Abstract
Purpose
We have previously shown that within tumors, recombinant interleukin-2 (rIL-2, Aldesleukin) consistently activates tumor-associated macrophages and up-regulates interferon stimulated genes (ISGs) while inducing minimal migration, activation or proliferation of T-cells. These effects are independent of tumor response to treatment. Here, we prospectively evaluated transcriptional alterations induced by rIL-2 in peripheral blood mononuclear cells (PBMCs) and within melanoma metastases.
Experimental Design
We evaluated gene expression changes by serially comparing pre- to post-treatment samples in 14 patients and also compared transcriptional differences among lesions displaying different responsiveness to therapy, focusing on 2 lesions decreasing in size and 2 remaining stable (responding lesions) compared to non-responding ones.
Results
As previously described, the effects of rIL-2 were dramatic within PBMC, while effects within the tumor microenvironment were lesion-specific and limited. However, distinct signatures specific to response could be observed in responding lesions pre-treatment that were amplified following rIL-2 administration. These signatures match the functional profile observed in other human or experimental models in which immune-mediated tissue-specific destruction (TSD) occurs underlying a common pathways leading to rejection. Moreover, the signatures observed in pre-treatment lesions were qualitatively similar to those associated with TSD underlining a determinism to immune responsiveness that depends upon the genetic background of the host or the intrinsic genetic makeup of individual tumors.
Conclusions
This is the first prospectively collected insight on global transcriptional events occurring during high-dose rIL-2 therapy in melanoma metastases responding to treatment.
Background
High-dose recombinant interleukin-2 (rIL-2, aldesleukin, Proleukin, rIL-2) was approved by the U.S. Food and Drug Administration in 1993 for the treatment of metastatic renal cell carcinoma and later for metastatic melanoma. Approval was based upon evidence of objective anticancer response in 15% and durable complete remissions in 7% of patients (1, 2). However, these results are achieved at a significant price in patient toxicity since rIL-2 induces a “capillary leak syndrome” with numerous hemodynamic consequences including fluid retention, acute renal insufficiency, and hypotension. For these reasons, rIL-2 has not been widely adopted and is delivered only at medical centers experienced in the management of its toxicities. Better understanding of the mechanisms of action of systemically administered of rIL-2 relevant to successful therapy could help in patient selection and/or in the strategy for administration of the drug by enhancing its therapeutic window.
The identification of parameters predictive of response remains elusive. Clinical parameters are only modestly predictive of the likelihood of response and most often are related to post-treatment observations such as development of vitiligo or alterations in blood counts that are of no use for predictive purposes (3). The expression of carbonic anhydrase 9 may predict response to rIL-2 therapy in patients with renal cell cancer (4). However, carbonic anhydrase 9 is not expressed by melanomas nor is a predictor of response (5). Using a limited array platform, we observed that response of melanoma metastases to treatment with antigen-specific vaccination plus the systemic administration of r-IL2 was associated with the expression in pre-treatment metastases of genes related to chronic inflammation (6). Similar finding were reported by others in the context of r-IL-2 therapy (7) or antigen-specific immunization (8, 9), suggesting that responsiveness is in part dependent upon the genetic predisposition of tumors to be susceptible to immune manipulation. Interestingly, the same signatures associated with responsiveness to immunotherapy are observed in other cancers including colon and breast carcinoma and are predictive of good prognosis independent of treatment (10–13). Moreover, similar signatures mark a broader aspect of immune biology which we recently defined as the immunologic constant of rejection (ICR), as they are observed in various types of immune-mediated tissue-specific destruction (TSD) including tumor rejection in response to immunotherapy, allograft rejection, graft versus host disease, flares of autoimmunity and clearance of pathogen during acute infection (14, 15).
We previously described how minimally invasive biopsies such as fine needle aspiration biopsy (FNA) consistently draw cells from tumor sites providing sufficient material to permit high fidelity gene expression analysis while allowing serial sampling of the same lesion (16, 17). Because FNA can be obtained immediately without ischemia or surgical trauma, the findings represent the tumor microenvironment more accurately than specimens harvested surgically. Moreover, serial sampling of the same lesion allows direct comparison of biological findings with the natural history of the same lesion, obviating correction for lesion-specific variability within a given patient (16). Although transcriptional changes due to the tissue disruption caused by the FNA can be expected, these are predictable and can be accounted for during the interpretation of the results (18–20). Thus, in this pilot study, we set out to compare, using an enhanced whole transcriptome platform, the systemic effects of high-dose r-IL-2 to its effects within the tumor tissue. PBMC and FNA biopsies from melanoma metastases were collected prior to and during rIL-2 administration.
Methods
Patients
Patients at least 18 years of age with cytologically- or histologically-confirmed metastatic malignant melanoma and with one or more metastases amenable to FNA were candidates for this study. Patients were required to have sufficient health predictive of tolerance of high-dose rIL-2 and an ECOG performance status of 0 or 1 (Table 1). Within 2 weeks prior to treatment, patients were required to have an hematocrit ≥ 25%, platelet count ≥ 50,000/cubic mm, prothrombin time INR ≤ 1.5 and to have the ability and willingness to provide written informed consent approved by our institutional review board (University of Virginia). Exclusion criteria included pregnancy, ischemic heart disease (negative stress test required), brain metastasis, or requirement for corticosteroids.
Table 1.
Patient Characteristics and Response to Therapy
| Patient # | Sex | Age | Tumor Site | Prior Therapy | Best Response |
|---|---|---|---|---|---|
| 1 | Male | 47 | ST, LN | S, L | PD |
| 2 | Female | 50 | ST,LV,LG,BO | S, L, R | PD |
| 3 | Female | 35 | ST,LN,LV,LG | S | PR |
| 4 | Male | 48 | ST,LG | S, R | PD |
| 5 | Male | 54 | LN | S | SD |
| 6 | Male | 64 | ST | S | CR |
| 7 | Female | 49 | ST,LG | S | PD |
| 8 | Female | 70 | ST,LN,LV | S, L, C | PD |
| 9 | Male | 66 | ST,LG | S, L | PD |
| 10 | Male | 57 | ST,LG | S, L | PD |
| 11 | Male | 68 | ST,LG,LV | S | SD |
| 12 | Female | 63 | ST,LG,BO | S, C, V | PD |
| 13 | Male | 45 | ST | S,R,V | PD |
| 14 | Female | 49 | ST,BO | None | PD |
ST=soft tissue, LN=lymph node, LG=lung, LV=liver, BO=bone, S=Excision, L=lymph node dissection, R=radiotherapy, C=chemotherapy, V=vaccine, CR=complete response, PR=partial response, SD=stable disease, PD=progressive disease. In bold are the lesions that were biopsied in patients who responded to treatment or remained stable.
Interleukin-2 administration
Patients were hospitalized in a monitored hematology-oncology unit at the University of Virginia Medical Center and received 600,000 International Units/kg (0.037 mg/kg) of rIL-2 (Proleukin,® Prometheus Laboratories, San Diego, CA) every 8 hours by a 15-minute intravenous infusion for a maximum of 14 doses. Following 9 days of rest, the schedule was repeated for up to 14 doses, for a maximum of 28 doses per course, as tolerated. During administration of therapy, doses were frequently withheld for severe toxicity. Patients were medicated with cephalexin, famotidine, ibuprofen and acetaminophen on a scheduled basis to reduce the risk of infection and cytokine-release symptoms.
