Abstract
Human parvovirus 4 (PARV4) is an emerging human virus, and little is known about the molecular aspects of PARV4 apart from its incomplete genome sequence, which lacks information of the termini. We analyzed the gene expression profile of PARV4 using a nearly full-length HPV4 genome in a replication competent system in 293 cells. We found that PARV4 utilizes two promoters to transcribe non-structural protein- and structural protein-encoding mRNAs, respectively, which were polyadenylated at the right end of the genome. Three major proteins, including the large non-structural protein NS1a, whose mRNA is spliced, and capsid proteins VP1 and VP2, were detected. Additional functional analysis of the NS1a revealed its capability to induce cell cycle arrest at G2/M phase in ex vivo-generated human hematopoietic stem cells. Taken together, our characterization of the molecular features of PARV4 suggests that PARV4 represents a new genus in the family Parvoviridae.
Introduction
Parvoviruses are small non–enveloped, icosahedral DNA viruses with a diameter of 18-26 nm that encapsidate a single-stranded genome of approximately (~)5-6 kb (Tattersall, 2006). To date, there are a number of parvoviruses known to infect humans, including adeno-associated viruses (AAVs), parvovirus B19 (B19V), and two newly identified human parvoviruses, which are the human bocavirus (HBoV) (Allander et al., 2005) and human parvovirus 4 (PARV4) (Jones et al., 2005). B19V, a prototype member in the genus Erythrovirus of family Parvoviridae, causes a number of human diseases ranging from mild erythema infectiosum and polyarthropathy syndrome to severe manifestations of transient aplastic crisis, pure red cell aplasia and hydrops fetalis (Young and Brown, 2004). In contrast, AAVs, members of the genus Dependovirus, are non-pathogenic to humans, and have been widely used as therapeutic vectors in human gene therapy (Brown, 2010; Mingozzi and High, 2011).
HBoV was firstly identified in respiratory samples from children with lower respiratory tract infections (Allander et al., 2005), and subsequently proven epidemiologically to be associated with the diseases (Manning et al., 2006). PARV4 was initially found in a blood sample from an intravenous drug user with acute viral infection syndrome (Jones et al., 2005). Subsequently, the PARV4 genome was detected in human plasma pools at a low titer (Schneider et al., 2008a). PARV4 had also been found in the livers of hepatitis C virus-positive individuals (Schneider et al., 2008b) and the bone marrow of HIV-positive individuals (Manning et al., 2007; Longhi et al., 2007). Recently, PARV4 DNA was detected in cerebrospinal fluid of two children with encephalitis of unknown etiology (Benjamin et al., 2011). However, the disease association of PARV4 remains unclear (Brown, 2010; Simmons et al., 2011).
HBoV has been classified as a member in the genus Bocavirus based on the similarity of its genome sequence with those of the two animal bocaviruses, minute virus of canines (MVC) and bovine parvovirus type 1 (BPV1) (Allander et al., 2005; Tijssen et al., 2011). However, the known PARV4 incomplete genome, which lacks information of the terminal repeats, does not show a close relationship to any of the known parvoviruses in the genera of family Parvoviridae that have been classified to date, but represents a deeply rooted lineage between avian dependoviruses and bovine parvovirus type 3 (Jones et al., 2005). Notably, PARV4-like viruses were detected in animals in Hong Kong, which were subsequently named as porcine and bovine hokoviruses (Lau et al., 2008). Later, a related porcine hokovirus was identified in wild boars (Adlhoch et al., 2010), and PARV4-like viral DNA was detected in plasma samples from chimpanzees and gorillas in Cameroon (Sharp et al., 2010). Phylogenetic analysis of the PARV4 genome together with those of hokoviruses has shown a close resemblance in nucleotide sequences, with an identity of 61.5-63% (Lau et al., 2008). These findings have led to the proposed classification of the PARV4 and PARV4-like viruses as members in a new genus called Partetravirus in the family Parvoviridae by the International Committee on Taxonomy of Viruses (ICTV) (Tijssen et al., 2011).
Little is known about the gene expression of PARV4 and the function of PARV4 proteins. Since the PARV4 has not been cultured in vitro, and the full-length genome with terminal repeats has not been sequenced, we profiled the gene expression of PARV4 by transfecting a replication-competent PARV4 genome, which harbors AAV5-inverted terminal repeats (ITRs) at two ends, in 293 cells expressing AAV5 Rep78 protein and the necessary adenovirus gene products. The function of the large nonstructural protein NS1 was studied in detail for its role in inducing cytopathic effects in ex vivo-generated human hematopoietic stem cells (CD34+ HSCs). In addition, the VP1 unique region (VP1u) was purified and tested in vitro for its hypothesized phospholipase A2 (PLA2)-like activity.
Results
Constructing a replication-competent clone of PARV4 in the context of AAV5 ITRs
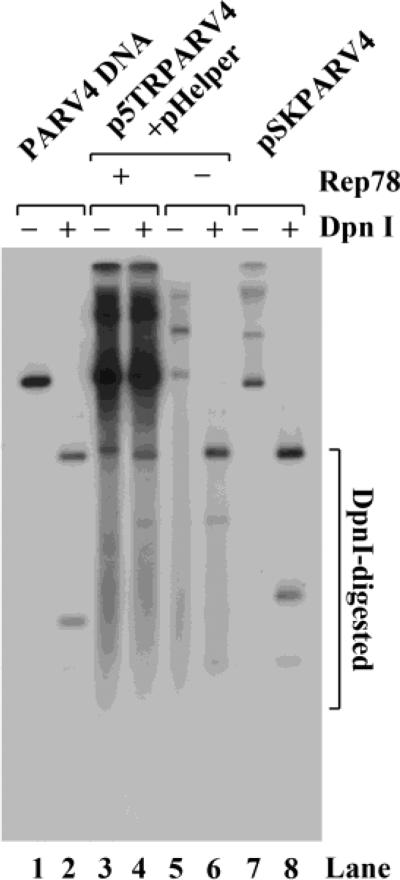
We first cloned nucleotide nt 127-5268 of the PARV4 genome (GenBank accession no.: NC_007018) into vector pCR2.1, which encompasses a coding region with two large open reading frames (ORF) and a portion of the terminal repeats at the 5' and 3' ends (Jones et al., 2005). This incomplete PARV4 genome lacks the terminal repeats at two ends, and therefore will not replicate in any cell culture system. To determine the transcriptional profile of PARV4 in a replication-competent system, we incorporated the incomplete PARV4 genome into the context of AAV5 ITRs, which resulted in plasmid p5TRPARV4. It replicated efficiently in 293 cells that expressed AAV5 Rep78 protein and necessary Ad5 gene products (i.e., E2, E4orf6 and VA RNA from the pHelper plasmid) (Guan et al., 2008) (Fig. 1, lane 4). In contrast, the pSKPARV4 clone, which did not contain the AAV5 ITRs, did not replicate in 293 cells (Fig. 1, lane 8). Thus, we used the p5TRPARV4 construct for further analysis of PARV4 gene expression.
Figure 1. Replication of the construct p5TRPARV4 in 293 cells.
293 cells were transfected with p5TRPARV4, pAV5Rep78 and pHelper, or pSKPARV4 alone. Hirt DNA was extracted from the cells 48 hrs post-transfection and digested with DpnI (DpnI +) or without DpnI (DpnI -). The DNA samples were then subjected to Southern blot analysis using the PARV4 NSCap probe (nt 127-5268). Ten ng of the probe template DNA was treated in parallel as a DpnI digestion control (lanes 1& 2).
