Abstract
CASPASE 8 initiates apoptosis downstream of TNF death receptors by undergoing autocleavage and processing the executioner CASPASE 31. However, the dominant function of CASPASE 8 is to transmit a pro-survival signal that suppresses programmed necrosis (or necroptosis) mediated by RIPK1 and RIPK32–6 during embryogenesis and hematopoiesis7–9. Suppression of necrotic cell death by CASPASE 8 requires its catalytic activity but not the autocleavage essential for apoptosis10, however, the key substrate processed by CASPASE 8 to block necrosis has been elusive. A key substrate must meet three criteria: (1) it must be essential for programmed necrosis; (2) it must be cleaved by CASPASE 8 in situations where CASPASE 8 is blocking necrosis; and (3) mutation of the CASPASE 8 processing site on the substrate should convert a pro-survival response to necrotic death without the need for CASPASE 8 inhibition. We now identify CYLD as a novel substrate for CASPASE 8 that satisfies these criteria. Upon TNF stimulation, CASPASE 8 cleaves CYLD to generate a survival signal. In contrast, loss of CASPASE 8 prevented CYLD degradation resulting in necrotic death. A CYLD substitution mutation at D215 that cannot be cleaved by CASPASE 8 switches cell survival to necrotic cell death in response to TNF.
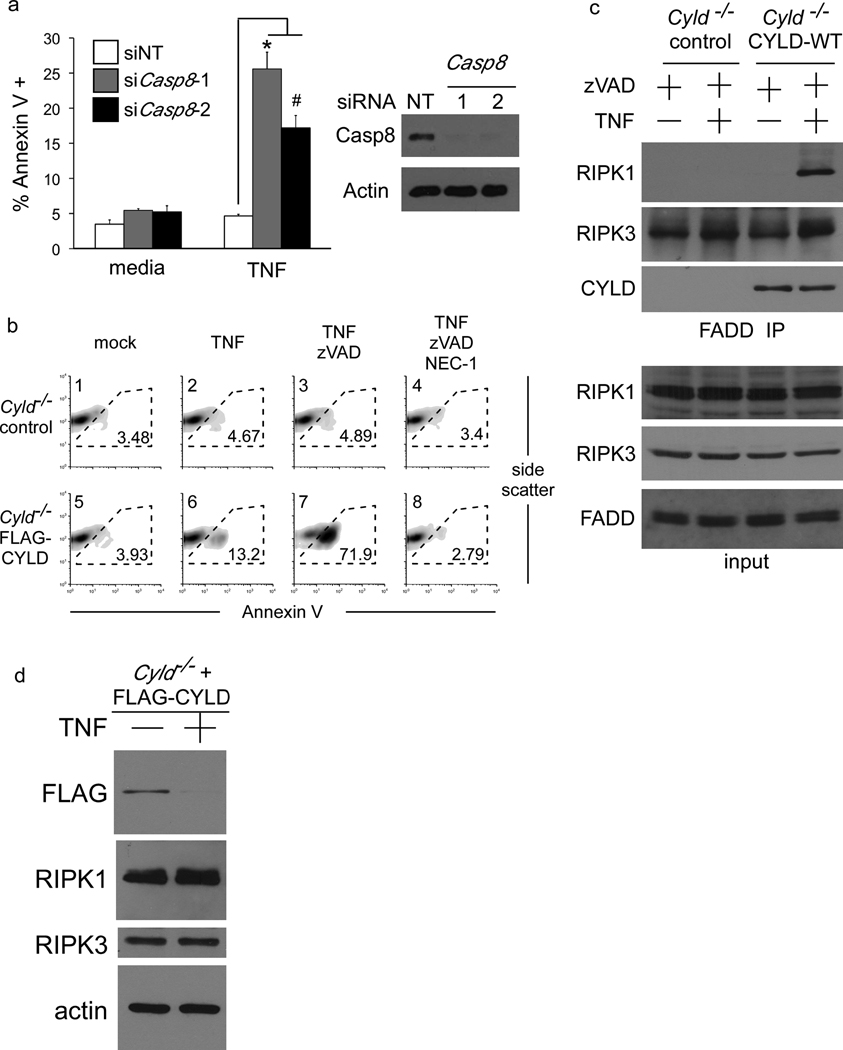
In mouse embryonic fibroblasts (MEFs), knockdown of CASPASE 8 sensitises cells to programmed necrosis upon TNF treatment, which confirms that endogenous CASPASE 8 functions as a pro-survival molecule in this cell-type (Figure 1a). CYLD was pinpointed as a key requirement for necrosis of L929 mouse fibrosarcoma cells by siRNA screen11. We observed that Cyld−/− MEFs remained viable when stimulated with TNF in the presence of the pan-caspase inhibitor zVAD-fmk, whereas Cyld−/− MEFs complemented with exogenous FLAG-CYLD rapidly died by programmed necrosis when caspase activity was blocked (Figure 1b), confirming that CYLD is essential for necrotic cell death (CRITERIA #1). Immunoprecipitation of FADD from CYLD-expressing and control Cyld−/− MEFs treated with TNF in the presence of zVAD-fmk revealed that recruitment of RIPK1 to the FADD necrosome is strictly dependent on CYLD (Figure 1c). To our surprise, immunoblotting to detect the ectopic CYLD in the reconstituted MEFs revealed that CYLD protein was rapidly lost upon TNF stimulation (Figure 1d). In contrast, protein levels of RIPK1 and RIPK3 were relatively unchanged suggesting that removal of CYLD may regulate necrosis.
Figure 1. CYLD is essential for necrosis.
(a) Wild-type MEFs transfected with two different Casp8 targeting RNAi oligos were stimulated with TNF for 24 hours and necrotic cell death quantified by Annexin V staining and flow cytometry. The mean percentage of cells that are Annexin V + is shown and the error bars display the standard deviation of each group (non-targetting n=3, siCasp8-1 n=4, siCasp8-2 n=5) and * denotes p=0.00035 and # denotes p=0.00061. (b) Cyld−/− MEFs reconstituted with a vector control or FLAG-CYLD were stimulated with TNF in the presence of zVAD-fmk or Necrostatin-1 (NEC-1). The percentage of cells undergoing necrosis (Annexin V +) after 24 hours is shown. (c) FADD was immunoprecipitated from MEFs described in (b) after stimulation with TNF for 90 minutes in the presence of zVAD-fmk and the isolated FADD complexes were immunoblotted for RIPK1, RIPK3 and FLAG-CYLD in the upper 3 panels. The lower 4 panels show immunoblots of the corresponding whole cell lysates. (d) Immunoblot of lysates from MEFs described in (b) 6 hours after TNF stimulation.