Response criteria
The minimal period for defining stable disease was 1 month. Following treatment with high-dose aldesleukin, serial surveillance CT scans were obtained 1 month and 2 months after completion of therapy. Response and stable disease assignment was based on RECIST criteria.
Blood sampling
Blood specimens were collected in CPT tubes containing sodium heparin, gel and density gradient media (8 ml) (BD bioscience) pre-therapy and 3 hours after the sixth dose of rIL-2 to separate PBMC for RNA preparation. PBMC were pelleted by centrifugation, lysed in 700 µl of Trizol reagent (Invitrogen) and stored at −80 till use.
Tumor biopsies
FNA of melanoma metastases were obtained pre-therapy and 3 hours after the sixth dose of rIL-2, as this was judged to be the most informative time point based on previous time-course studies, timing of blood sampling was secondarily chosen to directly compare differences between systemic and tumor effects (6, 17, 18). A 23 Gauge hypodermic needle was inserted into the tumor. After insertion, multiple passes at changing angles of insertion were made through all quadrants of the tumor. The serial sampling method had been previously validated for its intra-lesion, intra-patient and inter-patient reproducibility by several studies (6, 18); gene signatures are predominantly patient-specific independent of tissue in which the metastatic lesion is hosted. Serial sampling of the same lesion within a short time frame such as the one adopted in the study cause non treatment-specific alterations that are, however, limited and predictable (18, 20). Gene signatures portray a cumulative representation of functional interactions between cancer and host cells and are not a mere reflection of the number of immune cells in the sample (6).
Tissue collected into the needle was suspended in 10 ml cold RPMI and centrifuged at 1500 rpm for 5 minutes at 4°C. The cell pellet was harvested and red blood cells were eliminated with ACK lysis buffer. Cells were resuspended in 1 ml cold RPMI, transferred to a 1.5 ml Eppendorf tube, spun at 1500 rpm for 5 minutes and the supernatant was discarded. The cell pellet was resuspended in 500 µl of Trizol (Gibco BRL) and mixed thoroughly in the lysis buffer to ensure lysis of all cells. Lysates were frozen at −80°C until use.
Target preparation and hybridization to microarrays
Total RNA was processed by a two-cycle amplification as described elsewhere (17). Patient samples were labeled with Cy5 and mixed with Cy3 labeled reference consisting of pooled PBMC from 5 normal donors obtained from aphereses and hybridized to in house printed whole-genome human 36K oligo arrays, representing 25,100 unique human genes (Operon Human Genome Array-Ready Oligo Set version 4.0).
Sample Size considerations
This is an exploratory analysis, where sample size was based upon historical targets goals and no statistical precision. Based upon prior experience, it was estimated that 10 responders would be needed to assess the biomarkers in the melanoma population. Assuming subcutaneous metastases will represent the bulk of the melanoma patients where response rates tend to be around 20%, tissue samples from at least 50 melanoma patients and blood samples from at least 50 melanoma patients are required. As the accrual has progressed slower than anticipated the interim analysis (scheduled after the 25th patient accrual) was anticipated after the first 14 cases to test whether the study should proceed. This interim analysis represents a first step to assess the validity of this approach and support its continuation; however, due to the critical relevance of this approach in modeling immunotherapy monitoring of upcoming trials including those supported by the Cancer Immunotherapy Trial Network, it was deemed useful to publish these preliminary observations as a proof of concept.
Statistical Analysis
Statistical analyses were all performed using the NCI BRB-Array tool (21). Class comparison was based on paired or unpaired t test adopting various stringency cutoff criteria for gene identification or gene enrichment. Multivariate permutation test was applied to test whether the number of genes identified by each class comparison could be due to chance. Unsupervised sample differences were visualized as multidimensional scale (MDS) plots based on the complete data set. Functional interpretation was based on Ingenuity Pathway Analysis (IPA) Software (Ingenuity® Systems, Redwood City, CA). IPA computes a score to build functional networks according to the fit of the user’s set of significant genes. The score is derived from a p-value indicating the likelihood that genes in a network are together due to random chance. A score of ≥ 2 (the log10 value) indicates that there is a 1 in 100 or less probability that the genes are together due to random chance. Biological functions are then calculated and assigned to each network.
The primary scope of class comparison was to identify genes differentially expressed according to univariate comparison (though not always supported by multivariate analysis) that could be used for functional interpretation; the significance of the findings resulted from the non-random enrichment of immune pathways as described in the Results.
Results
This is a prospective study of the mechanism(s) of action of systemically administered rIL-2. Although the study has been open for approximately 5 years, accrual has been limited by the requirement for tissue biopsies. Fourteen patients with metastatic malignant melanoma have been enrolled so far. Patient characteristics are listed in Table 1. Most patients received rIL-2 as the first systemic therapy for metastatic disease. Three patients received either chemotherapy or multi-peptide vaccine therapy prior to rIL-2. The best antitumor responses (RECIST criteria) observed among this group included 1 complete response (CR), 1 partial response (PR) and 2 stable diseases (SD).
Overall effects of r-IL2 therapy
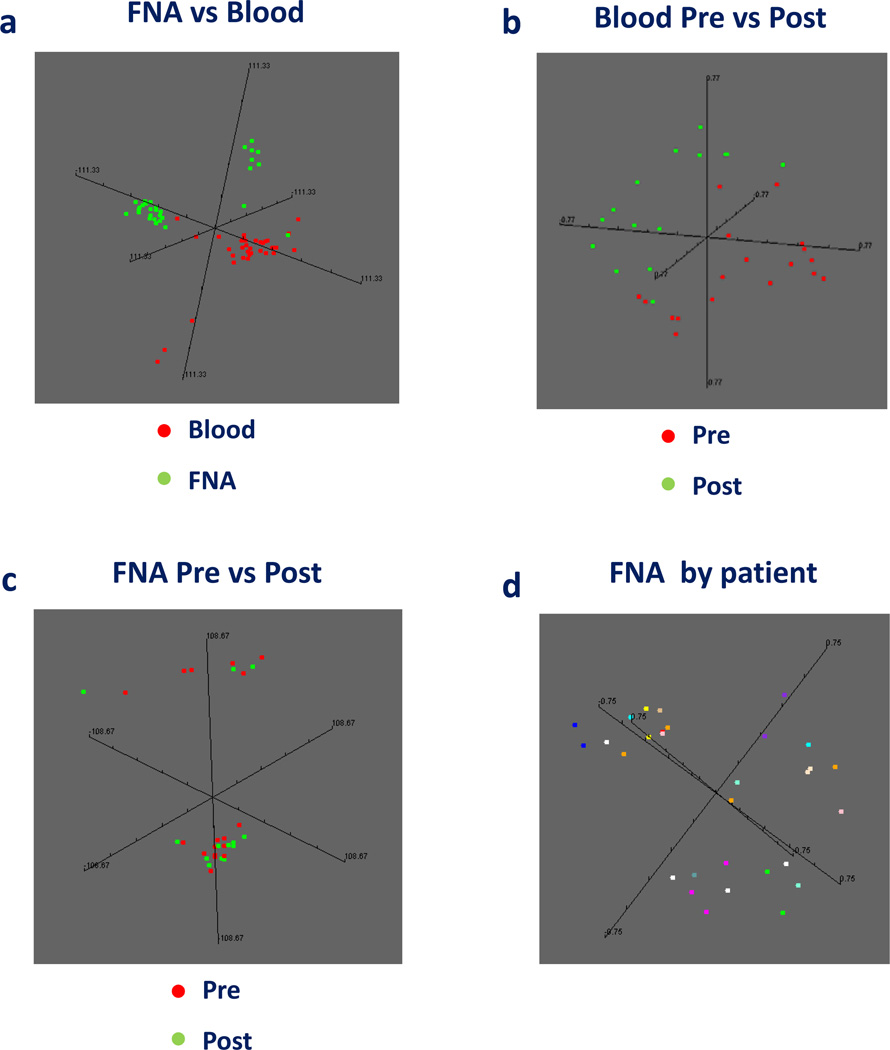
As expected, there were broad differences between the transcriptional profile of PBMC and FNA independent of treatment (Figure 1a). When PBMC were compared separately (Figure 1b), there was a dramatic separation between pre- and post-rIL-2 administration. The same separation was not identified between pre- and post-rIL-2 FNA samples (Figure 1c) confirming the previously noted limited effects of rIL-2 therapy within the tumor microenvironment compared to its systemic effects (18). In fact, for most patients, tumor expression profiles from autologous lesions pre- and post-IL-2 tended to group together independent of treatment (Figure 1d) demonstrating that the intrinsic genetic makeup of individual tumors overrides the effect of rIL-2. Moreover, no differences were noted among tumors from patients who received different previous treatments.