5'/3' rapid amplification of cDNA ends (RACE) and reverse transcription (RT)-PCR identification of the transcription units of the PARV4 genome
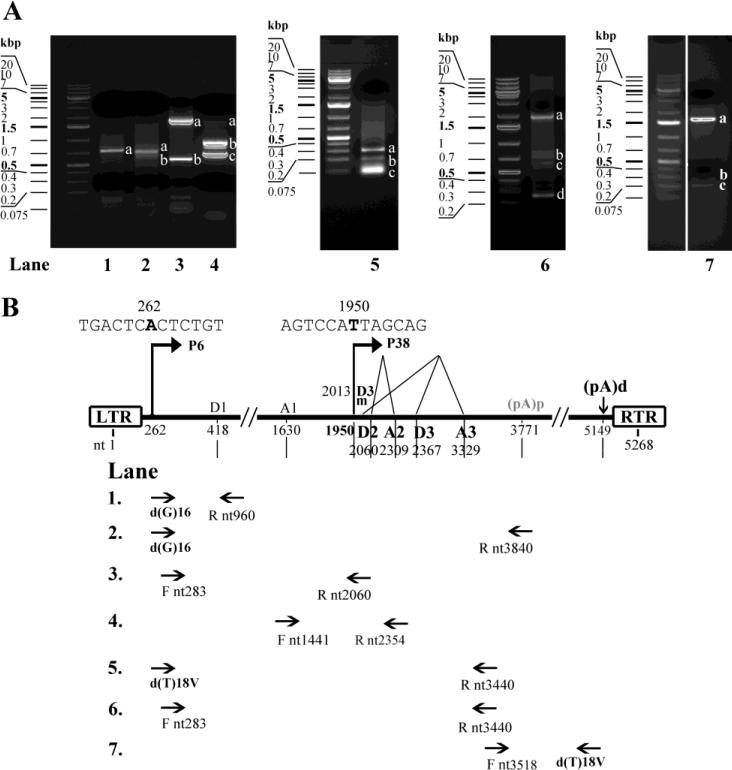
We used total RNA extracted from 293 cells that had been co-transfected with p5TRPARV4, pAAV5Rep78, and pHelper to analyze PARV4 mRNA transcripts. The 5' RACE using the primer Rnt960 generated a predominant band of ~690 nts (Fig. 2A, lane 1). Sequence analysis of this band revealed that mRNA transcripts spanning the region from the 5’ end to nt 960 initiated at nt 262. The 5' RACE with primers Rnt3840 and Rnt3440 produced major bands of ~700 nts and ~300 nts, and minor bands of ~600 nts and ~200 nts, respectively (Fig. 2A, lanes 2&5, respectively). Sequence analysis of the major bands revealed the presence of a 5’ end of PARV4 mRNAs at nt 1950, and splicing sites of D2 donor at nt 2060, A2 acceptor at nt 2309, D3 donor at nt 2367, and A3 acceptor at nt 3329. In addition, sequence analysis of the minor bands confirmed the 5’ end of PARV4 mRNAs at nt 1950, and revealed the splicing sites of D3m donor at nt 2013 and A3 acceptor at nt 3329. Taken together, these results demonstrated the existence of two promoters at map unit 6 and 38, namely P6 and P38, which transcribed PARV4 mRNAs of ORFs at the left and right hands of the PARV4 genome, respectively. The 3' RACE with primer Fnt3518 produced two predominant bands of ~1,500 nts and ~300 nts (Fig. 2A, lane 7). Sequence analysis of these two bands revealed that they terminated at nt 5149 and nt 3771, respectively, suggesting that PARV4 mRNAs are polyadenylated at either nt 5149 or nt 3771.
Figure 2. 5’/3’ RACE and RT-PCR analyses of PARV4 mRNAs.
Total RNA was isolated from 293 cells co-transfected with p5TRPARV4, pAV5Rep78, and pHelper and subjected to reverse transcription. The cDNA was then subjected to 5’/3’ RACE analysis or amplified with PARV4-specific primers. (A) Amplified DNA fragments were electrophoresed on 2% agarose gel and visualized using ethidium bromide staining. (B) Primer sets used for PCR amplification are depicted, and the transcription units identified, with the promoters (P6 and P38), splice donor (D1, D2, D3 and D3m) and acceptor (A1, A2 and A3) sites, and polyadenylation signals [(pA)p and (pA)d ], are shown. LTR, left terminal repeat; RTR, right terminal repeat.
More extensive RT-PCR analysis using different sets of primers followed by sequencing also showed the same results as those obtained by 5'/3' RACE, which confirmed the donor and acceptor sites for alternative splicing of PARV4 mRNAs generated from the P6 and P38 promoters (Fig. 2A, lanes 3, 4, and 6). Sequence analysis of the DNA fragments amplified by PCR using primers Fnt283 and Rnt2060 revealed an intron that lies in the NS1-encoding region from the D1 donor (nt 418) to the A1 acceptor sites (nt 1630).
Taken together, these data provide the locations of all transcription units in the PARV4 genome by 5’/3’ RACE and RT-PCR, which are diagramed in Fig. 2B. The sequence results of the amplified fragments are shown in detail in Table 1.
Table 1.
Sequencing results of the DNA bands obtained from 5'/3' RACE and RT-PCR
| Lane | Analysis | Primers* | Bands | Identified transcripts* |
|---|---|---|---|---|
| 1 | 5' RACE | d(G)16-R nt960 | a | P6 nt262-960 |
| 2 | 5' RACE | d(G)16-R nt3840 | a | P38 nt1950-2060Δ2309-2367Δ3329-3840 |
| b | P38 nt1950-2013Δ3329-3840 | |||
| 3 | RT-PCR | F nt283-R nt2060 | a | P6 nt283-2060 |
| b | P6 nt283-418Δ1630-2060 | |||
| 4 | RT-PCR | F nt1441-R nt2354 | a | non-specific |
| b | P6 nt1441-2354 | |||
| c | P6 nt1441-2060Δ2309-2354 | |||
| 5 | 5' RACE | d(T)18V-R nt3440 | a | P38 nt1950-2060Δ2309-2367Δ3329-3440 |
| b | P38 nt1950-2013Δ3329-3440 | |||
| c | Primer dimers | |||
| 6 | RT-PCR | F nt283-R nt3440 | a | P6 nt283-2060Δ2309-2367Δ3329-3440 |
| b | P6 nt283-418Δ1630-2060Δ2309-2367Δ3329-3440 | |||
| c | P6 nt283-418Δ1630-2013Δ3329-3440 | |||
| d | P6 nt283-418Δ3329-3440 | |||
| 7 | 3' RACE | F nt3518-d(T)18V | a | (pA)d at nt 5149 |
| b | (pA)p at nt 3771 | |||
| c | non-specific |
Nucleotide numbers shown refers to Genbank accession no.: NC_007018.
“Δ” denotes intron.
RNase protection assay (RPA) analysis of PARV4 mRNA transcripts
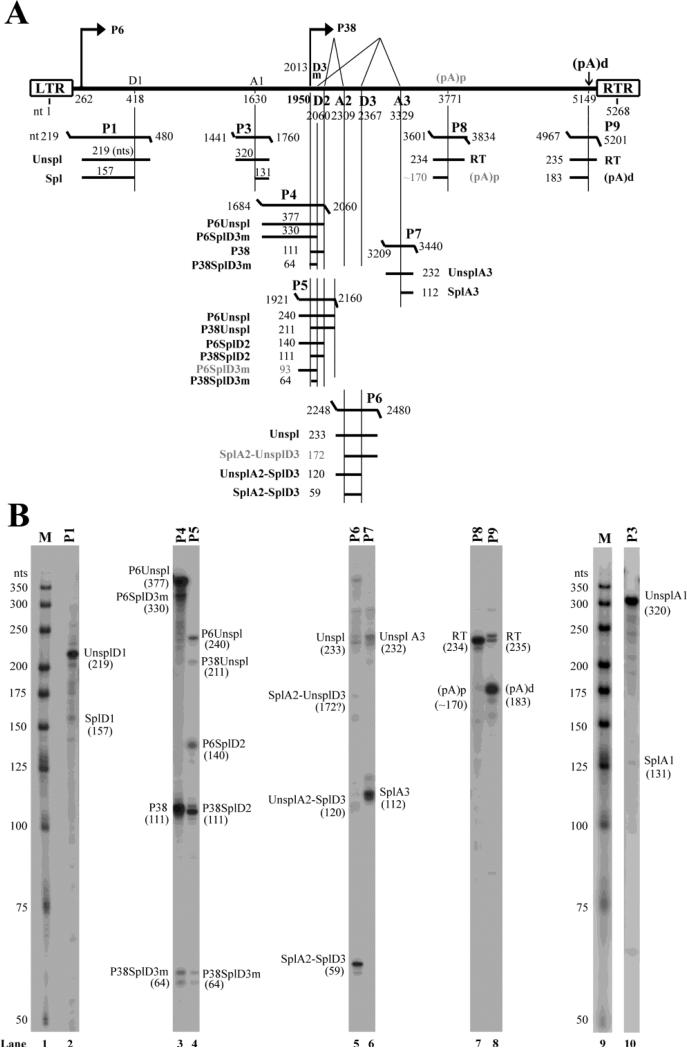
To determine the relative abundance of each PARV4 mRNA transcript, we used 8 anti-sense probes to protect individual transcript. A schematic diagram of the probes with their putatively protected bands and respective sizes is shown in Fig. 3A.
Figure 3. Transcription mapping by RNase protection assays (RPAs).
(A) Schematic diagram of the PARV4 genome and the probes used for the RPA. The PARV4 genome is depicted with the transcription units. Probes used for the RPAs are shown with their respective nucleotide numbers, together with the bands that they are expected to protect and the predicted sizes. Spl, spliced mRNAs; Unspl, unspliced mRNAs; RT, mRNAs read through the polyadenylation site. (B) Mapping of the PARV4 transcripts by RPA. Ten μg of total RNA was isolated from 293 cells transfected with p5TRPARV4, pAV5Rep78, and pHelper, and were protected by PARV4 probes P1 and P3-9. Lanes 1&9, 32P-labeled RNA markers with sizes indicated. Bands detected are designated in each lane.
