Abstract
Purpose
The dual modality of TGFβ, both as a potent tumor suppressor and a stimulator of tumor progression, invasion, and metastasis, make it a critical target for therapeutic intervention in human cancers. The ability to perform real-time, noninvasive imaging of TGFβ-activated Smad signaling in live cells and animal models would significantly improve our understanding of the regulation of this unique signaling cascade. To advance these efforts, we developed a highly sensitive molecular imaging tool that repetitively, non-invasively and dynamically reports on TGFBR1 kinase activity.
Experimental Design
The bioluminescent TGFβR1 reporter construct was developed using a split firefly luciferase gene containing a functional sensor of Smad2 phosphorylation wherein inhibition of TGFβ receptor1 kinase activity leads to an increase in reporter signaling. The reporter was stably transfected into mammalian cells and used to image in vivo and in vitro bioluminescent activity as a surrogate for monitoring TGFBR1 kinase activity.
Results
The reporter was successfully used to monitor direct and indirect inhibitors of TGFβ-induced Smad2 and SMAD3 phosphorylation in live-cells and tumor xenografts and adapted for high throughput screening to identify a role for receptor tyrosine kinase-inhibitors as modulators of TGFβ signaling.
Conclusion
The reporter is a dynamic, non-invasive imaging modality for monitoring TGFβ-induced Smad2 signaling in live cells and tumor xenografts. It has immense potential for identifying novel effectors of R-Smad phosphorylation; for validating drug-target interaction; and for studying TGFβ signaling in different metastasis models.
Keywords: bioluminescence, kinase activity, non-invasive molecular imaging, receptor-regulated Smads, TGFβ
Introduction
The transforming growth factor-β (TGFβ) signaling pathway regulates growth inhibition and tumor suppression through control of cell proliferation, differentiation, apoptosis, migration, and adhesion (1). Consequently, when the TGFβ signaling pathway is disrupted, either through genetic mutations of key signaling components or due to loss of its growth inhibition capabilities, tumorigenesis occurs. It has been demonstrated that TGFβ plays a dual role in cancer development (2, 3). During early stages of tumorigenesis, when the tumor cells are premalignant, TGFβ suppresses cellular outgrowth whereas at later stages, following alteration of TGFβ-responsiveness and increased ligand expression, it promotes tumor progression, invasion, and metastasis. Tumor cells that have lost TGFβ-mediated tumor suppression but maintain intact components of its core signaling pathway are especially aggressive. These cells exploit TGFβ for pro-oncogenic activities: epithelial-to-mesenchymal transition (EMT), tumor invasion, mitogen production, metastatic dissemination, and immune evasion (2, 3).
TGFβ cellular activity is mediated through a receptor serine-threonine kinase (RSK) complex. TGFβ initiates the interaction of TGFβ Type-1 and Type-2 receptors (TGFBR1 and TGFBR2, respectively) which phosphorylate the C-terminal SXS motif of receptor-regulated Smads (R-Smads), Smad2 and Smad3. The phosphorylated R-Smads complex with Co-Smad4, translocate into the nucleus, and, along with cell-specific transcriptional co-factors, interact with Smad-binding elements (SBE) in the DNA to regulate transcription of target genes. The C-terminal phosphorylation of R-Smads is a critical event in TGFβ signaling and transcriptional regulation. It is necessary for Smad complex assembly and disassembly, nuclear translocation, and transcriptional activity and stability required for Smad-mediated cellular responses (4).
Despite the importance of TGFβ-mediated Smad signaling in the progression of human cancers, methods for monitoring in vivo and in vitro Smad phosphorylation are limited. Indirect molecular imaging techniques have been developed using fluorescent-tagged proteins in the TGFβ-Smad signaling pathway to study cellular localization of Smad proteins in breast cancer cells (5, 6) and protein-protein interaction of critical elements of TGFβ signaling in xenopus embryogenesis (7). More direct measures of TGFβ signaling, have been designed using a firefly luciferase reporter linked to Smad responsive elements (SBE) to measure transcription of target genes in breast cancer metastasis(8, 9), skin wound healing (10), neurodegeneration, (11), and response to injury (12). These imaging methods are limited in that they do not measure the key upstream event in TGFβ signaling, the phosphorylation of R-Smads.
In this study, we developed a molecular imaging reporter for non-invasive, repetitive monitoring of TGFβ activity using the critical R-Smad phosphorylation step in the TGFβ signaling pathway. The reporter showed higher sensitivity to changes in Smad phosphorylation than measured by conventional western analysis and provided new insight into pharmacodynamics of TGFβ inhibitor SB431542 in vivo. It was successfully adapted for high throughput screening and utilized to delineate key modulators of Smad2 signaling. The reporter demonstrates immense potential for identifying novel effectors of R-Smad phosphorylation; for validating drug-target interactions; and for studying TGFβ signaling in different metastasis models.
Materials and Methods
Construction of the Bioluminescent TGFβR reporter (BTR)
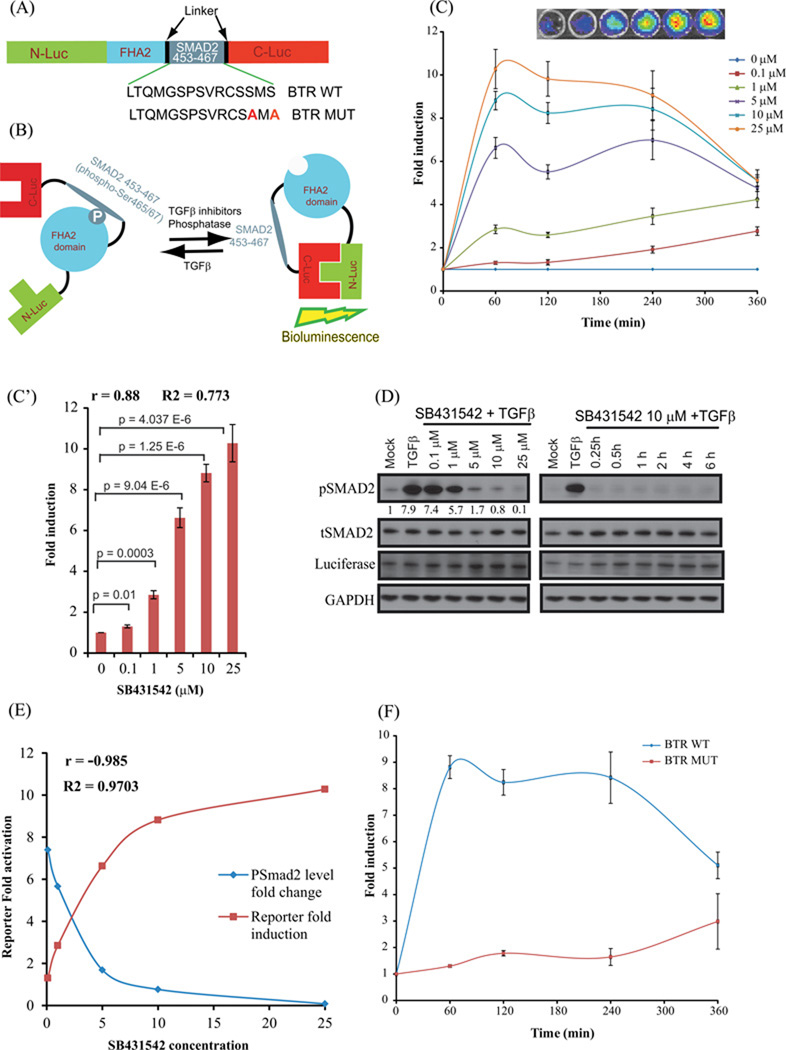
To image TGFBR1 kinase activity, we constructed a recombinant chimeric reporter (Fig. 1A, B) consisting of a 15 amino acid target peptide sequence derived from the C-terminus of Smad2 containing the SXS motif, aa 453–467, a TGFBR1 substrate (13). The BTR reporter was constructed through fusion of the Smad2 peptide sequence and the FHA2 phospho-serine-threonine binding domain of Rad53, which were flanked by the amino- (N-Luc) and carboxyl-(C-Luc) terminal domains of the firefly luciferase reporter molecule (14). The cloning primers were designed with a linker sequence (GlyGlyAlaGlyGly) on both ends and XbaI/XmaI precut sites. The BTR-WT was constructed using the appropriate primers (CTA GAG GAG GAA GTG GAG GGT TAA CTC AGA TGG GAT CCC CTT CAG TGC GTT GCT CAA GCA TGT CAC and CCG GGT GAC ATG CTT GAG CAA CGC ACT GAA GGG GAT CCC ATC TGA GTT AAC CCT CCA CTT CCT CCT). The mutant reporter, BTR-MUT, was generated by substituting both serines of the SXS motif with alanine using a single primer mutagenesis protocol, with minor modifications as described earlier (15), using the following primer: CCT TCA GTG CGT TGC TCA GCC ATG GCA CCC GGG GGA GGA GCT GGA GGA TCC GGT TAT GT. All the clones were sequence verified.
Figure 1. Construction and Validation of bioluminescent TGFβ reporter (BTR) with TGFBR1 kinase inhibitor, SB431542.