Figure 1.
Multi dimensional scaling (MDS) based on the complete data set comparing all samples (a), pre-versus during-rIL-2 PBMC (b) or FNA (c) samples. FNA sample distribution is also shown as MDS according to patient derivation for each sample (d).
Systemic effects of rIL-2 on PBMCs
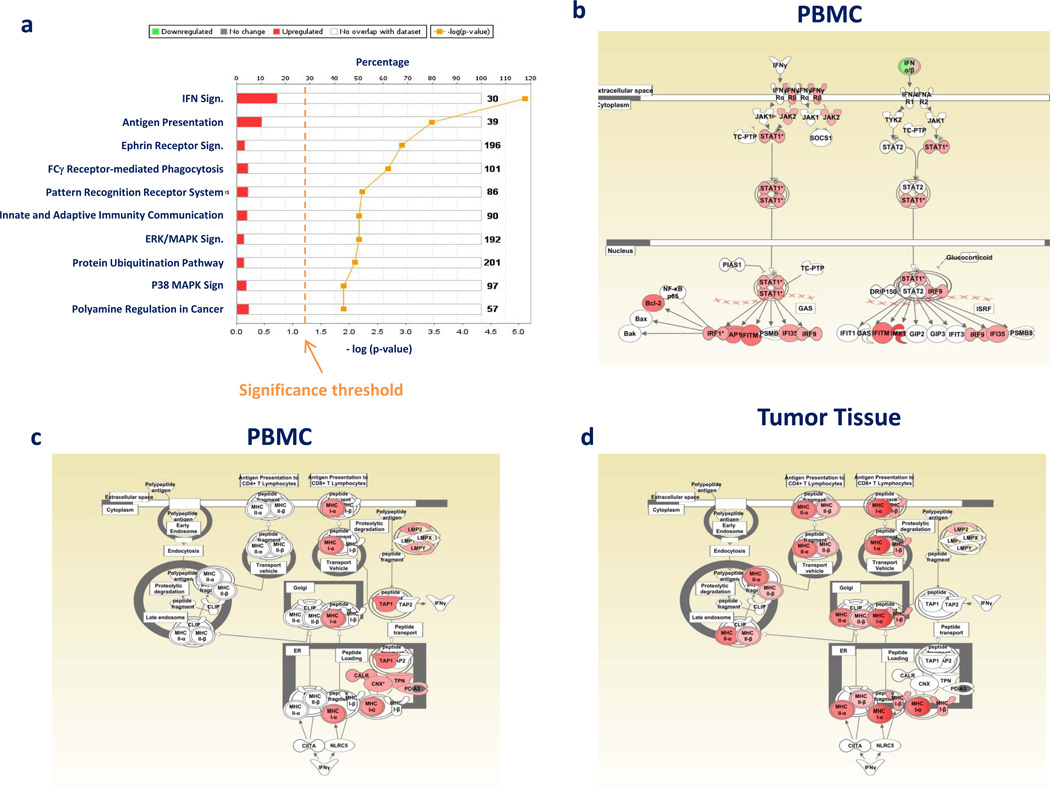
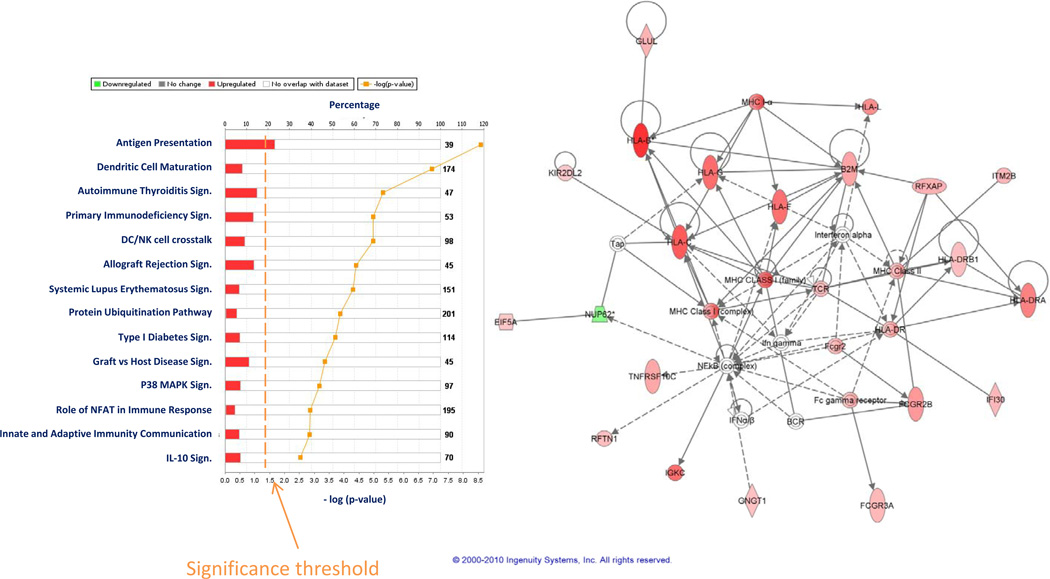
The effect of rIL-2 on gene expression in PBMC was measured by comparing blood drawn after the 6th dose of IL2 to pre-therapy, with a paired t test setting as a threshold a two tailed p2-value of < 0.001. This analysis identified 211 genes highly differentially expressed (permutation t test p-value < 0.001). The functional interpretation of the identified treatment-induced genes was assessed by IPA. The top biological processes affected by the treatment included innate and adaptive immunity. These findings resonated with a previous analysis performed on T cell subsets in vitro (22). Similarly, the ten top canonical pathways affected by IL-2 treatment included increases in interferon (IFN) and antigen presentation signaling (Figure 2a). To enrich for genes of potential relevance, a second less stringent analysis was performed using as a cutoff p2-value < 0.05. This analysis identified 1,257 genes (permutation analysis p-value = 0.04) that were then applied for IPA. Similarly, the top pathways included immune mechanisms such as IFN signaling (Figure 2b and antigen presentation (Figure 2c). These activated pathways are of particular relevance since they are a primary component of the ICR and their expression is a prerequisite for TSD. In fact, as later shown by studying tumor tissue, antigen presentation was strongly and specifically enhanced also in responding and stable lesions compared with non-responding ones (Figure 2d, see later). Comparison of the transcriptional profile of pre-treatment PBMC between responding and non-responding patients did not identify any candidate predictor of immune responsiveness suggesting that PBMC from different patients have similar potential to eradicate tumors while responsiveness to rIL-2 therapy is likely dependent upon the genetic makeup of tumors.