Probe P1, which spanned the P6 promoter and D1 donor site, protected bands of 219 and 157 nts in sizes. These bands confirmed the mRNA initiation site at nt 262 and the D1 donor site at nt 418. Notably, the spliced band (157 nts) at the D1 donor site accounted for less than 5% of the total protected bands (Fig. 3B, lane 2), suggesting that mRNA splicing at the D1 site is rather poor.
Probe P3, which spanned the A1 acceptor site of the first intron, protected bands of 320 and 131 nts in sizes (Fig. 3B, lane 10). The protected band of 131 nts confirmed the position of the A1 acceptor site at nt 1630. Similar to what observed at the D1 donor site, only a few PARV4 mRNAs were spliced at the A1 acceptor site, suggesting the weakness of the splicing capability of the first intron spanning D1-A1 sites.
Probe 4, which spanned the P38 promoter and D3m donor site, protected bands of 377, 330, 111 and 64 nts in sizes (Fig. 3B, lane 3). Probe 5, which spanned the P38 promoter, D3m and D2 donor sites, protected bands of 240, 211, 140, 111 and 64 nts in sizes (Fig. 3B, lane 4). These bands confirmed the locations of the P38 promoter as well as the D3m and D2 donor sites at nt 1950, 2013 and 2060, respectively. Only ~5% of mRNAs generated from the P38 promoter were spliced at the D3m donor site, which resulted in a band of 64 nts. The majority of the P38-generated mRNAs were spliced at the D2 site, which produced a band of 111 nts.
Probe P6, which spanned the A2 acceptor and the D3 donor sites, protected bands of 233, 120, and 59 nts in sizes. These bands confirmed the presence of the PARV4 mRNAs spliced at the A2 acceptor site (nt 2309) and the D3 donor site (nt 2367) (Fig. 3B, lane 5). A putative band of 172 nts, which refers to the transcripts that are spliced at the A2 site but not at the D3 donor site, was not detected, suggesting that the PARV4 mRNAs that were spliced at the A2 site were spliced concurrently at the D3 donor site,
Probe P7, which spanned the A3 acceptor site, protected two bands of 232 and 112 nts in sizes, respectively. Thus, the A3 acceptor site was mapped to nt 3329 (Fig. 3B, lane 6). The ratio of unspliced vs. spliced PARV4 mRNA transcripts across this region was ~1:4.
Probe P8 was used to confirm the internal poly A signal (pA)p site (Fig. 3B, lane 7). No band of ~170 nts in size was apparent, which suggests that the (pA)p site at nt 3771 detected by 3’ RACE is not significantly used for processing PARV4 pre-mRNA. Probe P9 protected a major band at ~183 nts, which was produced when PARV4 mRNAs were terminated at the (pA)d site, confirming the predominant polyadenylation site of PARV4 mRNA transcripts at nt 5149 (Fig. 3B, lane 8).
Northern blot analysis of PARV4 mRNA transcripts
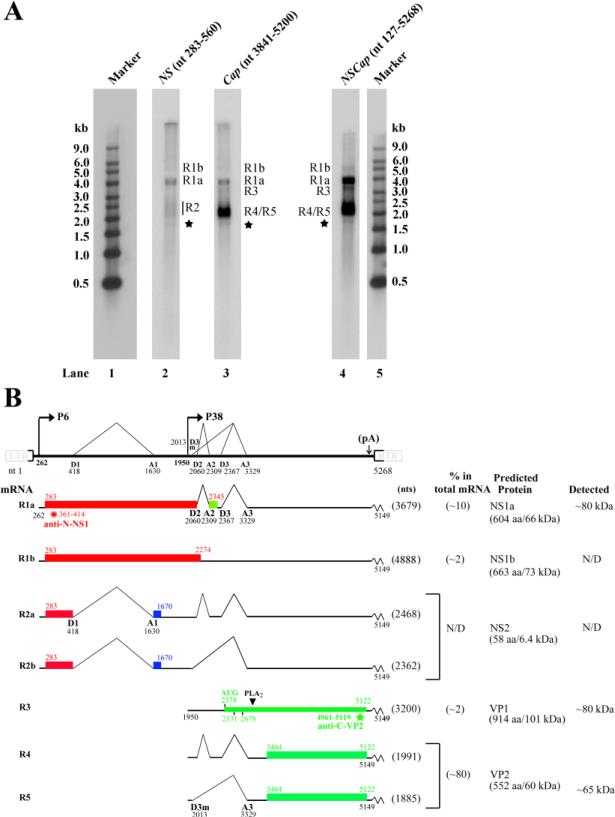
Hybridization of PARV4 mRNAs with the Cap probe (nt 3,841-5,200) detected four major bands (Fig. 4A, lane 3), which were putatively NS1a-encoding R1a mRNA (~4.0 kb), NS1b-encoding R1b mRNA (~5.0 kb), VP1-encoding R3 mRNA (~3.2 kb), and VP2-encoding R4 and R5 mRNAs (~2.0 kb). The putative NS2-encoding R2 mRNA (~2.4 kb) had a much lower expression level and was not clearly visualized in either lane 2, where the NS probe (nt 283-560) was supposed to detect the R2 mRNA, or in lane 3, where the Cap probe was used, in Fig. 4A. The NS probe was included to detect all transcripts generated from the P6 promoter; however, this probe only revealed a single major band, the R1a mRNA at ~4.0 kb. There were two smear areas above and below the R1a band, which potentially correspond to the R1b mRNA at ~5.0 kb and R2 (R2a+R2b) mRNAs at ~2.4 kb, respectively (Fig. 4A, lane 2).
Figure 4. Transcription profile of PARV4.
(A) Northern blot analysis. Total RNA was isolated from 293 cells transfected with p5TRPARV4, pAV5Rep78, and pHelper, and used for Northern blot analysis. The blots were hybridized to three PARV4 DNA probes (NSCap, NS, and Cap), respectively. Bands of mRNA detected by each probe are designated in each lane. The RNA marker ladder is shown in lanes 1 and 5. The asterisk denotes a likely non-specific band detected by all three probes. (B) Transcription map of PARV4. The PARV4 genome is shown to scale, with transcription landmarks indicated. Major mRNA species detected in RT-PCR, RPA, and Northern blot assays are diagrammed to display their identities and respective sizes in the absence of the polyA tail. The putative ORFs that each can encode are also diagramed in boxes, and the predicted sizes (kDa) of translated proteins are indicated on the right. The relative abundance of each mRNA and proteins detected by transfection are also shown on the right. Potential AUG usage for VP1 translation and peptides used for production of antibodies are depicted. N/D, not determined or not detected.
Analysis with the PARV4 genome probe (NSCap) showed an overall RNA profile generated from transfection of p5TRPARV4 (Fig. 4A, lane 4). Five major species of mRNA transcripts were observed, which were consistent in size with those detected when using the Cap probe and predicted to encode NS1 (R1a and R1b), VP1 (R3), and VP2 (R4 and R5). The R4/R5 mRNAs were the most abundant transcripts and constituted ~80% of the total viral mRNAs.
Accordingly, we generated a genetic map of PARV4 as a summary of the results obtained from 5’/3’ RACE, RT-PCR, RPA, and Northern blot analyses (Fig. 4B). The five major PARV4 mRNA transcripts and two minor transcripts (R2a and R2b) are depicted with their respective sizes, abundance by percentage, and encoded proteins.
Expression strategy of PARV4 proteins
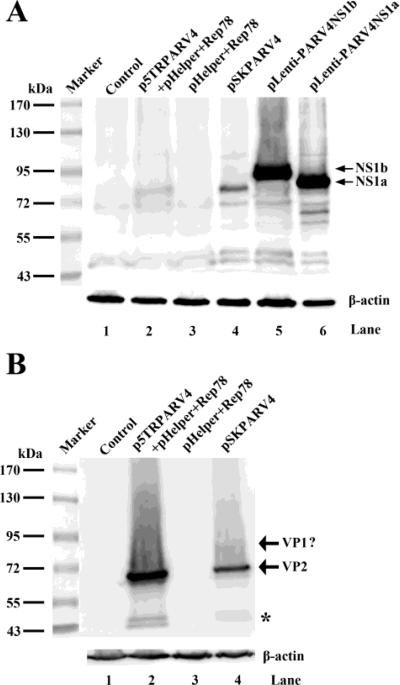
We next examined protein expression of PARV4 by transfecting. To determine whether the NS1a or NS1b protein is expressed after transfection of p5TRPARV4 and pSKPARV4 in 293 cells, we expressed NS1a and NS1b individually as size controls (Fig. 5A, lanes 5&6). We found that NS1a was clearly detected, but not NS1b, in both p5TRPARV4- and pSKPARV4-transfected cells (Fig. 5A, lanes 2&4). The bands of both NS1a were slightly lower than the one as the size control, which are likely due to the uneven migrations of NS1a in each lane (Fig. 5A). Although the predicted size of the NS1a protein is 66 kDa (Fig. 4B), it appeared larger than 72 kDa (~80 kDa) in the blot, which was likely due to a modification of the NS1 protein (e.g., phosphorylation) (Nuesch et al., 1998).
