Abstract
Repression of human papillomavirus (HPV) E6 and E7 oncogenes in established cervical carcinoma cell lines causes senescence due to reactivation of cellular tumor suppressor pathways. Here, we determined whether ongoing expression of HPV16 or HPV18 oncogenes is required for the proliferation of primary human cervical carcinoma cells in serum-free conditions at low passage number after isolation from patients. We used an SV40 viral vector expressing the bovine papillomavirus E2 protein to repress E6 and E7 in these cells. To enable efficient SV40 infection and E2 gene delivery, we first incubated the primary cervical cancer cells with the ganglioside GM1, a cell-surface receptor for SV40 limiting in these cells. Repression of HPV in primary cervical carcinoma cells caused them to undergo senescence, but the E2 protein had little effect on HPV-negative primary cells. These data suggest that E6 and E7 dependence is an inherent property of human cervical cancer cells.
Keywords: HPV; HeLa cells; GM1; viral tropism; oncogene addiction; cervical cancer, SV40
Introduction
Cervical carcinoma is initiated by infection with high-risk human papillomaviruses (HPV) such as HPV16 or 18 (Bosch et al., 2002). Expression of the HPV E6 and E7 oncogenes can immortalize primary, cultured human keratinocytes, the cells that give rise to cervical carcinomas (Durst et al., 1987; Hawley-Nelson et al., 1989; Hudson et al., 1990; Munger et al., 1989; Pirisi et al., 1987), and estrogen-treated transgenic mice expressing HPV16 E6 and E7 develop cervical carcinomas (Arbeit, Howley, and Hanahan, 1996). High-risk HPV E6 binds to p53, a tumor suppressor, and stimulates its ubiquitin-mediated degradation (Howie, Katzenellenbogen, and Galloway, 2009). High-risk HPV E7 binds to the p105Rb and other members of the retinoblastoma (Rb) family, causing their dissociation from E2F proteins and degradation of the active hypophosphorylated form of p105Rb, releasing E2F to activate genes required for cell cycle progression (Moody and Laimins, 2010). Transcription of E6 and E7 can be repressed by the papillomavirus E2 protein, which binds to the HPV E6/E7 promoter in the long control region of the viral genome (Thierry and Yaniv, 1987). Viral DNA integration or mutations that interfere with expression or activity of the E2 protein frequently occur during cervical carcinogenesis, leading to elevated expression of E6 and E7 (May et al., 1994; Schneider-Maunoury, Croissant, and Orth, 1987), which are continuously expressed in cervical carcinoma cell lines (Baker et al., 1987; Schwarz et al., 1985).
The proliferation and survival of cervical cancer cell lines requires ongoing expression of E6 and E7. Introduction of the HPV or bovine papillomavirus (BPV) E2 gene or siRNAs against HPV E6 and E7 inhibits the proliferation of cervical carcinoma cell lines or causes apoptosis (Desaintes et al., 1997; Dowhanick, McBride, and Howley, 1995; Hwang et al., 1993; Jiang and Milner, 2002; McBride, Romanczuk, and Howley, 1991; Parish et al., 2006). Repression of HPV18 E6 and E7 by BPV-E2 in HeLa cervical cancer cells results in growth arrest and cellular senescence due to reactivation of the p53 and Rb tumor suppressor pathways (Goodwin and DiMaio, 2000; Goodwin et al., 2000; Wells et al., 2000). Similarly, in a transgenic mouse model, repression of E7 expression driven by a tetracycline-regulated promoter leads to regression of high-grade cervical dysplasia and tumors (Jabbar et al., 2009). However, murine keratinocytes differ in some regards from their human counterparts (Balmain and Harris, 2000; Chaturvedi et al., 2004; Dotto, 1998), and cervical carcinogenesis in mice differs in certain aspects from human carcinogenesis. For example, the latency period of tumor formation in these mice is measured in months, whereas decades typically pass in women between HPV infection and the development of invasive cancer. It is thought that additional cellular mutations accumulate in infected human cells during this protracted latency period. Nevertheless, the fact that human cervical carcinoma cell lines and tumors in transgenic mice depend on HPV oncogene expression suggests that the E6 and E7 genes are potential targets for treatment of human cervical carcinoma.
It is not known whether dependence on E6 and E7 is an inherent property of human cervical carcinoma cells or whether this trait is acquired during the establishment or propagation of continuous cell lines. Continuous cell lines can be established from only a minority of primary tumors, and several pieces of evidence indicate that established cancer cell lines are not necessarily representative of the tumor of origin (Bignotti et al., 2006; Dairkee et al., 2004; Ertel et al., 2006; Lee et al., 2006; Santin et al., 2004; van Staveren et al., 2007; van Staveren et al., 2009). For example, analysis of gene expression profiles of the 60 cancer cells lines in the NC60 panel revealed only a 5 percent overlap in up-regulated genes compared to their corresponding tumor tissue (Sandberg and Ernberg, 2005). In addition, a comprehensive microarray-based analysis of cervical cancers and cervical cancer cell lines demonstrated that cervical cancers were more similar to normal primary cervical epithelial cells than they were to cervical cancer cell lines, indicating that the transcriptional changes that occur during the transition of normal cells to cancer cells are less pronounced than the changes that occur during the conversion of cancer cells into established cell lines (Carlson, Iyer, and Marcotte, 2007). These problems are a particular concern for cervical cancer cell lines, most of which have been in culture for decades (60 years in the case of HeLa cells). Thus, if a small population of cells that were dependent upon E6 and E7 possessed even a slight growth advantage in vitro, or if E6/E7-dependent cells were more likely to accumulate mutations that conferred a growth advantage during in vitro culture, these cells would eventually overgrow the culture, giving rise to a cell line that has diverged from its tumor of origin in terms of its oncogene dependence. In fact, HPV E6 and E7 engender a state of genomic instability and increased mutagenesis, which provides an accelerated path for cell lines to diverge from the original cancer (Moody and Laimins, 2010).
It might be expected that cells transformed by HPV in infected women would initially depend on the E6 and E7 oncogenes, and that the accumulation of mutations as the lesions progress in vivo or the cells are passaged in culture is likely to gradually render the cells independent of growth regulatory signals, so that they lose a requirement for E6 and E7. On the other hand, it is also possible that cancer cells do not require a particular oncogene for continued proliferation in culture until they are passaged in vitro. For example, in studies of a lung cancer cell line dependent on a mutant epidermal growth factor receptor, Turke et al. showed that in vitro culture of these cells in the presence of hepatocyte growth factor can generate cells that require continued signaling by c-Met, the hepatocyte growth factor receptor, as well as the EGF receptor (Turke et al., 2010). Published reports suggest a mechanism by which HPV E6/E7 could confer a growth advantage to cervical cancer cell lines cultured in vitro. Cervical carcinoma cell lines are typically maintained in high concentrations of calcium and fetal bovine serum (FBS). These conditions induce terminal differentiation of primary human keratinocytes and growth arrest. However, high-risk HPV E6 and E7 expression inhibits terminal differentiation of primary human keratinocytes and allows a small fraction of cells to proliferate in the presence of calcium and FBS (Munger et al., 1989; Pei et al., 1998; Schlegel et al., 1988; Sherman and Schlegel, 1996). Therefore, the standard conditions used to culture cell lines may have selected for expression of E6 and E7 to prevent terminal differentiation. If this were the case, the demonstrated requirement for E6/E7 in cell lines might reflect the ongoing need of these genes to continuously block terminal differentiation in response to serum and calcium in vitro, rather than an intrinsic growth property of the cells.
To minimize the risk of selection or genetic drift in testing the requirement for E6 and E7 in cervical cancer cells, we decided to assess the requirement for E6 and E7 in primary cultures of human cervical carcinoma cells. In work published by us and by others, gene expression profiling showed that various types of unmanipulated tumors are more similar to low passage primary cancer cells than they are to established cell lines derived from the same tumors (Bignotti et al., 2006; Dairkee et al., 2004; Lee et al., 2006; Santin et al., 2004). Thus, not surprisingly, primary cervical cancer cells are more representative of the original tumors than are long-term, established cell lines. There is a previous report that expression of BPV-E2 inhibited DNA synthesis in three early passage cervical carcinoma cell lines containing HPV16 DNA (Moon et al., 2001), but these cells were established and maintained in high calcium and FBS, so may have been subjected to selective pressure for E6/E7 dependence as described above.
In the experiments reported here, we isolated primary cancer cells from tumor biopsies of four patients with cervical carcinoma and cultured them for a minimal number of passages in low concentrations of calcium and in the absence of serum to prevent terminal differentiation and in vitro selection for E6/E7 dependence. For these experiments, we wished to use an SV40 recombinant virus vector to express the BPV-E2 protein, because this is an extremely efficient system to repress E6 and E7 in HeLa cells. However, most cells we tested infected poorly with this vector. By exploiting our understanding of factors that dictate SV40 infection, we were able to markedly enhance infection by the SV40 vector by treating the cells with the ganglioside GM1, a cellular receptor of SV40 (Tsai et al., 2003), prior to infection. GM1 added to the culture medium is incorporated into the plasma membrane (Schwarzmann, 2001), thus increasing the cell surface GM1 that can be used by SV40 for infection. By using this approach, we demonstrated that primary cervical carcinoma cells are indeed dependent on E6 and E7 for continued growth.
Results
Isolation of primary human cervical carcinoma cells
Primary cervical carcinoma cells were isolated from surgical biopsies as described previously (Santin et al., 2005) and cultured in serum-free keratinocyte medium, which contains low concentrations of calcium to prevent differentiation. Characteristics of these cells including the resident HPV DNA type are summarized in Table 1. Passage 0 is the point at which cells were initially placed in culture, and one passage number was recorded each time a 70–80 % confluent dish of cells was passed. The passage number at which each isolate was analyzed in various experiments is indicated where relevant in the figures. All primary cervical carcinoma isolates were confirmed to express dramatically elevated levels of p16ink4a mRNA, a marker of cervical cancer (Sano et al., 1998), when compared to primary keratinocytes and fibroblasts (data not shown).
Table 1.
Characteristics of primary cervical carcinoma cells.
| Cell Name | HPV Type |
Passage Numbera |
Histologyb | Stageb,c | Disease Siteb |
|---|---|---|---|---|---|
| HeLa | 18 | -d | AD | -e | cervix |
| SiHa | 16 | -d | SCC | II | cervix |
| CaSki | 16 | -d | SCC | REC | metastatic SBM |
| 101 | 16 | 39 | SCC | IB | cervix |
| 102 | 16 | 16 | SCC | IB | cervix |
| 104 | 18 | 33 | AD | IB | cervix |
| 106 | 16 | 3 | SCC | IB | cervix |
A passage number indicates the earliest passage number at which the effect of E2 expression on growth was measured
SCC, squamous cell carcinoma; AD, adenocarcinoma; REC, recurrent disease; SBM, small bowel mesentery
Staging for primary isolates was done according to the Figo staging system
The passage number for cell lines is not known. Cell lines were isolated in 1951(HeLa), 1970(SiHa), and 1977 (CaSki)
The cancer stage is not known
SV40 vector expressing BPV-E2 induces growth arrest preferentially in HeLa-Sen2 cells
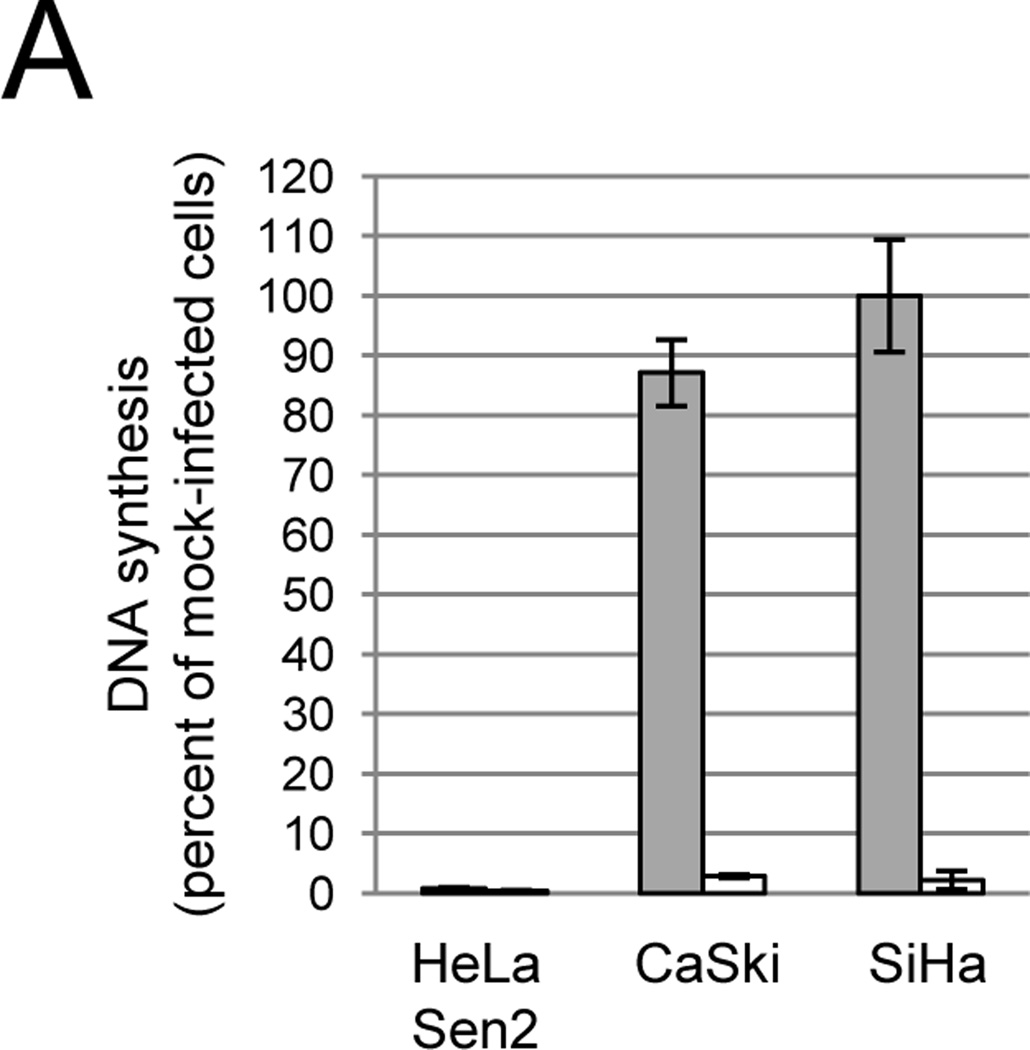
Exogenous expression of the BPV E2 gene (designated E2) in several different established cervical carcinoma cell lines causes irreversible growth arrest and senescence due to transcriptional repression of the HPV E6 and E7 genes (Goodwin et al., 2000; Wells et al., 2000). Previously we used an SV40-based recombinant virus vector (designated Pava) to express the wild-type (WT) E2 gene in HeLa-Sen2 cells, a strain of HeLa cells that is efficiently infected with this virus (Goodwin et al., 2000; Hwang et al., 1993). In this vector, the SV40 large T antigen gene is replaced with the E2 gene, but the SV40 capsid protein genes are retained. Replication-defective Pava virus particles containing the E2 gene packaged in an SV40 capsid are generated in permissive monkey CMT4 cells, which supply SV40 replication functions in trans (Settleman and DiMaio, 1988). Infection of HeLa-Sen2 cells with Pava delivers the E2 gene into essentially every cell, which leads to profound repression of E6/E7 and a rapid induction of senescence (Goodwin et al., 2000). Strikingly, although Pava caused dramatic growth arrest in HeLa-Sen2 cells, it had little or no effect on DNA synthesis of CaSki and SiHa cells, two cervical carcinoma cell lines that contain HPV16 DNA (Fig. 1A, grey bars). Pava infection also failed to inhibit DNA synthesis of all primary cervical carcinoma isolates tested, except CVX-102 (data not shown).