(A) The domain structure of the BTR construct: Two versions of the BTR construct were developed; the BTR-WT construct contains the wild-type Smad2 C-terminus, aa 463–467, and the BTR-MUT construct in which alanines were substituted for both serines of the SXS motif. (B) The mechanism of action of the BTR reporter involves TGFBR1 kinase dependent phosphorylation of the Smad2 target peptide which results in its interaction with the FHA2 domain. In this form, the reporter has minimal bioluminescence activity. In the absence of the kinase activity, the target peptide no longer interacts with the FHA2 domain allowing the N-Luc and C-Luc domains to re-associate, restoring bioluminescence activity. (C) A549 cells stably expressing the BTR-WT reporter were treated with increasing concentrations of SB431542 (0.1, 1, 5, 10, and 25µM) in presence of TGFβ (10ng/mL) and bioluminescence was measured serially from baseline through 6 h. Insert displays a representative bioluminescent image of BTR reporter response with increasing concentrations of SB431542 treated cells. The change in bioluminescence activity over mock treatment (DMSO) levels was calculated and plotted as fold induction. Bioluminescent measurements are in triplicates, error bars denote SEM. (C’) Reporter fold induction with SB431542 concentration is plotted to illustrate dose-dependent increase in the reporter activity. Pearson correlation coefficient (r), goodness of fit (R2) and statistical significance (p) is calculated and shown on the plot. (D) Cell lysates were prepared and analyzed by Western blot using anti-phospho-(S465–S467) Smad2, total Smad2, luciferase, and GAPDH antibodies. Level of pSmad2 was measured using imageJ. (E) Pearson correlation coefficient was calculated between pSmad2 level change and reporter fold induction. A very strong negative correlation (r=−0.985) was observed. (F) A549 cells stably expressing BTR-WT and BTR-MUT reporters were treated in triplicate with 10 µM SB431542 and bioluminescence was measured serially for 6 h. The change in bioluminescence activity over vehicle control was calculated and plotted as fold change.
Cell Culture and Transfection
The human lung carcinoma cell line A549 (American Type Culture collection, ATCC) was maintained in RPMI-1620 media supplemented with 10% heat-inactivated fetal bovine serum, 1% glutamine, and 0.1% penicillin/streptomycin/gentamycin (GIBCO-InVitrogen, Grand Island, NY). Cell cultures were grown in a humidified incubator at 37°C and 5% CO2. Breast cancer cell line 1833 derived from MDA-MB-431 (16) was kindly provided by Dr Joan Massague and maintained in DMEM media in the same conditions mentioned above. Cell cultures were grown in a humidified incubator at 37°C and 5% CO2. ATCC cell line was tested routinely for mycoplasma and purity. All ATCC lines were expanded immediately upon receipt and multiple vials of low passage cells were maintained in liquid N2. No vial of cells was cultured for more than 1–2 months. Stable cell lines were developed by transfecting the BTR reporter plasmids into A549 and 1833 cells using Fugene (Roche Diagnostics) and resulting clones were selected using 500 µg/ml (A549) or 250 µg/ml (1833) G418 (InVitrogen) containing media. Twenty-four stable clones were selected for both wild-type and mutant cell lines. The BTR-WT expressing clones were analyzed by bioluminescent imaging following treatment with 10 µM SB431542 (Cayman Chemical, Ann Arbor, MI). Single cell subclones were generated and maintained in 250 µg/ml G418 containing media.
Antibodies and Reagents
Rabbit polyclonal antibodies to phospho(S465/467)Smad2, Smad2, p(S423/425)Smad3, Smad6, TGFΒR1/ALK5, TGFΒRII, C-Myc, C-Jun, p(Y845)EGFR, EGFR, p(Y1248)HER2, p(T180/Y182)P38MAPK, P38MAPK, p(P85-Y458/P55-Y199)PI3-K, PI3-K, p(T183/Y185)JNK, JNK and GAPDH were obtained from Cell Signaling Technology (Beverly, MA); anti-E-cadherin and N-cadherin from BD Biosciences (San Jose, CA); anti-pSer, Smad3, and SMAD7 from Invitrogen (Camarillo, CA); Smad4 (SantaCruz Biotechnology, Santa Cruz, CA); and anti-HER2 and anti-firefly luciferase antibodies from Millipore (Billerica, MA). The HRP-conjugated secondary antibodies were from Jackson Laboratories (West Grove, PA).
TGFBR1/ALK5 inhibitor SB431542, AG1296, Tyrphostin AG1478, and SP600125 were obtained from Cayman Chemical (Ann Arbor, MI); pan-RTK inhibitor PP2 from Calbiochem-EMD chemicals (Gibbstown, NY); and SB203580 (Invivogen, San Diego, CA). Lapatinib in tablet form (Tykerb) was purchased from the University of Michigan pharmacy. Erlotinib was a kind gift from Genentech (San Francisco, CA). D-Luciferin was purchased from Promega Corp. (Madison, WI). An 84-compound kinase-specific library was obtained from Timtek Drug Discovery (Newark, DE). SBE4-Luc reporter plasmid (17) was a kind gift of Dr Bert Vogelstein (Addgene plasmid #16495).
Bioluminescence reporter assay and live-cell imaging
For the reporter assay, A549 and 1833 BTR cell lines were seeded in 96-well (5×103 cells/well), clear bottom, white-walled plates (Corning, Inc., Corning, NY) 48-hours prior to assaying. Cells were treated in serum-free media with varied concentrations of test compounds for indicated time periods. Luminescence was read with an Envision 2104 multi-label plate reader (Perkin Elmer, Waltham, MA) for 5 repeats after addition of D-Luciferin (100µg/ml final concentration) to the cell medium; each experiment was done in triplicate. For the high throughput screen, 10µl of an intermediate DMSO stock of each compound in the kinase inhibitor library was added to the cell medium using a Beckman Biomek NXP Laboratory Automation Workstation (Beckman Coulter Inc.), yielding a final assay concentration of 10 µM for each compound; all appropriate controls were included and cells incubated for 4 h before D-luciferin was added and bioluminescence measured.
Live-cell luminescent imaging was achieved by adding D-Luciferin (100µg/ml final concentration) to the growth medium on cells seeded in black-walled, clear-bottom, 96-well plates. Photon counts for each condition were acquired 5 minutes after incubation with D-luciferin using an IVIS 200 imaging system (Xenogen, Alameda, CA).
Small interfering RNA transfection analysis
TGFBR1 and TGFBR2 small interfering RNA (siRNA), an siGENOME Smart Pool, and nonsilencing siRNA (NSS) were synthesized by Dharmacon Research (Lafayette, CO).Knockdown experiments were performed by transfecting A549 and 1833 BTR-WT reporter cells with 100nM siRNA for TGFBR1 and TGFBR2, as well as for non-targeted siRNA (NSS) as a negative control using Dharmafect1 (Dharmacon Research, Lafayette, CO). The transfected cells were incubated for 60 h, serum starved overnight, and 10ng/ml TGFβ added 1 h prior to evaluating reporter activity with bioluminescent imaging and Western analysis. Fold change in reporter activity was calculated over change in activity in NSS treated cells.
Western analysis and immunoprecipitation
Western analysis was carried out using standard protocols. A549 and 1833 BTR cell lines were grown in culture dishes, treated with select compounds for designated time periods, and cell lysates resolved on SDS-PAGE gels and transferred to PVDF membranes. Membranes were probed against specific primary antibodies followed by HRP conjugated secondary antibodies then visualized using the Enhanced Chemiluminescence (ECL) Western Blotting System (GE Healthcare, Piscataway, NJ).
Immunoprecipitation of the BTR reporter molecule was carried out by incubating cell lysate (400µg protein) with 5µg luciferase specific antibody for 1 h. The immune complex was captured using 20µl slurry of protein A/G-coupled Sepharose beads (GE Healthcare) for 1 h, washed three times with RIPA buffer. The resulting pellet was resolved by SDS-PAGE and transferred to PVDF membrane for Western analysis.
To study the effect of various inhibitors on Smad2/3 complex formation with Smad4, co-immunoprecipiation was carried out using a Smad3 antibody. A549 cells were serum starved and treated with 10µM inhibitors in the presence of 10ng/ml TGFβ for 1hr. Lysates were made in native lysis buffer (50mM Tris PH 7.4, 1% NP40, 0.25% Deoxycholate sodium salt, 150mM NaCl, 10% Glycerol, and 1mM EDTA) supplemented with 1X PhosStop (Roche), 1X Protease inhibitor cocktail (Roche), Sodium Ortho Vanadate, Sodium fluoride, PMSF, and β-Glycerol phosphate. The samples were gently rocked for 2h at 4°C. Immune complexes were captured using 40µl slurry of protein A/G-coupled Sepharose beads (GE Healthcare) for 1h. The resulting pellet was washed and resolved by SDS-PAGE and transferred to PVDF membrane and probed using a human anti-Smad4 antibody.
Transcriptional response assay
A549 cells were transiently transfected with SBE4-Luc reporter plasmid along with Gaussia luciferase plasmid (pEF-GLuc) as internal control. Transfected cells were incubated with 10µM inhibitors in the presence or absence of 10ng/ml TGFβ in media containing 1% FBS. SBE4-Luc reporter activity was measured 22hr post treatment. Each condition was done in triplicate.