Figure 5. Western blot analysis of PARV4 proteins.
293 cells were transfected with plasmids as indicated. At 48 hrs post-transfection, the cells were harvested and lysed for Western blotting using polyclonal antibodies anti-N-NS1 (A) and anti-C-VP2 (B), respectively. The blots were reprobed using an anti-β-actin antibody. The identities of detected proteins are shown to the right on the blot. Untransfected cells were used as a control. The asterisk indicates bands that were likely degraded from VP2.
The anti-N-NS1 antibody used for Western blot was produced against the N-terminus (aa 27-44) of the NS1 protein (Fig. 4B), which is potentially able to detect both NS1 and NS2 proteins. However, we did not detect specific bands around 7 kDa, which should have corresponded to the putative NS2 protein, using a high percentage (15%) PAGE gel (data not shown). These results indicated that the NS2 protein is likely expressed at a very low level.
Using the anti-C-VP2 antibody raised against the C-terminus (aa 500-552) of the VP2 protein, we detected a major band at a molecular weight of ~65 kDa, which corresponded to the VP2 protein, in both p5TRPARV4 and pSKPARV4-transfected cells (Fig. 5B, lanes 2&4). Notably, VP1, which has a predicted molecular weight of 101 kDa, was not detected. However, we observed a minor band around 90 kDa above the VP2 band, which could be the VP1 protein translated from the second AUG (nt 2531) or the third AUG (nt 2678) on the VP1-encoding R3 mRNA (Fig. 4B). However, it is also possible that the band of ~90 kDa was a cleaved product of the VP1 protein of 101 kDa, since the capsid proteins of other parvoviruses have been reported to be cleaved (Farr et al., 2006; Cheng et al., 2009).
Both the PARV4 NS1a and NS1b proteins induce cell cycle arrest at G2/M phase
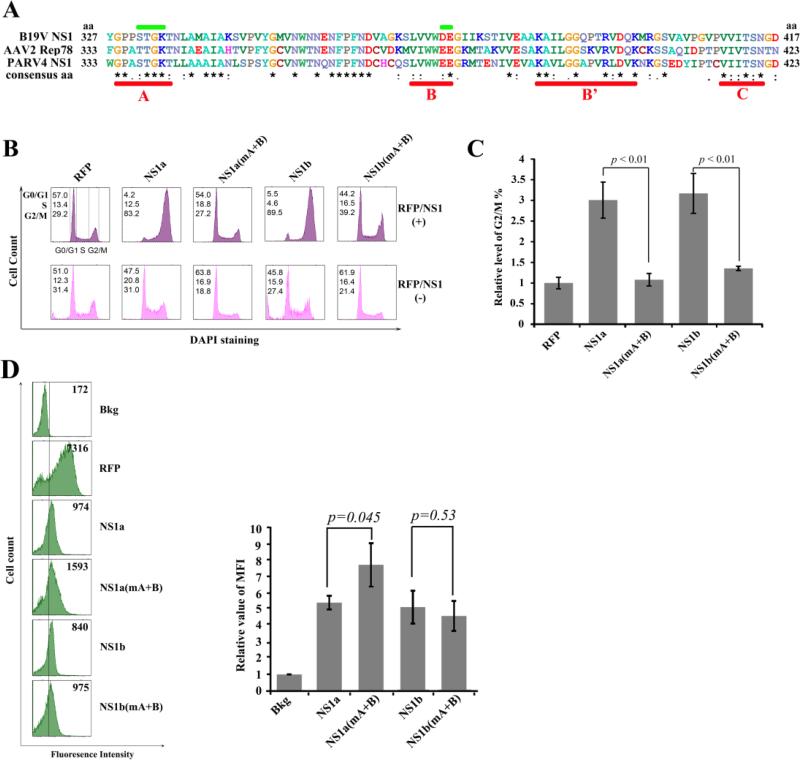
As shown in other genera of parvoviruses, the large nonstructural protein NS1 or Rep78 protein is able to induce cell cycle arrest (Chen and Qiu, 2010), which is also a hallmark of B19V infection of primary erythroid progenitor cells (Wan et al., 2010). Since PARV4 was previously detected in human bone marrow (Manning et al., 2007; Longhi et al., 2007), we examined the effect of PARV4 NS1 on the cell cycle progression of CD34+ HSCs. We first performed a sequence alignment among PARV4 NS1, B19V NS1, and AAV2 Rep78 proteins (Fig. 6A). Highly conserved regions were identified in the helicase motifs (the four sites of Walker boxes, A, B, B’ and C sites (Jindal et al., 1994; Walker et al., 1997; Momoeda et al., 1994)). Therefore, we introduced mutations in boxes A and B of both NS1a and NS1b to explore the potential role of the helicase motifs of NS1 in disturbing cell cycle progression.
Figure 6. The helicase motifs of the PARV4 NS1 are responsible for G2/M arrest induced by PARV4 NS1 protein.
(A) Comparison of the NS1 helicase motifs. The amino acid sequences of B19V NS1, AAV2 Rep78, and PARV4 NS1 were aligned using ClustalW2 (Larkin et al., 2007). The alignment of aa 327-417 of B19V NS1, aa 333-423 of AAV2 Rep78, and aa 333-423 of PARV4 NS1 is depicted with identical amino acids shown by the asterisk, while homologous amino acids are shown as two dots under the amino acid. Conserved motifs (Walker boxes A, B, B’, and C) are underlined in red, while the mutated regions of boxes A and B are marked in green. (B&C) PARV4 NS1 protein induces G2/M arrest. (B) CD34+ HSCs were transduced with lentiviruses to express proteins as indicted. At 48 hrs post-transduction, RFP- or NS1-expressing cells were selectively gated for anti-Flag staining, followed by DAPI staining for cell cycle analysis. The cell cycle patterns of both NS1/RFP-positive and NS1/RFP-negative cells are shown in parallel. In each panel, the percentages of cells in the G1, S, and G2/M phases are indicated, respectively (top left). A representative experiment is shown. (C) The percentage of RFP-expressing cells in G2/M was arbitrarily set as 1. Relative values are shown with average and standard deviation from at least three independent experiments. P values were determined using a student's t test. (D) Expression levels of PARV4 NS1 proteins. The expression levels of RFP and NS1 as mean fluorescence intensity (MFI) were analyzed by flow cytometry in lentivirus-transduced (NS1/RFP+) cells at 48 hrs post-transduction. The reference line is selected arbitrarily to show the relative position of the NS1/RFP-positive and negative peaks, and Bkg (background) represents the secondary antibody only control. A representative experiment is shown to the left. Relative MFI values were determined as fold changes of MFI of NS1/RFP+ cells compared to that of Bkg, and are shown to the right with average and standard deviation.
We transduced lentiviruses in CD34+ HSCs in order to express NS1a and NS1b. At 48 hrs post-transduction, we analyzed the cell cycle progression using flow cytometry. As shown in Fig. 6B, an obvious G2/M arrest (p < 0.01) was observed in both NS1a and NS1b-expressing CD34+ HSCs compared to RFP-expressing cells. Notably, both NS1a(mA+B) and NS1b(mA+B), which bear the mutations in boxes A+B, significantly rescued the G2/M arrest (p < 0.01, Fig. 6C). The expression levels of NS1a, NS1b and their mutants in transduced cells were quantified at similar levels as shown with mean fluorescence intensity in Fig. 6D. These results demonstrated that the PARV4 NS1a/b proteins are capable of inducing a G2/M arrest in NS1-expressing cells. Thus, these data reveal a remarkable feature of the PARV4 NS1, which is similar to that seen with B19V infection of primary erythroid progenitor cells (Wan et al., 2010) and clearly involves the predicted helicase motifs.