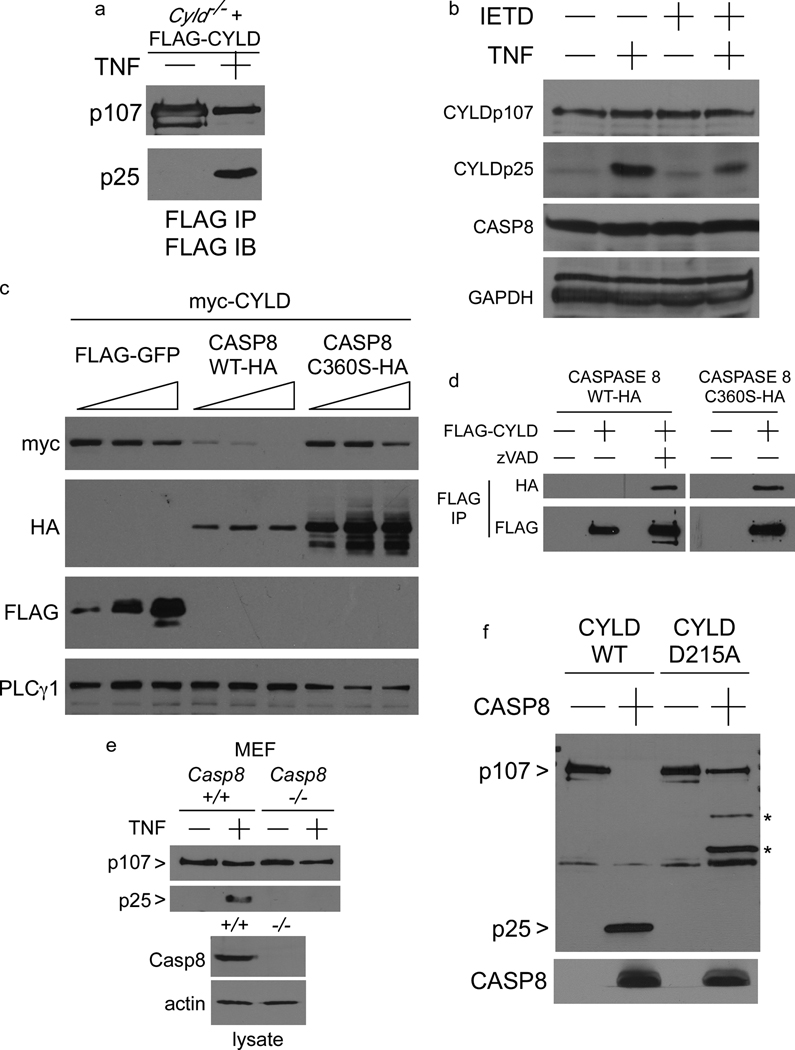
In order to examine whether degradation of CYLD observed in TNF stimulated MEFs was due to proteolytic cleavage, FLAG-CYLD was immunoprecipitated from the reconstituted Cyld−/− MEFs and blotted with the same antibody. A FLAG-tagged product from CYLD of approximately 25kDa (CYLDp25) was detected upon TNF stimulation (Figure 2a) suggesting that CYLD undergoes cleavage. Furthermore, the 25kDa cleavage product from endogenous CYLD was similarly detected in untransfected wildtype MEFs (Figure 2b). We hypothesised that CYLD protein might be regulated by active CASPASE 8, particularly since computational analysis also indicated a relationship between CASPASE 8 and CYLD gene expression levels (Supplementary Figure 1), especially in lymphoid cells. Consistent with this hypothesis, the CASPASE 8 inhibitor IETD-fmk reduced the level of the CYLDp25 fragment. Co-transfection of HEK 293 cells revealed that over-expression of wild-type CASPASE 8, but not the catalytically inactive mutant CASPASE 8-C360S, causes degradation of CYLD protein (Figure 2c). Interaction between transfected CYLD and CASPASE 8 by co-immunoprecipitation was observed only when the activity of CASPASE 8 was blocked by the pan-caspase inhibitor zVAD-fmk, or by mutation of the CASPASE 8 active site, suggesting that CYLD is a substrate for proteolytic cleavage by CASPASE 8 (Figure 2d). To provide genetic evidence that CASPASE 8 is cleaving CYLD, FLAG-CYLD was stably expressed in Casp8+/+ and Casp8−/− MEFs and the cells were stimulated with TNF. The p25 fragment was not detected in the Casp8−/− MEFs (Figure 2e) indicating that CYLDp25 is a product of the proteolytic cleavage of CYLD by CASPASE 8, under conditions where active CASPASE 8 suppresses necrosis7, 12 (CRITERIA #2).
Figure 2. CYLD is a substrate for proteolysis by CASPASE 8.
(a) FLAG immunoprecipitations from Cyld−/− MEFs reconstituted with a vector control or FLAG-CYLD stimulated for 6 hours with TNF were blotted with anti-FLAG. Full-length unprocessed CYLD is indicated by CYLDp107, cleaved CYLD is shown as CYLDp25. (b) Immunoblot of lysates from wild-type MEFs 5 hours after stimulation with TNF, in the presence of the CASPASE 8 inhibitor IETD-fmk, as indicated. The anti-CYLD used was raised against the N-terminus of CYLD to detect the endogenous CYLDp25 fragment. (c) Immunoblot of HEK 293EBNA lysates 24 hours after transfection with the indicated plasmids. (d) Immunoblot of HA-CASPASE 8 proteins co-immunoprecipitating with FLAG-CYLD 24 hours post-transfection in the presence of vehicle or zVAD-fmk, as indicated. (e) FLAG-CYLD stably-expressed in Casp8+/+ and Casp8−/− MEFs was immunoprecipitated after stimulation with TNF for 5 hours and immunoblotted with FLAG-specific antibody to detect unprocessed CYLD (p107) and cleaved CYLD (p25). (f) FLAG-CYLD-WT and D215A proteins stably expressed in Cyld−/− MEFs were purified by anti-FLAG immunoaffinity, eluted with FLAG peptide, and then incubated with recombinant CASPASE 8 for 2 hours. The reaction mixture was immunoblotted with FLAG-specific antibody to detect full length CYLDp107 and the cleavage product CYLDp25, followed by CASPASE 8-specific antibody to detect the active CASPASE 8 p18 fragment. The bands indicated with * correspond to cryptic cleavage products generated from the CYLD-D215A protein.