Fig. 1.
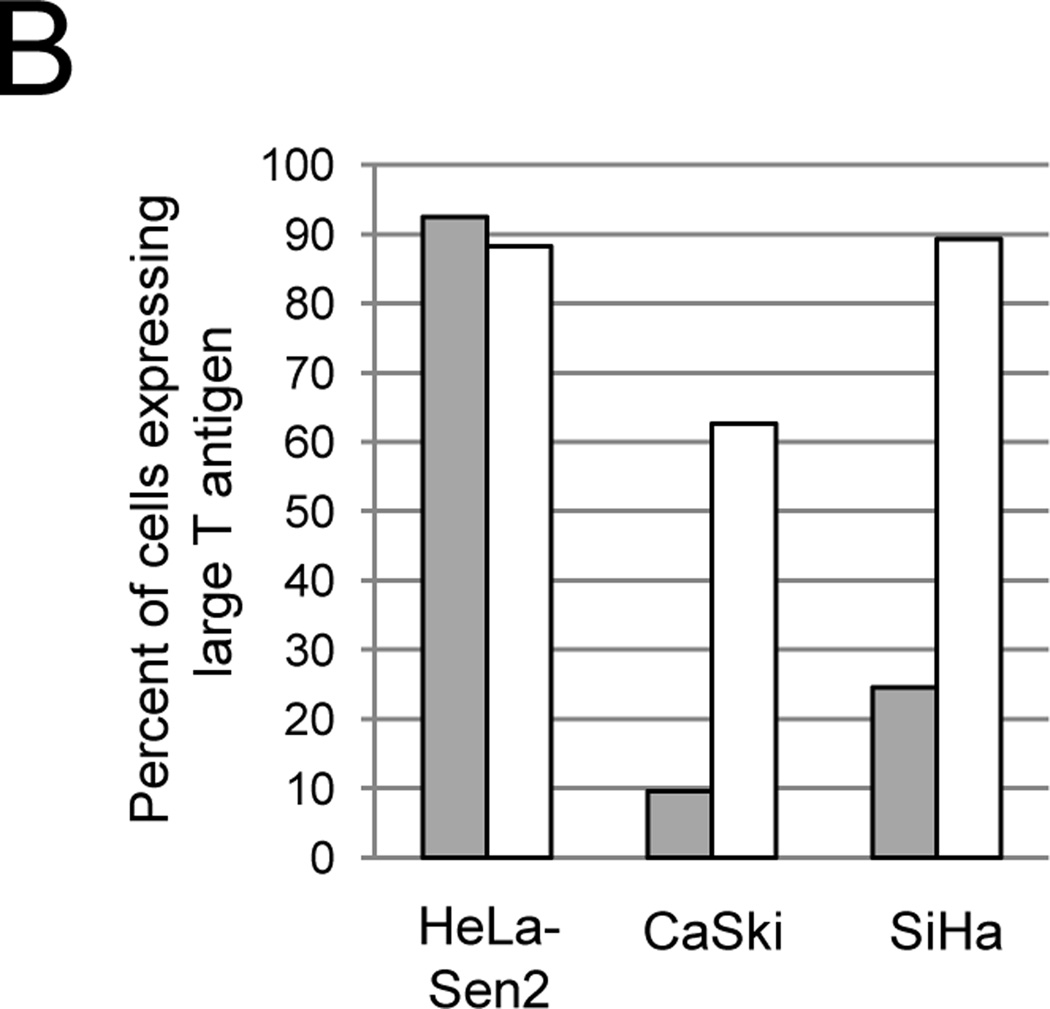
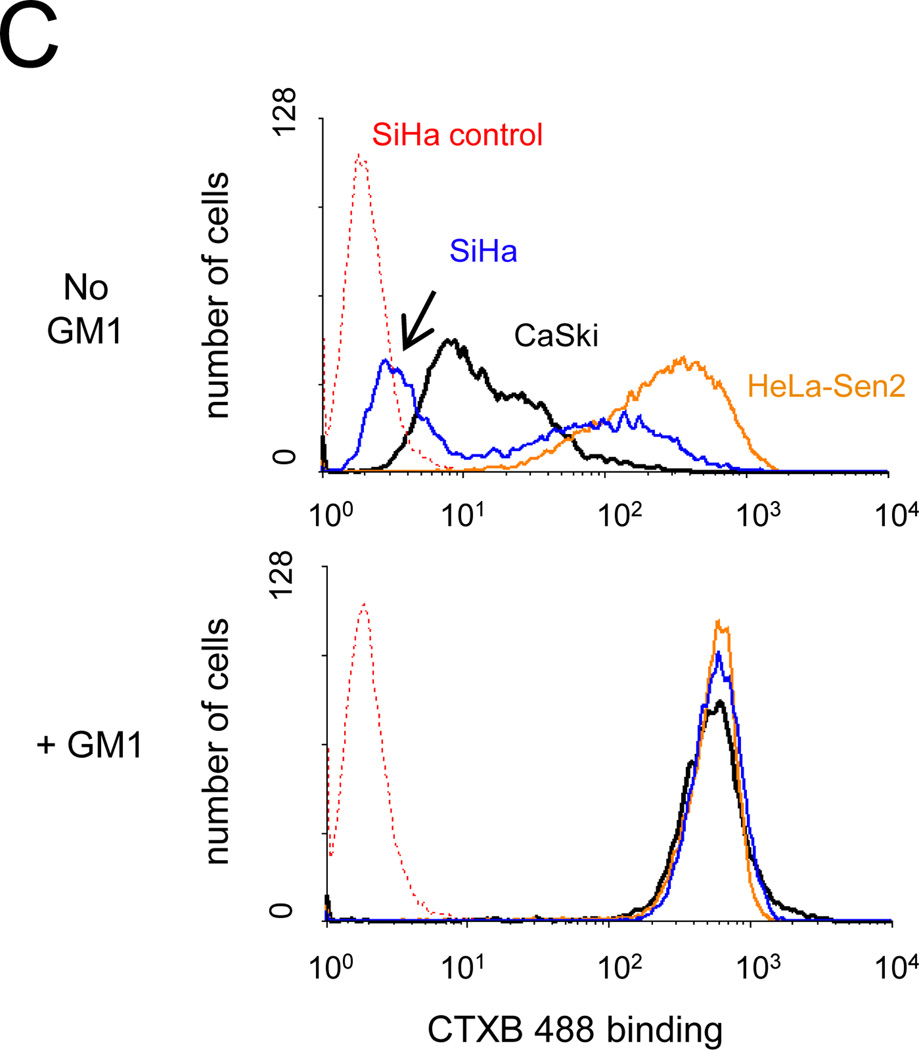
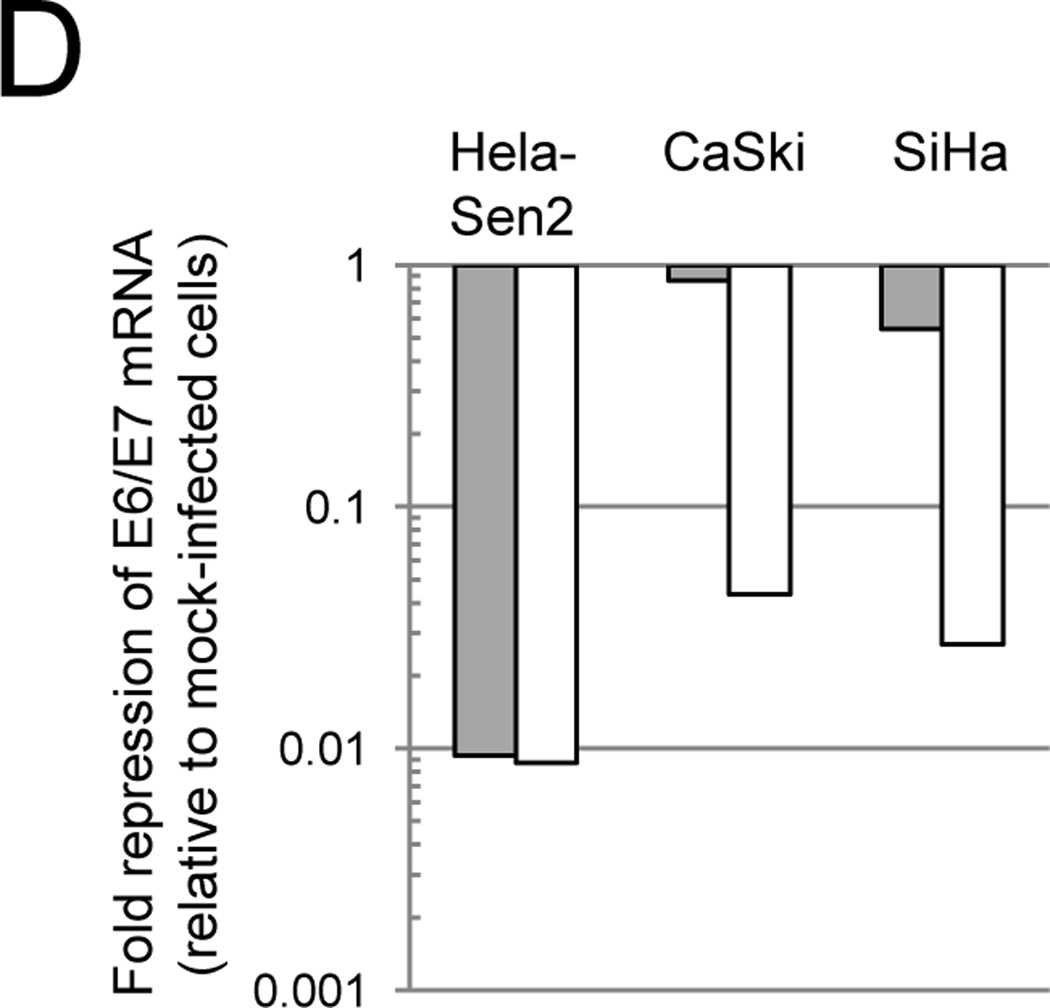
Effect of GM1 on SV40 infection of cervical carcinoma cell lines. A) Cells were left untreated (grey bars) or treated with GM1 overnight (white bars) and then either mock-infected or infected with Pava at an MOI of 20. At 48 hrs post-infection, DNA synthesis was measured by incorporation of tritiated thymidine and is presented as the percent of tritiated thymidine incorporation in infected cells relative to mock-infected cells. The results of a typical experiment are presented, and represent the average of triplicate samples. Similar results were obtained in at least three independent experiments. B) Cells were left untreated (grey bars) or treated with GM1 overnight (white bars) and infected with SV40 at an MOI of 10. The fraction of cells expressing large T antigen was measured by immunostaining and flow cytometry at 48 hrs post-infection. Similar results were obtained in three independent experiments. C) Cells were left untreated (top panel) or treated with GM1 overnight (bottom panel), and GM1 surface levels were measured by flow cytometry of binding of fluorescently-labeled CTXB. The dotted red lines represent SiHa cells incubated in the absence of CTXB. Similar results were obtained in at least three independent experiments. D) Cells were left untreated (grey bars) or treated with GM1 overnight (white bars), and either mock-infected or infected with Pava at an MOI of 20. At 48 hrs post-infection, HPV18 E6/E7 mRNA levels in HeLa-Sen2 cells and HPV16 E6/E7 mRNA levels in SiHa and CaSki cells were measured by qRT-PCR and normalized to GAPDH mRNA levels. E7 expression is reported as fold-reduction in the level of mRNA in infected cells relative to mock-infected cells. The results of a typical experiment are presented, and represent the average of triplicate samples. Similar results were obtained in two independent experiments.
WT SV40 and Pava infection of established cervical carcinoma cell lines
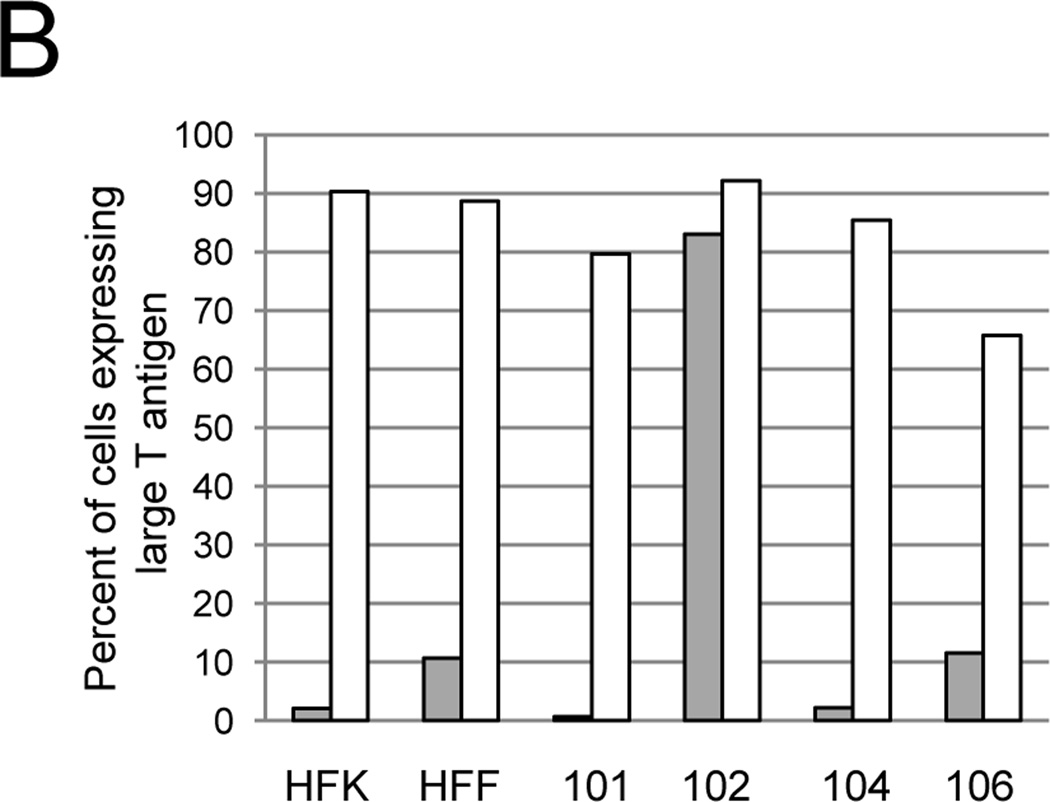
Because the introduction of the E2 gene into CaSki and SiHa cells by transfection or adenovirus vectors leads to growth arrest (Lee et al., 2002; Wells et al., 2000), we hypothesized that the SV40-based vector used to deliver the E2 gene did not efficiently infect these cells. In order to test this hypothesis, we measured the ability of WT SV40 to infect the three established cervical cancer cell lines. HeLa-Sen2, CaSki, and SiHa cells were infected with SV40 at an MOI of 10 (titer determined in permissive monkey cells), and infection was measured by immunostaining and flow cytometry of large T antigen expression. Typical histograms of flow cytometry data are presented in supplementary Figure S1. As predicted, more than 90 percent of HeLa-Sen2 cells expressed large T antigen (Fig. 1B, grey bars). However, less than 25 percent of CaSki or SiHa cells expressed large T antigen at this MOI. The inability of SV40 to efficiently infect SiHa and CaSki cells suggests that these cells also infect poorly with Pava, providing an explanation to their resistance to Pava-induced growth arrest.