The PARV4 VP1 unique region does not exhibit phospholipase A2-like activity in vitro
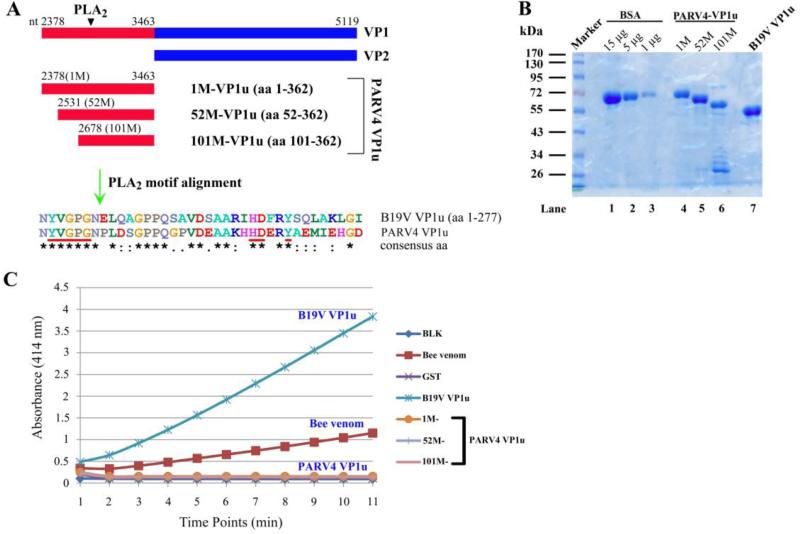
The VP1 unique region (VP1u) of parvoviruses functions like PLA2 with conserved motifs, which is critical for virus infection (Zadori et al., 2001). We first aligned the PARV4 VP1u with that of B19V, and identified conserved PLA2 motifs (Fig. 7A). To examine the PLA2-like activity of the PARV4 VP1u, three forms of VP1u (1M-VP1u, 52M-VP1u, and 101M-VP1u) were purified (Fig. 7B), which originated from three different methionines of the putative VP1u. This approach was taken because we had not determined the start AUG for translation of PARV4 VP1. Purified B19V VP1u and GST proteins were used as positive and negative controls, respectively. As seen in Fig. 7C, both B19V VP1u and bee venom exhibited significant PLA2-like activity (Dorsch et al., 2002); however, none of the PARV4 VP1u proteins possessed PLA2-like function in vitro, implying that either the PARV4 VP1u requires a structural conformation for PLA2-like activity or PLA2-like activity is not required for PARV4 infection.
Figure 7. The PARV4 VP1 unique region does not exhibit phospholipase A2-like activity in vitro.
(A) Comparison of B19V VP1u and PARV4 VP1u. The PARV4 VP1 and potential unique regions are diagramed. The alignment of PLA2-conserved motifs of B19V VP1u and three forms of PARV4 VP1u is depicted, with identical amino acids shown as an asterisk and homologous amino acids shown as two dots. (B) SDS-PAGE analysis of purified PARV4-VP1u and B19V VP1u proteins. Five microliter of each purified protein and BSA standards was analyzed on SDS-10%PAGE gel. The gel was stained using Coomassie blue. A protein maker is shown. (C) PARV4 VP1u does not exhibit PLA2-like activity. An in vitro sPLA2 assay kit was used to test PLA2-like activity of various substrates as indicated on the right. The PLA2-like activity is shown as a value of the absorbance at 414 nm at various time points. BLK, blank.
Discussion
With a recurrent pandemic of newly emerging viruses worldwide, strong interest has been drawn on uncovering novel viruses that can potentially infect humans. The newly identified human parvoviruses HBoV and PARV4 have been widely found to persist in humans (Allander et al., 2005; Jones et al., 2005; Brown, 2010). In this study, we revealed the basic gene expression profile of PARV4, which not only has important clinical significance, but also identified its position as a member in the newly formed genus Partetravirus in family Parvoviridae (Tijssen et al., 2011).
The characterized transcription map of PARV4, which resulted from transfection of a replication-competent PARV4 clone, is similar to those of the parvoviruses in the genus Parvovirus (Qiu et al., 2006b). Two promoters, P6 and P38, were used to transcribe NS-encoding and VP-encoding mRNAs, respectively. In contrast to the parvoviruses in genera Amdovirus, Bocavrius, and Erythrovirus, PARV4 does not employ internal polyadenylation for pre-mRNA processing (Qiu et al., 2006a; Qiu et al., 2007a; Sun et al., 2009; Ozawa et al., 1987). Although NS2 protein was not detected after transfection, R2 mRNAs, which are transcribed from the P6 promoter and spliced in the NS1-encoding region, likely encode NS2 protein during PARV4 infection similar to members of the genus Parvovirus, such as the minute virus of mice (MVM) and canine parvovirus (CPV). PARV4-like parvoviruses have emerged in animals worldwide (Lau et al., 2008; Adlhoch et al., 2010; Sharp et al., 2010); however, like PARV4, none of the PARV4-like parvoviruses has been cultured in vitro as yet, and infectious clones have not been established. We have employed an AAV5 ITRs-supported replication system to study gene expression of parvoviruses B19V and HBoV, which closely reflects the profiles during virus infection (Guan et al., 2008; Chen et al., 2010a). We realize that the transcription profile of PARV4 generated from this AAV5 ITRs-supported replication system might not fully depict the profile during PARV4 infection; however, our characterization of the gene expression profile of PARV4 within this artificial replication system will lay a fundamental guidance for future study of PARV4 gene expression during infection.
The large nonstructural protein NS1 or Rep78 of parvoviruses is essential for viral DNA replication (Cotmore and Tattersall, 2005a), transcriptional transactivation (Legendre and Rommelaere, 1994; Ahn et al., 1992), and encapsidation of the viral genome (Cotmore and Tattersall, 2007). In addition, it causes cytopathic effects of infected cells by inducing cell cycle arrest and apoptosis (Chen and Qiu, 2010). The PARV4 NS1a protein is encoded from a spliced form of R1 mRNA (R1a, Fig. 4B) and was clearly detected after transfection. Although the NS1b protein was not detectable in PARV4-transfetced cells, the R1b mRNA was clearly detected (Fig. 4A). Therefore, PARV4 NS1a is likely the major NS1 protein expressed during infection, while NS1b is produced at a relatively low level. Parvoviruses use different strategies to express their NS1 proteins. Some NS1 proteins are expressed from unspliced mRNAs (e.g., B19V and AAVs) (Ozawa et al., 1987; Qiu et al., 2002). Some parvoviruses only express one form of NS1 (e.g., B19V and MVM) (Ozawa et al., 1987; Cotmore and Tattersall, 2005b), but some others express multiple isoforms that possess different functions (Di and Chiorini, 2003). NS1a and NS1b proteins differ at the C-termini (Fig. 4B). The NS1b C-terminus-encoding region lies in the intron that spans the D2 and A2 splice sites, which is only used to encode the NS1b C-terminus. Since the NS1 C-terminus of parvoviruses has transcriptional transactivation function (Legendre and Rommelaere, 1994), we hypothesize that both NS1a and NS1b proteins are expressed, and that the NS1b is also required for the life cycle of PARV4 during infection. But one possibility still could not be ruled out that the R1b transcript could be a nuclear precursor mRNA that may not be used for translation of NS1b protein. If a virus infection system is available, the existence of NS1b protein could be easier to be proved.
We found that both NS1a and NS1b induced G2/M arrest in NS1-expressing hematopoietic stem cells (Fig. 6). This characteristic is quite similar to that of B19V NS1 (Wan et al., 2010). Amino acid alignment showed that PARV4 NS1 contains conserved helicase motifs (Jindal et al., 1994; Walker et al., 1997; Momoeda et al., 1994). Mutations of the helicase motifs abolished the G2/M arrest, suggesting that the helicase motifs of NS1 are involved in the G2/M arrest induced by PARV4 NS1. It has been shown that the helicase motifs of B19V NS1 are critical for the induction of apoptosis (Momoeda et al., 1994). Besides, the nuclear localization signal (NLS) has been shown to be important for B19V NS1 to induce G2/M arrest in primary erythroid progenitor cells (Wan et al., 2010). It is interesting to hypothesize that the mechanism underlying PARV4 NS1-induced G2/M arrest may differ from that of B19V NS1. We also found that PARV4 NS1 induced G2/M arrest in 293T and HeLa cells (data not shown). Thus we believe that PARV4 NS1 induces G2/M arrest in PARV4-infected cells. Further studies are needed to prove that NS1 can disturb the cell cycle and elucidate its mechanism during PARV4 infection.
Parvovirus is non-enveloped, and has an icosahedral capsid formed by the main structural VP2 protein and the minor structural VP1 protein with PLA2-like activity, which is a lipolytic enzyme lying within the capsid (Tsao et al., 1991). The PLA2-like activity is required for efficient penetration of virions through the cellular membrane (Farr et al., 2005) and for the transfer of the viral genome from the endosomes or lysosomes into the nucleus (Zadori et al., 2001). However, the structural conformation of VP1u in the virions among different genera of parvoviruses could be different (Rosenfeld et al., 1992; Anderson et al., 1995; Cotmore et al., 1999). In our study, none of the three forms of purified PARV4 VP1u appeared to exhibit PLA2-like activity in vitro, while B19V VP1u exhibited a strong activity (Dorsch et al., 2002), which was somewhat unexpected. This result could be due to either the requirement of a specific structural conformation of PARV4 VP1u or the possibility that PARV4 utilizes certain novel mechanisms to penetrate through cellular membranes without the need of PLA2-like activity.
In conclusion, as a representative member in the new genus Partetravirus in family Parvoviridae, PARV4 shares characteristics of transcription and protein expression with the parvoviruses in the genus Parvovirus. PARV4 NS1 protein shares features with B19V NS1 protein and can induce G2/M arrest in primary hematopoietic cells and certain cell lines. Therefore, our study has shown for the first time the detailed transcription map of PARV4, which can be beneficial for subsequent study of PARV4 infection.
