2002;99:190–195. doi: 10.1073/pnas.012485299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Talloczy Z, Virgin HW, IV, Levine B. PKR-fependent autophagic degradation of herpes simplex virus type 1. Autophagy. 2006;2:24–29. doi: 10.4161/auto.2176. [DOI] [PubMed] [Google Scholar]
- 18.Orvedahl A, et al. HSV-1 ICP34.5 confers neurovirulence by targeting the Beclin 1 autophagy protein. Cell Host Microbe. 2007;1:23–35. doi: 10.1016/j.chom.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 19.Geisler S, et al. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat Cell Biol. 2010;12:119–131. doi: 10.1038/ncb2012. [DOI] [PubMed] [Google Scholar]
- 20.Narendra D, Kane LA, Hauser DN, Fearnley IM, Youle RJ. p62/SQSTM1 is required for Parkin-induced mitochondrial clustering but not mitophagy; VDAC1 is dispensable for both. Autophagy. 2010;6:1090–1106. doi: 10.4161/auto.6.8.13426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yamashita M, et al. Ubiquitin ligase Smurf1 controls osteoblast activity and bone homeostasis by targeting MEKK2 for degradation. Cell. 2005;121:101–113. doi: 10.1016/j.cell.2005.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cho W, Stahelin RV. Membrane binding and subcellular targeting of C2 domains. Biochim Biophys Acta. 2006;1761:838–849. doi: 10.1016/j.bbalip.2006.06.014. [DOI] [PubMed] [Google Scholar]
- 23.Lu K, et al. Pivotal role of the C2 domain of the Smurf1 ubiquitin ligase in substrate selection. J Biol Chem. 2011;286:16861–16870. doi: 10.1074/jbc.M110.211979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Singh R, et al. Autophagy regulates lipid metabolism. Nature. 2009;458:1131–1135. doi: 10.1038/nature07976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mizushima N, Levine B. Autophagy in mammalian development and differentiation. Nat Cell Biol. 2010;12:823–830. doi: 10.1038/ncb0910-823. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.