In the MEF model, CASPASE 8 generates a survival signal. Since CASPASE 8 is also known to initiate apoptosis, which antagonizes programmed necrosis, we examined whether CYLD is similarly cleaved by CASPASE 8 during apoptosis in the human Jurkat T cell model. Levels of endogenous CYLD were decreased upon TNF stimulation of SMAC mimetic13-treated parental Jurkat T cells (clone A3) but not in CASPASE 8-deficient mutant clone I9.2 (Supplementary Figure 2a). Likewise, endogenous CYLD was cleaved to generate CYLDp25 when CASPASE 8 was activated following TNF stimulation of NEMO-deficient Jurkat T cells (clone 832114, Supplementary Figure 2c). CYLD was not cleaved in CASPASE 8-deficient Jurkat T cells treated with TNF and SMAC mimetic (Supplementary Figure 2a, 2d), which failed to undergo apoptosis but instead died by programmed necrosis5 (Supplementary Figure 2b). Similar to the MEF model, a correlation between the stabilisation of CYLD and necrosis is also observed in the Jurkat T cell model. Finally, we examined CYLD cleavage in a third model using murine L929 cells. Cleavage of endogenous CYLD (Supplementary Figure 3a) or transfected FLAG-tagged CYLD (Supplementary Figure 3b) could be detected in TNF-treated L929 cells in a caspase-dependent manner. When Caspase 8 activity was inhibited, L929 cells died by necrosis, which was blocked by knockdown of either CYLD or RIPK3 (Supplementary Figure 3c). Processing of CYLD to CYLDp25 was also observed in SMAC mimetic and TNF-treated MCF7 human breast cancer cells (Supplementary Figure 4a), which are deficient in CASPASE 315, indicating that CASPASE 3 is not required for CYLD cleavage. Therefore CYLD cleavage by CASPASE 8 is observed whether it is generating a survival or an apoptotic death signal, and CYLD stabilisation coincided with cells undergoing necrosis. To provide further corroboration that CASPASE 8 activity was required for CYLD cleavage, we stably expressed CrmA, a viral inhibitor of CASPASE 8 and a known trigger of programmed necrosis2, 4, in FLAG-CYLD-expressing MEFs. CrmA prevented processing of CYLD to CYLDp25 (Supplementary Figure 4b) whereas an aspartate to alanine mutation that abrogates the ability of CrmA to inhibit CASPASE 8 (CrmA-D303A) was unable to prevent cleavage of CYLD. Likewise, wildtype CrmA expression predisposed MEFs to programmed necrosis upon TNF treatment whereas mutant CrmA did not (Supplementary Figure 4c). Therefore, the genetic data from MEFs and Jurkat T cells indicate that the cleavage of CYLD upon TNF treatment requires CASPASE 8. The data with the pharmacological and viral inhibitors of CASPASE 8 activity suggest that proteolytically active CASPASE 8 is required for processing of CYLD. To test whether CASPASE 8 could directly cleave CYLD, affinity-purified FLAG-tagged CYLD protein was incubated with recombinant CASPASE 8. Purified CYLD protein was processed by recombinant CASPASE 8 to generate the N-terminal CYLDp25 fragment (Figure 2f). Therefore, CASPASE 8 directly cleaves CYLD.
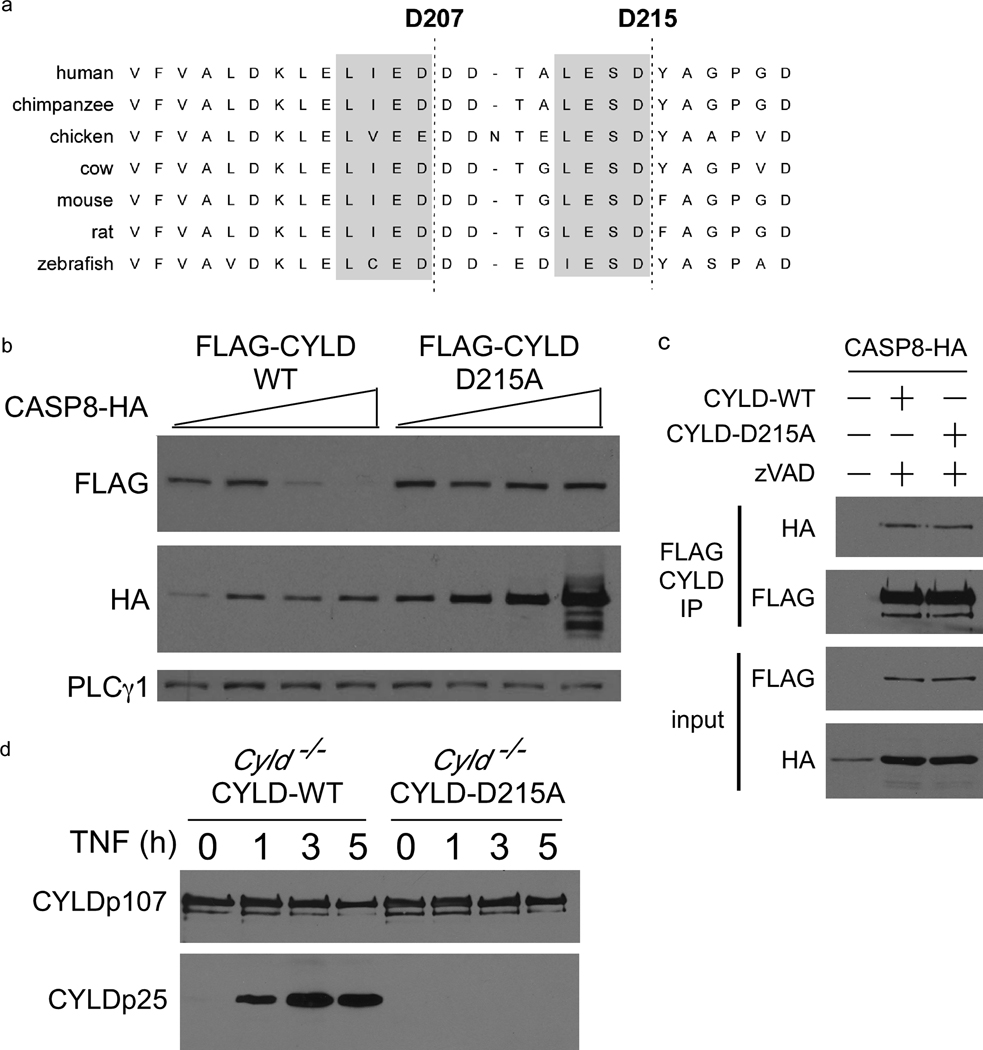
Amino acid sequences of CYLD from several species were aligned to identify a conserved CASPASE 8 cleavage site motif (LxxD) that would produce an N-terminal 25kDa fragment (Figure 3a). Two potential motifs were identified and mutation of D215 was sufficient to prevent proteolysis of CYLD by CASPASE 8. Upon co-expression in HEK 293 cells, CYLD-D215A was more resistant to degradation by CASPASE 8 (Figure 3b) even though both CYLD-WT and CYLD-D215A co-precipitated with CASPASE 8 (Figure 3c) to the same extent. The D215A mutation abrogated the appearance of CYLDp25 in TNF-stimulated reconstituted Cyld−/− MEFs (Figure 3d) or in Jurkat T cells and L929 cells (Supplementary Figure 2e and 3b, respectively), suggesting that CASPASE 8 cleaves CYLD at D215. Recombinant CASPASE 8 was unable to generate the CYLDp25 fragment from purified CYLD-D215A in vitro (Figure 2f), confirming that CASPASE 8 directly cleaves CYLD after D215.
Figure 3. CASPASE 8 processes CYLD at aspartate 215.
(a) Alignment of CYLD amino acid sequences from multiple species. The conserved CASPASE 8 motifs are highlighted in gray shading with the potential CASPASE 8 cleavage sites indicated by a dashed line. (b) Immunoblot of HEK 293EBNA lysates 24 hours after transfection with the indicated plasmids. (c) Immunoblot of HA-CASPASE 8 co-immunoprecipitating with FLAG-CYLD-WT or FLAG-CYLD-D215A, 24 hours post-transfection of HEK 293EBNA cells. (d) Cyld−/− MEFs reconstituted with FLAG-CYLD-WT or FLAG-CYLD-D215A were subjected to anti-FLAG immunoprecipitation after TNF stimulation for the indicated times followed by anti-FLAG blotting.