In vivo bioluminescence imaging of mouse tumor models
Tumor xenografts expressing BTR-WT were established by implanting 2.5×106 stably-transfected BTR-WT A549 cells on each flank of four to six week old female athymic mice of the genotype CD-1 nu/nu (Charles River Laboratories, Wilmington, MA). When tumors reached a volume of approximately 40–60 mm3, in about four weeks, bioluminescence activity was monitored. The mice were anesthetized with 2% isofluorane/air mixture and given a single intraperitoneal injection (i.p.) of 150 mg/kg luciferin in normal saline. Image acquisition was initiated after injection of luciferin and serial background bioluminescence images acquired for four hours prior to drug administration. Mice were injected with a single i.p. dose of 10mg/kg body weight with SB431542, a dosing regimen previously reported to inhibit TGFβ signaling in vivo (19, 20), or vehicle control (DMSO) and bioluminescent images performed before treatment as well as, at 1, 4, 8, and 24 h.
In parallel, tumor xenografts were generated in mice using A549 BTR WT and MUT reporter expressing cells. Mice were treated as described and bioluminescence was measured 4hr after treatment. All mice experiments were approved by the University Committee on the Use and Care of Animals (UCUCA) of the University of Michigan.
Immunohistochemistry
Tumors from mice treated with SB431542 (10mg/kg) or vehicle, for 4 h, were excised and fixed in formalin. The tumors were processed at the UMCCC Tissue Core Facility and stained with anti-pSmad2 antibody (1:100); micrographs were taken using an Olympus microscope fitted with an Olympus DP-70 high resolution digital camera. Cell nuclei stained positive for pSmad2 were counted in three random fields for vehicle (n=3) and SB431542 (n=3) treated tumors. A two sided student’s t-test was performed to assess statistical significance. Slides were adjusted for brightness and contrast with Adobe Photoshop CS2 (Adobe Inc., San Jose, CA) but the micrographs underwent no other manipulations.
Data analyses
Western blot signal intensity was measured using the image processing and analysis program, ImageJ v1.45 (18). Pearson correlation coefficient (r) was estimated to confirm how well the SB431542 doses correlate to BTR reporter fold induction and pSmad2 levels. Analysis of statistical significance (student’s t test, p values) was performed to estimate the significance of reporter fold induction with SB431542 doses. Additionally, regression analysis (goodness of fit; R2) was carried out to confirm relationships between SB431542 dose, reporter fold induction and pSmad2 levels. All the statistical analyses were performed on Microsoft Excel 2010. GraphPad Prism v5 (GraphPad Software) nonlinear regression analyses and sigmoidal dose-response (variable slope) was used to generate EC50 values.
Results
Construction and mechanism of the bioluminescent TGFβR1 reporter
The key step in TGFβ signaling is the phosphorylation of receptor-regulated Smads by active TGFΒR1 receptor. To study this essential step, we designed a bioluminescent TGFβR1 reporter using a split firefly luciferase construct (Fig. 1A), consisting of the critical TGFβ substrate, the C-terminal SXS motif of Smad2, linked to the phospho-peptide-binding domain FHA2, and flanked by the N-terminal (N-Luc) and C-terminal (C-Luc) domains of the firefly luciferase reporter molecule. The functional basis of the reporter is shown schematically in Figure 1B. The reporter exhibits low level luminescence when the Smad2 substrate is phosphorylated allowing it to bind to the FHA2 domain thereby sterically preventing reconstitution of the luciferase reporter molecule. A reduction in Smad2 phosphorylation through inhibition of TGFΒR1 kinase activity, or other downstream modulators of Smad2 phosphorylation, releases the stearic inhibition allowing reconstitution of the luciferase reporter molecule whose activity can be detected by bioluminescence imaging.
In addition to the wild-type reporter (BTR-WT), we constructed a BTR mutant (BTR-MUT) reporter based on reports that mutations of the SXS motif abolish Smad2 phosphorylation by TGFΒR1 (13). The Smad2 C-terminal serine residues 465 and 467 were mutated to the neutral amino acid alanine (Fig. 1A).
Increased BTR reporter activity following treatment with TGFβR1 inhibitor SB431542
To evaluate whether the BTR reporter provides a sensitive and robust response to inhibition of TGFβ-mediated Smad2 phosphorylation, we imaged A549 and 1833 BTR-WT cells following treatment with varied concentrations of SB431542 (21–24) in the presence of 10ng/ml TGFβ. Bioluminescence activity was measured sequentially for 6 hours revealing both concentration-and time-dependent increases in reporter activity, reaching peak luminescence within 60 minutes post-treatment (Fig. 1C, Suppli Fig. 2A). The BTR reporter activity increased with greater concentrations of inhibitor, reaching a maximum expression of more than 10-fold with 25µM SB431542 in A549 (Fig. 1C) and 8-fold in 1833 (Suppli Fig. 2A) maintaining a steady, although lower, bioluminescence for approximately 4 hours followed by a gradual decline in signal. EC50 values from the same datasets (reporter fold induction at 1h) were estimated to be 2.09 µM and 17.67 µM for A549 and 1833 cell lines respectively. The increased BTR reporter activity correlates with a concomitant decrease in TGFΒR1-dependent levels of phosphorylated Smad2 and with increasing concentrations of SB431542 (Fig. 1D, Suppli Fig. 2B). EC50 values (pSmad2 levels at 1h) were calculated to be 1.6 µM and 0.36 µM for A549 and 1833 cell lines respectively. A very strong positive correlation (Pearson correlation coefficient r=0.88 in Fig 1C’and r= 0.998 in Suppli Fig 2A’) was found between reporter fold induction values and SB431542 concentration. In addition, we also estimated R2 values (goodness of fit) which determines how closely our data fit linear regression. R2 values were estimated to be 0.773 (Fig 1C’) and 0.996 (Suppli Fig2A’). An analysis of statistical significance between reporter fold induction and SB431542 concentration was performed. P values were found to be statistically significant. Similarly, a strong negative correlation (r=−0.985 in Fig 1E and r=−0.5308 in Suppli. Fig 2B bottom panel) between reporter fold induction and pSmad2 levels was found which further confirms the robustness of the reporter. A549 BTR-MUT cells treated with SB431542 (10 µM) in the presence of TGFβ demonstrate only a slight increase in reporter activity which was not sustained throughout the duration of the experiment (Fig. 1F) confirming the specificity of the BTR-WT reporter activity to TGFBR1 kinase phosphorylation of the Smad2 substrate. These changes in BTR reporter activity were not the result of changes in levels of total Smad2 or reporter (Fig. 1D, Suppli Fig. 2B–C).
BTR reporter sensitivity to indirect inhibition of Smad2 phosphorylation
Studies have shown that active src-kinase is a downstream mediator of TGFβ-induced cellular responses (25). To study the sensitivity of the BTR reporter assay to indirect effectors of Smad2 phosphorylation, we treated A549 BTR-WT cells with the src-kinase inhibitor, PP2 (26), in the presence of 10ng/ml TGFβ. Inhibition of src-kinase activity results in both a concentration- and time-dependent increase in reporter response, with peak bioluminescence occurring approximately two hours post-treatment (Suppli. Fig. 1A). Western blot analysis confirms that increase in reporter activity corresponds to decrease in pSmad2 levels, two hours post-PP2 treatment (Suppli. Fig. 1B) as well as a concentration-dependent decrease in pSmad2 with increasing PP2 concentration (Suppli. Fig. 1B). Identical experiments using the BTR-MUT expressing A549 cell line show minimal effect of PP2 on reporter activity (Suppli. Fig. 1C). Similar to SB431542 treatment, these results were not due to any significant change in levels of total Smad2 (Suppli. Fig. 1B).