Materials and methods
Cells and transfection
HEK-293 (ATCC-1573) cells were maintained in Dulbecco's modified Eagle's medium with 10% fetal calf serum (FCS) in 5% CO2 at 37°C. Human hematopoietic stem cells were purchased from the National Disease Research Interchange (NDRI, Philadelphia, PA). CD34+ HSCs were cultured in StemSpan serum-free expansion medium (SFEM; StemCell Technologies Inc., Vancouver, Canada) as previously described (Chen et al., 2010b), and stored in liquid nitrogen on day 4 of the culture. The day 4 stock of CD34+ HSCs was then thawed and expanded ex vivo in SFEM until day 6 for lentivirus transduction.
Transfection was performed using the Lipofectamine™ and Plus™ reagent (Invitrogen, Carlsbad, CA) or LipoD293 transfection reagent (SigmaGen Laboratories, Rockville, MD) following the manufacturers’ instructions.
Plasmid construction
(i) Clones of the PARV4 genome
A primer based on the common repeated sequence in both the 5' and 3' terminal repeats of the PARV4 genome was used to amplify the full encoding sequence. DNA extracted from a patient's serum was amplified with primer HP4-FL (5’-WCG TAT TTC CGC TTC CGG TC-3’), and ExTaq DNA polymerase (Takara Mirius Bio., Madison, WI) using the following amplification parameters: 94 °C for 2 min, and then 30 cycles of 92 °C for 40s, 52 °C for 40 s, and 68 °C for 4 min. A final extension was performed at 75 °C for 5 min. The PCR product was cloned into pCR2.1 TOPO vector (Invitrogen, Carlsbad, CA), which resulted in pCR2.1-PARV4. The PARV4 sequence was confirmed to be from nt 127 to 5268 (GenBank accession no.: NC_007018).
The p5TRPARV4 plasmid, which contained the PARV4 sequence flanked by AAV5 ITRs at two ends, was constructed by ligating PARV4 nt 127-5268 into the EcoRV-digested AAV5 ITRs-containing plasmid pAV5ITR (Guan et al., 2008). The PARV4 sequence was also cloned into a KpnI/EcoRI-digested pBluescript SK(+) vector (Agilent, Santa Clara CA), which created the plasmid pSKPARV4.
(ii) Lentiviral vectors
Two PARV4 NS1 sequences, NS1a (nt 283-2060/2309-2345) and NS1b (nt 283-2271), as well as an RFP-coding sequence (Chen et al., 2010c), were tagged with the Flag-encoding sequence at the C-terminus and cloned into a BamHI/BsrG-digested pLenti-CMV-IRES-GFP-WPRE vector (Chen et al., 2011), which resulted in pLent-NS1a, pLenti-NS1b, and pLenti-RFP, respectively. In addition, two helicase motifs (box A+B) of the NS1 were mutated from amino acid number (aa) 337STGK340 to AAAA and 378EE379 to AA, resulting in plasmids pLenti-NS1a(mA+B) and pLenti-NS1b(mA+B), respectively.
(iii) Probe constructs for RNase protection assay
RPA probe clones, P1 and P3-9, were constructed by ligating the following regions of PARV4 into pGEM3Z (Promega, Madison, WI) at BamHI/HindIII sites: nt 219-480 (P1), nt 1441-1760 (P3), nt 1684-2060 (P4), nt 1921-2160 (P5), nt 2248-2480 (P6), nt 3209-3440 (P7), nt 3601-3834 (P8), and nt 4967-5201 (P9).
(ii) Constructs for glutathione S-transferase (GST)-fusion protein expression
The PARV4 NS1 N-terminus (aa 27-44) and the VP2 C-terminus (aa 500-552) were cloned into BamHI/XhoI-digested pGEX4T3 (GE Healthcare, Piscataway, NJ) as pGEX-N-NS1 and pGEXC-VP2, respectively.
PARV4 VP1u-encoding sequences, which originate from the methionine (M) at three different positions, nt 2378 (1M-VP1u), nt 2531 (52M-VP1u) and nt 2678 (101M-VP1u), respectively, were cloned into a BamHI/XhoI-digested pGEX4T3 vector as pGEX-1M-VP1u, pGEX-52M-VP1u and pGEX-101M-VP1u, respectively. As a control, B19V VP1u (nt 2624-3304, accession no: AY386330) was also cloned into pGEX4T3 to create pGEX-B19VVP1u.
The sequences of all the clones were confirmed at MCLAB (South San Francisco, CA).
Southern blot analysis
Hirt DNA was extracted from p5TRPARV4- and pSKPARV4-transfected 293 cells, and DpnI digestion and Southern blotting were performed as previously described (Guan et al., 2008) using the PARV4 NSCap probe template (nt 127-5268). Signals were developed by exposing the blot to an X-ray film.
RNA isolation and RNA analysis
p5TRPARV4 was co-transfected with pAV5Rep78 and pHelper plasmids into 293 cells as previously described (Guan et al., 2008). Total RNA was isolated 48 hrs later using Trizol® reagent (Invitrogen).
RNase protection assay
Probes were generated from EcoRI-digested templates by in vitro transcription with T7 polymerase, using a MAXIscript kit (Ambion, Austin, TX) following the manufacturer's instruction. RPAs were performed as previously described (Naeger et al., 1992; Schoborg and Pintel, 1991). RPA signals were quantified with a Typhoon 9700 PhosphorImager and ImageQuant TL software v2005 (GE Healthcare). Relative molar ratios for individual species of mRNAs were determined after adjusting the number of 32P-labeled uridines (U) in each protected fragment, as previously described (Schoborg and Pintel, 1991).
Northern blot analysis
Northern blot analysis was performed with an input of 5 μg of total RNA as previously described (Pintel et al., 1983; Qiu et al., 2002). Blots were hybridized with the PARV4 NS, Cap and NSCap probes, which were amplified PARV4 DNA fragments that spanned nt 283-560, nt 3841-5200 and nt 127-5268, respectively. Signals were developed and analyzed with a Typhoon 9700 PhosphorImager.
5′/3’ RACE
The 5’ and 3’ RACE was performed following the instructions of the RACE kit (Roche, Nutley, NJ). The primers for 5′ RACE were as follows: forward primers: d(G)16 (5’-GGC CAC GCG TCG ACT AGT ACG GGG GGG GGG GGG GGG-3’), or d(T)18V (5’-CGC GGA TCC TTT TTT TTT TTT TTT TTT A/C/G-3’); reverse primers: Rnt960 (5’-CAT GAA TCT TTG TGT TCC-3’), Rnt3440 (5’-GCA AGG CCG GGT CAG ACT CG-3’), and Rnt3840 (5’-CTT GCA GAA TCA GAA ACA G-3’). The primers for 3′ RACE were as follows: Fnt3518 (5’-ATA GGT GGA ACT AGT TTT TCT GAT TCT GTA GTT-3’) and d(T)18V.
RT-PCR
The cDNA was synthesized as previously described (Guan et al., 2008). Primers used for RT-PCR are diagramed in Figure 2B. The sequences were as follows: Fnt283 (5’-ATG GAC GCT CCT GCC TGG ATT-3’), Fnt1441 (5’-GTT GAA GTA GCC AAA GCT GTG-3’), Rnt2060 (5’- CTT TCA GCA GTT TCG TCA-3’), and Rnt2354 (5’-TGA AGC TTG TTA GTC TTC TGC-3’). PCR was performed with Vent®Taq DNA polymerase (NEB, Ipswich, MA) using the following amplification parameters: 94°C for 2 min, followed by 30 cycles of 92 °C for 45s, 52°C for 60 s, and 72°C for 1 min. A final extension was performed at 72 °C for 10 min. The amplified DNA fragments were separated on a 2% agarose gel, and all bands were excised and purified by using a gel extraction kit (Qiagen, Valencia, CA). The final products were sequenced at MCLAB.
Production of antibodies against PARV4 proteins
GST-N-NS1 and GST-C-VP2 proteins were expressed in Escherichia coli BL21 cells transformed with pGEX-N-NS1 and pGEX-C-VP2, respectively, and were purified using a GSTrap FF 1-ml column (GE Healthcare) on the Biologic LP system (Bio-Rad, Hercules, CA). Purified proteins were analyzed by SDS-PAGE and appeared as two major bands with approximate sizes of 28 kDa and 32 kDa, respectively. Purified GST-N-NS1 and GST-C-VP2 proteins (500 μg/ml) were emulsified with TiterMax® Gold adjuvant (TiterMax® USA, Norcross, GA) and then used to immunize animals as previously described (Sun et al., 2009). All animal procedures were approved by the KUMC Institutional Animal Care and Use Committee.
SDS-PAGE and Western blot analysis
Sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) was performed as previously described (Chen et al., 2010c). For Western blotting, cell lysates were prepared 48 hrs post-transfection and analyzed as previously described (Qiu et al., 2000; Qiu et al., 2007b).