Figure 2.
Ingenuity pathway analysis (IPA) representation of the top most significantly enriched canonical pathways among approximately 400 included in the software that were activated in PBMC by rIL-2 administration (a) based on 200 genes differentially expressed between pre- and post-rIL-2 PBMC according to an unpaired t test (cut off p2-value < 0.001). IFN signaling (b) and antigen presentation (c) enrichment analysis based on 1,257 genes differentially expressed at a cutoff p2value <0.05 (unpaired Student t test) between pre- and post treatment PBMC; antigen presentation enrichment analysis based on genes differentially expressed between responding and stable disease metastases compared with non responding ones at the same cutoff level (d).
Effect of rIL-2 therapy on melanoma metastases
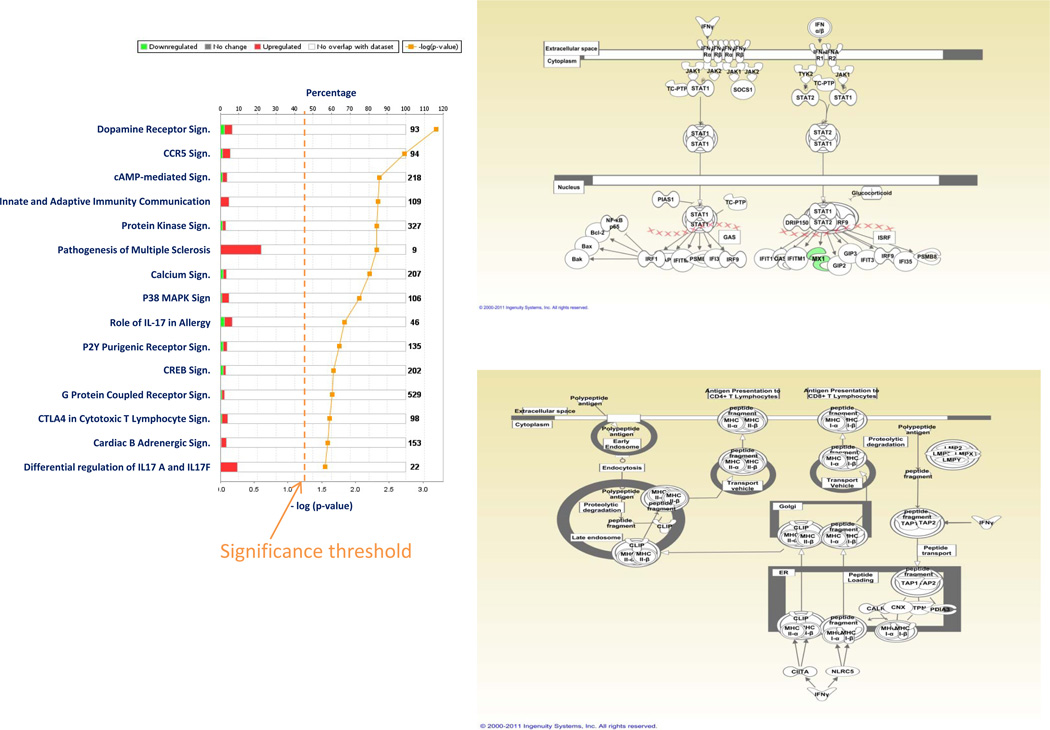
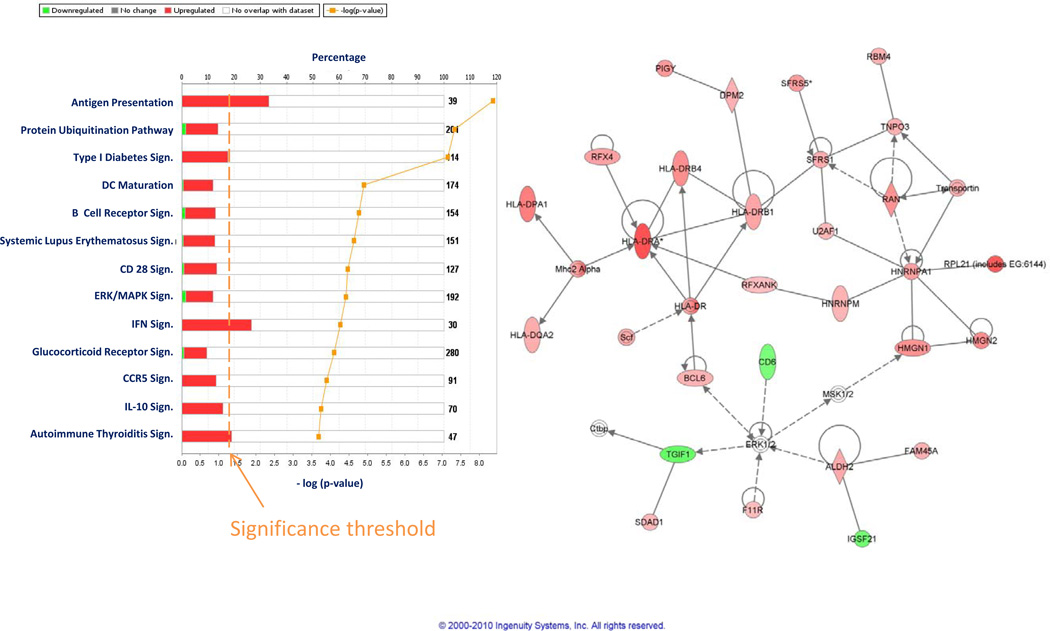
As previously observed (18), the effects of rIL-2 therapy at the tumor site were minimal compared to those observed in PBMC; a paired analysis of all pre- vs post- treatment biopsies independent of clinical outcome did not identify any gene differentially expressed at the stringent cutoff p2-value of < 0.001 and only 350 genes differentially expressed at the cutoff p2 value of < 0.05 (permutation test non significant). Of the 350 transcripts, 325 had functional annotations and were analyzed by IPA. This analysis demonstrated activation of immunologic pathways related to macrophage function confirming previous observations (18). Moreover, we identified pathways associated with IL-17 signaling that were probably missed by the previous analyses due to an incomplete representation of the transcriptome by those earlier platforms (Figure 3a). IFN signaling (Figure 3b) and antigen presentation (Figure 3c) strongly activated in PBMC at the same time points were not represented within the tumor microenvironment. This data demonstrates that the effects of the systemic administration of rIL-2 are very limited within the tumor microenvironment in most circumstances and this may explain in quantitative terms the lack of responsiveness of most tumors to this therapy.