In vitro A549 BTR-WT cellular responses to TGFβ stimulus
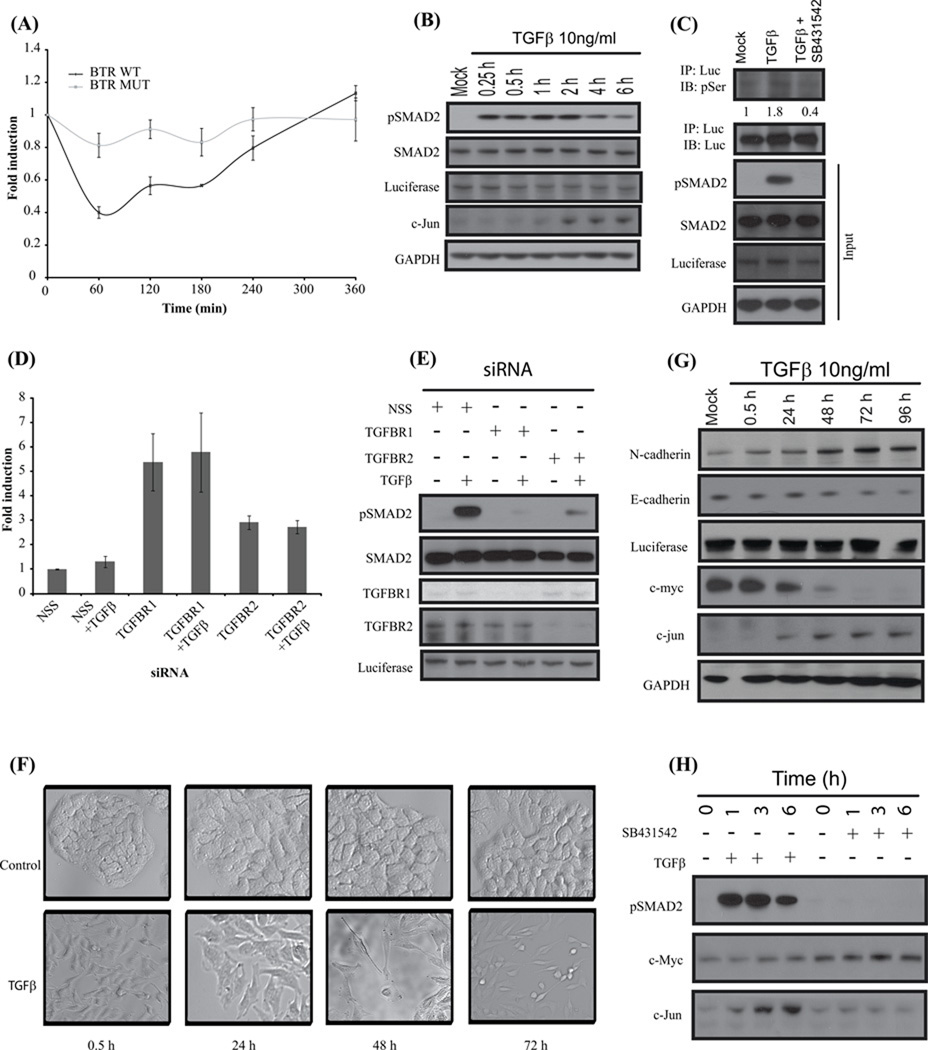
To monitor the effect of the biological transducer on BTR reporter activity, we treated A549 BTR-WT cells with 10ng/ml TGFβ, revealing a nearly 60% decrease in reporter activity at 1h post treatment, whereas treatment of A549 cells expressing the mutant reporter show minimal effect on reporter response (Fig. 2A). Western blot analysis of similarly treated A549 BTR-WT cells indicate a corresponding increase in pSmad2 at 15 minutes that remained at maximal levels for 2 h, followed by slightly lower levels at 4 and 6 hours (Fig. 2B).
Figure 2. BTR reporter activity is dependent on reporter substrate phosphorylation through TGFβ signaling.
(A) A549 cells stably expressing BTR-WT reporters and BTR-MUT treated with TGFβ (10ng/ml) and bioluminescence measured serially for 6 h. (B) Cell lysates prepared and analyzed by Western blot against anti-phospho-(S465–S467) Smad2, total Smad2, luciferase, C-jun and GAPDH antibodies. (C) BTR-WT reporter expressing A549 cells were treated with TGFβ (10ng/ml) or TGFβ and SB431542 (10 µM) for 1 h, cell lysates were prepared and immunoprecipitated with luciferase specific antibody. The level of substrate phosphorylation was determined by Western blot analysis with phospho-Ser antibody. The immunoblot was probed with luciferase specific antibody as a control and the input lysate was probed against phospho-(S465–S467) Smad2, total Smad2, luciferase and GAPDH. (D) A549 cells stably expressing the BTR-WT reporter were transfected with 100nM siRNA for 72 h, TGFβ was added for an additional 1 h, and reporter activity was measured. Fold change in reporter activity was calculated from change in non-targeted siRNA (NSS). Error bars denote SEM. (E) Cell lysates were prepared and analyzed by Western blot against anti-phospho-(S465–S467) Smad2, total Smad2, TGFBR1, TGFBR2, and luciferase. (F) A549 BTR-WT cells were serum-starved and treated with TGFβ (10ng/ml) for 72 h demonstrating epithelial-to-mesenchymal transition. (G) Cell lysates of A549-BTR WT cells treated with TGFβ for 0.5, 24, 48, 72, and 96 hours were probed with antibodies against N-cadherin, E-cadherin, luciferase, c-jun, c-myc and GAPDH. (H) Western blots analysis of A549 BTR-WT cells treated with 10µM SB431542 or 10ng/ml TGFβ for 0, 1, 3, 6 h with antibodies against phospho-(S465–S467) Smad2, c-myc and c-jun.
Verifying that TGFβ-induction phosphorylates the target peptide, the reporter molecule was immunoprecipitated from cell extracts with anti-luciferase antibody and immuno-blotted with a phospho-serine-specific antibody. The BTR protein had a substantial increase in phosphoserine levels following TGFβ treatment as compared to controls (Fig.2C, Suppli Fig. 2D). Following treatment with SB431542, phosphoserine levels are significantly lower than control levels (Fig. 2C, Suppli Fig. 2D) corresponding with increased BTR reporter activity as determined directly by bioluminescence imaging.
To further confirm the link between reporter activity and TGFβ signaling, we suppressed the expression of the upstream kinases by transiently transfecting A549 BTR-WT cells with TGFΒR1 and TGFΒR2 specific siRNA resulting in approximately 6-fold and 3-fold increases in reporter activity, respectively (Fig. 2D). Cells transfected with non-target siRNA (NSS) did not affect reporter activity (Fig. 2D). Similar observations were made in 1833 BTR WT cells which exhibited 2.5 fold increase in reporter activity with TGFBR1siRNA (Suppli Fig. 2E) although no significant activation of the reporter was observed with siRNA to TGFBR2. Cell extracts prepared from parallel experiments and probed with antibodies against TGFΒR1 and TGFBR2 confirm the siRNA mediated suppression of these proteins. A substantial increase in pSmad2 was observed in NSS transfected TGFβ-treated cells while theTGFBR1 and TGFBR2 knockdown cells, with or without TGFβ stimulation, have minimal levels of pSmad2 (Fig. 2E, Suppli Fig. 2F) which closely correlates with reporter bioluminescence activity (Fig. 2D, Suppli Fig. 2E).
To demonstrate that A549 BTR WT cells undergo epithelial-to-mesenchymal (EMT) transition and exhibit the characteristic mesenchymal phenotype with TGFβ treatment (3, 27, 28), we treated serum starved cells with 10ng/ml TGFβ for up to 96 h and studied the changes in cell morphology every 24 h post-treatment. TGFβ treated cells lost cell-to-cell contact and exhibited characteristic elongated, spindle shaped morphology (Fig. 2F). These samples were prepared for Western blot analysis and probed against epithelial marker E-cadherin, and mesenchymal marker N-cadherin. An increase in levels of N-cadherin and corresponding decrease of E-cadherin was observed (Fig. 2G) which is a characteristic of cells undergoing EMT.
Two early regulatory genes shown to be downstream targets of R-Smad activation are the oncogenes c-jun and c-myc (29, 30). Expression levels of c-myc decreased over 96 hours and c-jun levels increased during the same time period (Fig. 2G). We further elucidated the connection of c-myc and c-jun expression to TGFβ signaling in A549 cells by treating with TGFβ alone and in the presence of SB431542. With TGFβ, c-myc levels remain constant for up to 6h but when TGFβ signaling was inhibited, c-myc levels increased (Fig. 2H). Conversely, c-jun expression levels increase within 1 h after TGFβ treatment and are abolished in the presence of SB431542 (Fig. 2H).
In vivo imaging and analysis of TGFβ signaling in BTR expressing tumor xenografts
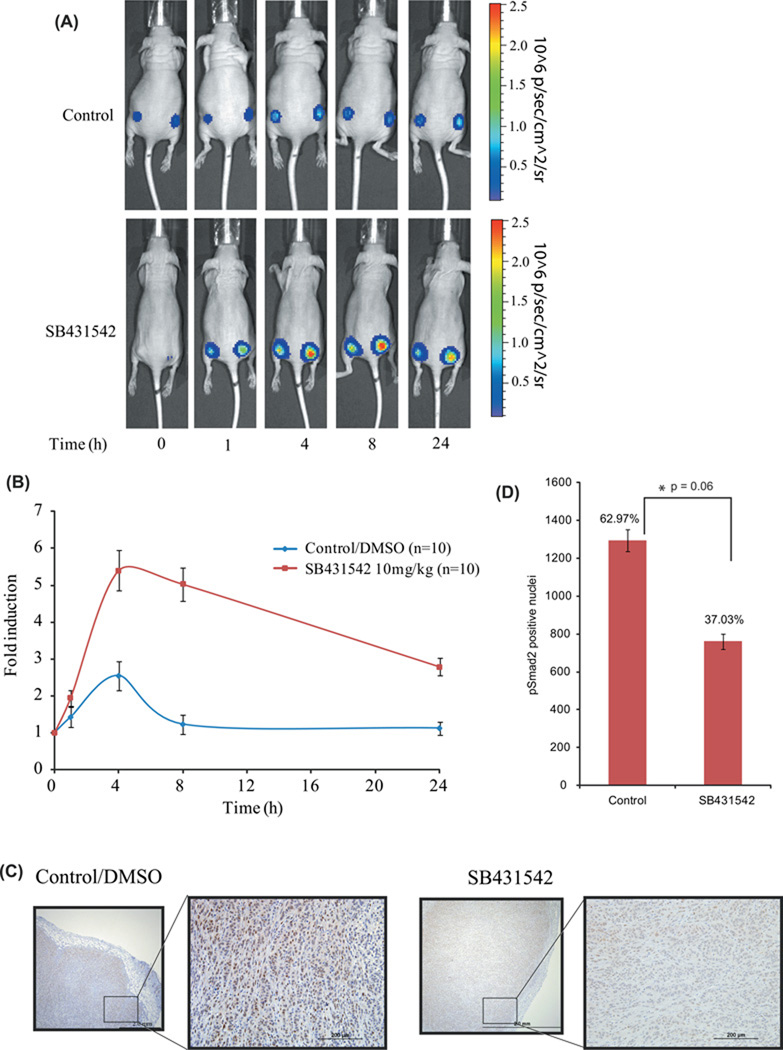
Having established the specificity and sensitivity of the BTR reporter in vitro, we next investigated reporter utility for monitoring changes in TGFβ signaling in a tumor mouse model. We implanted A549 BTR-WT cells into the flanks of nude mice to establish tumors. When the tumors reached a volume of 40–60mm3, we monitored bioluminescence over time in mice treated with TGFBR1 kinase inhibitor or vehicle control. Representative images of control and SB431542 treated mice are shown in Figure 3A. Bioluminescence activity in mice treated with SB431542 increased in the first hour and reached peak activity of greater than 5-fold within 4 hours post-treatment. Reporter activity remained elevated through 8 hours followed by lower, but sustained, reporter activity at 24 h (Fig. 3A–B). Bioluminescence was unchanged over the 24-hour period in vehicle-treated mice (Fig. 3A and 3B).