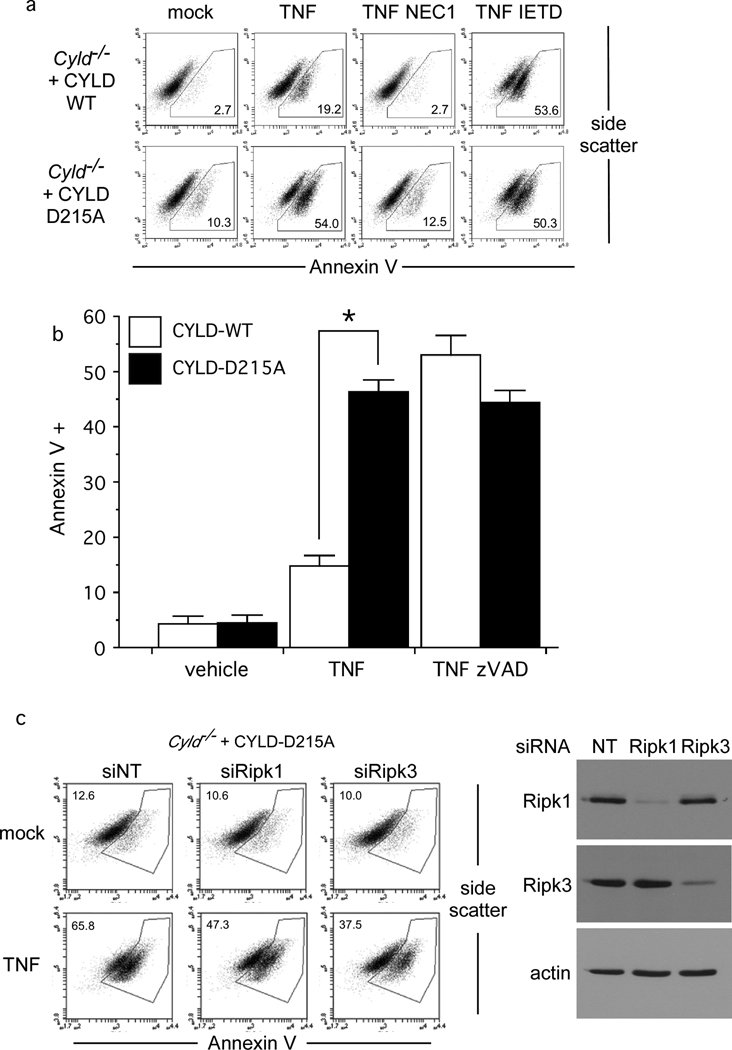
In both the MEF and Jurkat T cell models, there was a strict correlation between necrosis and stabilisation of CYLD. CYLD was rapidly lost in TNF-treated CASPASE 8-sufficient Jurkat T cells dying by apoptosis but remained unchanged in the CASPASE 8-deficient cells dying by necrosis (Supplementary Figure 2a). Cell death in CASPASE 8-deficient Jurkat T cells was blocked by Necrostatin-12, 16, confirming that CASPASE 8 represses programmed necrosis (Supplementary Figure 2b). Similarly, blocking CYLD cleavage in MEFs with caspase inhibitors correlated with entry into necrotic death (Figures 1b & 2b). While dramatic down-regulation of CYLD was observed in TNF-treated cells, no detectable change occurred for RIPK1 and RIPK3 in either model (Figure 1d and Supplementary Figure 2a). Therefore, we hypothesised that CYLD is the key substrate cleaved by CASPASE 8 to prevent necrosis. To test this hypothesis, Cyld−/− MEFs stably reconstituted with CYLD-WT or CYLD-D215A were treated with TNF. CYLD-D215A-expressing MEFs quickly died by necrosis, without the need for a caspase inhibitor (Figure 4a). In contrast, MEFs expressing CYLD-WT only underwent significant amounts of necrosis if treated with TNF in the presence of IETD-fmk (Figure 4a) or zVAD-fmk (Figure 4b). Cell death initiated by TNF in Cyld−/− MEFs expressing CYLD-D215A was blocked by the RIPK1 inhibitor Necrostatin-117 (Figure 4a, compare panel 6 and panel 7), or by specific knockdown of Ripk1 or Ripk3 (Figure 4c), demonstrating that RIPK1 and RIPK3 are downstream of CYLD. Neither RIPK1 nor RIPK3 was required for the processing of CYLD to CYLDp25 in MEFs (Supplementary Figure 4d), indicating that CASPASE 8 inactivation of CYLD occurs upstream of these necrosome components. Recruitment of RIPK1 to the necrosome is strictly dependent on CYLD (Figure 1c) and CASPASE 8 removal of CYLD would be expected to prevent the recruitment of RIPK1. Therefore, stabilising CYLD by mutating the CASPASE 8 cleavage site at D215 is sufficient to permit TNF-induced necrosis even in the presence of CASPASE 8 (CRITERIA #3).
Figure 4. CYLD-D215A triggers programmed necrosis in the absence of caspase inhibitors.
(a) CYLD-WT and CYLD-D215A MEFs from Figure 3d undergoing necrosis 24 hours after stimulation with TNF in the presence of IETD-fmk or Necrostatin-1 (NEC1) was quantified by Annexin V staining. (b) Bar chart displays the mean percentage of CYLD-WT and CYLD-D215A cells cultured in triplicate undergoing necrosis in the presence of vehicle or zVAD-fmk after TNF stimulation for 24 hours. Error bars display standard deviation and * denotes p = 0.0000446 (n=3). (c) Cyld−/− MEFs reconstituted with CYLD-D215A were depleted of RIPK1 and RIPK3 for 48h prior to TNF stimulation for 24 hours to induce necrosis. Immunoblot confirms knockdown of RIPK1 and RIPK3.