Lentivirus production and transduction
Lentiviruses were generated and concentrated following the instructions provided by Addgene (Cambridge, MA). Cells were transduced at a multiplicity of infection (MOI) of ~10 as previously described (Chen et al., 2010b; Chen et al., 2011).
Flow cytometry analysis
Lentivirus-transduced CD34+ HSCs were collected 48 hrs post-transduction. The cells were fixed with 1% paraformaldehyde-phosphate buffered saline (PBS) at room temperature for 30 min and then permeabilized in 2%FCS-PBS containing 0.3% Tween-20 (PBST) for 20 min. The cells were then stained with a 1:100-diluted anti-Flag antibody (Agilent, Santa Clara, CA) for 45 min followed by staining with a Cy5-conjugated anti-mouse antibody for 30 min. After incubation, the cells were washed and incubated with 2 μg/ml 4'-6-Diamidino-2-phenylindole (DAPI) in PBST for 30 min at 37 °C, and then analyzed on a three-laser flow cytometry (LSR II, BD Biosciences, San Jose, CA) within one hour. Flow cytometry data were analyzed using FACS DIVA software (BD Biosciences).
Phospholipase A2-like activity assessment
Concentrations of the purified GST, GST-1M-, 52M-, and 101M-VP1u proteins as well as GST-B19V-VP1u protein were determined by using the CB-X assay kit (GBioscience, Maryland Heights, MO). The proteins were aliquoted and stored at -80 °C. PLA2-like activity was assayed following the protocol provided with the sPLA2 assay kit (Cayman, Ann Arbor, MI). The input of GST-fused proteins was adjusted to 0.2 nmol per well during analysis. A wavelength at 414 nm was used to detect the absorbance.
Acknowledgements
This work was supported by PHS grant R01 AI070723 from NIAID. We thank members in Qiu lab for valuable discussions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adlhoch C, Kaiser M, Ellerbrok H, Pauli G. High prevalence of porcine Hokovirus in German wild boar populations. Virol. J. 2010;7:171. doi: 10.1186/1743-422X-7-171. 171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn JK, Pitluk ZW, Ward DC. The GC box and TATA transcription control elements in the P38 promoter of the minute virus of mice are necessary and sufficient for transactivation by the nonstructural protein NS1. J. Virol. 1992;66:3776–3783. doi: 10.1128/jvi.66.6.3776-3783.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allander T, Tammi MT, Eriksson M, Bjerkner A, Tiveljung-Lindell A, Andersson B. Cloning of a human parvovirus by molecular screening of respiratory tract samples. Proc. Natl. Acad. Sci. U. S. A. 2005;102:12891–12896. doi: 10.1073/pnas.0504666102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson S, Momoeda M, Kawase M, Kajigaya S, Young NS. Peptides derived from the unique region of B19 parvovirus minor capsid protein elicit neutralizing antibodies in rabbits. Virology. 1995;206:626–632. doi: 10.1016/s0042-6822(95)80079-4. [DOI] [PubMed] [Google Scholar]
- Benjamin LA, Lewthwaite P, Vasanthapuram R, Zhao G, Sharp C, Simmonds P, Wang D, Solomon T. Human parvovirus 4 as potential cause of encephalitis in children, India. Emerg. Infect. Dis. 2011;17:1484–1487. doi: 10.3201/eid1708.110165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown KE. The expanding range of parvoviruses which infect humans. Rev. Med. Virol. 2010;20:231–244. doi: 10.1002/rmv.648. [DOI] [PubMed] [Google Scholar]
- Chen AY, Cheng F, Lou S, Luo Y, Liu Z, Delwart E, Pintel D, Qiu J. Characterization of the gene expression profile of human bocavirus. Virology. 2010a;403:145–154. doi: 10.1016/j.virol.2010.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen AY, Guan W, Lou S, Liu Z, Kleiboeker S, Qiu J. Role of Erythropoietin Receptor Signaling in Parvovirus B19 Replication in Human Erythroid Progenitor Cells. J. Virol. 2010b;84:12385–12396. doi: 10.1128/JVI.01229-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen AY, Kleiboeker S, Qiu J. Productive Parvovirus B19 Infection of Primary Human Erythroid Progenitor Cells at Hypoxia is Regulated by STAT5A and MEK Signaling but not HIF alpha. PLoS. Pathog. 2011;7:e1002088. doi: 10.1371/journal.ppat.1002088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen AY, Qiu J. Parvovirus infetcion-induced cell detah and cell cycel arrest. Future Virology. 2010;5:731–741. doi: 10.2217/fvl.10.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen AY, Zhang EY, Guan W, Cheng F, Kleiboeker S, Yankee TM, Qiu J. The small 11kDa non-structural protein of human parvovirus B19 plays a key role in inducing apoptosis during B19 virus infection of primary erythroid progenitor cells. Blood. 2010c;115:1070–1080. doi: 10.1182/blood-2009-04-215756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng F, Chen AY, Best SM, Bloom ME, Pintel D, Qiu J. The capsid proteins of Aleutian mink disease virus (AMDV) activate caspases and are specifically cleaved during infection. J. Virol. 2009;84:2687–2696. doi: 10.1128/JVI.01917-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotmore SF, D'Abramo AM, Jr., Ticknor CM, Tattersall P. Controlled conformational transitions in the MVM virion expose the VP1 N-terminus and viral genome without particle disassembly. Virology. 1999;254:169–181. doi: 10.1006/viro.1998.9520. [DOI] [PubMed] [Google Scholar]
- Cotmore SF, Tattersall P. A rolling-haipin strategy: basic mechanisms of DNA replication in the parvoviruses. In: Kerr J, Cotmore SF, Bloom ME, Linden RM, Parrish CR, editors. Parvoviruses. Hoddler Arond; London: 2005a. pp. 171–181. [Google Scholar]
- Cotmore SF, Tattersall P. Structure and Organization of the Viral Genome. In: Kerr J, Cotmore SF, Bloom ME, Linden RM, Parrish CR, editors. Parvoviruses. Hodder Arnold; London: 2005b. pp. 73–94. [Google Scholar]
- Cotmore SF, Tattersall P. Parvoviral host range and cell entry mechanisms. Adv. Virus Res. 2007;70:183–232. doi: 10.1016/S0065-3527(07)70005-2. 183-232. [DOI] [PubMed] [Google Scholar]
- Di PG, Chiorini JA. PKA/PrKX activity is a modulator of AAV/adenovirus interaction. EMBO J. 2003;22:1716–1724. doi: 10.1093/emboj/cdg153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorsch S, Liebisch G, Kaufmann B, von LP, Hoffmann JH, Drobnik W, Modrow S. The VP1 unique region of parvovirus B19 and its constituent phospholipase A2-like activity. J. Virol. 2002;76:2014–2018. doi: 10.1128/JVI.76.4.2014-2018.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farr GA, Cotmore SF, Tattersall P. VP2 cleavage and the leucine ring at the base of the fivefold cylinder control pH-dependent externalization of both the VP1 N terminus and the genome of minute virus of mice. J. Virol. 2006;80:161–171. doi: 10.1128/JVI.80.1.161-171.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farr GA, Zhang LG, Tattersall P. Parvoviral virions deploy a capsid-tethered lipolytic enzyme to breach the endosomal membrane during cell entry. Proc. Natl. Acad. Sci. U. S. A. 2005;102:17148–17153. doi: 10.1073/pnas.0508477102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan W, Cheng F, Yoto Y, Kleiboeker S, Wong S, Zhi N, Pintel DJ, Qiu J. Block to the production of full-length B19 virus transcripts by internal polyadenylation is overcome by replication of the viral genome. J. Virol. 2008;82:9951–9963. doi: 10.1128/JVI.01162-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jindal HK, Yong CB, Wilson GM, Tam P, Astell CR. Mutations in the NTP-binding motif of minute virus of mice (MVM) NS-1 protein uncouple ATPase and DNA helicase functions. J. Biol. Chem. 1994;269:3283–3289. [PubMed] [Google Scholar]
- Jones MS, Kapoor A, Lukashov VV, Simmonds P, Hecht F, Delwart E. New DNA viruses identified in patients with acute viral infection syndrome. J. Virol. 2005;79:8230–8236. doi: 10.1128/JVI.79.13.8230-8236.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Lau SK, Woo PC, Tse H, Fu CT, Au WK, Chen XC, Tsoi HW, Tsang TH, Chan JS, Tsang DN, Li KS, Tse CW, Ng TK, Tsang OT, Zheng BJ, Tam S, Chan KH, Zhou B, Yuen KY. Identification of novel porcine and bovine parvoviruses closely related to human parvovirus 4. J. Gen. Virol. 2008;89:1840–1848. doi: 10.1099/vir.0.2008/000380-0. [DOI] [PubMed] [Google Scholar]