Figure 3. Molecular imaging of TGFβ signaling in vivo.
(A) Tumor xenografts expressing BTR-WT were established on each flank of four to six week old female athymic CD-1 nu/nu mice. Images of in vivo BTR reporter activity are shown following intraperitoneal injection of SB431542 (10mg/kg body weight) or vehicle control (DMSO) at 1, 4, 8, and 24 h. (B) Bioluminescence activity in tumor-bearing mice before treatment and in response to treatment were measured and expressed as fold induction. Values represent the mean ± SEM.
(C) Tumor xenografts were sectioned 4h post-treatment with SB431542 (10mg/kg body weight) or vehicle control (DMSO) and immunohistochemical analysis was performed using pSmad2(S465–S467) specific antibody. The scale bar is 2 mm and a10-fold magnification of selected section. (D) Quantification of number of cells staining positive for nuclear pSmad2 for control (n=3) and SB431542 treated (n=3) tumors in three random fields. A decrease in pSmad2 stained nuclei in SB431542 treated tumors (761 nuclei, 37.03%) was observed when compared to DMSO treated tumors (1294 nuclei, 62.97%). Statistical analysis: *p= 0.06 calculated by a two-sided Student’s t-test.
In a similar experiment mice (n=5) bearing xenografts of A549 BTR WT and MUT reporters were treated with 10 mg/kg SB431542 and DMSO control. Bioluminescence was acquired 4hr after treatment and plotted as fold change over control (Suppli. Fig 3). BTR WT expressing tumors showed around 8 fold induction in bioluminescence activity with SB431542 while MUT tumors exhibited an attenuated response (around 3.5 fold).
Since BTR reporter bioluminescence represents decreased levels of pSmad2, we next evaluated pSmad2 levels in tumor xenografts harvested 4h post treatment when peak bioluminescence was observed. Immunohistological staining was performed using pSmad2 specific antibody (Fig. 3C–D). Total numbers of pSmad2 positive nuclei were counted for each group and plotted, revealing a significant decrease (p = 0.06) in levels of pSmad2 stained nuclei in SB431542 treated tumors (761 nuclei, 37.03%) as compared to control tumors (1294 nuclei, 62.97%), (Fig. 3D).
High throughput screening of BTR reporter assay against a kinase-inhibitor compound library reveals crosstalk with multiple kinase signaling pathways
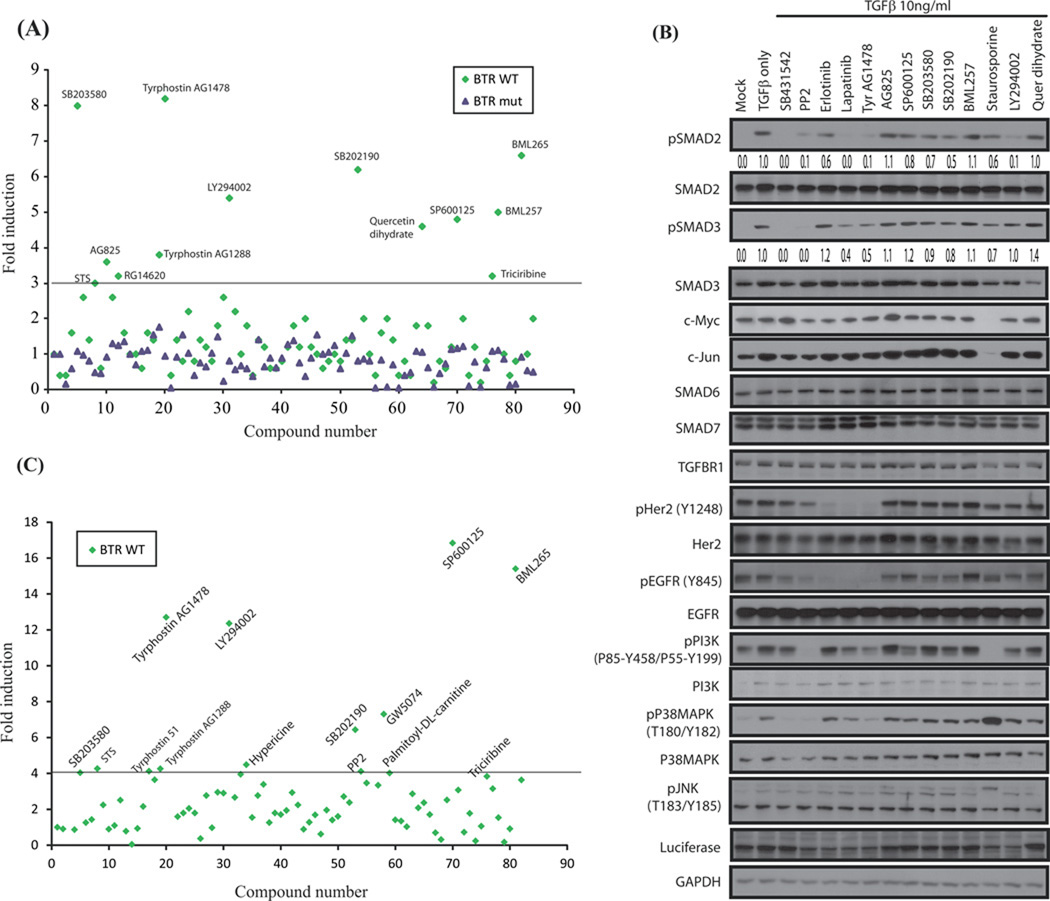
Having confirmed the sensitivity of the BTR reporter to both direct and indirect inhibition of TGFβ-stimulated phosphorylation of the C-terminal serines of Smad2, we examined its suitability for high throughput screening. Cellular activity of the BTR reporter was screened against a kinase-inhibitor library in the presence of TGFβ. Of the 84 small molecular weight kinase inhibitors screened, 13 compounds increased reporter activity. The active compounds included five protein tyrosine kinase (PTK) inhibitors, (Tyrphostin analogs, AG1478, AG 1288, AG 825, and RG14620, and an Erlotinib analog, BML-265); a selective JNK inhibitor (SP600125); two p38 MAP kinase inhibitors (SB203580, SB202190); a PI3K inhibitors (LY294002); two inhibitors of Akt signaling (BML257, triciribine); and finally, Staurosporine, an apoptosis inducer and potent pan-specific protein kinase inhibitor and Quercetin, a common dietary flavanoid believed to have antioxidant and anticancer properties. No significant change in reporter activity was observed when the screen was repeated using A549 BTR-MUT cells, confirming assay specificity for phosphorylation of the C-terminal SXS motif (Fig. 4A). Additionally, the HTS screen was repeated using the 1833 BTR WT reporter cell line, to independently validate the obtained hits (Fig. 4C). Most of the hits between two cell lines were similar although the reporter fold induction varied (Fig. 4A and 4C, Table 1). The EGFR directed inhibitor AG1478 was a hit in A549 and 1833 cells (8.2 fold and 12.7 fold). Not surprisingly, the Akt inhibitor (BML257) and the PI3K inhibitor (LY294002) were also hits in the A549 cells (5.0 fold and 5.4 fold, respectively). In 1833 cells LY294002 yielded a 12.3 fold increase in bioluminescence, while BML257 yielded 3.1 fold increase. A JNK inhibitor (SP600125) and p38MAPK inhibitors (SB203580 and SB202190), important regulators of TGFβ signaling were also hits in both cell lines (see Table 1). Interestingly, GW5074 was a hit in 1833 cells but not in the A549 cells.
Figure 4. High throughput screening of BTR reporter assay against a small molecular weight library of kinase-inhibitors reveals TGFβ-signaling crosstalk with multiple kinase signaling pathways.