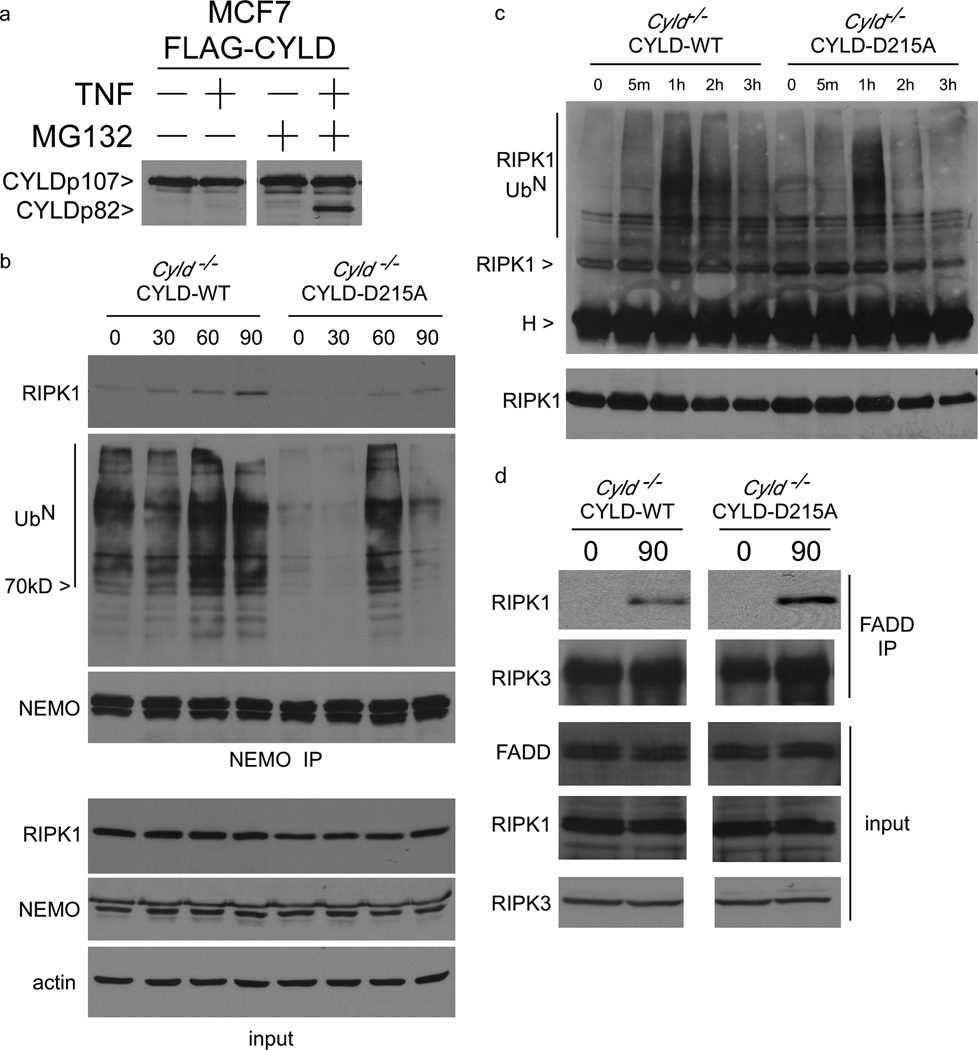
CYLD is a deubiquitinase that removes K63-polyubiquitin chains from RIP118; these ubiquitin chains are required for recruitment of signaling molecules such as NEMO19, 20 that prevent RIPK1 from functioning as a pro-death molecule21, 22, in part by preventing RIPK1 from associating with downstream death signaling molecules and in part by mediating activation of pro-survival NFκB. Consistent with an inhibitory role for CYLD during IKK activation, Cyld−/− MEFs display faster kinetics of IκBα phosphorylation when compared to Cyld−/− MEFs reconstituted with CYLD-WT (Supplementary Figure 5a). However, the kinetics of IκBα phosphorylation appears to be fairly similar between the Cyld−/− cells reconstituted with CYLD-WT or CYLD-D215A with the D215A cells displaying a modest reduction in IκBα phosphorylation at 60 minutes. In a control experiment, mutation of D215 did not affect the ability of CYLD to bind NEMO in HEK 293 cells (Supplementary Figure 5b). We confirmed that the level of CYLD-WT was reduced following TNF stimulation whereas the CYLD-D215A mutant was not (Supplementary Figure 6a), coincident with survival in the former and necrotic cell death in the latter. Thus while the CYLD D215A mutation had a minimal effect on NF-κB signaling, it had a striking effect on necrosis suggesting that CYLD deubiquitination of downstream molecules such as RIPK1 controls entry into this death pathway. To test this hypothesis, we first examined the fate of the C-terminal p82 fragment containing the deubiquitinase domain resulting from the cleavage. In experiments up to this point, this C-terminal fragment was not detectable suggesting that it may be unstable. Sequence analysis of CYLD indicated a PEST motif from amino acids 397 to 415. Addition of the proteosome inhibitor MG132 to TNF-stimulated MCF7 cells (Figure 5a) or MEFs (Supplementary Figure 6b) lead to the stabilisation of the p82 fragment. Therefore when TNF is promoting survival, CASPASE 8 cleaves CYLD to remove the deubiquitinase domain, which is likely to affect RIPK1 ubiquitination and its interaction with signaling partners. We examined this by immunoprecipitating the pro-survival NEMO complex from TNF-treated CYLD-WT and CYLD-D215A-expressing MEFs because NEMO binds ubiquitinated RIPK119, 20. Therefore, the amount of RIPK1 in the NEMO complex can be regarded as an indirect measure of RIPK1 ubiquitination. Furthermore, the NEMO-RIPK1 complex has been proposed to be a survival complex that functions by preventing RIPK1 from interacting with downstream death signaling molecules22–24. More RIPK1 protein co-precipitated with NEMO from CYLD-WT than CYLD-D215A-expressing MEFs suggesting that RIPK1 ubiquitination may be reduced in the D215A cells (Figure 5b). Interestingly, the level of ubiquitinated proteins in the NEMO complex was reduced in CYLD-D215A MEFs at both steady state and upon TNF stimulation consistent with the idea that CYLD stabilisation resulted in enhanced deubiquitinase activity (Figure 5b). The ubiquitination level of RIPK1 was directly examined by RIPK1 immunoprecipitation followed by blotting with anti-ubiquitin. RIPK1 isolated from the CYLD-D215A MEFs exhibited lower levels of ubiquitination after TNF stimulation, which is most discernible at the 2 and 3 hour time points (Figure 5c), suggesting that stabilisation of the CYLD protein leads to accelerated deubiquitination of RIPK1 concurrent with reduced interaction of RIPK1 with the pro-survival NEMO protein. Disruption of the interaction of RIPK1 with NEMO would be predicted to lead to increased association of RIPK1 with the downstream necrotic death apparatus. Indeed, more RIPK1 was recruited to the necrosome complex after 90 minutes of TNF treatment of CYLD-D215A-expressing MEFs (Figure 5d). Therefore, removal of CYLD-WT by CASPASE 8 prolongs the ubiquitination state of RIPK1 and maintains RIPK1 in a pro-survival complex with NEMO. In contrast, the resistance of CYLD-D215A to proteolytic cleavage by CASPASE 8 resulted in less ubiquitination of RIPK1 (Figure 5), which enhanced RIPK1 interaction with the necrosome to initiate death (Figure 4). These observations demonstrate that CASPASE 8 cleavage of CYLD functions as a pro-survival event by preventing CYLD from deubiquitinating RIPK1.
Figure 5. CYLD-D215A switches pro-survival NEMO-RIPK1 complex to pro-necrotic RIPK1-FADD complex.