We hypothesized that inefficient infection of CaSki and SiHa cells was a consequence of low levels of GM1, a cellular receptor used by SV40 (Tsai et al., 2003), on the surface of these cells. To test the levels of GM1 at the surface of cervical carcinoma cell lines, GM1 surface expression was measured with fluorescently-labeled cholera toxin B (CTXB), a bacterial toxin subunit which binds to GM1 (Chinnapen et al., 2007). Binding to intact cells was conducted at 4°C to prevent internalization and was measured by flow cytometry. SiHa and CaSki cells showed markedly lower levels of CTXB binding than HeLa-Sen2 cells (Fig. 1C, top), consistent with our suggestion that low levels of surface GM1 may limit SV40 infection in SiHa and CaSki cells. Addition of GM1 to the culture medium of murine cells that lack endogenous GM1 results in the incorporation of GM1 into the plasma membrane and a dramatic increase in the number of cells susceptible to SV40 infection (Schwarzmann, 2001; Tsai et al., 2003). As shown in Fig. 1C, bottom, addition of GM1 increased CTXB binding in Hela-Sen2, CaSki, and SiHa cells so that the level of CTXB binding was high in all cell lines.
We then tested whether elevating GM1 surface levels increased the ability of SV40 to infect CaSki and SiHa cells. Cells were treated with GM1 or left untreated. After washing away unincorporated GM1, cells were infected with SV40 at an MOI of 10, and infection was measured by assessing large T antigen expression by flow cytometry. As shown in Figure 1B (white bars), increasing the level of cell surface GM1 dramatically increased the ability of SV40 to infect CaSki and SiHa cells. In order to determine whether incorporation of GM1 also increased Pava infection, we measured E2 mRNA levels in untreated or GM1-treated cells. Pava-infected CaSki and SiHa cells displayed increased E2 mRNA expression after treatment with GM1, indicating that GM1 addition to cells increased Pava infection (Fig. S2.)
We also measured the effect of increased Pava infection on cell physiology. E2 expression in GM1-treated cells led to a 6- to 20-fold repression of HPV16 E6/E7 mRNA in CaSki and SiHa cells when compared to infected, untreated cells (Fig. 1D). We next tested whether Pava infection inhibited the DNA synthesis of GM1-treated cells. In contrast to the minimal effects of Pava infection in native CaSki and SiHa cells, infection of these cells with Pava following GM1 treatment led to a substantial repression of DNA synthesis to less than 3% of the mock-infected cells (Fig. 1A, white bars). Pava infection of GM1-treated CaSki and SiHa cells also led to a loss of the inactive hyperphosphorylated form of p105Rb and an increase in the active hypophosphorylated form (Fig. S3). Pava infection also increased total levels of p53 protein in CaSki cells (Fig. S3). The effects are more pronounced after GM1 addition. Although E2 expression did not appear to increase p53 levels in SiHa cells, an increase in p53 may occur prior to the 48hr time point at which protein was harvested (Goodwin and DiMaio, 2000). Taken together, these results indicated that the range of cells susceptible to efficient infection by SV40 and SV40 vectors can be expanded by addition of GM1. Therefore, we tested whether we could use this approach to express E2 in primary cervical carcinoma cells to determine the effect of E6 and E7 repression on cell growth.
SV40 infection of primary cervical carcinoma cells
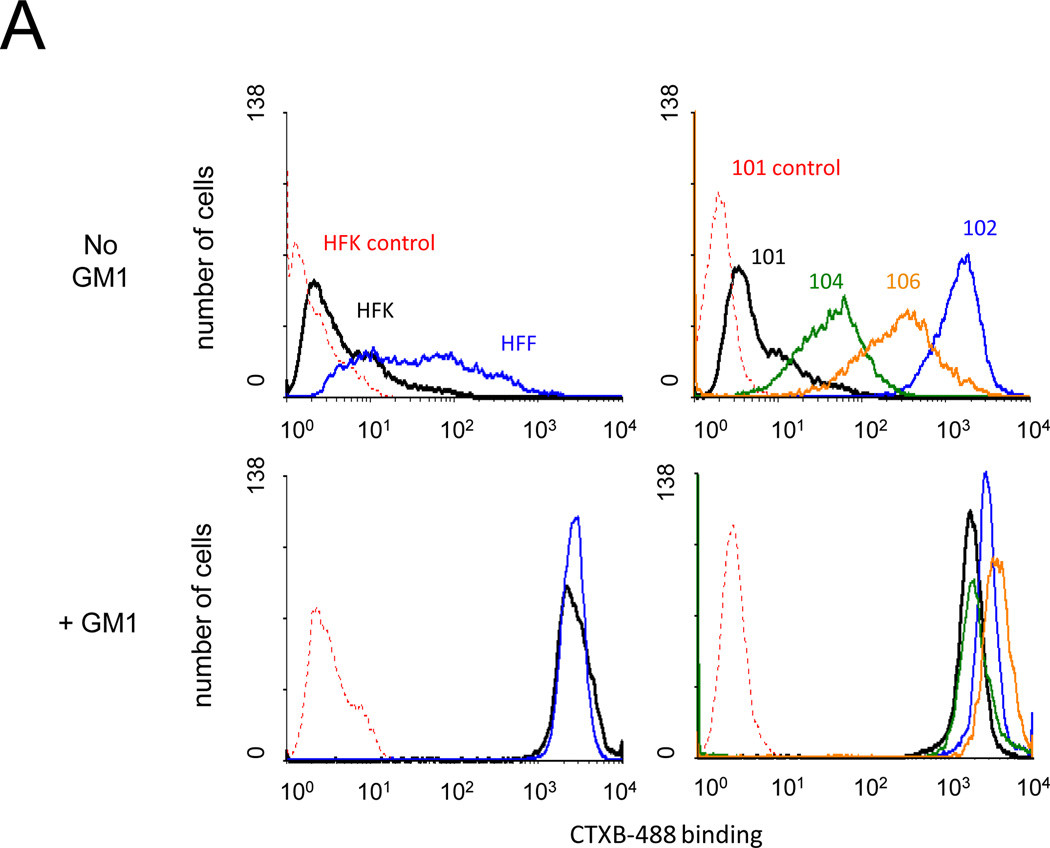
We first used CTXB binding to measure GM1 surface expression in four HPV-positive primary human cervical carcinoma cell isolates and two primary cell isolates that lack HPV DNA, human foreskin fibroblasts (HFF) and keratinocytes (HFK). As shown in the top panels of Figure 2A, these cells displayed a wide range of GM1 surface expression. Addition of GM1 to the culture medium caused a marked increase in CTXB binding in all cells tested except CVX-102 cells, which displayed high basal CTXB binding (Fig. 2A). SV40 infection was measured as described above in untreated cells or cells treated with GM1. The high levels of endogenous GM1 in CVX-102 cells allowed efficient SV40 infection even without GM1 addition. In all other cell isolates, addition of GM1 to cells caused a dramatic increase in the fraction of cells expressing large T antigen after infection, approaching 90% in most cases (Fig. 2B).
Fig. 2.
Effect of GM1 on SV40 infection of primary cervical carcinoma cells. A) The indicated cells were left untreated (top panels) or treated with GM1 overnight (bottom panels), and GM1 surface expression was measured by flow cytometry of binding of fluorescently-labeled CTXB. Dotted red lines represent fluorescence of HFK (left panels) or CVX-101 cells (right panels) incubated in the absence of CTXB. Similar results were obtained in two independent experiments. B) The indicated cell strains were left untreated (grey bars) or treated with GM1 overnight (white bars) and infected with SV40 at an MOI of 10. The fraction of cells expressing large T antigen was measured as is described in Figure 1B. Similar results were obtained in two independent experiments.
Effect of E2 expression on DNA synthesis of primary cervical carcinoma cells
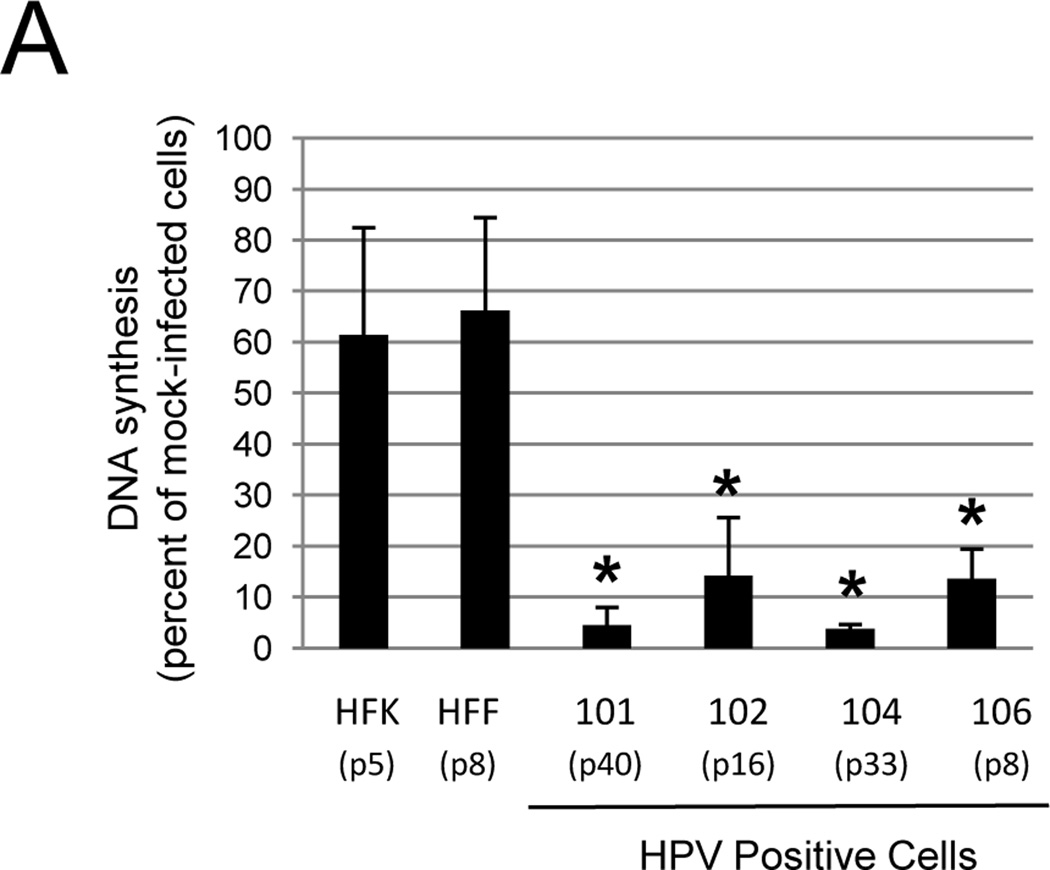
Improved SV40 infection allowed us to investigate the effect of E2 expression on the proliferation of primary cervical carcinoma cells. HPV negative HFFs and HFKs were infected in parallel as controls. Cells were first treated with GM1 overnight to maximize Pava infection and delivery of the E2 gene. After removing unincorporated GM1, cells were infected with Pava, and DNA synthesis was measured 48 hrs post-infection. Expression of E2 caused a 6–20 fold reduction in DNA synthesis in the primary carcinoma cell lines tested, but had a minimal effect on DNA synthesis in HPV-negative cells (Fig. 3A). qRT-PCR of E2 mRNA levels demonstrated that differences in E2 expression did not dictate differences in DNA synthesis among the cell lines, because HFKs expressed at least as much E2 mRNA as any of the primary carcinoma cells (Fig. S4). These results indicated that growth arrest induced by E2 expression is specific to HPV-positive cells.
Fig. 3.