Figure 3.
IPA based on 325 genes with functional annotations differentially expressed (t test p2-value cutoff < 0.05) between pre- and post-rIL-2 administration FNA samples independent of treatment outcome (a). IFN signaling (b) and antigen presentation (c) pathways based on the same gene set (compare to Figure 2 b and c)
Analysis of Immune responsiveness
Pre-treatment samples
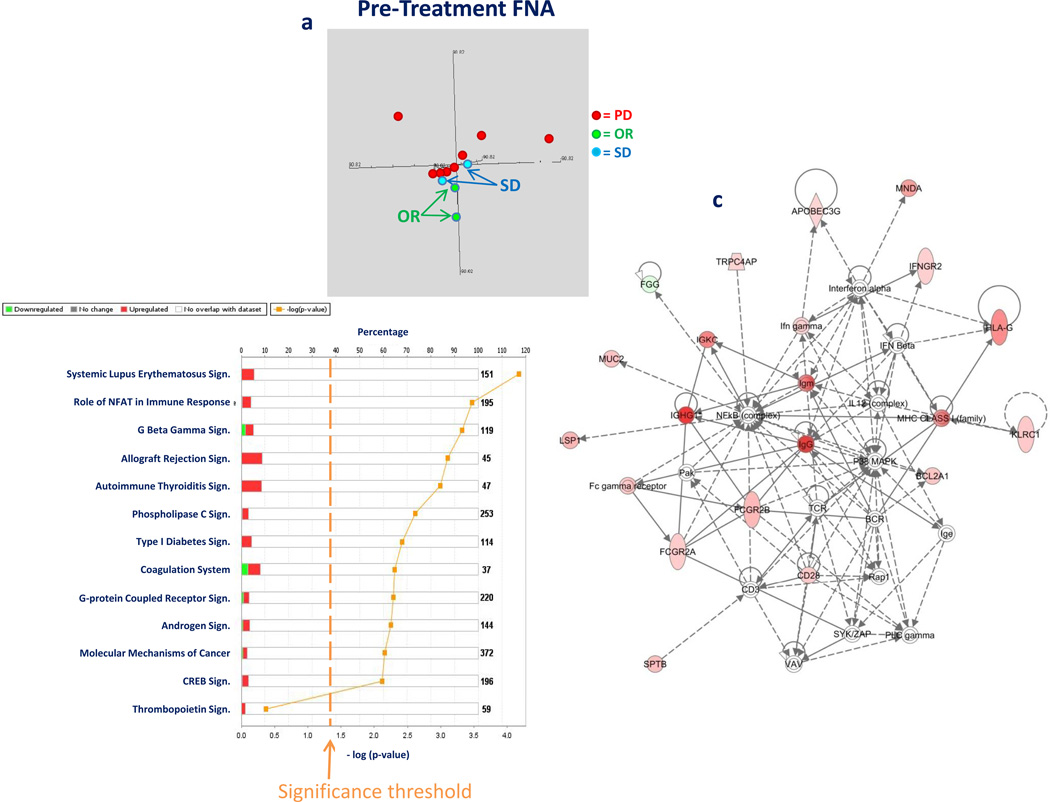
FNA were compared between patients with clinical response (CR+PR) or SD and those with tumor progression. MDS of pre-treatment lesions based on the complete data set suggested that lesions destined to respond to rIL-2 therapy segregate separately from those that are not, with tumors from two patients with SD positioning in between (Figure 4a). Transcriptional patterns from CR, PR and SD were compared to PD based on a non-paired t test using low stringency criteria (p2 value < 0.05), which identified 262 genes; the robustness of this finding was not supported by permutation analysis. Nevertheless, the data set was applied to IPA to test whether response-specific pathways could be identified. The results were striking: ranking of the top canonical pathways identified 12 significantly (Fisher test) involved in determining the differences between the two phenotypes (Figure 4b); among them, the top 6 were associated with TSD (14) and good prognostic significance in various cancers (10, 12, 13, 23). The enrichment in top ranking immune-related pathways was highly significant by Fisher test (p2-value = 0) and could not be attributed to random chance. In fact, the top functional network identified by IPA (Figure 4c) was clearly associated with immune effector activation, centered on IFN-γ signaling, and including enhanced expression of Human Leukocyte Antigen (HLA) molecules. Furthermore, B cell signatures (i.e. IgG mRNA expression) were detected which are increasingly recognized as harbingers of better prognosis and markers of rejection (24).
Figure 4.
MDS based on the complete gene data set (a) comparing pre treatment transcriptional patterns of lesions from patients who subsequently responded (CR and PR referred as overall response = OR), experienced stable (SD) or progression of disease (PD). IPA canonical pathway analysis comparing transcriptional patterns cumulative CR, PR and SD with PD in pre-treatment lesions (unpaired t test cutoff p2-value < 0.05, 262 genes identified); in red is the proportion of genes up regulated in responding lesions compared with non-responding ones (b). First self-organizing IPA network based on the same data set (in red are genes up regulated in responding lesions, intensity of color is proportional to the value of the ratio of expression between the two phenotypes (c)
Post Treatment Samples
Transcriptional differences following rIL-2 administration between congregated CR, PR and SD lesions compared with PD lesions were strikingly consistent with those observed pre-treatment. However they were magnified, suggesting that rejection of tumors, when it occurs, represents an enhancement of a pre-existing biology. Applying the same statistical criteria used for pre-treatment lesions, 393 genes were differentially expressed between the two phenotypes. Although the permutation test was not significant, IPA demonstrated that the largest majority was associated with effector immune responses associated with TSD. Ten out of ten top ranking canonical pathways (Figure 5a) were associated with immune rejection (Fisher p2 value = 0) confirming that tumor rejection follows pathways common to other forms of rejection (14). Interestingly, the self-organizing network descriptive of immune activation during therapy (Figure 5b) strongly reflected patterns seen in the pre-treatment network (Figure 4c). However, a striking broadening of the repertoire of HLA molecules was observed including HLA class II strongly dependent upon IFN-γ activation and more specifically associated with rejection (14).
Figure 5.
IPA canonical pathway analysis comparing cumulative OR, PR and SD differences compared to PD transcriptional patterns in post-treatment lesions (unpaired t test with cutoff p2-value < 0.05, 393 genes identified); in red is the proportion of genes up regulated in responding lesions compared with non responding ones (a). First organizing IPA network based on the same data set (in red are genes up regulated in responding lesions, intensity of color is proportional to the value of the ratio of expression between the two phenotypes, b).