(a) MCF7 cells stably expressing FLAG-CYLD-WT were treated with vehicle or 25µM MG132 for one hour and then stimulated with 100 ng/ml TNF for 5 hours. Cytoplasmic lysates were blotted with antibody against the C-terminus of CYLD. (b) NEMO was immunoprecipitated from Cyld−/− MEFs reconstituted with CYLD-WT or CYLD-D215A after TNF stimulation for different periods of time. The NEMO protein complex was sequentially immunoblotted with anti-RIPK1 to detect the pro-survival NEMO-RIPK1 complex, and with anti-ubiquitin to detect the level of ubiquitinated protein in the NEMO complex. Corresponding whole cell lysates were blotted with for RIPK1, NEMO and actin in the lower three panels. (c) RIPK1 was immunoprecipitated from CYLD-WT and CYLD-D215A-expressing MEFs after stimulation with TNF for different periods of time in high stringency buffer. The immunoprecipitates were blotted with anti-ubiquitin to detect ubiquitinated RIPK1. (d) FADD immunoprecipitates from CYLD-WT and CYLD-D215A-expressing MEFs stimulated for 90 minutes with TNF were immunoblotted for the presence of RIPK1. Samples of the lysates were also immunoblotted for RIPK1, RIPK3 and FADD.
In conclusion, we provide evidence that CYLD is cleaved by CASPASE 8 to promote survival. Significantly, the cleavage fragment containing the deubiquitinase domain is unstable and undergoes degradation by the proteosome. CYLD level diminished in a time-dependent manner after TNF stimulation suggesting that CYLD may be continuously recruited to and degraded by the CASPASE 8 signaling complex to maintain the suppression of necrosis. CASPASE 8-dependent CYLD degradation is also observed in cells undergoing apoptosis, which antagonizes necrosis. Therefore proteolytic degradation of CYLD may serve to disable the necrosis machinery in multiple settings. More significantly, mutation of the cleavage site on CYLD drives MEFs into necrotic cell death with no requirement for caspase inhibition. Cleavage of CYLD by CASPASE 8 is both necessary and sufficient for CASPASE 8 to repress programmed necrosis, indicating that processing of CYLD is the critical determinant of cell survival versus death by necrosis in MEFs. Processing of CYLD by CASPASE 8 does not require RIPK1 or RIPK3, which suggests that this cleavage event occurs early after TNFR1 ligation in order to prevent recruitment of RIPK1 to the necrosome. When the removal of CYLD by CASPASE 8 is prevented by mutation of D215, we observe accelerated deubiquitination of RIPK1 and the dismantling of the pro-survival RIPK1 and NEMO complex, with the concomitant formation of the RIPK1 and FADD necrosome complex. Since CYLD is a tumour-suppressor25 and can be removed by CASPASE 8, this study should motivate a re-examination of the controversial role of CASPASE 8 in tumourigenesis. CYLD is a known regulator of other non-apoptotic functions attributed to active CASPASE 8 such as proliferation, cell migration and metastasis26; therefore, processing of CYLD by CASPASE 8 is likely to have important biological functions that extend beyond the repression of programmed necrosis.
Materials and Methods
Reagents and antibodies
Phycoerythrin-conjugated Annexin V (Pharmingen), propidum iodide, Necrostatin-1, MG132 and zVAD-fmk (Calbiochem), zVAD-fmk and IETD-fmk (Imgenex), recombinant human TNF (PeproTech) were purchased from the indicated sources. Antibodies against CASPASE 3, CASPASE 8, phospho-IκBα (Cell Signaling), IκBα (Imgenex) CYLD C-terminus, FADD and ubiquitin (Santa Cruz), CYLD N-terminus (Invitrogen), mouse RIPK3 (ProScience), HA and myc (Roche), RIPK1 (Pharmingen), actin and FLAG antibody and beads (Sigma) were purchased from the indicated sources. The human RIPK3 antibody and SMAC mimetics5 were generously provided by Dr. Xiaodong Wang (UT Southwestern, Dallas, TX). All western blots were probed with primary antibodies at 1 µg/ml, except human RIPK3 (rabbit serum was diluted 1:2000), horse radish peroxidase-conjugated secondary antibodies (Jackson Laboratories) were used at the following concentrations: anti-mouse 0.08 µg/ml, anti-rabbit 0.026 µg/ml.
Cell lines
The HEK 293 derivative cell line 293EBNA was obtained from Invitrogen. Wildtype (clone A3) and CASPASE 8-deficient (clone I9.2) Jurkat cell lines were obtained from ATCC. Casp8+/+ and Casp8−/− MEFs have been described previously7. Cyld−/− MEFs have been previously described25.
Cell viability assays
Cells undergoing programmed necrosis expose phosphatidyl serine on the outer face of the plasma membrane27–30, therefore Annnexin V staining is a valid method to detect necrotic cells. Annexin V staining of fibroblasts: subconfluent MEFs or L929 cells were counted and plated to give approximately 30 to 50% cell density. 24 h after treatment with TNF, detached dead cells were removed with the tissue culture supernatant. The remaining adherent cells were briefly trypsinized and combined with the detached cells. MEFs were washed once with Annexin V Binding buffer (10 mM HEPES, 140 mM NaCl, 2.5 mM CaCl2, pH7.5), resuspended in 100 µl binding buffer and incubated with 1 mg/L Annexin V-PE for 5 minutes before immediate analysis on a Becton Dickinson FACScan flow cytometer or an Accuri C6 flow cytometer. Annexin V staining Jurkat T cells: approximately 5×105 T cells per sample were stimulated as described and then washed once with Annexin V binding buffer, resuspended in binding buffer and incubated with 1 mg/L Annexin V-PE for 5 minutes before immediate analysis on a Becton Dickinson FACScan flow cytometer. Flow cytometry data was analysed using CellQuest (Becton Dickinson) FlowJo or CFlow (Accuri) software. Samples were gated to exclude cellular debris according to the forward and side scatter. Cells were typically treated with between 10–100 pg/ml (MEFs) and 10 ng/ml (Jurkat T cells) human TNF to induce programmed necrosis. Cells were pre-incubated with 10 µM zVAD-fmk, 50 µM IETD-fmk, 30 µM Necrostatin-1 or 100 nM SMAC mimetic, as indicated.
Bioinformatics Analysis
Analysis of gene expression across 79 tissues was carried out as described previously31. Briefly, microarray data files were obtained from the Novartis GNF human expression atlas version 2 resource32, and expression values of probe sets from the HG-U133A (Affymetrix, Santa Clara, CA, USA) platform and the GNF1H custom chip were analyzed. The data set was normalized by using global median scaling and we filtered the data by excluding from the analysis probe sets with 100% ‘absent’ calls (MAS 5.0 algorithm) across all 79 tissues. Z-score transformation was applied to each probe set across all arrays before generating ‘heatmaps’ for visualization using TreeView33.