(A) A549 cells stably expressing the BTR-WT and BTR-MUT reporters were incubated with compounds from an 84-compound small molecular-weight kinase inhibitor library (10 µM) for 4 h, TGFβ (10ng/ml) was added for 1 h, and reporter activity measured and plotted as fold change over DMSO controls. Compounds which exhibit 3 folds or higher induction in the reporter activity are marked in the plot. All assays were repeated at least 3 times. Values from one representative experiment are shown. (B) Cell lysates from similarly treated A549 BTR-WT cells were probed with antibodies against phospho-(S465–S467) Smad2, total Smad2, phospho-(S423–425) Smad3, total Smad3, c-Myc, c-Jun, Smad6, Smad7, TGFBR1, p(Y1248)-HER2, HER2, p(Y845)-EGFR, EGFR, p-(P85-Y458/P55-Y199)-PI3K, total PI3K, p(T180-Y182)-P38MAPK, total P38MAPK, p(T183/Y185)-JNK, luciferase, and GAPDH. Densitometric analysis of pSmad2 level for all treatments were measured and calculated as fold change over TGFβ-treated samples. (C) Kinase inhibitor library screen as described in Fig. 4A was carried out with 1833 cells stably expressing the BTR WT reporter.
Table 1.
Summary of kinase-inhibitors and their effects on bioluminescent TGFBR1 reporter activity and phosphorylated Smad2 levels
| Kinase- inhibitors (10µM) |
Reporter bioluminescence (Fold response) |
1833 (Fold response) |
pSmad2 levels |
Kinase-inhibitor target(s)* |
References: inhibitors and C-terminal pSmad2/3 |
|
|---|---|---|---|---|---|---|
| BTR- WT |
BTR- MUT |
BTR- WT |
||||
| Vehicle (DMSO) | 1.0 | 1.0 | 1 | 0.0 | ||
| TGFβ only | 0.4 | 1.0 | - | 1.0 | ||
| SB431542 | 10.0 | 2.0 | 3.9 | 0.0 | TGFBR1 kinase | (21, 22, 41, 50) |
| PP2 | 4.0 | 1.0 | 4.1 | 0.1 | Src family non-receptor tyrosine kinases | (25) |
| AG1478 | 8.2 | 0.9 | 12.7 | 0.1 | PTK inhibitor selective for EGFR | (37) |
| AG825 | 3.6 | 0.9 | 0.89 | 1.1 | PTK inhibitor selective for EGFR | (37) |
| SP600125 | 4.8 | 1.2 | 16.8 | 0.8 | c-Jun N-terminal kinase (JNK) | (41, 42, 45) |
| SB203580 | 8.0 | 1.1 | 4.0 | 0.7 | p38 MAP kinase activity | (41, 42, 44) |
| SB202190 | 6.2 | 0.8 | 6.4 | 0.5 | p38 MAP Kinase | (43) |
| BML257 | 5.0 | 1.1 | 3.1 | 1.1 | Akt1/2/3 serine-threonine protein kinases | |
| Staurosporine | 3.0 | 0.5 | 4.2 | 0.6 | Pan-specific | (32) |
| LY294002 | 5.4 | 0.8 | 12.3 | 0.1 | Phosphoinositide 3-kinase (PI3K) | (41, 47) |
| Quercetin | 4.6 | 1.1 | 2.0 | 1.0 | EGFR and Her-2 tyrosine kinase, PI3K, | (34) |
| Erlotinib | n/a | n/a | n/a | 0.6 | EGFR tyrosine kinase | (37) |
| Lapatinib | n/a | n/a | n/a | 0.0 | EGFR and Her-2 tyrosine kinase | (37) |
target information from product inserts
To evaluate the effect of these inhibitors on TGFβ-induced Smad2 phosphorylation, select active compounds, along with two clinically important receptor kinase inhibitors, Erlotinib and Lapatinib, underwent further analysis. A549 BTR-WT cells were treated with inhibitors in the presence of TGFβ, lysates were prepared, and Western blots probed with pSmad2 antibodies. The results (summarized in Table 1) confirm the previously described inhibition of TGFβ-activated Smad2 phosphorylation by the direct TGFΒR1 inhibitor, SB431542, and indirect inhibition with src-kinase inhibitor, PP2. Of the selected kinase inhibitors, Lapatinib, AG1478 and LY294002 completely block TGFβ-induced Smad2 phosphorylation while Erlotinib, AG825, SP600125, SB203580, SB202190 and Staurosporine show only moderate reduction in measurable pSmad2 levels. BML257 and Quercetin treatments resulted in no measurable reduction in pSmad2 levels despite increased reporter activity (Fig. 4B). Additionally western blots were probed with pSmad3 antibody to access the potential role of non-TGFβ receptor kinase inhibitors in inhibiting TGFβ mediated Smad3 phosphorylation. Lapatinib and AG1478 moderately block TGFβ-induced Smad3 phosphorylation while SB203580, SB202190 and Staurosporine show only marginal reductions in measurable pSmad3 levels.
To delineate other signaling events affecting TGFβ receptor activity we probed the Western blot with antibodies to target proteins (see Table 1 for references). The PTK inhibitors Erlotinib, Lapatinib, and Tyrphostin AG1478 reduce levels of pHER2 and pEGFR while increasing the expression of the inhibitory-Smad, Smad7. The Tyrphostin analog, AG825, had no measurable effect on activation of HER2 or EGFR kinase. AG825 is an ATP-competitive inhibitor of HER2 and EGFR kinase activity in cell-free assays but, due to high intracellular ATP levels, it has minimal activity in whole cells (31). Treatment with JNK inhibitor, SP600125, resulted in a moderate decrease in TGFβ mediated Smad2 phosphorylation but levels of pJNK, as measured with pJNK (T180/Y182) antibody, remain unchanged with TGFβ or SP600125 treatment. TGFβ treatment increases phosphorylated p38 MAP kinase (p-p38MAPK) levels which is unchanged following treatment with the p38 MAPK inhibitors SB203580 and SB202190; p-p38MAPK levels are attenuated by Tyrphostin AG1478, SB431542, and PP2 treatment. Levels of phosphorylated PI3K were partly reduced by LY294002, a PI3K inhibitor, as well as, with Lapatinib, Tyrphostin AG1478, and SP600125. A slight decrease in luciferase expression was observed with PP2, Erlotinib, Lapatinib, Tyrphostin AG1478, Staurosporine, and LY294002 treatments with no corresponding decrease in GAPDH levels suggesting results are not due to differences in protein loading (Fig. 4B).
Staurosporine treatment caused only a small increase in BTR activity and a moderate decrease in Smad2 phosphorylation while completely blocking activation of PI3K and causing a marked increase in p-p38 MAPK along with reduced expression of most of the other proteins measured including luciferase. It has been reported that Staurosporine treatment of Mv1Lu cells, 100ng/ml, increased pSmad2 and enhanced TGFβ-stimulated apoptosis (32) suggesting that a 5-hour exposure to a 10-fold higher concentration of Staurosporine, as seen in our experiment, most likely initiated cell death thus decreasing protein expression (Fig. 4B).
Quercetin, a common dietary flavanoid believed to have antioxidant and anticancer properties, was observed to decrease levels of HER2 protein and inhibit downstream PI3K signaling in breast cancer cells (33) and suppress basal expression of TGFBR1/2, Smad2/3/4, and pSmad2/3 after 72 hour treatment of keloid fibroblasts (34). In this study, quercetin treatment stimulated BTR reporter bioluminescence and showed a moderate inhibition of TGFBR1 and pEGFR levels but did not demonstrate any measureable inhibition of pSmad2 levels, PI3K signaling, or increased expression of inhibitory Smad7. These results suggest that the reporter response may be a result of an immeasurable change in pSmad2 level due to moderate changes in TGFBR1 and pEGFR expression (Fig. 4B). Lysates were also probed with c-Jun and c-Myc antibodies to evaluate the effect of each inhibitor on TGFβ/Smad-mediated transcription.