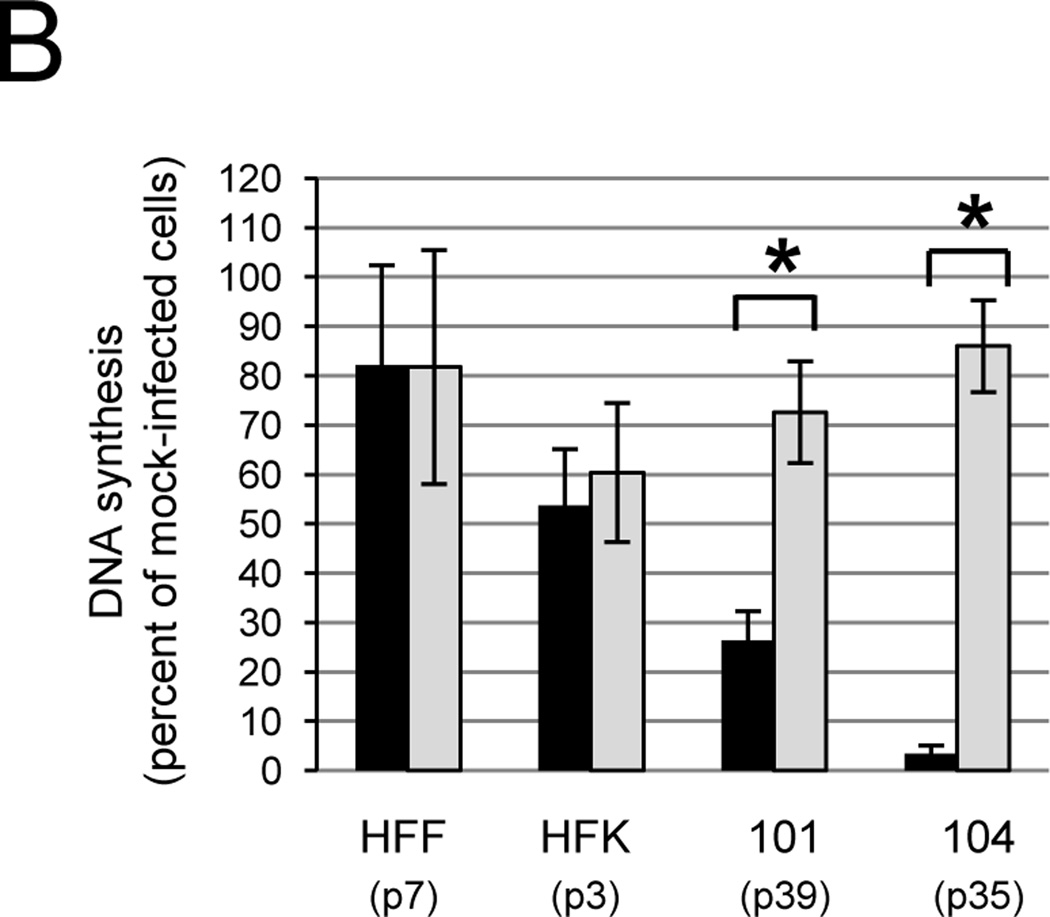
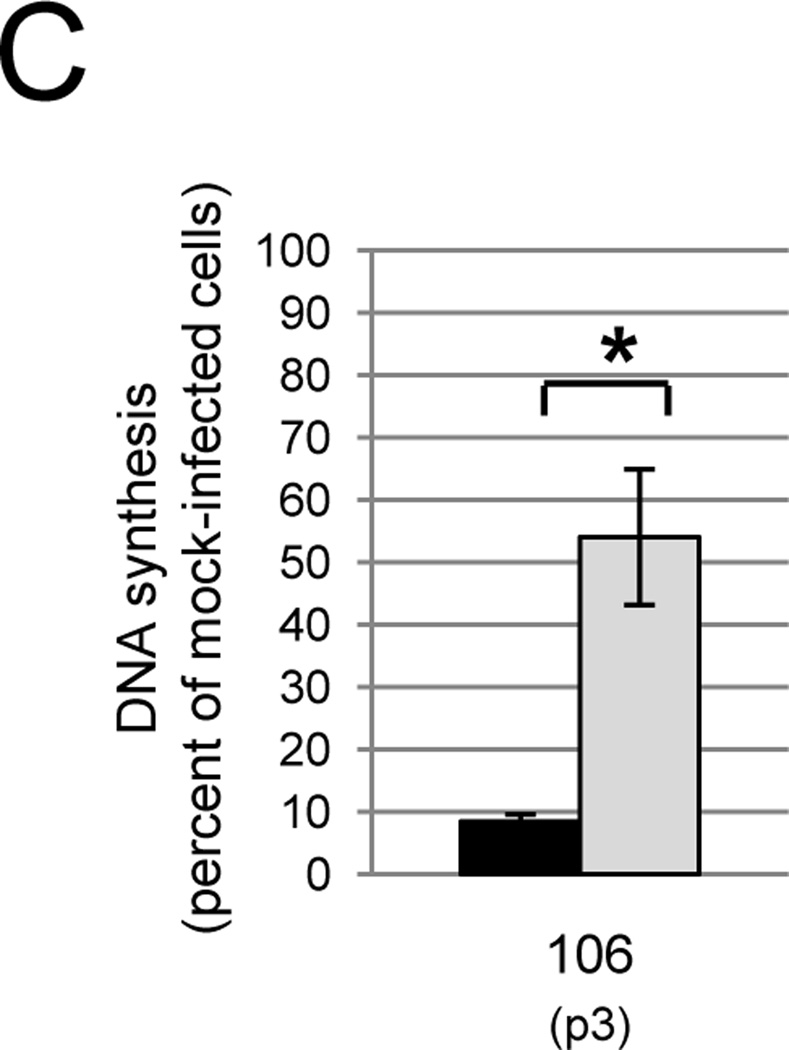
The WT E2 protein inhibits DNA synthesis in primary carcinoma cells. A) The indicated cell strains were treated with GM1 overnight and either mock-infected or infected with Pava at an MOI of 10. DNA synthesis was measured as in Figure 1A and is presented as the percent of tritiated thymidine incorporated in infected cells relative to mock-infected cells. The numbers in parentheses indicate the passage at which the cells were tested. The results of a typical experiment are shown, and represent the average of triplicate samples. Asterisks indicate a significant difference between DNA synthesis of mock and infected cells (*p <.05). Similar results were obtained in at least four independent experiments. B) Cells were treated with GM1 in panel A and infected with Pava encoding WT (black bars) or K339M (grey bars) E2 at an MOI of 10. DNA synthesis was measured and is presented as in panel A. The brackets indicate a significant difference between DNA synthesis of cells infected with WT or K339M-E2 (* p<.005). Similar results were obtained in at least four independent experiments. C) Cells were treated with GM1 and infected with WT or mutant Pava as in panel A. DNA synthesis was measured 72 hrs post-infection and is presented as in panel A. The brackets indicate a significant difference between DNA synthesis of cells infected with WT or K339M E2 (* p <.02). Similar results were obtained in two independent experiments.
To begin to determine whether the observed repression of DNA synthesis was the result of E6 and E7 repression in primary cervical carcinoma cells, CVX-101, CVX-104, HFF, and HFK were treated with GM1 and infected with Pava expressing the WT E2 protein or an E2 mutant, K339M, defective for DNA binding. Reduced DNA binding by K339M E2 inhibits transcriptional repression of E6 and E7 (Goodwin et al., 1998; Prakash et al., 1992). The K339M E2 protein had little effect on DNA synthesis in CVX-101 and 104 cells when compared to the WT E2 protein, suggesting that the DNA binding activity of E2 is required for its ability to induce growth arrest (Fig. 3B). Similar results were also obtained with CVX-102 cells (data not shown). Neither WT nor K339M E2 caused significant inhibition of DNA synthesis in HPV-negative cells. The minor inhibition observed in HFKs suggested that Pava infection and/or E2 expression caused a minor, non-specific inhibition of DNA synthesis in HPV-negative cells, independent of the DNA binding activity of the E2 protein (Desaintes et al., 1999) (Fig. 3B). qRT-PCR for E2 mRNA expression confirmed that differences in growth inhibition between cells infected with WT or K339M Pava were not the result of differences in E2 mRNA levels, because K339M E2 mRNA levels were approximately equal to or greater than that of WT E2 (Fig. S5). Taken together, these data imply that the growth repression caused by E2 expression in primary cervical carcinoma cells is dependent upon the transcriptional repressor activity of the E2 protein and is likely the result of E6 and E7 repression.
In order to test whether earlier passage cells were also dependent on E6 and E7 for growth, we measured the effect of the E2 protein on CVX-106 cells at passage 3. Cells were treated with GM1 and infected with Pava expressing the WT or K339M E2 protein as described above. Due to the limiting number and slow growth of these early passage cells, DNA synthesis was measured at 72 hrs post infection in order to maximize the effect of E2 expression on growth. Even at these very early passages, WT E2 repressed DNA synthesis to a substantially greater degree than K339M E2 (Fig. 3C). These data indicated that extremely early passage primary cervical carcinoma cells are also dependent on E6 and E7 for continued growth.
Effect of the E2 protein on E6 and E7 expression and activity in primary cervical carcinoma cells
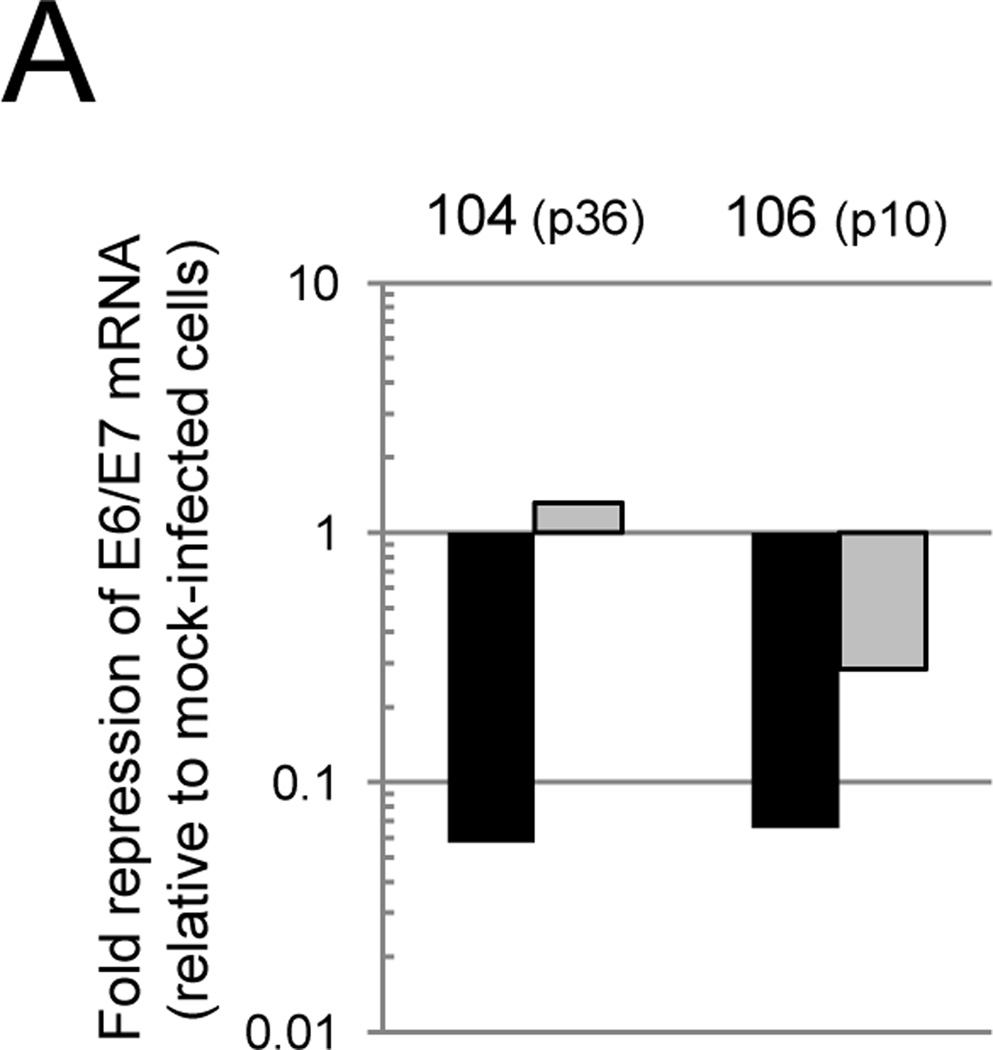
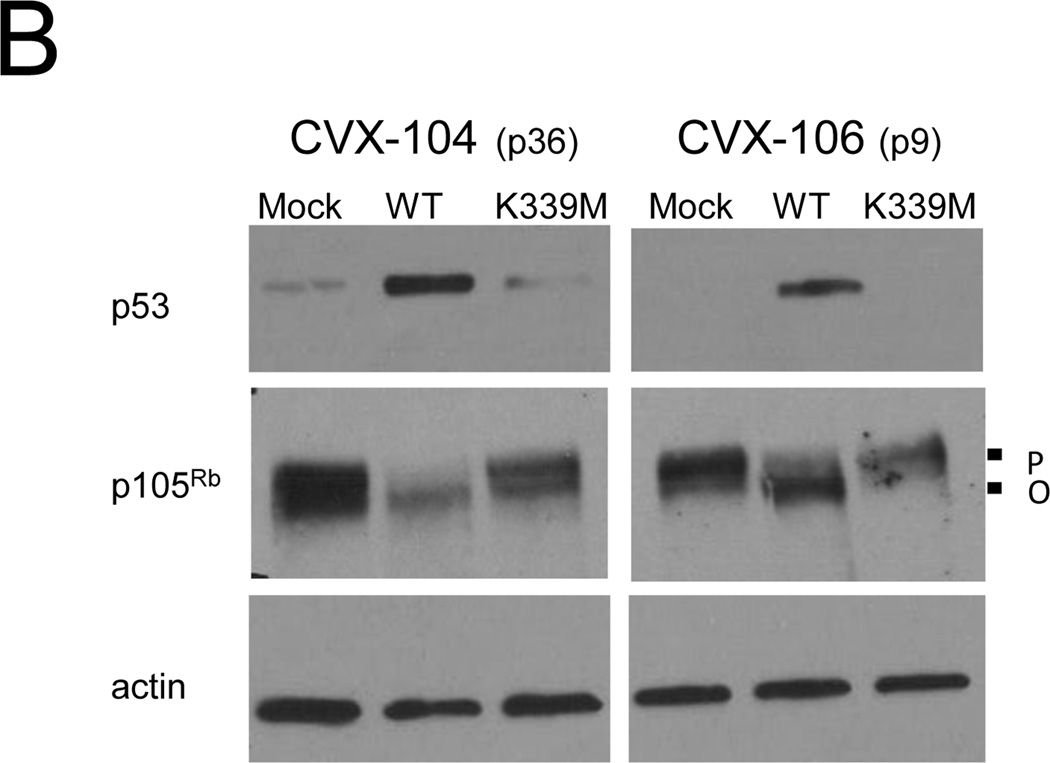
To determine whether the WT E2 protein repressed E6 and E7 mRNA expression in primary carcinoma cells, qRT-PCR was used to measure endogenous HPV mRNA levels in two representative cervical carcinoma cell strains, CVX-104 and CVX-106. WT E2 repressed endogenous levels of HPV18 E6/E7 17-fold in CVX-104 cells and HPV16 E6/E7 levels 15-fold in CVX-106 cells, where as K339M E2 failed to repress 18E6/E7 in CVX-104 cells and repressed 16E6/E7 only three-fold in CVX-106 cells (Fig. 4A). In order to determine whether repression of E6 and E7 by E2 led to the reactivation of the p105Rb and p53 tumor suppressor pathways in primary carcinoma cells, protein was isolated from infected and mock-infected CVX-104 and CVX-106 cells and analyzed by immunoblotting. WT E2 caused an increase in p53 levels and the hypophosphorylation/activation of p105Rb in CVX-104 and CVX-106 cells, but K339M had no effect on p53 levels and/or the phosphorylation state of p105Rb (Fig. 4B). These results indicated that repression of E6 and E7 by the E2 protein led to the reactivation of p53 and p105Rb pathways in primary cervical carcinoma cells.
Fig. 4.
The WT E2 protein represses E6 and E7 and activates p53 and p105Rb in primary cervical carcinoma cells. A) Cells were treated with GM1 and either mock-infected or infected with Pava encoding WT (black bars) or K339M (grey bars) E2 at an MOI of 10. At 40 hrs post-infection, levels of HPV18 E6/E7 (CVX-104) and HPV16 E6/E7 (CVX-106) mRNA were measured and reported as described in Figure 1D. The passage numbers at which the cells were tested are indicated in parentheses. Similar results were obtained in three independent experiments. B) Cells were treated with GM1 and either mock-infected or infected as in panel A. Protein was harvested at 48 hrs post-infection and analyzed by SDS-PAGE and immunoblotting for p105Rb, p53, and actin (loading control). P indicates hyperphosphorylated p105Rb, while O indicates active, hypophosphorylated p105Rb. Similar results were obtained in three independent experiments.