- Legendre D, Rommelaere J. Targeting of promoters for trans activation by a carboxy-terminal domain of the NS-1 protein of the parvovirus minute virus of mice. J. Virol. 1994;68:7974–7985. doi: 10.1128/jvi.68.12.7974-7985.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longhi E, Bestetti G, Acquaviva V, Foschi A, Piolini R, Meroni L, Magni C, Antinori S, Parravicini C, Corbellino M. Human parvovirus 4 in the bone marrow of Italian patients with AIDS. AIDS. 2007;21:1481–1483. doi: 10.1097/QAD.0b013e3281e38558. [DOI] [PubMed] [Google Scholar]
- Manning A, Russell V, Eastick K, Leadbetter GH, Hallam N, Templeton K, Simmonds P. Epidemiological profile and clinical associations of human bocavirus and other human parvoviruses. J. Infect. Dis. 2006;194:1283–1290. doi: 10.1086/508219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning A, Willey SJ, Bell JE, Simmonds P. Comparison of tissue distribution, persistence, and molecular epidemiology of parvovirus B19 and novel human parvoviruses PARV4 and human bocavirus. J. Infect. Dis. 2007;195:1345–1352. doi: 10.1086/513280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mingozzi F, High KA. Therapeutic in vivo gene transfer for genetic disease using AAV: progress and challenges. Nat. Rev. Genet. 2011;12:341–355. doi: 10.1038/nrg2988. [DOI] [PubMed] [Google Scholar]
- Momoeda M, Wong S, Kawase M, Young NS, Kajigaya S. A putative nucleoside triphosphate-binding domain in the nonstructural protein of B19 parvovirus is required for cytotoxicity. J. Virol. 1994;68:8443–8446. doi: 10.1128/jvi.68.12.8443-8446.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naeger LK, Schoborg RV, Zhao Q, Tullis GE, Pintel DJ. Nonsense mutations inhibit splicing of MVM RNA in cis when they interrupt the reading frame of either exon of the final spliced product. Genes Dev. 1992;6:1107–1119. doi: 10.1101/gad.6.6.1107. [DOI] [PubMed] [Google Scholar]
- Nuesch JP, Corbau R, Tattersall P, Rommelaere J. Biochemical activities of minute virus of mice nonstructural protein NS1 are modulated In vitro by the phosphorylation state of the polypeptide. J. Virol. 1998;72:8002–8012. doi: 10.1128/jvi.72.10.8002-8012.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozawa K, Ayub J, Hao YS, Kurtzman G, Shimada T, Young N. Novel transcription map for the B19 (human) pathogenic parvovirus. J. Virol. 1987;61:2395–2406. doi: 10.1128/jvi.61.8.2395-2406.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pintel D, Dadachanji D, Astell CR, Ward DC. The genome of minute virus of mice, an autonomous parvovirus, encodes two overlapping transcription units. Nucleic Acids Res. 1983;11:1019–1038. doi: 10.1093/nar/11.4.1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu J, Cheng F, Burger LR, Pintel D. The transcription profile of Aleutian Mink Disease Virus (AMDV) in CRFK cells is generated by alternative processing of pre-mRNAs produced from a single promoter. J. Virol. 2006a;80:654–662. doi: 10.1128/JVI.80.2.654-662.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu J, Cheng F, Johnson FB, Pintel D. The transcription profile of the bocavirus bovine parvovirus is unlike those of previously characterized parvoviruses. J. Virol. 2007a;81:12080–12085. doi: 10.1128/JVI.00815-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu J, Cheng F, Pintel D. The abundant R2 mRNA generated by aleutian mink disease parvovirus is tricistronic, encoding NS2, VP1, and VP2. J. Virol. 2007b;81:6993–7000. doi: 10.1128/JVI.00244-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu J, Handa A, Kirby M, Brown KE. The interaction of heparin sulfate and adeno- associated virus 2. Virology. 2000;269:137–147. doi: 10.1006/viro.2000.0205. [DOI] [PubMed] [Google Scholar]
- Qiu J, Nayak R, Tullis GE, Pintel DJ. Characterization of the transcription profile of adeno-associated virus type 5 reveals a number of unique features compared to previously characterized adeno-associated viruses. J. Virol. 2002;76:12435–12447. doi: 10.1128/JVI.76.24.12435-12447.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu J, Yoto Y, Tullis GE, Pintel D. Parvovirus RNA processing strategies. In: Kerr JR, Cotmore SF, Bloom ME, Linden ME, Parish CR, editors. Parvoviruses. Hodder Arnold; London, UK: 2006b. pp. 253–274. [Google Scholar]
- Rosenfeld SJ, Yoshimoto K, Kajigaya S, Anderson S, Young NS, Field A, Warrener P, Bansal G, Collett MS. Unique region of the minor capsid protein of human parvovirus B19 is exposed on the virion surface. J. Clin. Invest. 1992;89:2023–2029. doi: 10.1172/JCI115812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider B, Fryer JF, Oldenburg J, Brackmann HH, Baylis SA, Eis-Hubinger AM. Frequency of contamination of coagulation factor concentrates with novel human parvovirus PARV4. Haemophilia. 2008a;14:978–986. doi: 10.1111/j.1365-2516.2008.01800.x. [DOI] [PubMed] [Google Scholar]
- Schneider B, Fryer JF, Reber U, Fischer HP, Tolba RH, Baylis SA, Eis-Hubinger AM. Persistence of novel human parvovirus PARV4 in liver tissue of adults. J. Med. Virol. 2008b;80:345–351. doi: 10.1002/jmv.21069. [DOI] [PubMed] [Google Scholar]
- Schoborg RV, Pintel DJ. Accumulation of MVM gene products is differentially regulated by transcription initiation, RNA processing and protein stability. Virology. 1991;181:22–34. doi: 10.1016/0042-6822(91)90466-o. [DOI] [PubMed] [Google Scholar]
- Sharp CP, LeBreton M, Kantola K, Nana A, Diffo JD, Djoko CF, Tamoufe U, Kiyang JA, Babila TG, Ngole EM, Pybus OG, Delwart E, Delaporte E, Peeters M, Soderlund-Venermo M, Hedman K, Wolfe ND, Simmonds P. Widespread infection with homologues of human parvoviruses B19, PARV4, and human bocavirus of chimpanzees and gorillas in the wild. J. Virol. 2010;84:10289–10296. doi: 10.1128/JVI.01304-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons R, Sharp C, Sims S, Kloverpris H, Goulder P, Simmonds P, Bowness P, Klenerman P. High frequency, sustained T cell responses to PARV4 suggest viral persistence in vivo. J. Infect. Dis. 2011;203:1378–1387. doi: 10.1093/infdis/jir036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Chen AY, Cheng F, Guan W, Johnson FB, Qiu J. Molecular characterization of infectious clones of the minute virus of canines reveals unique features of bocaviruses. J. Virol. 2009;83:3956–3967. doi: 10.1128/JVI.02569-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tattersall P. The evolution of parvovirus taxonomy. In: Kerr J, Cotmore SF, Bloom ME, Linden RM, Parrish CR, editors. Parvoviruses. Hodder Arond; London: 2006. pp. 5–14. [Google Scholar]
- Tijssen P, Agbandje-McKenna M, Almendral JM, Bergoin M, Flegel TW, Hedman K, Kleinschmidt JA, Li D, Pintel D, Tattersall P. Parvoviridae. In: King MQ, Adams MJ, Carstens E, Lefkowitz EJ, editors. Virus taxonomy: classification and nomenclature of viruses: Ninth Report of the International Committee on Taxonomy of Viruses. Elsevier; San Diego: 2011. [Google Scholar]
- Tsao J, Chapman MS, Agbandje M, Keller W, Smith K, Wu H, Luo M, Smith TJ, Rossmann MG, Compans RW. The three-dimensional structure of canine parvovirus and its functional implications. Science. 1991;251:1456–1464. doi: 10.1126/science.2006420. [DOI] [PubMed] [Google Scholar]
- Walker SL, Wonderling RS, Owens RA. Mutational analysis of the adeno-associated virus type 2 Rep68 protein helicase motifs. J. Virol. 1997;71:6996–7004. doi: 10.1128/jvi.71.9.6996-7004.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Z, Zhi N, Wong S, Keyvanfar K, Liu D, Raghavachari N, Munson PJ, Su S, Malide D, Kajigaya S, Young NS. Human parvovirus B19 causes cell cycle arrest of human erythroid progenitors via deregulation of the E2F family of transcription factors. J. Clin. Invest. 2010;120:3530–3544. doi: 10.1172/JCI41805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young NS, Brown KE. Parvovirus B19. N. Engl. J. Med. 2004;350:586–597. doi: 10.1056/NEJMra030840. [DOI] [PubMed] [Google Scholar]
- Zadori Z, Szelei J, Lacoste MC, Li Y, Gariepy S, Raymond P, Allaire M, Nabi IR, Tijssen P. A viral phospholipase A2 is required for parvovirus infectivity. Dev. Cell. 2001;1:291–302. doi: 10.1016/s1534-5807(01)00031-4. [DOI] [PubMed] [Google Scholar]