Immunoprecipitation
Immunoprecipitation of FLAG-tagged proteins: approximately 10 x106 HEK 293 cells or 3×106 MEFs were lysed in 1500 µl triton-X-100 lysis buffer on ice for 20 min as previously described31. Samples were centrifuged at 20000×g for 10 min and the supernatants incubated with M2 FLAG beads (Sigma) for 2h to overnight. Beads were washed 5 times with 1000 µl triton-X lysis buffer supplemented to 250 mM NaCl and once with peptide elution buffer. Immunoprecipitated proteins were eluted in 300 µg/ml 3X FLAG peptide (Sigma) in peptide elution buffer for 1h on ice before SDS-PAGE. Cells were treated with human TNF for 5 hours, in the presence of 100 nM SMAC mimetic when appropriate, to induce cleavage of FLAG-CYLD: 10 ng/ml human TNF was used for Cyld−/− MEFs, 50 ng/ml human TNF for for Casp8−/− MEFs, 50 ng/ml human TNF + SMAC mimetic for Jurkat T cells and MCF7 cells. To isolate NEMO and FADD complexes, approximately 30×106 MEFs were lysed in NP-40 lysis buffer (0.2% NP-40, 20 mM Tris, 150mM NaCl, 10% glycerol, pH7.5) for 30 minutes on ice. Samples were centrifuged at 20000×g for 10 min and the supernatants incubated with NEMO or FADD-specific antibody (Santa Cruz) overnight. Protein A/G beads (Santa Cruz) were added for a further two hours at 4°C and the beads were washed 5 times with lysis buffer. NEMO and FADD-associated proteins were eluted by incubating the beads in SDS-PAGE loading buffer at 70°C for 20 minutes. RIPK1 immunoprecipitations were performed similarly with anti-RIPK1 but the beads were washed 5 times with RIPA buffer (50mM Tris, 500mM NaCl, 1% Triton-X-100, 1% Na deoxycholate, 0.1% SDS, pH7.4) prior to SDS-PAGE.
Recombinant CASPASE 8 cleavage assay
FLAG-tagged CYLD-WT and CYLD-D215A were purified from reconstituted Cyld−/− MEFs by FLAG immunoprecipitation as described above except the beads were washed with buffer containing 500mM NaCl. FLAG-tagged proteins were eluted with FLAG peptide and then combined with 3U of recombinant CASPASE 8 (Millipore) in a reaction buffer (50 mM HEPES, 50 mM NaCl, 0.1% CHAPS, 10 mM EDTA, 5% Glycerol, and 10 mM DTT). Samples were incubated at 37°C for two hours and the entire reaction was subject to SDS-PAGE followed by immunoblotting with anti-FLAG and anti-CASPASE 8.
Whole cell lysates
Cells were washed once with PBS and lysed with 0.5% SDS. Lysates were vortexed for 1 min, boiled for 5 min and vortexed again to shear genomic DNA. Protein concentration was determined by BCA assay (Pierce) and between 25 and 75 µg of protein was typically subject to SDS-PAGE.
RNA interference
Transient knockdown: cells were plated in media with no antibiotics and once attached were transfected with siRNA duplexes targeting Casp8, Cyld, Ripk1 and Ripk3 or with a non-silencing control. MEFs were transfected on two consecutive days with 150 nM non-targeting RNA duplexes or duplexes targeting Ripk1 or Ripk3 using the DharmaFECT1 reagent. Forty-eight hours after the second transfection, cells were re-plated and used in experiments the following day. siRNA duplex oligos were obtained from Dharmacon: On-targetplus mouse Casp8-1(GUGAAUGGAACCUGGUAUA), On-targetplus mouse Casp8-2 (GUCACGGACUUCAGACAAA) On-targetplus smartpool mouse Ripk1 (CAACCGCGCUGAGUACAAU, GAAGGCAUGUGCUACUUAC, CCUCGUUGAUCGUGACUUU, UCACCAAUGUUGCAGGAUA) and On-targetplus smartpool mouse Ripk3 (ACACGGCACUCCUUGGUAU, GGUAAAGCAUUAUCUGUCU, CAAGUUCGGCCAAGUAUGA, UCAAGAUCGUGAACUCGAA). Stable knockdown: L929 cells were transduced with pSUPERretro-puromycin encoding scrambled or RIPK3 targeting short hairpins as described previously7.
Plasmids
Constructs encoding FLAG-GFP, myc-CYLD, FLAG-CYLD-WT, FLAG-CYLD-D215A, CASPASE 8-WT-HA, CASPASE 8-C360S-HA, HA-CrmA-WT and HA-CrmA-D303A were constructed using standard recombinant DNA techniques. Point mutations were generated with QuikChangeXL site-directed mutagenesis kit (Stratagene). For transient expression, genes were cloned into the EF1α-driven PEAK8 or CMV-driven pcDNA3 vectors. For stable expression, genes were inserted into a MMLV-based retroviral vector upstream of an IRES-puromycin resistance cassette described previously21, 31. VSV-G-pseudotyped viruses were generated and infection carried out as previously described21.
Statistical analysis
Bar charts display mean values and the error bars indicate the s.d. of the sample sizes described in the figure legends. Two-tailed student’s t-test (assuming equal variance) was used to determine statistical significance and the p values are shown in the figure legends.
Supplementary Material
Acknowledgements
We are indebted to a number of colleagues for their generosity in providing reagents utilized in these studies. We thank Dr. Xiaodong Wang (UT Southwestern, Dallas, TX) for his generous gift of SMAC mimetics and human RIPK3 antibody, Dr Michelle Kelliher (UMASS, Worcester, MA) for Ripk1+/+ and Ripk1−/− MEFs, Dr. Jerry Chipuk (Mount Sinai School of Medicine, New York, NY) for suggesting the use of MCF7 cells and Dr. Stuart Aaronson (Mount Sinai School of Medicine, New York, NY) for providing these cells. This work was supported by National Institutes of Health grants AI052417 (ATT) and AI44828 (DRG). M.A.O’D. is a recipient of a Research Fellowship Award from the Crohn’s and Colitis Foundation of America. A.T.T. is a recipient of the Irma T. Hirschl Career Scientist Award.
Footnotes
Author contributions
M.A.O’D. and E.P.-J. designed and conducted the experiments. A.O. designed and performed the siCasp8 and siCyld experiments in L929 cells. A.N. and R.X. performed the bioinformatics analyses and wrote the corresponding section of the manuscript. A.O. and D.R.G. provided Casp8+/+ and Casp8−/− MEFs on both Ripk3+/+ or Ripk3−/− background and helpful discussions of the manuscript. R.M. provided the Cyld−/− MEFs. M.A.O’D. and A.T.T. wrote the manuscript. A.T.T. directed the studies.