Repression of E6 and E7 in primary cervical carcinoma cells induces senescence
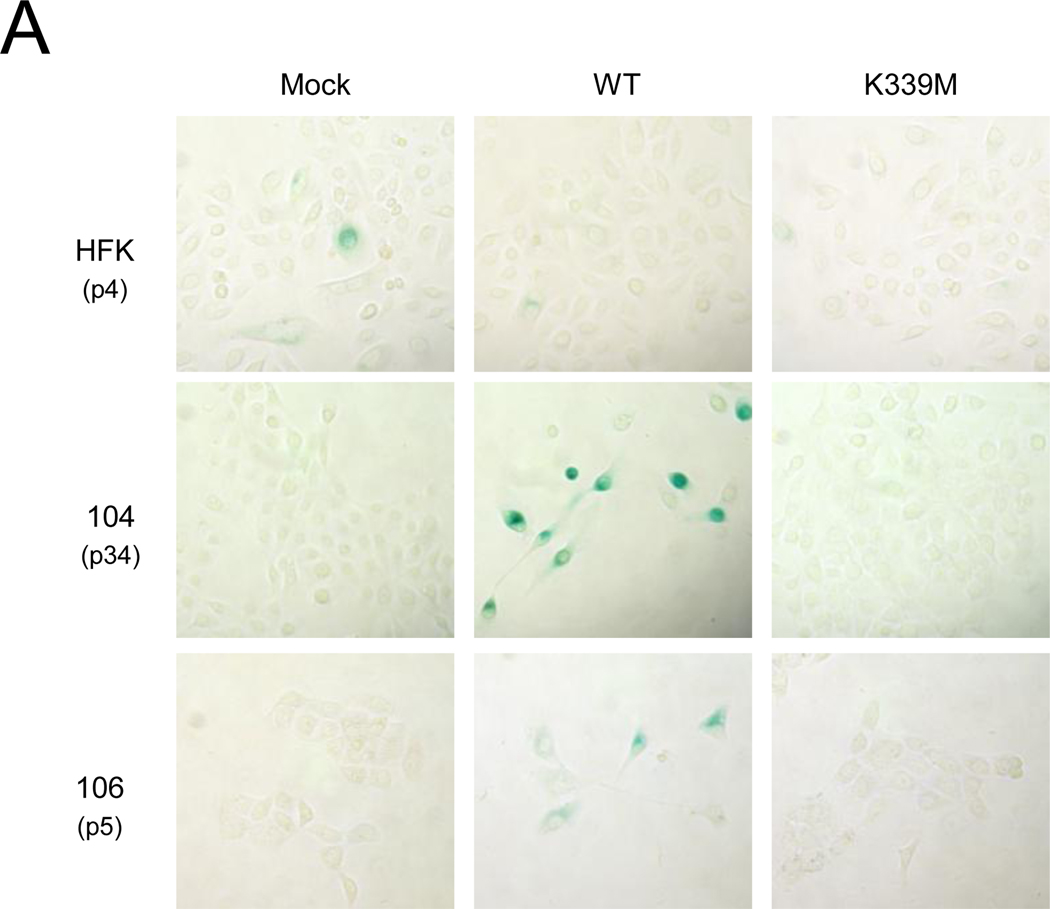
Repression of HPV E6 and E7 in HeLa and other established cervical carcinoma cell lines induces senescence, a form of irreversible growth arrest (Campisi and d'Adda di Fagagna, 2007; Goodwin et al., 2000; Wells et al., 2000). One biomarker used to distinguish senescence from other forms of growth arrest is an increase in senescence-associated (SA)-β-galactosidase activity (Dimri et al., 1995). In order to determine whether repression of E6 and E7 induced senescence in primary cervical carcinoma cells, CVX-104, CVX-106, and HFK cells were treated with GM1 and infected with Pava. To decrease the number of cells that escaped infection, cells were treated with GM1 and infected with Pava a second time. Nine days after the initial Pava infection, SA-β-galactosidase activity was assessed by staining with a colorimetric substrate. WT E2 expression increased SA-β-galactocidase staining in CVX-104 and 106 cells but not in HPV-negative HFK cells (Fig. 5A). Cells that expressed SA-β-galactocidase also displayed an enlarged and flattened morphology, which is characteristic of senescent cells (Fig. 5A). As expected, K339M E2 did not induce SA-β-galactosidase activity or these morphological changes, suggesting that these effects required repression of E6 and E7.
Fig. 5.
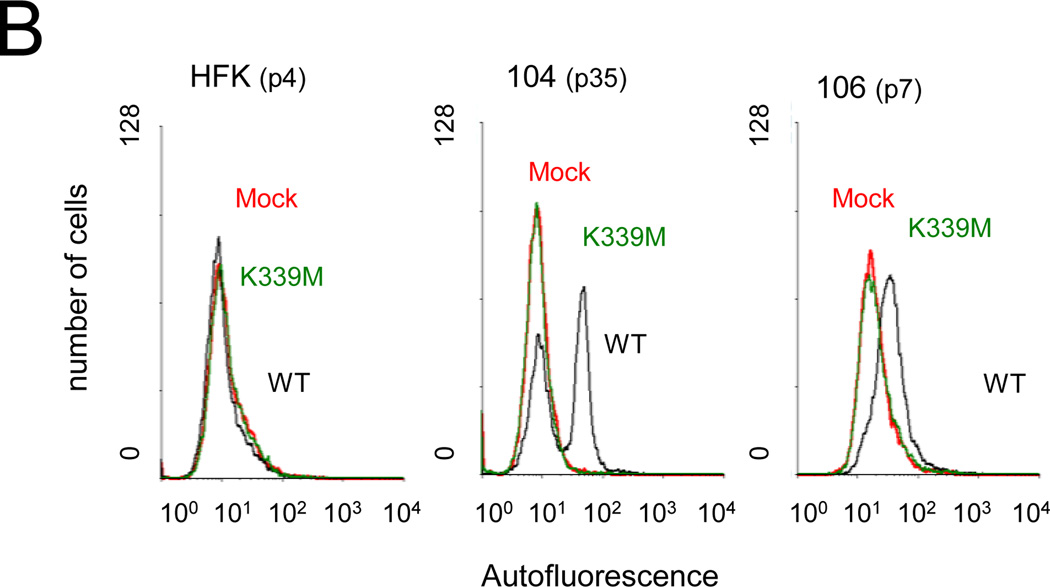
Repression of E6 and E7 induces senescence in primary cervical carcinoma cells. A) HFK, CVX-104, and CVX-106 cells at the indicated passage number were twice treated with GM1 and infected with Pava encoding WT or K339M E2 at an MOI of 10. At nine days post-infection, cells were fixed and stained for SA-β-galactosidase activity. Similar results were obtained for each cell type in at least two independent experiments. B) Cells were infected as in panel A. Autofluorescence was measured by flow cytometry at day seven post-infection. Similar results were obtained in three independent experiments. Red, mock-infected; black, WT E2; green, K339M E2.
In addition to increased SA-β-galactosidase activity, senescence is also characterized by an increase in autofluorescence due to the accumulation of the fluorescent pigment lipofuscin (von Zglinicki et al., 1995). Following two rounds of GM1 treatment and infection with Pava, autofluorescence was measured by flow cytometry seven days after the initial infection with Pava (Fig. 5B). WT E2 and K339M E2 expression had no effect on autofluorescence in HFK cells, which continued to proliferate. Expression of the WT E2 protein, but not K339M E2, induced an increase in autofluorescence in a large fraction of CVX-104 and CVX-106 cells. Although a subpopulation of CVX-104 cells displayed low autofluorescence, we believe that these are cells that escaped infection with Pava and continued to proliferate during the seven day course of the experiment. This proliferating population is not seen in CVX-106 cells. However, since CVX-106 cells grow significantly slower than CVX-104 cells (data not shown), cells that escaped infection would still be a very minor fraction at day seven post-infection. Taken together, these data demonstrated that repression of E6 and E7 induces senescence in primary cervical carcinoma cells.
Exogenous expression of E6 and E7 in CVX-104 cells impairs growth arrest induced by E2 expression
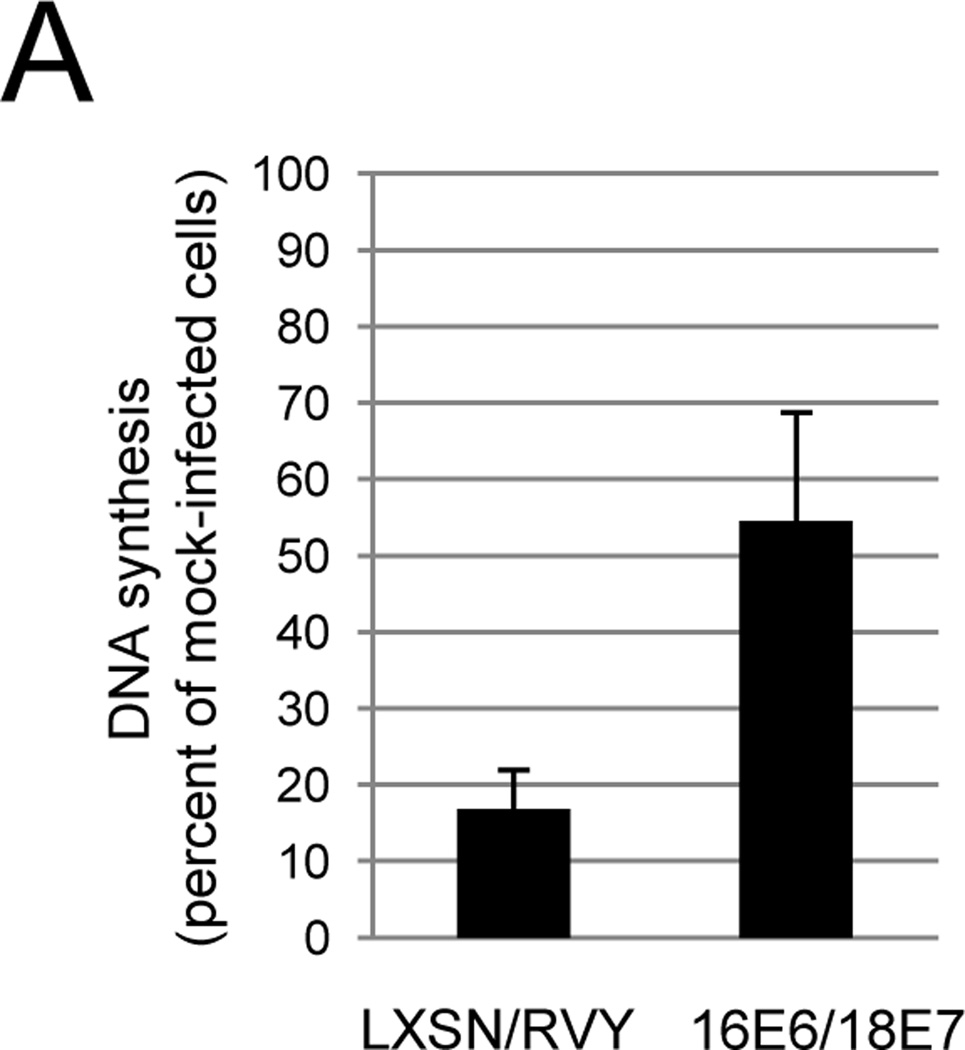
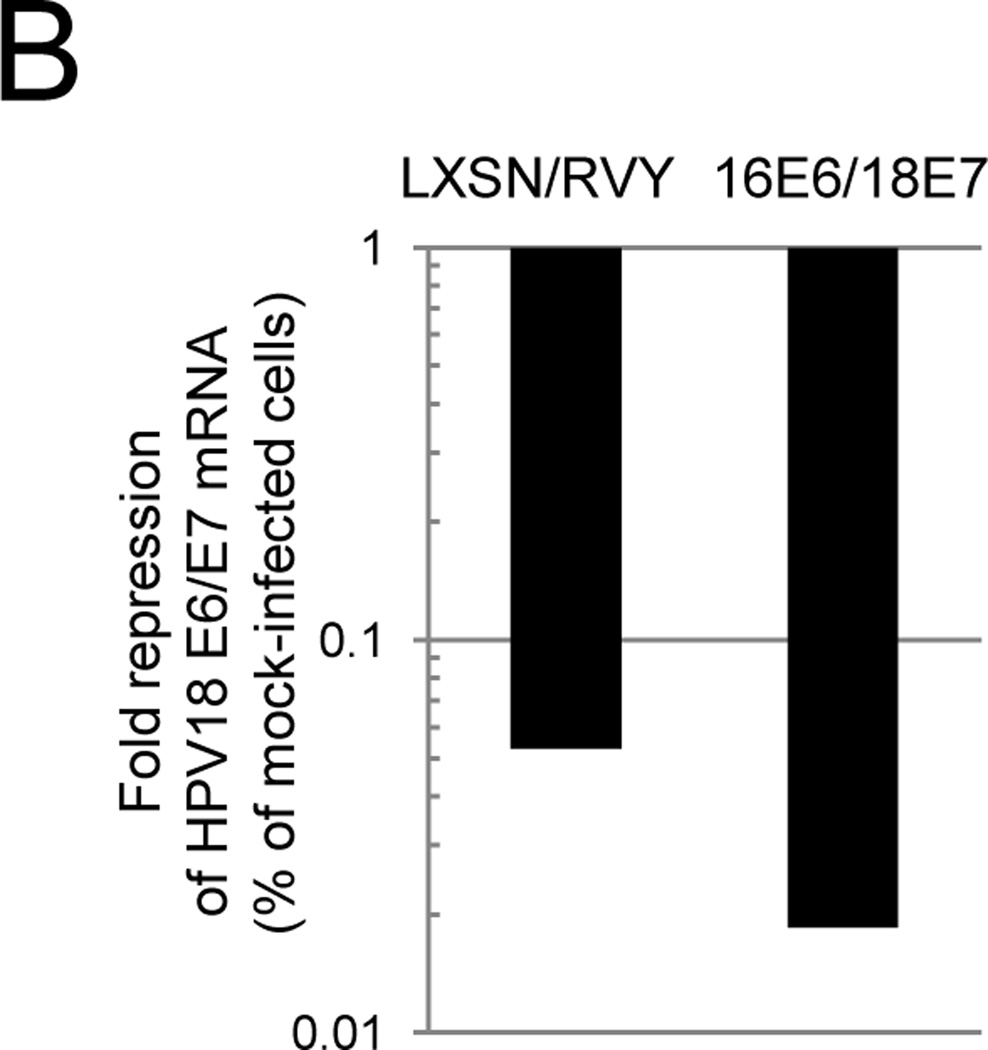
We performed a gene rescue experiment to confirm that the phenotypes elicited by Pava infection in primary cervical carcinoma cells were due to E6/E7 repression. CVX-104 cells were infected either with an empty retroviral vector or the vector encoding HPV16 E6 and HPV18 E7. Since these genes are expressed from the retroviral LTR, E2 does not affect their expression. After confirming expression of these genes by qRT-PCR (data not shown), the cells were treated with GM1 and infected with Pava. Growth arrest was then measured by DNA synthesis. As shown in Figure 6A, expression of exogenous HPV16 E6 and 18 E7 largely abrogated the inhibition of DNA synthesis caused by E2 expression in these cells. qRT-PCR confirmed that Pava-infected 16E6/18E7 expressing cells contained at least the same amount of E2 mRNA as infected control cells (data not shown). qRT-PCR using primers specific for the endogenous or exogenous E6/E7 transcripts confirmed that expression of the endogenous E6 and E7 genes in 16E6/18E7 cells was repressed by Pava infection to at least the same level seen in control cells (Fig. 6B), but expression of the exogenous E6/E7 genes was not affected (data not shown). Similarly, as measured by autofluorescence and granularity, which are both increased during senescence, there was a four- to five-fold reduction in the number of CXV-104 16E6/18E7 cells that senesced following Pava infection compared to infected control CVX-104 cells (Fig. S6). These data showed that E2-induced growth arrest and senescence in CVX-104 cells was the result of E6 and E7 repression.
Fig. 6.
Growth inhibition of primary cervical carcinoma cells is dependent upon repression of E6 and E7. A) CVX-104 cells infected with either empty retroviral vectors (LXSN/RVY) or vectors encoding E2-resistant HPV16 E6 and HPV18 E7 genes (16E6/18E7) were treated with GM1 and either mock-infected or infected with Pava as described in Figure 3A. DNA synthesis was measured as in Figure 3A and is presented as the average of three independent experiments. The difference between DNA synthesis of infected LXSN/RVY cells as a % of mock-infected cells and 16E6/18E7 infected cells as a % of mock-infected cells was statistically significant (* p <.05). B) Vector only or 16E6/18E7-expressing CVX-104 cells were treated with GM1 and infected with Pava as described in Figure 4A. Endogenous HPV18 E6 mRNA levels were measured by qRT-PCR as described in Figure 4A. Similar results were obtained in two independent experiments.