Cumulative Differences between CR, PR and SD compared to PD lesions before and during rIL-2 administration
We then compared CR, PR and two SD with PD lesions independent of time point to identify the most consistent patterns associated with better responsiveness to rIL-2 therapy. This analysis identified 1,267 genes differentially expressed (permutation test p-value < 0.001). IPA (Figure 6a) demonstrated highly significant enrichment of pathways associated with TSD and the top functional network (Figure 6b) identified HLA class II activation as a key component. Overall, the large majority of pathways and transcripts identified by this study reflected previous analyses, strongly supporting the hypothesis (14, 15) that immune rejection represents a continuum from a chronic inflammatory process insufficient to eliminate target tissues to an acute one under the orchestration of IFN-γ.
Figure 6.
IPA canonical pathway analysis comparing cumulative OR, PR and SD differences compared to PD transcriptional patterns in all lesions independent of treatment (unpaired t test with cutoff p2-value < 0.05, 393 genes identified); in red is the proportion of genes up regulated in responding lesions compared with non responding ones (a). First organizing IPA network based on the same data set (in red are genes up regulated in responding lesions, intensity of color is proportional to the value of the ratio of expression between the two phenotypes, b).
Discussion
It has been shown that IFN-γ is required for tumor immune surveillance as mice impaired in the secretion of this cytokine are prone to develop spontaneous tumors (25). More recently, it was observed that an IFN-γ dependent signature and a Th1 immune environment are associated with better prognosis in cancer (10, 12, 13, 23, 26, 27). For instance, good prognosis of ovarian cancer is associated with enhanced Th1 infiltrates and expression of IFN-γ, IL-2 and HLA-class I molecules (27, 28). Similarly, a coordinated cytotoxic Th1 immune phenotype expressing the transcription factor T-box protein 21 (T-bet), interferon regulatory factor 1 (IRF-1) and IFN-γ is associated with good prognosis in colorectal cancer (10, 23). A similar phenomenon was recently reported by our group in breast cancer (13). Interestingly, similar signatures associated with activated IFN-γ signaling are predictive of immune responsiveness to vaccine therapy. Gajewski et al (8, 29) observed that pre-treatment melanoma metastases likely to respond to active specific immunization express interferon stimulated genes. Similar findings were observed by GSK-Biologics in non small cell lung cancer and melanoma patients undergoing vaccination with MAGE-A3 protein (9, 30). A more broadly defined inflammatory phenotype also characterizes lesions likely to respond to rIL-2 (7), anti-CTLA-4 mAb (31) or IFN-α therapy (32).
Applying serial sampling of the same lesion, we reported a decade ago that melanoma metastases likely to respond to systemic rIL-2 administration displayed a pre-existing status of immune activation (6). Besides limitations in the array platform utilized at that time, the study was hampered by the heterogeneity of the treatment received by the patients who underwent conceptually similar yet different active-specific vaccination protocols in combination with rIL-2. Moreover, we previously analyzed the transcriptional patterns of tumors undergoing immunotherapy such as melanoma metastases in 6 patients treated with high dose systemic rIL-2 administration (18) or basal cell carcinomas treated topically with the Toll-like receptor 7 agonist Imiquimod (19). In addition, we evaluated in a mouse model the immunologic effects of viral oncolytic therapy that leads to tumor elimination through activation of effector innate immune responses (33). Comparison of these and others’ studies, led to the identification of a convergent pathway leading to TSD that was independent of animal species, disease model and treatment applied. This pathway includes the expression of a group of genes that we defined as the “immunologic constant of rejection” (ICR). Interestingly, the ICR signature qualitatively resembles patterns predictive of good prognosis or immune responsiveness, leading to the hypothesis that immune manipulation of the cancer-bearing host with the purpose of inducing tumor rejection leads to an exaggeration of a natural susceptibility to immune surveillance (15).
This study was, therefore, designed to validate previous attempts to identify predictors of immune responsiveness. Moreover, the analysis was extended to the study of lesions during treatment to test whether changes specific to tumor rejection represented an enhancement in a continuum scale of baseline conditions. It should be emphasized that although this study was proposed a decade ago (16, 34) it took extensive planning confronting conceptual, ethical and regulatory hurdles to be approved. Although the planned accrual is larger than what achieved so far, an interim analysis presented sufficient novel information in line with previous hypotheses.
This study confirmed that the effects of systemically administered rIL-2 are exponentially higher in the peripheral circulation compared to the tumor site as postulated in a previous study of 6 patients receiving rIL-2 (18). A time course parallel analysis of simultaneously collected PBMC and FNA samples demonstrated that the most informative time point to study the effects of rIL-2 was three hours after the bolus administration of the drug and the effects were proportionately limited within the tumor site. Moreover, the effects at the tumor site could not be ascribed to a direct activation of immune cells by rIL-2 but rather to an indirect activation of macrophages by soluble factors secreted systemically by circulating IL-2 receptor-bearing cells (35–37).
This study also confirms that immune responsiveness of tumors is pre-determined and the determinant can be identified within the functional genome of individual tumors (12). Remarkably, signatures associated with immune responsiveness are quite similar independent of tumor type and treatment received; the findings observed here are comparable to those observed in melanoma or lung cancer patients receiving active-specific vaccination with tumor antigens (29, 38) (9). Experimental animal models provide a mechanistic interpretation of these findings, suggesting a role for IFN-dependent chemokine release guiding the homing of effector immune cells to the tumor site (39, 40). This study also proposes that the status of immune responsiveness of growing tumors is turned into tumor rejection through an enhancement of the same phenomenology; responding lesions studied during rIL-2 administration were qualitatively similar but quantitatively enhanced confirming what we term the “enhancement theory of tumor immune rejection”. Analysis of lesions post-rIL-2 administration confirmed the ICR theory, identifying activation of canonical pathways that are not restricted to tumor rejection but are shared by other phenomenologies related to TSD including allograft rejection, graft versus host disease, acute clearance of pathogen through elimination of infected cells and flares of autoimmunity, a phenomenon originally postulated by Jonas Salk as the “delayed allergy reaction” (41) which lead to the definition of the ICR theory (42).
We had recently the opportunity to study two rare cases of mixed response to vaccine therapy plus systemic cytokine administration in patients with metastatic melanoma (43). In these 2 patients some metastases decreased in size while others simultaneously grew; these observations eliminate the genetic background of the host or environmental influences as determinants of response since the regression or lack thereof occurred in the same individual at the same time. The mixed response study identified in the responding lesions of both patients’ signatures identical to those observed in responding lesions in the current study.
In summary, this interim report provides insight into the mechanism of action of the systemic administration of rIL-2 relevant to its anti-cancer function. The study suggests that the transcriptional program of tumors is likely responsible for immune responsiveness. This information may lead to better patient stratification. However, because of the small sample size, it would be difficult to justify patient selection in any future immunotherapy study on the basis of this gene signature although the results may justify the inclusion of FNA and gene signature analysis in future studies to confirm the present findings. This approach may have broader implications than the understanding of how melanoma metastases respond to rIL-2 as the same transcriptional patterns are progressively more recognized as biomarkers of good clinical outcome in solid tumors in relation to their natural history or responsiveness to therapy (12). We recognize that the information provided here is limited, however, this preliminary report may galvanize a hesitant research environment currently comfortable with more standard approaches (44).
Statement of Translational Relevance.