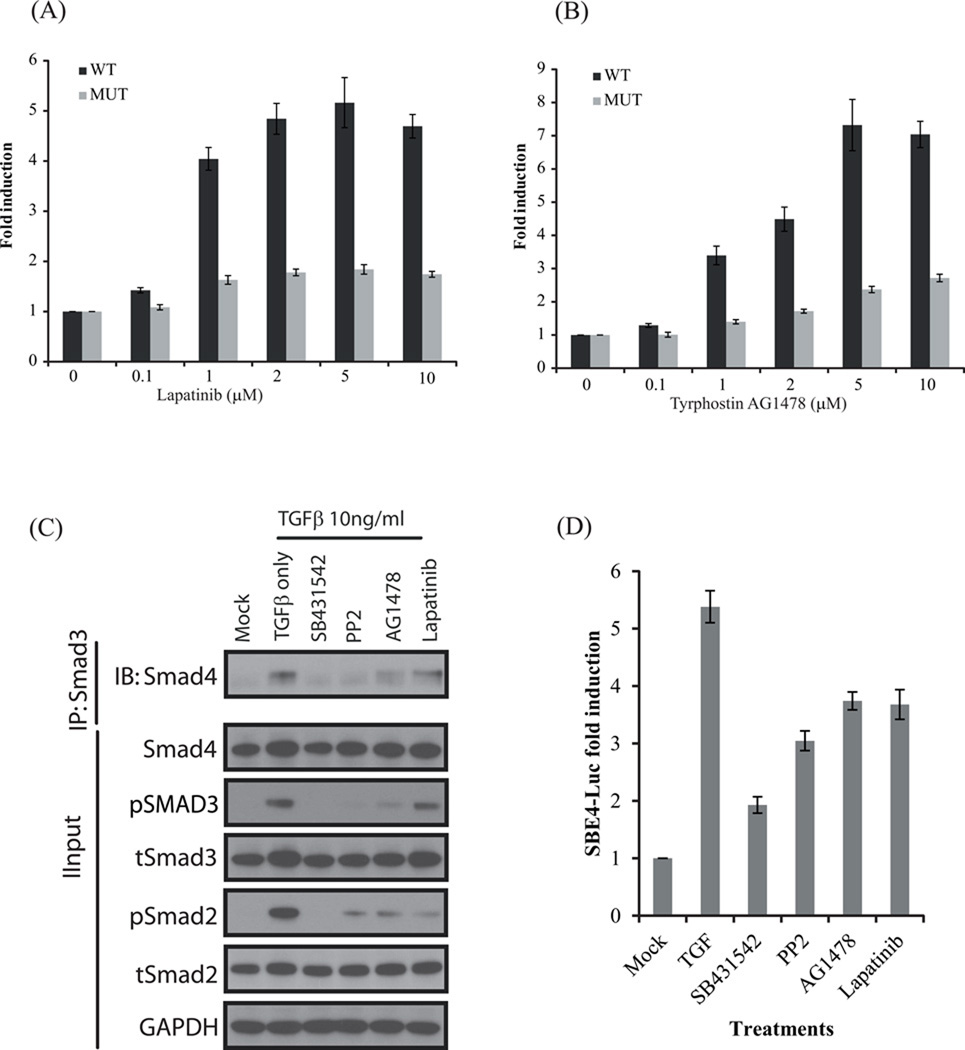
Two RTK inhibitors which abrogated TGFβ-induced activation of Smad2 (Lapatinib and AG1478) were selected for further analysis. Lapatinib activated the BTR-WT reporter at a dose of 1µM (~4 fold), but failed to activate the mutant reporter significantly at doses as high as 10µM (Fig 5 A). Tyrphostin AG1478 also activated the WT reporter similarly, and did not alter the activity of the MUT reporter substantially (Fig 5 A). These data demonstrate that Lapatanib and AG1478 were valid hits in the screen.
Figure 5.
(A) A549 cells stably expressing the BTR WT and MUT reporter were treated with increasing concentrations of RTK inhibitor Lapatinib (0.1, 1, 2, 5, and 10µM) and Tyrphostin AG1478 (B). Bioluminescence was measured after 1hr. The change in bioluminescence activity over mock treatment (DMSO) levels was calculated and plotted as fold induction. Bioluminescent measurements were done in 8 wells. (C) A549 cells were treated with TGFβ (10ng/ml) or TGFβ and SB431542, PP2, AG1478 and Lapatinib (10 µM) for 1 h, cell lysates were prepared and immunoprecipitated with Smad3 specific antibody. The level of Smad2/3–Smad4 complex formation was determined by Western blot analysis with Smad4 antibody. The input lysate was probed against pSmad2, total Smad2, pSmad3, total Smad3, Smad4 and GAPDH. (D) A549 cells were transiently transfected with SBE4-Luc reporter plasmid and GLuc plasmid as internal control. Cells were treated with 10 µM SB431542, PP2, AG1478 and Lapatinib in presence of 10ng/ml TGFβ for 22h. Firefly luciferase activity was normalized to Gaussia luciferase and fold induction (over mock /DMSO treated) is expressed as the mean±SEM of triplicate measurements.
To further evaluate the validity of the two RTK-inhibitors as modulators of TGFβ signaling, their impact on Smad2/3-Smad4 complex formation was evaluated. Formation of a Smad2/3-Smad4 complex is a critical event for nuclear translocation of Smad2/3. Lysates from A549 cells treated with SB431542, PP2, AG1478 and Lapatinib in the presence of TGFβ were immunoprecipitated using a Smad3 antibody and co-immunoprecipitation of Smad4 was evaluated (Fig 5C). SB431542 and PP2 completely blocked Smad3-Smad4 complex formation compared to TGFβ treated cells, while AG1478 substantially and Lapatinib marginally blocked complex formation between Smad3-Smad4 in A549 cells. The reduced Smad complex formation was due to blockade of TGFβ-induced phosphorylation and activation of Smad2/3 as seen in the input lysates (Fig 5C). Additionally to elucidate if inhibition of Smad2/3–4 complex formation by PP2 and RTK-inhibitors have any effect on downstream transcriptional responses mediated by TGFβ, we carried out transcriptional response assay using a TGFβ responsive reporter SBE4-Luc (17) (Fig 5D). TGFβ strongly activated the SBE4-Luc reporter in A549 cells which was significantly blocked by the ALK5 inhibitor SB431542 and moderately blocked by PP2, AG1478 and Lapatinib. These results suggest that PP2, AG1478 and Lapatinib partially inhibit the activation of Smad pathway resulting in inhibition of TGFβ-induced transcriptional activity.
Discussion
The dual modality of TGFβ, both as a potent tumor suppressor and a stimulator of tumor progression, invasion and metastasis, make it a critical target for therapeutic intervention in human cancers. The ability to perform real-time, non-invasive imaging of TGFβ-activated Smad signaling in live cells and animal models would significantly improve our ability to develop targeted therapeutics for cancer and to monitor their effects in vivo. To study TGFβ-mediated phosphorylation of the C-terminal serines of R-Smads, we constructed a TGFBR1 bioluminescent molecular imaging reporter, described in Figure 1A–B, based on split-luciferase technology (14). We validated the specificity and responsiveness of the BTR reporter through direct inhibition of TGFBR1 kinase activity, and indirectly, through inhibition of src-kinase, a downstream modulator of TGFβ-stimulated Smad2 phosphorylation (25). Reporter bioluminescence activity shows a robust response following inhibition of TGFBR1 kinase activity with SB431542. Time- and concentration-dependence of SB431542 inhibition of TGFBR1 kinase was confirmed by increased reporter activity and a concomitant decrease in levels of pSmad2 (Fig. 1C–E, Suppli. Fig. 2).
The EC50 derived using the reporter (for SB431542) was estimated to be 2.09 µM, while the pSmad2 westerns yielded an EC50 of 1.6 µM in A549 cells. In 1833 cells, the values were 15.68 µM using the reporter and 0.36 µM using pSmad2 westerns. Strong positive correlation (Fig. 1C’; Suppli. Fig. 2A’), goodness of fit, and statistical significance between reporter fold induction and SB431542 concentration clearly demonstrate the robustness nature of the BTR reporter. A strong negative correlation between reporter fold induction and pSmad2 levels (Fig. 1E; Suppli. Fig. 2B bottom panel) further substantiated the effectiveness of the reporter. Inhibition of downstream modulation of pSmad2 levels by src-kinase also demonstrated timeand concentration-dependent increases in reporter bioluminescence along with a decrease in measurable levels of pSmad2 (Suppli. Fig. 1). These results strongly support the relationship between increased reporter activity and both direct and indirect inhibition of Smad2 phosphorylation.
Direct confirmation that TGFβ is a biological transducer of BTR reporter activity is demonstrated by the reduction of reporter bioluminescence following TGFβ treatment of A549 BTR-WT cells (Fig. 2A) and the corresponding increase in pSmad2 (Fig. 2B). The sustained decrease in reporter activity and prolonged elevation of pSmad2 are consistent with continuous shuttling of Smad proteins between cell nucleus and cytoplasm shown to exist during TGFβ signaling (23). Immunoprecipitation of the reporter, followed by analysis of phosphoserine content, confirms that the reporter was phosphorylated in a TGFβ-dependent manner (Fig. 2C; Suppli. Fig. 2D). In addition, suppression of the upstream kinases by siRNA knockdown of TGFΒR1 and TGFΒR2 verifies the direct link between reporter activity and TGFβ receptor signaling. Knockdown TGFBR1 and TGFBR2 cells have reduced levels of pSmad2 and show a significant increase in reporter bioluminescence (Fig. 2D–E; Suppli. Fig. 2E–F).
TGFβ signaling contributes to tumor progression and metastasis through activation of epithelialto-mesenchymal transition (EMT), a process in which epithelial cells convert to fibroblastoid-like cells which generate an invasive and metastatic phenotype associated with poor clinical outcomes for cancer patients (35). TGFβ-activation of Smad2 signaling induces EMT in A549 BTR-WT cells, resulting in the loss of cell-to-cell contact and the cellular conversion to an elongated, spindle-shaped morphology (Fig. 2F). Protein analysis reveals the characteristic EMT downregulation of E-cadherin expression, a critical caretaker of epithelial integrity, and a corresponding upregulation of N-cadherin (Fig. 2G) (35). Activated Smad2 has also been shown to regulate expression of the proto-oncogenes c-myc (30) and c-jun (29). TGFβ downregulation of c-myc is critical to growth inhibition (30) where the upregulation of c-jun is important for cell survival (29). TGFβ stimulation of A549 BTR-WT cells suppresses c-myc expression with a corresponding upregulation of c-jun (Fig. 2G). Cell treatment with TGFBR1 kinase inhibitor SB431542 results in increased expression of c-myc and eliminates c-jun expression (Fig. 2H). A TGFβ increase in total c-jun levels is consistent with previous findings demonstrating an effective downstream signaling cascade initiated by activated R-Smads in response to TGFβ (29).