References
- 1.Brenner D, Mak TW. Mitochondrial cell death effectors. Curr Opin Cell Biol. 2009;21:871–877. doi: 10.1016/j.ceb.2009.09.004. [DOI] [PubMed] [Google Scholar]
- 2.Chan FK, et al. A role for tumor necrosis factor receptor-2 and receptor-interacting protein in programmed necrosis and antiviral responses. J Biol Chem. 2003;278:51613–51621. doi: 10.1074/jbc.M305633200. [DOI] [PubMed] [Google Scholar]
- 3.Degterev A, et al. Chemical inhibitor of nonapoptotic cell death with therapeutic potential for ischemic brain injury. Nat Chem Biol. 2005;1:112–119. doi: 10.1038/nchembio711. [DOI] [PubMed] [Google Scholar]
- 4.Cho YS, et al. Phosphorylation-driven assembly of the RIP1-RIP3 complex regulates programmed necrosis and virus-induced inflammation. Cell. 2009;137:1112–1123. doi: 10.1016/j.cell.2009.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.He S, et al. Receptor interacting protein kinase-3 determines cellular necrotic response to TNF-alpha. Cell. 2009;137:1100–1111. doi: 10.1016/j.cell.2009.05.021. [DOI] [PubMed] [Google Scholar]
- 6.Zhang DW, et al. RIP3, an energy metabolism regulator that switches TNF-induced cell death from apoptosis to necrosis. Science. 2009;325:332–336. doi: 10.1126/science.1172308. [DOI] [PubMed] [Google Scholar]
- 7.Oberst A, et al. Catalytic activity of the caspase-8-FLIP(L) complex inhibits RIPK3-dependent necrosis. Nature. 2011;471:363–367. doi: 10.1038/nature09852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ch'en IL, Tsau JS, Molkentin JD, Komatsu M, Hedrick SM. Mechanisms of necroptosis in T cells. J Exp Med. 2011;208:633–641. doi: 10.1084/jem.20110251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang H, et al. Functional complementation between FADD and RIP1 in embryos and lymphocytes. Nature. 2011;471:373–376. doi: 10.1038/nature09878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kang TB, et al. Mutation of a self-processing site in caspase-8 compromises its apoptotic but not its nonapoptotic functions in bacterial artificial chromosome-transgenic mice. J Immunol. 2008;181:2522–2532. doi: 10.4049/jimmunol.181.4.2522. [DOI] [PubMed] [Google Scholar]
- 11.Hitomi J, et al. Identification of a molecular signaling network that regulates a cellular necrotic cell death pathway. Cell. 2008;135:1311–1323. doi: 10.1016/j.cell.2008.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li M, Beg AA. Induction of necrotic-like cell death by tumor necrosis factor alpha and caspase inhibitors: novel mechanism for killing virus-infected cells. J Virol. 2000;74:7470–7477. doi: 10.1128/jvi.74.16.7470-7477.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang L, Du F, Wang X. TNF-alpha induces two distinct caspase-8 activation pathways. Cell. 2008;133:693–703. doi: 10.1016/j.cell.2008.03.036. [DOI] [PubMed] [Google Scholar]
- 14.He KL, Ting AT. Essential role for IKKgamma/NEMO in TCR-induced IL-2 expression in Jurkat T cells. Eur J Immunol. 2003;33:1917–1924. doi: 10.1002/eji.200323650. [DOI] [PubMed] [Google Scholar]
- 15.Wolf BB, Schuler M, Echeverri F, Green DR. Caspase-3 is the primary activator of apoptotic DNA fragmentation via DNA fragmentation factor-45/inhibitor of caspase-activated DNase inactivation. J Biol Chem. 1999;274:30651–30656. doi: 10.1074/jbc.274.43.30651. [DOI] [PubMed] [Google Scholar]
- 16.Holler N, et al. Fas triggers an alternative, caspase-8-independent cell death pathway using the kinase RIP as effector molecule. Nat Immunol. 2000;1:489–495. doi: 10.1038/82732. [DOI] [PubMed] [Google Scholar]
- 17.Degterev A, et al. Identification of RIP1 kinase as a specific cellular target of necrostatins. Nat Chem Biol. 2008;4:313–321. doi: 10.1038/nchembio.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wright A, et al. Regulation of early wave of germ cell apoptosis and spermatogenesis by deubiquitinating enzyme CYLD. Dev Cell. 2007;13:705–716. doi: 10.1016/j.devcel.2007.09.007. [DOI] [PubMed] [Google Scholar]
- 19.Ea CK, Deng L, Xia ZP, Pineda G, Chen ZJ. Activation of IKK by TNFalpha requires site-specific ubiquitination of RIP1 and polyubiquitin binding by NEMO. Mol Cell. 2006;22:245–257. doi: 10.1016/j.molcel.2006.03.026. [DOI] [PubMed] [Google Scholar]
- 20.Wu CJ, Conze DB, Li T, Srinivasula SM, Ashwell JD. Sensing of Lys 63-linked polyubiquitination by NEMO is a key event in NF-kappaB activation [corrected] Nat Cell Biol. 2006;8:398–406. doi: 10.1038/ncb1384. [DOI] [PubMed] [Google Scholar]
- 21.O'Donnell MA, Legarda D, Skountzos P, Yeh WC, Ting AT. Ubiquitination of RIP1 regulates an NF-kappaB-independent cell-death switch in TNF signaling. Curr Biol. 2007;17:418–424. doi: 10.1016/j.cub.2007.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Legarda-Addison D, Hase H, O'Donnell MA, Ting AT. NEMO/IKKgamma regulates an early NF-kappaB-independent cell-death checkpoint during TNF signaling. Cell Death Differ. 2009;16:1279–1288. doi: 10.1038/cdd.2009.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.O'Donnell MA, Ting AT. Chronicles of a death foretold: Dual sequential cell death checkpoints in TNF signaling. Cell Cycle. 2010;9 doi: 10.4161/cc.9.6.10982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.O'Donnell MA, Ting AT. RIP1 comes back to life as a cell death regulator in TNFR1 signaling. FEBS J. 2011;278:877–887. doi: 10.1111/j.1742-4658.2011.08016.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Massoumi R, Chmielarska K, Hennecke K, Pfeifer A, Fassler R. Cyld inhibits tumor cell proliferation by blocking Bcl-3-dependent NF-kappaB signaling. Cell. 2006;125:665–677. doi: 10.1016/j.cell.2006.03.041. [DOI] [PubMed] [Google Scholar]
- 26.Peters ME. Programmed cell death: Apoptosis meets necrosis. Nature. 2011;471:310–312. doi: 10.1038/471310a. [DOI] [PubMed] [Google Scholar]
- 27.Brouckaert G, et al. Phagocytosis of necrotic cells by macrophages is phosphatidylserine dependent and does not induce inflammatory cytokine production. Mol Biol Cell. 2004;15:1089–1100. doi: 10.1091/mbc.E03-09-0668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hirt UA, Leist M. Rapid, noninflammatory and PS-dependent phagocytic clearance of necrotic cells. Cell Death Differ. 2003;10:1156–1164. doi: 10.1038/sj.cdd.4401286. [DOI] [PubMed] [Google Scholar]
- 29.van Delft MF, Smith DP, Lahoud MH, Huang DC, Adams JM. Apoptosis and non-inflammatory phagocytosis can be induced by mitochondrial damage without caspases. Cell Death Differ. 2010;17:821–832. doi: 10.1038/cdd.2009.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Waring P, Lambert D, Sjaarda A, Hurne A, Beaver J. Increased cell surface exposure of phosphatidylserine on propidium iodide negative thymocytes undergoing death by necrosis. Cell Death Differ. 1999;6:624–637. doi: 10.1038/sj.cdd.4400540. [DOI] [PubMed] [Google Scholar]
- 31.Friedman CS, et al. The tumour suppressor CYLD is a negative regulator of RIG-I-mediated antiviral response. EMBO Rep. 2008;9:930–936. doi: 10.1038/embor.2008.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Su AI, et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc Natl Acad Sci U S A. 2004;101:6062–6067. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saldanha AJ. Java Treeview--extensible visualization of microarray data. Bioinformatics. 2004;20:3246–3248. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.