This is the first translational attempt to prospectively gain mechanistic insights about the events occurring during an immunotherapy trial (systemic administration of recombinant interleukin-2). This study was designed to address how rIL-2 treatment affects the host at the systemic level and the target tissue. It is the culmination of a decade of work by our groups defining the strategy and addressing its pitfalls. We believe the findings reported here are critical for the promotion of a paradigm shift in clinical research in the field of tumor immunology. They will give credibility to this approach to study the kinetics of tumor behavior during therapy and provide a proof of principle and a guideline for immune monitoring.
Acknowledgements
Funding support - This study was funded by: the University of Virginia Cancer Center Support Grant (NIH/NCI P30 CA44579: Clinical research support, Biorepository and Tissue Research Facility, Flow Cytometry Core, and Biomolecular Core Facility); the UVA General Clinical Research Center (NIH M01 RR00847), and philanthropic support from the Commonwealth Foundation for Cancer Research and Alice and Bill Goodwin, the James and Rebecca Craig Foundation, George S. Suddock, Richard and Sherry Sharp, and the Patients and Friends Research Fund of the University of Virginia Cancer Center. No corporate funding support was provided for this study.
We appreciate the work of Joshua Judge in specimen collection, Alison Gaucher in patient enrollment and study compliance and Gina Petroni for expert technical assistance in the statistical design of the clinical portion of the study.
References
- 1.Atkins MB, Lotze MT, Dutcher JP, Fisher RI, Weiss G, Margolin K, et al. High-dose recombinant interleukin-2 therapy for patients with metastatic melanoma: analysis of 270 patients treated between 1985 and 1993. J Clin Oncol. 1998;17:2105–2116. doi: 10.1200/JCO.1999.17.7.2105. [DOI] [PubMed] [Google Scholar]
- 2.Atkins MB, Regan M, McDermott D. Update on the role of interleukin 2 and other cytokines in the treatment of patients with stage IV renal carcinoma. Clin Cancer Res. 2004;10:6342S–6346S. doi: 10.1158/1078-0432.CCR-040029. [DOI] [PubMed] [Google Scholar]
- 3.Phan GQ, Attia P, Steinberg SM, White DE, Rosenberg SA. Factors associated with response to high-dose interleukin-2 in patients with metastatic melanoma. J Clin Oncol. 2001;19:3477–3482. doi: 10.1200/JCO.2001.19.15.3477. [DOI] [PubMed] [Google Scholar]
- 4.Atkins MB, Regan M, McDermott D, Mier J, Stanbridge E, Youmans A, et al. Carbonic anhydrase IX expression predicts outcome in interleukin-2 therapy of renal cancer. Clin Cancer Res. 2005;11:3714–3721. doi: 10.1158/1078-0432.CCR-04-2019. [DOI] [PubMed] [Google Scholar]
- 5.Panelli MC, Wang E, Marincola FM. The pathway to biomarker discovery: carbonic anhydrase IX and the prediction of immune responsiveness. Clin Cancer Res. 2005;11:3601–3603. doi: 10.1158/1078-0432.CCR-05-0475. [DOI] [PubMed] [Google Scholar]
- 6.Wang E, Miller LD, Ohnmacht GA, Mocellin S, Petersen D, Zhao Y, et al. Prospective molecular profiling of subcutaneous melanoma metastases suggests classifiers of immune responsiveness. Cancer Res. 2001;62:3581–3586. [PMC free article] [PubMed] [Google Scholar]
- 7.Sullivan RJ, Hoshida Y, Brunet J, Tahan S, Aldridge J, Kwab C, et al. A single center experience with high-dose IL-2 treatment of patients with advanced melanoma and pilot investigation of novel gene expression signature as a predictor of response. J Clin Oncol. 2009;27:A9003. [Google Scholar]
- 8.Gajewski TF, Zha Y, Thurner B, Schuler G. Association of gene expression profile in melanoma and survival to a dentritic cell-based vaccine. J Clin Oncol. 2009;27:A9002. [Google Scholar]
- 9.Louahed J, Grusell O, Gaulis S, Coche T, Eggermont AM, Kruit W, et al. Expression of defined genes indentifed by pre-treatment tumor profiling: association with clinical response to GSK MAGE A-3 immunetherapeutic in metastatic melanoma patients. J Clin Oncol. 2008;26:A9045. [Google Scholar]
- 10.Galon J, Costes A, Sanchez-Cabo F, Kirilovsky A, Mlecnik B, Lagorce-Pages C, Tosolini M, et al. Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science. 2006;313:1960–1964. doi: 10.1126/science.1129139. [DOI] [PubMed] [Google Scholar]
- 11.Pages F, Galon J, Dieu-Nosjean MC, Tartour E, Sautes-Fridman C, Fridman WH. Immune infiltration in human tumors, a prognostic factor that should not be ignored. Oncogene. 2009;29:1093–1102. doi: 10.1038/onc.2009.416. [DOI] [PubMed] [Google Scholar]
- 12.Ascierto ML, De Giorgi V, Liu Q, Bedognetti D, Murtas D, Chouchane L, et al. An immunologic portrait of cancer. J Transl Med. 2011;9:146. doi: 10.1186/1479-5876-9-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ascierto ML, Kmieciak M, Idowo MO, Manjili R, Zhao Y, Grimes M', et al. A signature of immune function genes associated with recurrence-free survival in breast cancer patients. Breast Cancer Res Treat. 2011 doi: 10.1007/s10549-011-1470-x. Epub ahead of print:2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang E, Worschech A, Marincola FM. The immunologic constant of rejection. Trends Immunol. 2008;29:256–262. doi: 10.1016/j.it.2008.03.002. [DOI] [PubMed] [Google Scholar]
- 15.Wang E, Marincola FM, editors. Immunologic signatures of rejection. 1st ed. New York, NY: Springer; 2010. [Google Scholar]
- 16.Wang E, Marincola FM. A natural history of melanoma: serial gene expression analysis. Immunol Today. 2000;21:619–623. doi: 10.1016/s0167-5699(00)01724-2. [DOI] [PubMed] [Google Scholar]
- 17.Wang E, Miller L, Ohnmacht GA, Liu E, Marincola FM. High fidelity mRNA amplification for gene profiling using cDNA microarrays. Nature Biotech. 2000;17:457–459. doi: 10.1038/74546. [DOI] [PubMed] [Google Scholar]
- 18.Panelli MC, Wang E, Phan G, Puhlman M, Miller L, Ohnmacht GA, et al. Gene-expression profiling of the response of peripheral blood mononuclear cells and melanoma metastases to systemic IL-2 administration. Genome Biol. 2002;3:R35. doi: 10.1186/gb-2002-3-7-research0035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Panelli MC, Stashower M, Slade HB, Smith K, Norwood C, Abati A, et al. Sequential gene profiling of basal cell carcinomas treated with Imiquimod in a placebo-controlled study defines the requirements for tissue rejection. Genome Biol. 2006;8:R8. doi: 10.1186/gb-2007-8-1-r8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Deonarine K, Panelli MC, Stashower ME, Jin P, Smith K, Slade HB, et al. Gene expression profiling of cutaneous wound healing. J Transl Med. 2007;5:11. doi: 10.1186/1479-5876-5-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Simon R, Lam A, LI MC, Ngan M, Menenzes S, Zhao Y. Analysis of Gene Expression Data Using BRB-Array Tools. Cancer Inform. 2007;3:11–17. [PMC free article] [PubMed] [Google Scholar]