Discussion
The experiments reported here demonstrate that HPV oncogene expression is required for the proliferation of primary human cervical cancer cells in culture and provide insight into factors that control tropism of SV40 and SV40-based vectors. Numerous laboratories have demonstrated that established human cervical carcinoma cells lines depend on HPV E6 and E7 for growth or survival (Francis, Schmid, and Howley, 2000; Goodwin and DiMaio, 2000; Hwang et al., 1993; Jiang and Milner, 2002; Parish et al., 2006), but these previous experiments did not determine whether this requirement was an intrinsic property of cervical cancer cells or whether it was acquired during the establishment or propagation of permanent cell lines. Because established cancer cell lines, especially HeLa cells, diverge widely from tumors and primary cancer cells (Carlson, Iyer, and Marcotte, 2007; Sandberg and Ernberg, 2005), it seemed prudent to examine the requirement of E6/E7 in low passage human cancer cells. Furthermore, E6/E7 expression induces human keratinocytes to become resistant to terminal differentiation caused by high calcium and FBS (Munger et al., 1989; Pei et al., 1998; Schlegel et al., 1988; Sherman and Schlegel, 1996), the conditions typically used to culture established cell lines. Therefore, these conditions may allow the proliferation of only those cell that express E6 and E7. If this were the case, E6/E7 dependence is not an intrinsic property of cervical cancer cells, but rather an artifact of the cell culture conditions.
To determine whether cervical cancer cells have an intrinsic requirement for HPV E6 and E7, we examined the behavior of low passage cervical cancer cells cultured under conditions that do not select for E6 and E7 expression. We isolated cells from primary cervical cancers, cultured them in serum-free keratinocyte media with low calcium, and, after a minimal number of passages, analyzed their dependence on HPV E6 and E7. We believe that the early passage numbers at which these cells were tested and the conditions used to maintain them provide a better model for studying E6 and E7 dependence than established cell lines or cells immortalized in vitro. Unfortunately, the original tumors from which our primary cervical cancer strains were isolated no longer exist, so we cannot directly compare them to the primary cell strains analyzed here.
Expression of BPV E2 in all four primary cervical carcinoma cell strains repressed E6 and E7 and caused a significant inhibition of DNA synthesis (6- to 20-fold). Cancer cells as early as three passages after isolation displayed this response. E2 expression had only a slight effect on DNA synthesis of HPV-negative, primary HFFs and HFKs, and a DNA binding-defective E2 mutant had a minimal effect on DNA synthesis in the cancer cells. Moreover, growth arrest was abrogated by exogenous copies of E6 and E7. These results demonstrate that the ability of the E2 protein to bind to and repress the E6/E7 promoter is essential for its ability to induce growth arrest in primary human cervical cancer cells. Taken together, our results indicate that these primary human cervical cancer cells are dependent on E6 and E7 as early as three passages after isolation from patients and suggest that some in vivo human tumors are also dependent on E6/E7 for growth. However, even though the earliest passage cells we tested were highly dependent on HPV E6/E7, we have not ruled out the possibility that time in culture affects the sensitivity of cervical cancer cells to E6/E7 repression.
Repression of the E6 and E7 genes by the E2 protein in CVX-104 and CVX-106 cells led to elevated levels of p53 and active, hypophosphorylated p105Rb. Levels of total p105Rb were not elevated in the primary cervical cancer cells, unlike the situation in the established cell lines, highlighting a biochemical difference between primary cells and established cancer cell lines. In addition, E6/E7 repression in the primary cells resulted in growth arrest, increased autofluorescence and SA-β-galactosidase activity, and cell enlargement and flattening, all markers of cellular senescence. These data imply that repression of E6 and E7 in these primary cells induces senescence as a result of reactivation of p53 and p105Rb, as is the case in HeLa cells (DeFilippis et al., 2003). Senescence in this system is not due to up-regulation of p16ink4a, which is constitutively up-regulated in proliferating cervical cancer cells (Sano et al., 1998), including the primary cells studied here (unpublished data). Since senescence is irreversible (Campisi and d'Adda di Fagagna, 2007), we infer that repression of E6 and E7 in primary cervical carcinoma cells induces permanent growth arrest.
Lambert and colleagues demonstrated that repression of HPV16 E7 in a transgenic mouse model of cervical cancer caused tumor regression (Jabbar et al., 2009). Our results extend these findings from mice to human cells from invasive cancers that had undergone the decades-long process of carcinogenesis in women. The cellular basis of tumor regression in the mouse system has not been established, but in human cervical cancer cells, HPV repression causes senescence, which, like cell transformation, is under markedly different control in human and mouse cells (Balmain and Harris, 2000; Chaturvedi et al., 2004; Dotto, 1998; Goodwin et al., 2000; Newbold, 1997; Rangarajan et al., 2004). Nevertheless, the dependence on HPV oncogenes in primary human cervical cancer cells, established cervical cancer cell lines (and HPV-associated head-and-neck cancer cell lines (Rampias et al., 2009)) and transgenic mice indicates that oncogene dependence is a fundamental feature of HPV-induced cancers.
It was previously shown that human keratinocytes immortalized in vitro with transfected HPV16 DNA and maintained in the absence of FBS and in low concentrations of calcium are dependent on E6 and E7 for continued growth (Lee et al., 2002). However, immortalized cells are very different from cervical carcinoma cells. For example, unlike cells derived from tumors, cells immortalized in vitro by HPV are not tumorigenic in experimental animals (Durst et al., 1987; Lee et al., 2002; Pirisi et al., 1987; Woodworth et al., 1988). Furthermore, the vast majority of cervical high-risk HPV infections do not progress to cancer, and when progression does occur, it typically takes many years. These findings indicate that E6/E7 expression is not sufficient for tumorigenesis and that additional factors and genetic alterations are required for tumor formation in women. Thus, while it is not surprising that cells recently immortalized with E6/E7 in vitro retain their dependence on the proximal immortalizing agent, the relevance of this finding to human cancer cells is unclear.
It is striking that cancer cells retain E6/E7 dependence despite the onslaught of mutagenesis engendered by the viral oncoproteins. Remarkably, this dependence on E6/E7 extends to established cervical cancer cell lines such as HeLa, SiHa, CaSki, which have been exposed to decades of stress and mutagenesis during time spent in culture. It is possible that the viral oncogenes are more effective than acquired mutations at inactivating cellular tumor suppressor pathways or activating mitogenic pathways, thereby eliminating selective pressure for mutations in these pathways. However, expression of dominant-negative p53 blocks p53 signaling more completely than expression of HPV E6 (Butz et al., 1995). Rather, we propose that sustained dependence on the viral oncogenes is a consequence of the multiplicity of pathways that are targeted by the viral oncoproteins. For example, in addition to the p53 pathway, the E6 protein targets telomerase, several PDZ domain-containing proteins, and components of the DNA repair and apoptosis machineries (Howie, Katzenellenbogen, and Galloway, 2009). Because these multiple pathways are unlikely to be activated or inactivated by mutations in one or a few cellular genes, viral proteins are more effective drivers of cell growth and survival than are random mutations occurring in cellular DNA. This implies that therapies directed against individual cellular pathways targeted by the viral oncoproteins are likely to be less effective than therapies against the viral oncogenes themselves.
We have been able to expand primary cultures with high purity from 20–30% of cervical cancer tumor biopsies. Although this rate of success is higher than in a previously reported attempt to culture cervical carcinoma cells (Ku et al., 1997), we do not know if cervical carcinomas that fail to grow in culture also depend on E6 and E7. Nevertheless, our data indicate that E6 and E7 dependence is an intrinsic property of cervical carcinoma cells from at least a substantial fraction of patients and is not a trait that is acquired during the establishment and propagation of permanent cell lines. These findings in turn imply that treatments that inhibit the expression or activity of E6 and E7 may have therapeutic benefit in cervical cancer patients. The cancer isolates we studied were from early stage carcinomas; additional studies should determine whether cells isolated from advanced cervical carcinomas are also dependent on E6 and E7 for growth.
Our ability to infect these primary cancer cells with our E2 vector was informed by an understanding of the factors that control SV40 infection. The efficiency of SV40 infection appears to be dictated in large part by levels of cell-surface GM1, which binds directly to the major viral capsid protein, VP1 (Campanero-Rhodes et al., 2007; Neu et al., 2008; Tsai et al., 2003). In our experiments, CVX-102 and HeLa-Sen2 cells, which displayed the highest level of GM1, were efficiently infected with SV40 and Pava, whereas cells with lower levels of GM1, such as SiHa and CaSki cells and most primary cervical cancer cells, were infected poorly unless supplemented with GM1. These results demonstrated that infection by SV40 vectors can be improved by addition of GM1 to human cells.
There are reports that recombinant SV40 vectors and in vitro assembled SV40 virus-like particles are capable of efficiently transducing many mammalian cell types (Kimchi-Sarfaty et al., 2004; Lund et al., 2005; Strayer et al., 2005). However, by using quantitative measures of early viral gene expression, we found considerable differences in the ability of different cells to be infected by SV40 and SV40-based vectors generated in permissive monkey cells. In fact, seven of nine cell strains tested in this study infected poorly in the absence of GM1 supplementation. Similar differences have been documented by other groups in studies of the mechanism of SV40 infection, which identified GM1 as a cell surface receptor for this virus (Campanero-Rhodes et al., 2007; Low et al., 2004; Tsai et al., 2003). Our demonstration that different cell isolates display marked differences in GM1 cell-surface levels provides an explanation, at least in part, for these differences in infectivity. The addition of GM1 to the culture medium provides a simple and inexpensive method to expand the tropism of SV40 and SV40-based vectors for gene transfer and gene therapy applications.
Materials and Methods
Cells
CV-1, 293-T, and SiHa cells were purchased from American Type Culture Collection (ATTC, Manassas, VA). CV-1 cells were cultured in Minimal Essential Medium (MEM) Eagle, with 10 % FBS, standard antibiotics, 10 mM L-glutamine, and 10 mM HEPES pH 7.2 (standard supplements). HeLa-Sen2 cells were described previously (Goodwin et al., 2000). Normal diploid human foreskin fibroblasts (HFF) were obtained from the Yale Skin Diseases Research Center (YSDRC). CMT4 cells were provided by Y. Gluzman. CMT-4, HeLa-Sen2, HFF, 293-T, and SiHa cells were cultured in Dulbecco’s MEM (DMEM) with standard supplements. CaSki cells were obtained from Michael Reiss (University of Medicine and Dentistry of New Jersey) and cultured in RPMI-1640 with standard supplements. Primary human foreskin keratinocytes (HFK) were obtained from YSDRC and cultured in Epilife medium (Invitrogen, Carlsbad, CA) with 60 µM calcium chloride, human keratinocyte growth supplement (Invitrogen, Carlsbad, CA), and standard antibiotics in the absence of serum. Hybridomas expressing PAb 108 mouse anti-large T antigen (purchased from ATTC) and PAb 597 mouse anti-VP1 (obtained from Edward Harlow, Harvard Medical School)) were cultured in DMEM plus 20% FBS and standard supplements.
CVX-101 to CVX-106 cells were isolated from primary tumors as described previously (Santin et al., 2005). Briefly, viable tumor tissue was harvested from solid tumor biopsies under sterile conditions at room temperature. Samples were mechanically minced, washed with RPMI-1640, and then incubated on a magnetic stirring apparatus in 0.14% collagenase type I (Sigma-Aldrich Corp., St. Louis, MO) and 0.01% DNAse (Sigma, 2000 kU/mg) in RPMI-1640 for 2 hrs at 37°C or overnight at 4°C. Single cell suspensions were generated by filtering enzymatically-dissociated tumor through 150 µM nylon mesh, washing in RPMI-1640 plus 10% human AB serum (Gemini Bioproducts, Calabasas, CA), and culturing in keratinocyte serum-free media (KSFM) (Invitrogen, Carlsbad,CA) supplemented with 35–50 µg/ml bovine pituitary extract, 5 ng/ml recombinant human EGF, 90 µM calcium chloride, and standard antibiotics (KSFM+). We succeeded in establishing primary cervical carcinoma cell strains in approximately 20–30% of attempts. Purity and homogeneity of fresh tumor cultures were tested by morphology, immunohistochemistry staining and/or flow cytometry with antibodies against cytokeratins and p16ink4a (CDKN2). Only primary cultures that had at least 95% viability and contained >99% epithelial cells expressing p16 were used in the experiments described below. HPV genotyping was conducted as described previously (Santin et al., 2005). Primary cell strains were shown to be free of HeLa cell contamination by using a PCR-based assay to detect a novel HeLa cell-specific retro-transposon (Rahbari et al., 2009). All cell isolates and cell lines were confirmed to have originated from different patients by amplifying and sequencing five genomic regions known to contain several single nucleotide polymorphisms (SNPs). The protocol and results for this analysis are available upon request. Cells were also found to be free of mycoplasma by using Mycoscope PCR Detection Kit (Genlantis, San Diego, CA) according to the manufacturer’s instructions. Passage 0 of primary cells is the point at which cells were placed in culture. A passage number was recorded each time a 70–80% confluent 10 cm dish of cells was passed 1:4. Cells were thawed, expanded, and analyzed at the earliest passage available.
To generate cells that stably express exogenous HPV16 E6 and HPV18 E7, CVX-104 cells were first infected with the concentrated LXSN or LXSN 16E6 retrovirus stocks (see below) in KSFM+ and selected with 35 µg/ml G418 for nine days. Cells were then infected with concentrated RVY or RVY 18E7 retrovirus and selected in 20 µg/ml hygromycin for 12 days. Following selection, cells were maintained in KSFM+ with 17.5 µg/ml G418 and 10 µg/ml hygromycin.
Reagents
Monosialoganglioside GM1 from bovine brain was purchased from Sigma-Aldrich Corp. Alexa Fluor 488 donkey anti-mouse IgG (H+L) and Alexa Fluor 488 Cholera Toxin B were purchased from Invitrogen. Mouse monoclonal anti-human p105Rb and mouse monoclonal anti-human p53 were purchased from BD Pharmingen (San Jose, CA). Goat polyclonal anti-humanβ-actin was purchased from Santa Cruz Biotechnology Inc (Santa Cruz, CA). RNeasy kits were purchased from Qiagen (Valencia, CA). iScript cDNA synthesis and iQ SYBR Green Supermix kits were purchased from BioRad (Hercules, CA). Tritiated thymidine was purchased from MP Biomedicals (Solon,OH).
Viruses
WT SV40 strain 776 was generated from pBRSV DNA (obtained from ATTC) in permissive CV-1 cells. A final high titer stock was generated using the Pava harvesting protocol as described previously (Goodwin et al., 1998; Naeger et al., 1999). Titers of WT SV40 stocks were determined by infecting CV-1 cells and measuring the fraction of cells expressing large T antigen (see below) at 24 hrs post infection by immunostaining and flow cytometry. Prior to the generation of SV40 virus from pBRSV DNA, a phenylalanine mutation at position 325 of VP1 found in pBRSV DNA was converted to WT serine by standard site-directed mutagenesis.