- 22.Jin P, Wang E, Provenzano M, Deola S, Selleri S, Jiaqiang R, Voiculescu S, et al. Molecular signatures induced by interleukin-2 on peripheral blood mononuclear cells and T cell subsets. J Transl Med. 2006;4:26. doi: 10.1186/1479-5876-4-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pages F, Berger A, Camus M, Sanchez-Cabo F, Costes A, Molidor R, et al. Effector memory T cells, early metastasis, and survival in colorectal cancer. N Engl J Med. 2005;353:2654–2666. doi: 10.1056/NEJMoa051424. [DOI] [PubMed] [Google Scholar]
- 24.Sarwal M, Chua MS, Kambham N, Hsieh SC, Satterwhite T, Masek M, et al. Molecular heterogeneity in acute renal allograft rejection identified by DNA microarray profiling. N Engl J Med. 2003;349:125–138. doi: 10.1056/NEJMoa035588. [DOI] [PubMed] [Google Scholar]
- 25.Shankaran V, Ikeda H, Bruce AT, White JM, Swanson PE, Old LJ, et al. a IFN-g and lymphocytes prevent primary tumour development and shape tumour immunogenicity. Nature. 2001;410:1107–1111. doi: 10.1038/35074122. [DOI] [PubMed] [Google Scholar]
- 26.Tosolini M, Kirilovsky A, Mlecnik B, Fredriksen T, Mauger S, Bindea G, et al. Clinical impact of different classes of infiltrating T cytotoxic and helper cells (Th1, th2, treg, th17) in patients with colorectal cancer. Cancer Res. 2011;71:1263–1271. doi: 10.1158/0008-5472.CAN-10-2907. [DOI] [PubMed] [Google Scholar]
- 27.Zhang L, Conejo-Garcia JR, Katsaros D, Gimotty PA, Massobrio M, Regnani G, et al. Intratumoral T cells, recurrence, and survival in epithelial ovarian cancer. N. Engl. J. Med. 2003;348:203–213. doi: 10.1056/NEJMoa020177. 2003. [DOI] [PubMed] [Google Scholar]
- 28.Kooi S, Zhang HZ, Patenia R, Edwards CL, Platsoucas CD, Freedman RS. HLA class I expression on human ovarian carcinoma cells correlates with T-cell infiltration in vivo and T-cell expansion in vitro in low concentrations of recombinant interleukin-2. Cell Immunol. 1996;174:116–128. doi: 10.1006/cimm.1996.0301. [DOI] [PubMed] [Google Scholar]
- 29.Gajewski TF, Fuertes M, Spaapen R, Zheng Y, Kline J. Molecular profiling to identify relevant immune resistance mechanisms in the tumor microenvironment. Curr Opin Immunol. 2011;23:286–292. doi: 10.1016/j.coi.2010.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brichard VG, Lejeune D. GSK's antigen-specific cancer immunotherapy programme: pilot results leading to Phase III clinical development. Vaccine. 2007;25 Suppl 2:B61–B71. doi: 10.1016/j.vaccine.2007.06.038. [DOI] [PubMed] [Google Scholar]
- 31.Hamid O, Chasalow SD, Tsuchihashi Z, Alaparthy S, Galbraith S, Berman D. Association of baseline and on-study tumor biopsy markers with clinical activity in patients with advanced melanoma treated with ipilimumab. J Clin Oncol. 2009;27:A9008. [Google Scholar]
- 32.Moschos SJ, Edington HD, Land SR, Rao UN, Jukic D, Shipe-Spotloe J, Kirkwood JM. Neoadjuvant treatment of regional stage IIIB melanoma with high-dose interferon alfa-2b induces objective tumor regression in association with modulation of tumor infiltrating host cellular immune responses. J Clin Oncol. 2006;24:3164–3171. doi: 10.1200/JCO.2005.05.2498. [DOI] [PubMed] [Google Scholar]
- 33.Worschech A, Chen N, Yu YA, Zhang Q, Pos Z, Weibel S, et al. Systemic treatment of xenografts with vaccinia virus GLV-1h68 reveals the immunologic facets of oncolytic therapy. BMC Genomics. 2009;10:301. doi: 10.1186/1471-2164-10-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang E, Marincola FM. cDNA microarrays and the enigma of melanoma immune responsiveness. Cancer J Sci Am. 2001;7:16–23. [PubMed] [Google Scholar]
- 35.Panelli MC, White RL, Jr, Foster M, Martin B, Wang E, et al. Forecasting the cytokine storm following systemic interleukin-2 administration. J Transl Med. 2004;2:17. doi: 10.1186/1479-5876-2-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Panelli MC, Martin B, Nagorsen D, Wang E, Smith K, Monsurro' V, et al. A genomic and proteomic-based hypothesis on the eclectic effects of systemic interleukin-2 administration in the context of melanoma-specific immunization. Cells Tissues Organs. 2003;177:124–131. doi: 10.1159/000079986. [DOI] [PubMed] [Google Scholar]
- 37.Rossi L, Martin B, Hortin G, White RL, Jr, Foster M, Stroncek D, et al. Inflammatory protein profile during systemic high dose interleukin-2 administration. Proteomics. 2006;6:709–720. doi: 10.1002/pmic.200500004. [DOI] [PubMed] [Google Scholar]
- 38.Gajewski TF. Transcriptional profiling of melanoma as a potential predictive biomarker for response to immunotherapy. In: Wang E, Marincola FM, editors. Signatures of rejection. New York: Springer; 2010. pp. 229–238. [Google Scholar]
- 39.Harlin H, Meng Y, Peterson AC, Zha Y, Tretiakova M, Slingluff C, et al. Chemokine expression in melanoma metastases associated with CD8+ T-cell recruitment. Cancer Res. 2009;69:3077–3085. doi: 10.1158/0008-5472.CAN-08-2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dengel LT, Norrod AG, Gregory BL, Clancy-Thompson E, Burdick MD, Strieter RM, et al. Interferons induce CXCR3-cognate chemokine production by human metastatic melanoma. J Immunother. 2010;33:965–974. doi: 10.1097/CJI.0b013e3181fb045d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Salk J. Immunological paradoxes: theoretical considerations in the rejection or retention of grafts, tumors, and normal tissue. Ann. N. Y. Acad. Sci. 1969;164:365–380. doi: 10.1111/j.1749-6632.1969.tb14051.x. [DOI] [PubMed] [Google Scholar]
- 42.Wang E, Marincola FM. From the "delayed allergy reaction" to the "immunologic constant of rejection". In: Wang E, Marincola FM, editors. Immunologic signatures of rejection. New York, NY: Springer; 2010. pp. 3–8. [Google Scholar]
- 43.Carretero R, Wang E, Rodriguez A, Reinboth J, Ascierto ML, Engle AM, et al. Regression of melanoma metastases is associated with activation of antigen presentation and immune-mediated rejection genes. Int J Cancer. 2001 doi: 10.1002/ijc.26471. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Marincola FM. The trouble with translational medicine. J Intern Med. 2011;270:123–127. doi: 10.1111/j.1365-2796.2011.02402.x. [DOI] [PubMed] [Google Scholar]