BPV1/SV40 recombinant viruses, designated Pava, which encode WT or K339M BPV-E2 were generated and harvested as described previously (Goodwin et al., 1998; Naeger et al., 1999). The E2 repressor and E5 genes were disrupted in WT Pava vector but remained intact in K339M Pava {described in (Goodwin et al., 1998)}. Virus stocks were titered by infecting CMT-4 cells with Pava and measuring the fraction of cells that expressed VP1 by immunostaining and flow cytometry. The protocol used for measuring VP1 expression is identical to the protocol used for measuring large T antigen expression (see below), except cells were fixed in 10% formaldehyde and permeabilized in 10% formaldehyde with 0.2% triton X-100. PAb 597 mouse anti-VP1 was used to measure VP1 expression.
Retroviruses expressing HPV16 E6 and HPV18 E7 were described previously (DeFilippis et al., 2003; Halbert, Demers, and Galloway, 1991). Retrovirus stocks were generated in 293-T cells as described previously (Yates et al., 2008). Virus was harvested at 48 hrs post transfection and concentrated with Peg-It (System Biosciences, Mountain View, CA) according to the manufacturer’s instructions. Briefly, one volume of Peg-It was added to four volumes of retrovirus at 4°C overnight. Precipitated retrovirus was pelleted by centrifugation, washed, and resuspended in KSFM+.
DNA synthesis
To measure DNA synthesis, an equal number of cells were plated in triplicate. Cells were treated with 10 (HeLa, SiHa, CasKi) or 15 µg/ml GM1 (all other cells) overnight in either the medium used to culture cells (keratinocytes and CVX-101 to CVX-106 cells) or DMEM with 1% FBS (all other cells). Cells were then washed with PBS (keratinocytes and CVX-101 to CVX-106 cells) or DMEM with 10% FBS (all other cells lines) and either mock-infected or infected with Pava. At the indicated time post-infection, cells were treated with media containing 1.5 uCi of [3H]thymidine/ml for five to six hours. After precipitating nucleic acids with 10% trichloroacetic acid, DNA synthesis was determined by measuring the incorporation of [3H]thymidine into acid-insoluble material using a liquid scintillation counter.
SDS-PAGE and immunoblotting
For immunoblotting, cells were harvested at the indicated times post-infection and lysed on ice in NP40 buffer (50 mM TRIS pH 8.0, 1 mM EDTA, 150 mM NaCl, 1% NP40, 0.1% SDS, standard protease inhibitors) for 12 minutes. Debris was cleared by centrifugation, and equal quantities of protein were electrophoresed on standard 7.5% or 12% polyacrylamide gels containing SDS. After electrophoresis, proteins were transferred to polyvinylidene fluoride membranes (Millipore, Billerica, MA) in transfer buffer (12.5 mM Tris, 0.1 M glycine, 20% methanol) and blocked in 5% milk/TBST buffer (5% nonfat dry milk, 25 mM Tris-HCl [pH 8.0], 125 mM NaCl, 0.1% Tween-20). The membranes were probed with the following specific antibodies: mouse monoclonal anti-human p105Rb, mouse monoclonal anti-human p53, or goat polyclonal anti-humanβ-actin.
Quantitative reverse transcriptase real-time PCR
Quantitative real-time PCR (qRT-PCR) and primer design were performed as described previously (Johung, Goodwin, and DiMaio, 2007). Briefly, total RNA was harvested using the RNeasy with DNAse treatment or RNeasy Plus kit at indicated times post-infection. 1 µg RNA was used as a template for cDNA synthesis using an iScript cDNA synthesis kit. qRT-PCR was performed by using iQ SYBR Green Supermix with 40 ng cDNA per 20 µl reaction and the BioRad MyiQ Single-color Real-time PCR detection system. GAPDH transcripts were detected using primers 5’-CAGCCTCAAGATCATCAGCA-3’ and 5’-TGTGGTCATGAGTCCTTCCA-3’. BPV-E2 transcripts were detected using 5’-GACGAGGCAGCCAGATTTAG-3’ and 5’GGGTCTCCTTCAGGTCCTTC-3’. HPV16 E6/E7 transcripts were detected using primers 5’-ACAAGCAGAACCGGACAGAG-3’ and 5’-GCCCATTAACAGGTCTTCCA-3’, which annealed to the E7 ORF present in both transcripts. HPV18 E6/E7 transcripts were detected using primers 5’-TGAAATTCCGGTTGACCTTC -3’and 5’-CACGGACACACAAAGGACAG-3’, which annealed to the E7 ORF present in both transcripts. Exogenous HPV18 E7 transcripts were detected using primers 5’-AAGCTCAGCAGACGACCTTC-3’ and 5’-CACAGCCGGATCAGCTTACT-3’, which annealed to the E7 ORF and the 3’UTR of the RVY vector. Endogenous HPV18 E6/E7 transcripts were distinguished from exogenous HPV18 E7 transcripts by using primers 5’-CCAGAAACCGTTGAATCCAG-3’and 5’-GTTGGAGTCGTTCCTGTCGT-3’, which annealed to the HPV18 E6 ORF present in both endogenous transcripts but absent from the exogenous E7 ORF.
SV40-infection and flow cytometry of large T antigen
Cells were treated with GM1 as described above and infected with SV40 at the indicated MOI. At 48 hrs post-infection, cells were trypsinized, neutralized, and washed once with PBS. Cells were then fixed by adding ice cold methanol dropwise to the cell pellet while vortexing gently, and the fixation continued for at least 20 minutes on ice. After centrifugation, methanol was removed, and cells were blocked for 5 min in PBS plus 0.5% bovine serum albumin (BSA). The cell pellet was then resuspended in 100 µl of a 1:1 mixture of 5% normal donkey serum in PBS (NDS-PBS) to PAb 108 monoclonal mouse anti-large T antigen supernatant and incubated for 1 hr at 37°C. The cells were then washed twice with PBS + 0.5% BSA and resuspended in 100 µl NDS-PBS containing a 1:500 dilution of Alexa Fluor 488 donkey anti-mouse IgG and incubated at 37°C for 30 minutes. Following two washes in PBS + 0.5% BSA, the cell pellet was resuspended in 200 µl PBS and kept on ice until analysis. The fraction of cells expressing large T antigen was measured using a 488 nm excitation and 530 nm emission filter on a FACS Calibur flow cytometer (BD Biosciences, San Jose, CA) and plotted on a 650 nm vs 530 nm emission filter 2D-density plot.
Cholera toxin B binding
Cells were mock-treated or treated with GM1 as described above. After removing unincorporated GM1, cells were trypsinized, neutralized, and washed once with ice cold PBS. Cells were then resuspended in 500 µl of ice cold DMEM with 1 µg/ml Alexa Fluor 488 labeled cholera toxin B (CTXB) and incubated at 4°C for 30 min. Cells were then washed twice in cold PBS, resuspended in PBS, and kept on ice until analysis. CTXB binding was analyzed using a 488 nm excitation and 530 nm emission filter on a FACS Calibur flow cytometer.
Senescence assays
To assess autofluoresence and/or granularity, cells were treated with GM1 and infected with Pava at the indicated MOI as described above. Where indicated, the process was repeated a second time to reduce the number of cells that escaped infection. At the indicated times after the first infection with Pava, autofluoresence was measured by flow cytometry of unstained cells using 488 nm excitation and a 650 nm emission filter on a FACS Calibur flow cytometer. Granularity was measured using a 488 nm laser and side scatter channel (SSC). For measuring senescence-associated (SA)-β-galactosidase, cells were treated with GM1 and infected with Pava twice as above. Nine days after the first infection, SA-β-galactosidase staining was performed as described previously (Dimri et al., 1995; Goodwin et al., 2000).
Statistical analysis
A two-tailed T-test was used for all statistical analysis of data.
Conclusions
Low passage human cervical carcinoma cell lines maintained in serum-free medium in low calcium require sustained expression of the human papillomavirus oncogenes for continuous proliferation.
Extinction of HPV oncogene expression in these cells causes cellular senescence. Incubation of primary human cells with the ganglioside GM1 is a simple method to improve infection efficiency by SV40 and SV40-based viral vectors.
Supplementary Material
Acknowledgements
We thank Ivan Martinez and Edward Goodwin for advice and reagents, Daniel Dykas for analysis of SNPs, and Jan Zulkeski for assistance in preparing this manuscript. T.G.M. was supported in part by a training grant from the National Institutes of Health. L.L.A. was supported in part by a training grant and an individual National Research Service Award from the National Institutes of Health. This work was supported by a grant from the National Cancer Institute to DD (P01 CA016038).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arbeit JM, Howley PM, Hanahan D. Chronic estrogen-induced cervical and vaginal squamous carcinogenesis in human papillomavirus type 16 transgenic mice. Proc Natl Acad Sci U S A. 1996;93(7):2930–2935. doi: 10.1073/pnas.93.7.2930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker CC, Phelps WC, Lindgren V, Braun MJ, Gonda MA, Howley PM. Structural and transcriptional analysis of human papillomavirus type 16 sequences in cervical carcinoma cell lines. J Virol. 1987;61(4):962–971. doi: 10.1128/jvi.61.4.962-971.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balmain A, Harris CC. Carcinogenesis in mouse and human cells: parallels and paradoxes. Carcinogenesis. 2000;21(3):371–377. doi: 10.1093/carcin/21.3.371. [DOI] [PubMed] [Google Scholar]
- Bignotti E, Tassi RA, Calza S, Ravaggi A, Romani C, Rossi E, Falchetti M, Odicino FE, Pecorelli S, Santin AD. Differential gene expression profiles between tumor biopsies and short-term primary cultures of ovarian serous carcinomas: identification of novel molecular biomarkers for early diagnosis and therapy. Gynecol Oncol. 2006;103(2):405–416. doi: 10.1016/j.ygyno.2006.03.056. [DOI] [PubMed] [Google Scholar]
- Bosch FX, Lorincz A, Munoz N, Meijer CJ, Shah KV. The causal relation between human papillomavirus and cervical cancer. J Clin Pathol. 2002;55(4):244–265. doi: 10.1136/jcp.55.4.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butz K, Shahabeddin L, Geisen C, Spitkovsky D, Ullmann A, Hoppe-Seyler F. Functional p53 protein in human papillomavirus-positive cancer cells. Oncogene. 1995;10(5):927–936. [PubMed] [Google Scholar]
- Campanero-Rhodes MA, Smith A, Chai W, Sonnino S, Mauri L, Childs RA, Zhang Y, Ewers H, Helenius A, Imberty A, Feizi T. N-glycolyl GM1 ganglioside as a receptor for simian virus 40. J Virol. 2007;81(23):12846–12858. doi: 10.1128/JVI.01311-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campisi J, d'Adda di Fagagna F. Cellular senescence: when bad things happen to good cells. Nat Rev Mol Cell Biol. 2007;8(9):729–740. doi: 10.1038/nrm2233. [DOI] [PubMed] [Google Scholar]
- Carlson MW, Iyer VR, Marcotte EM. Quantitative gene expression assessment identifies appropriate cell line models for individual cervical cancer pathways. BMC Genomics. 2007;8:117. doi: 10.1186/1471-2164-8-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaturvedi V, Bacon P, Bodner B, Nickoloff BJ. Proliferating cultured human keratinocytes are more susceptible to apoptosis compared with mouse keratinocytes. J Invest Dermatol. 2004;123(6):1200–1203. doi: 10.1111/j.0022-202X.2004.23514.x. [DOI] [PubMed] [Google Scholar]
- Chinnapen DJ, Chinnapen H, Saslowsky D, Lencer WI. Rafting with cholera toxin: endocytosis and trafficking from plasma membrane to ER. FEMS Microbiol Lett. 2007;266(2):129–137. doi: 10.1111/j.1574-6968.2006.00545.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dairkee SH, Ji Y, Ben Y, Moore DH, Meng Z, Jeffrey SS. A molecular 'signature' of primary breast cancer cultures; patterns resembling tumor tissue. BMC Genomics. 2004;5(1):47. doi: 10.1186/1471-2164-5-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeFilippis RA, Goodwin EC, Wu L, DiMaio D. Endogenous human papillomavirus E6 and E7 proteins differentially regulate proliferation, senescence, and apoptosis in HeLa cervical carcinoma cells. J. Virol. 2003;77:1551–1563. doi: 10.1128/JVI.77.2.1551-1563.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desaintes C, Demeret C, Goyat S, Yaniv M, Thierry F. Expression of the papillomavirus E2 protein in HeLa cells leads to apoptosis. EMBO J. 1997;16:504–514. doi: 10.1093/emboj/16.3.504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desaintes C, Goyat S, Garbay S, Yaniv M, Thierry F. Papillomavirus E2 induces p53- independent apoptosis in HeLa cells. Oncogene. 1999;18:4538–4545. doi: 10.1038/sj.onc.1202818. [DOI] [PubMed] [Google Scholar]
- Dimri GP, Lee X, Basile G, Acosta M, Scott G, Roskelley C, Medrano EE, Linskens M, Rubelj I, Pereira-Smith O, et al. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc Natl Acad Sci U S A. 1995;92(20):9363–9367. doi: 10.1073/pnas.92.20.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dotto P. The keratinocyte growth-differentiation switch. Front Biosci. 1998;3:d502–d508. doi: 10.2741/a297. [DOI] [PubMed] [Google Scholar]
- Dowhanick JJ, McBride AA, Howley PM. Suppression of cellular proliferation by the papillomavirus E2 protein. J. Virol. 1995;69:7791–7799. doi: 10.1128/jvi.69.12.7791-7799.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durst M, Dzarlieva-Petrusevska RT, Boukamp P, Fusenig NE, Gissmann L. Molecular and cytogenetic analysis of immortalized human primary keratinocytes obtained after transfection with human papillomavirus type 16 DNA. Oncogene. 1987;1(3):251–256. [PubMed] [Google Scholar]
- Ertel A, Verghese A, Byers SW, Ochs M, Tozeren A. Pathway-specific differences between tumor cell lines and normal and tumor tissue cells. Mol Cancer. 2006;5(1):55. doi: 10.1186/1476-4598-5-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis DA, Schmid SI, Howley PM. Repression of the integrated papillomavirus E6/E7 promoter is required for growth suppression of cervical cancer cells. J Virology. 2000;74:2679–2686. doi: 10.1128/jvi.74.6.2679-2686.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin EC, DiMaio D. Repression of human papillomavirus oncogenes in HeLa cervical carcinoma cells causes the orderly reactivation of dormant tumor suppressor pathways. Proc Natl Acad Sci USA. 2000;97:12513–12518. doi: 10.1073/pnas.97.23.12513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin EC, Naeger LK, Breiding DE, Androphy EJ, DiMaio D. Transactivation-competent bovine papillomavirus E2 protein is specifically required for efficient repression of human papillomavirus oncogene expression and for acute growth inhibition of cervical carcinoma cell lines. J. Virol. 1998;72:3925–3934. doi: 10.1128/jvi.72.5.3925-3934.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin EC, Yang E, Lee CJ, Lee HW, DiMaio D, Hwang ES. Rapid induction of senescence in human cervical carcinoma cells. Proc Natl Acad Sci U S A. 2000;97(20):10978–10983. doi: 10.1073/pnas.97.20.10978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halbert CL, Demers GW, Galloway DA. The E7 gene of human papillomavirus type 16 is sufficient for immortalization of human epithelial cells. J. Virol. 1991;65:473–478. doi: 10.1128/jvi.65.1.473-478.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawley-Nelson P, Vousden KH, Hubbert NL, Lowy DR, Schiller JT. HPV16 E6 and E7 proteins cooperate to immortalize human foreskin keratinocytes. EMBO J. 1989;8(12):3905–3910. doi: 10.1002/j.1460-2075.1989.tb08570.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howie HL, Katzenellenbogen RA, Galloway DA. Papillomavirus E6 proteins. Virology. 2009;384(2):324–334. doi: 10.1016/j.virol.2008.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson JB, Bedell MA, McCance DJ, Laiminis LA. Immortalization and altered differentiation of human keratinocytes in vitro by the E6 and E7 open reading frames of human papillomavirus type 18. J Virol. 1990;64(2):519–526. doi: 10.1128/jvi.64.2.519-526.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang ES, Riese DJ, 2nd, Settleman J, Nilson LA, Honig J, Flynn S, DiMaio D. Inhibition of cervical carcinoma cell line proliferation by the introduction of a bovine papillomavirus regulatory gene. J Virol. 1993;67(7):3720–3729. doi: 10.1128/jvi.67.7.3720-3729.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jabbar SF, Abrams L, Glick A, Lambert PF. Persistence of high-grade cervical dysplasia and cervical cancer requires the continuous expression of the human papillomavirus type 16 E7 oncogene. Cancer Res. 2009;69(10):4407–4414. doi: 10.1158/0008-5472.CAN-09-0023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang M, Milner J. Selective silencing of viral gene expression in HPV-positive human cervical carcinoma cells treated with siRNA, a primer of RNA interference. Oncogene. 2002;21:6041–6048. doi: 10.1038/sj.onc.1205878. [DOI] [PubMed] [Google Scholar]
- Johung K, Goodwin EC, DiMaio D. Human papillomavirus E7 repression in cervical carcinoma cells initiates a transcriptional cascade driven by the retinoblastoma family, resulting in senescence. J Virol. 2007;81(5):2102–2116. doi: 10.1128/JVI.02348-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimchi-Sarfaty C, Alexander NS, Brittain S, Ali S, Gottesman MM. Transduction of multiple cell types using improved conditions for gene delivery and expression of SV40 pseudovirions packaged in vitro. Biotechniques. 2004;37(2):270–275. doi: 10.2144/04372RR04. [DOI] [PubMed] [Google Scholar]
- Ku JL, Kim WH, Park HS, Kang SB, Park JG. Establishment and characterization of 12 uterine cervical-carcinoma cell lines: common sequence variation in the E7 gene of HPV1-16-positive cell lines. Int J Cancer. 1997;72(2):313–320. doi: 10.1002/(sici)1097-0215(19970717)72:2<313::aid-ijc19>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- Lee CJ, Suh EJ, Kang HT, Im JS, Um SJ, Park JS, Hwang ES. Induction of senescence-like state and suppression of telomerase activity through inhibition of HPV E6/E7 gene expression in cells immortalized by HPV16 DNA. Exp. Cell Res. 2002;277:173–182. doi: 10.1006/excr.2002.5554. [DOI] [PubMed] [Google Scholar]
- Lee J, Kotliarova S, Kotliarov Y, Li A, Su Q, Donin NM, Pastorino S, Purow BW, Christopher N, Zhang W, Park JK, Fine HA. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell. 2006;9(5):391–403. doi: 10.1016/j.ccr.2006.03.030. [DOI] [PubMed] [Google Scholar]
- Low J, Humes HD, Szczypka M, Imperiale M. BKV and SV40 infection of human kidney tubular epithelial cells in vitro. Virology. 2004;323(2):182–188. doi: 10.1016/j.virol.2004.03.027. [DOI] [PubMed] [Google Scholar]
- Lund PE, Hunt RC, Gottesman MM, Kimchi-Sarfaty C. Pseudovirions as vehicles for the delivery of siRNA. Pharm Res. 2005;27(3):400–420. doi: 10.1007/s11095-009-0012-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May M, Dong XP, Beyer-Finkler E, Stubenrauch F, Fuchs PG, Pfister H. The E6/E7 promoter of extrachromosomal HPV16 DNA in cervical cancers escapes from cellular repression by mutation of target sequences for YY1. EMBO J. 1994;13(6):1460–1466. doi: 10.1002/j.1460-2075.1994.tb06400.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride AA, Romanczuk H, Howley PM. The papillomavirus E2 regulatory proteins. J. Biol. Chem. 1991;266:18411–18414. [PubMed] [Google Scholar]
- Moody CA, Laimins LA. Human papillomavirus oncoproteins: pathways to transformation. Nat Rev Cancer. 2010;10(8):550–560. doi: 10.1038/nrc2886. [DOI] [PubMed] [Google Scholar]
- Moon MS, Lee CJ, Um SJ, Park JS, Yang JM, Hwang ES. Effect of BPV1 E2-mediated inhibition of E6/E7 expression in HPV16-positive cervical carcinoma cells. Gynecologic Oncology. 2001;80:168–175. doi: 10.1006/gyno.2000.6053. [DOI] [PubMed] [Google Scholar]
- Munger K, Phelps WC, Bubb V, Howley PM, Schlegel R. The E6 and E7 genes of the human papillomavirus type 16 together are necessary and sufficient for transformation of primary human keratinocytes. J Virol. 1989;63(10):4417–4421. doi: 10.1128/jvi.63.10.4417-4421.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naeger LK, Goodwin EC, Hwang ES, DeFilippis RA, Zhang H, DiMaio D. Bovine papillomavirus E2 protein activates a complex growth-inhibitory program in p53-negative HT-3 cervical carcinoma cells that includes repression of cyclin A and cdc25A phosphatase genes and accumulation of hypophosphorylated retinoblastoma protein. Cell Growth Differ. 1999;10:413–422. [PubMed] [Google Scholar]
- Neu U, Woellner K, Gauglitz G, Stehle T. Structural basis of GM1 ganglioside recognition by simian virus 40. Proc Natl Acad Sci U S A. 2008;105(13):5219–5224. doi: 10.1073/pnas.0710301105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newbold RF. Genetic control of telomerase and replicative senescence in human and rodent cells. Ciba Found Symp. 1997;211:177–189. doi: 10.1002/9780470515433.ch12. discussion 189-97. [DOI] [PubMed] [Google Scholar]
- Parish JL, Kowalczyk A, Chen HT, Roeder GE, Sessions R, Buckle M, Gaston K. E2 proteins from high- and low-risk human papillomavirus types differ in their ability to bind p53 and induce apoptotic cell death. J Virol. 2006;80(9):4580–4590. doi: 10.1128/JVI.80.9.4580-4590.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei XF, Sherman L, Sun YH, Schlegel R. HPV-16 E7 protein bypasses keratinocyte growth inhibition by serum and calcium. Carcinogenesis. 1998;19:1481–1486. doi: 10.1093/carcin/19.8.1481. [DOI] [PubMed] [Google Scholar]
- Pirisi L, Yasumoto S, Feller M, Doniger J, DiPaolo JA. Transformation of human fibroblasts and keratinocytes with human papillomavirus type 16 DNA. J Virol. 1987;61(4):1061–1066. doi: 10.1128/jvi.61.4.1061-1066.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash SS, Grossman SR, Pepinsky RB, Laimins LA, Androphy EJ. Amino acids necessary for DNA contact and dimerization imply novel motifs in the papillomavirus E2 trans-activator. Genes Dev. 1992;6(1):105–116. doi: 10.1101/gad.6.1.105. [DOI] [PubMed] [Google Scholar]
- Rahbari R, Sheahan T, Modes V, Collier P, Macfarlane C, Badge RM. A novel L1 retrotransposon marker for HeLa cell line identification. Biotechniques. 2009;46(4):277–284. doi: 10.2144/000113089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rampias T, Sasaki C, Weinberger P, Psyrri A. E6 and e7 gene silencing and transformed phenotype of human papillomavirus 16-positive oropharyngeal cancer cells. J Natl Cancer Inst. 2009;101(6):412–423. doi: 10.1093/jnci/djp017. [DOI] [PubMed] [Google Scholar]
- Rangarajan A, Hong SJ, Gifford A, Weinberg RA. Species- and cell type-specific requirements for cellular transformation. Cancer Cell. 2004;6(2):171–183. doi: 10.1016/j.ccr.2004.07.009. [DOI] [PubMed] [Google Scholar]
- Sandberg R, Ernberg I. Assessment of tumor characteristic gene expression in cell lines using a tissue similarity index (TSI) Proc Natl Acad Sci U S A. 2005;102(6):2052–2057. doi: 10.1073/pnas.0408105102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sano T, Oyama T, Kashiwabara K, Fukuda T, Nakajima T. Expression status of p16 protein is associated with human papillomavirus oncogenic potential in cervical and genital lesions. Am J Pathol. 1998;153(6):1741–1748. doi: 10.1016/S0002-9440(10)65689-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santin AD, Zhan F, Bellone S, Palmieri M, Cane S, Bignotti E, Anfossi S, Gokden M, Dunn D, Roman JJ, O'Brien TJ, Tian E, Cannon MJ, Shaughnessy J, Jr, Pecorelli S. Gene expression profiles in primary ovarian serous papillary tumors and normal ovarian epithelium: identification of candidate molecular markers for ovarian cancer diagnosis and therapy. Int J Cancer. 2004;112(1):14–25. doi: 10.1002/ijc.20408. [DOI] [PubMed] [Google Scholar]
- Santin AD, Zhan F, Bignotti E, Siegel ER, Cane S, Bellone S, Palmieri M, Anfossi S, Thomas M, Burnett A, Kay HH, Roman JJ, O'Brien TJ, Tian E, Cannon MJ, Shaughnessy J, Jr, Pecorelli S. Gene expression profiles of primary HPV16- and HPV18-infected early stage cervical cancers and normal cervical epithelium: identification of novel candidate molecular markers for cervical cancer diagnosis and therapy. Virology. 2005;331(2):269–291. doi: 10.1016/j.virol.2004.09.045. [DOI] [PubMed] [Google Scholar]
- Schlegel R, Phelps WC, Zhang YL, Barbosa M. Quantitative keratinocyte assay detects two biological activities of human papillomavirus DNA and identifies viral types associated with cervical carcinoma. EMBO J. 1988;7(10):3181–3187. doi: 10.1002/j.1460-2075.1988.tb03185.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider-Maunoury S, Croissant O, Orth G. Integration of human papillomavirus type 16 DNA sequences: a possible early event in the progression of genital tumors. J Virol. 1987;61(10):3295–3298. doi: 10.1128/jvi.61.10.3295-3298.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz E, Freese UK, Gissmann L, Mayer W, Roggenbuck B, Stremlau A, zur Hausen H. Structure and transcription of human papillomavirus sequences in cervical carcinoma cells. Nature. 1985;314(6006):111–114. doi: 10.1038/314111a0. [DOI] [PubMed] [Google Scholar]
- Schwarzmann G. Uptake and metabolism of exogenous glycosphingolipids by cultured cells. Semin Cell Dev Biol. 2001;12(2):163–171. doi: 10.1006/scdb.2000.0233. [DOI] [PubMed] [Google Scholar]
- Settleman J, DiMaio D. Efficient transactivation and morphologic transformation by bovine papillomavirus genes expressed from a bovine papillomavirus/simian virus 40 recombinant virus. Proc. Natl. Acad. Sci. USA. 1988;85:9007–9011. doi: 10.1073/pnas.85.23.9007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman L, Schlegel R. Serum- and calcium-induced differentiation of human keratinocytes is inhibited by the E6 oncoprotein of human papillomavirus type 16. J Virol. 1996;70(5):3269–3279. doi: 10.1128/jvi.70.5.3269-3279.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strayer DS, Cordelier P, Kondo R, Liu B, Matskevich AA, McKee HJ, Nichols CN, Mitchell CB, Geverd DA, White MK, Strayer MS. What they are, how they work and why they do what they do? The story of SV40-derived gene therapy vectors and what they have to offer. Curr Gene Ther. 2005;5(2):151–165. doi: 10.2174/1566523053544281. [DOI] [PubMed] [Google Scholar]
- Thierry F, Yaniv M. The BPV1-E2 trans-acting protein can be either an activator or a repressor of the HPV18 regulatory region. EMBO J. 1987;6:3391–3397. doi: 10.1002/j.1460-2075.1987.tb02662.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai B, Gilbert JM, Stehle T, Lencer W, Benjamin TL, Rapoport TA. Gangliosides are receptors for murine polyoma virus and SV40. EMBO J. 2003;22(17):4346–4355. doi: 10.1093/emboj/cdg439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turke AB, Zejnullahu K, Wu YL, Song Y, Dias-Santagata D, Lifshits E, Toschi L, Rogers A, Mok T, Sequist L, Lindeman NI, Murphy C, Akhavanfard S, Yeap BY, Xiao Y, Capelletti M, Iafrate AJ, Lee C, Christensen JG, Engelman JA, Janne PA. Preexistence and clonal selection of MET amplification in EGFR mutant NSCLC. Cancer Cell. 2010;17(1):77–88. doi: 10.1016/j.ccr.2009.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Staveren WC, Solis DW, Delys L, Duprez L, Andry G, Franc B, Thomas G, Libert F, Dumont JE, Detours V, Maenhaut C. Human thyroid tumor cell lines derived from different tumor types present a common dedifferentiated phenotype. Cancer Res. 2007;67(17):8113–8120. doi: 10.1158/0008-5472.CAN-06-4026. [DOI] [PubMed] [Google Scholar]
- van Staveren WC, Solis DY, Hebrant A, Detours V, Dumont JE, Maenhaut C. Human cancer cell lines: Experimental models for cancer cells in situ? For cancer stem cells? Biochim Biophys Acta. 2009;1795(2):92–103. doi: 10.1016/j.bbcan.2008.12.004. [DOI] [PubMed] [Google Scholar]
- von Zglinicki T, Nilsson E, Docke WD, Brunk UT. Lipofuscin accumulation and ageing of fibroblasts. Gerontology. 1995;41 Suppl 2:95–108. doi: 10.1159/000213728. [DOI] [PubMed] [Google Scholar]
- Wells SI, Francis DA, Karpova AY, Dowhanick JJ, Benson JD, Howley PM. Papillomavirus E2 induces senescence in HPV-positive cells via pRB- and p21CIP-dependent pathways. EMBO J. 2000;19:5762–5771. doi: 10.1093/emboj/19.21.5762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodworth CD, Bowden PE, Doniger J, Pirisi L, Barnes W, Lancaster WD, DiPaolo JA. Characterization of normal human exocervical epithelial cells immortalized in vitro by papillomavirus types 16 and 18 DNA. Cancer Res. 1988;48(16):4620–4628. [PubMed] [Google Scholar]
- Yates KE, Korbel GA, Shtutman M, Roninson IB, DiMaio D. Repression of the SUMO-specific protease Senp1 induces p53-dependent premature senescence in normal human fibroblasts. Aging Cell. 2008;7(5):609–621. doi: 10.1111/j.1474-9726.2008.00411.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.