Several imaging techniques have been designed to monitor TGFβ signaling in various in vivo models (5–12, 36) but none directly measure the phosphorylation of R-Smads, the key event in the TGFβ signaling pathway. Accordingly, high throughput screening of a kinase-inhibitor library identified the existence of a complex crosstalk between TGFβ signaling and tyrosine kinase receptors, as well as p38 MAPK, JNK, and PI3K pathways through attenuation of RSmad phosphorylation at the C-terminal serines (Summarized in Suppli. Fig. 4).
Research has shown growth factors which signal through receptor tyrosine kinases (RTKs) such as, hepatocyte growth factor (HGF) and epidermal growth factor (EGF), mediate Smad2 activity independent of TGFΒR1 activation, suggesting intricate crosstalk between RTK and RSK signaling pathways (37, 38). Our results demonstrate that EGF receptor (EGFR) and human epidermal growth factor receptor 2 (HER2) kinase inhibitors Erlotinib, Lapatinib, and Tyrphostin AG1478 block TGFβ-mediated increases in pSmad2 (Fig. 4B). Lapatinib, and Tyrphostin AG1478 marginally reduce TGFβ-mediated phosphorylation of Smad3 while Erlotinib had no significant effect (Fig. 4B).The reduction in pSmad2 levels coincides with an equally pronounced decrease in expression of the active kinases, phosphorylated HER2 (pHER2) and phosphorylated EGFR (pEGFR). Treatment of A549 BTR-WT cells with SB431542 also reduces basal levels of pEGFR, further supporting signaling crosstalk between RTK and RSK pathways.
An important component of crosstalk between TGFβ- and RTK-signaling is the increased expression of inhibitory-Smad (I-Smad) proteins (39). The I-Smads, Smad6 and Smad7, modulate signaling of TGFβ and, another TGFβ isoform, bone morphogenetic protein (BMP) by binding to the activated Type-1 receptor thereby, preventing recruitment and phosphorylation of R-Smads (38, 39). In some cell-types, TGFβ or BMP induce I-Smad expression resulting in a negative feedback mechanism to regulate further TGFβ- or BMP-signaling. Besides induction by TGFβ or BMP, I-Smad expression can also be induced in a cell-dependent manner by other cytokine signaling pathways and stress induction (39, 40). In A549 cells, expression levels of Smad6 and Smad7 did not increase following TGFβ treatment. However, treatment with EGFR and HER2 kinase inhibitors significantly increased expression of Smad7 in A549 BTR-WT cells resulting in interference with recruitment and phosphorylation of Smad2 by active TGFΒR1 (Fig. 4B). These data suggest that, at least in part, the RTK signaling pathways increase activation of R-Smads by suppressing Smad7 expression and subsequent interference with Smad2 phosphorylation, providing evidence of the importance of RTK and RSK crosstalk in regulating TGFβ-induced Smad2 signaling.
In addition to crosstalk between RTK and RSK signaling, other kinase pathways have been shown to modulate Smad2 phosphorylation. BTR reporter activity was increased by inhibitors of PI3K, p38 MAPK, and JNK signaling pathways. Martin, et al. (41) demonstrated that kinase inhibitors against PI3K (LY294002), p38 MAPK (SB203580), and JNK (SP6000125) activity attenuate TGFβ-mediated phosphorylation of Smad2 in lung fibroblasts where treatment with inhibitors for other kinase signaling pathways, MEK1 (PD98059), PKC (Ro31–8425), and Erk/MEK (U0126), had no effect. These observations are supported by our screening results, BTR reporter activity was increased for inhibitors shown to reduce R-Smad activation (LY294002, SB203580, and SP600125) (Fig. 4A) and was inactive when treated with inhibitors to kinase pathways that do not modulate TGFβ-stimulated phosphorylation of C-terminal serines of Smad2/3 (PD98059, Ro31–8425, and U0126) (Data not shown). A parallel screen using the 1833 cell line stably expressing the BTR WT reporter yielded hits that were overlapping with the hits obtained from the A549 cells (Fig 4C). Published studies support some of these hits, for example, inhibitors for JNK (SP600125), and p38 MAPK (SB203580) have been shown to downregulate phosphorylation of the C-terminal serines in rat mesangial cells (42) and the p38 MAPK inhibitor (SB202190) abolishes TGFβ-activation of Smad-dependent promoters and attenuate pSmad2 levels in glioblastoma cells (43).
However, the extent of signaling crosstalk between TGFβ and MAPK pathways has been shown to be dependent on cell-type and cell-context. Where the previous studies demonstrate JNK and p38MAPK modulation of pSmad, TGFβ-induced p38 MAPK activity in mouse mammary epithelial (NMuMG) cells was shown to mediate TGFβ responses independent of Smad phosphorylation (44). The cell-specific differences in crosstalk between MAPK and Smad pathways are also evident with the JNK pathway. Where active JNK enhances or maintains TGFβ-mediated expression in fibroblasts(41) and mesangial cells(42) through phosphorylation of C-terminal Smad2, inhibiting JNK activity in embryonic lung explants increases both endogenous and TGFβ-induced Smad phosphorylation (45) suggesting that active JNK antagonizes TGFβ-induced pSmad2. In yet another study, TGFβ-activated JNK was shown to only phosphorylate Smad3 outside the C-terminal SXS motif in Mv1Lu cells (46).
Besides crosstalk between p38 MAPK and JNK with TGFβ signaling pathways, our data suggests that the PI3K-Akt signaling pathway may also provide significant regulation of TGFβ-induced cellular responses. Treatment with the PI3K inhibitor, LY294002, results in a robust activation of the BTR reporter and a nearly complete inhibition of TGFβ-mediated phosphorylation of the C-terminal serines of Smad2 (Fig. 4A–B). Inhibition of Akt1/2/3, a PI3K target protein, with BML257 also activates the BTR reporter but did not yield a measurable decrease in pSmad. PI3K signaling has been shown to be critical for Smad2-dependent activity in fibroblasts (41) and transcriptional responses, EMT and cell migration of NMuMG cells (47).
Although C-terminal SXS phosphorylation by the TGFβ type-1 receptor is the key event in TGFβ-signaling, phosphorylation of the linker region and, to a lesser extent, the MH1 domain of Smad proteins by intracellular protein kinases can also positively and negatively regulate R-Smad activity (2). Inhibitors for many of these kinases were screened with the BTR reporter and, as expected, did not increase bioluminescence, confirming the specificity of the BTR reporter assay to phosphorylation of the critical SXS motif. MEK1 inhibitor, PD98059, which inhibits RTK phosphorylation of Smad2 through activation of ras proteins does not affect TGFβ-mediated Smad2 phosphorylation (37) and did not activate the BTR reporter in our kinase-inhibitor screen. Protein kinase inhibitors known to modulate pSmad in the linker region, were also inactive in the high throughput screen including, the Ca2+ calmodulin-dependent protein kinase II (CaMKII) specific inhibitor KN93 (48) and inhibitors of ROCK, PKA/PKG, and MLCK (Y027632, HA-1077, and ML-7, respectively) (49).
In summary, we have designed and validated a sensitive and highly specific bioluminescence imaging tool to monitor TGFβ-mediated phosphorylation of the C-terminal serines of R-Smads in live cells and tumor xenografts. The BTR reporter is adaptable to high throughput screening where it can be used to aide in identification and evaluation of therapeutic compounds targeting TGFβ-Smad2 signaling. Following compound identification, this bioluminescent imaging technique provides a repetitive, non-invasive, dynamic method to monitor in vivo TGFβ signaling to evaluate drug-target interaction and pharmacodynamics of therapeutic compounds that directly or indirectly effect TGFβ-stimulated Smad2 phosphorylation providing new insights into TGFβ signaling and help in identifying new drug therapeutics for cancer treatment.
Statement of Translational Relevance.
The response of cells to TGFβ stimulation is highly contextual in cancer. Loss or mutational inactivation of genes that mediate the TGFβ pathway early in carcinogenesis is indicative of its role as a tumor suppressor. Yet, paradoxically, TGFβ also modulates processes such as cell invasion and modifications of the tumor microenvironment that are critical for metastasis and tumor establishment at distant sites. Although therapeutic agents have been developed targeting this pathway, their rational use will require a better understanding of the regulation of its signaling. In this effort, we in this paper describe the development of a reporter molecule wherein TGFβ receptor I kinase activity can be imaged dynamically and non-invasively in living subjects. We also provide preliminary results that TGFβ signaling activity is impacted by seemingly unrelated signaling cascades, providing potential explanation for the contextual differences in TGFβ downstream signaling events in cancer.
Supplementary Material
Acknowledgements
We thank Barbara Olson for critical reading of the manuscript. We thank Dr J Massague for providing the 1833 cell line and Dr Bert Vogelstein for providing the SBE4-Luc reporter plasmid. We acknowledge kind gift of Erlotinib by Genentech. Light microscopy was performed at the Microscopy and Image Analysis Laboratory at the University of Michigan. The tumors were processed at the UMCCC Tissue Core Facility.
Grant support
This work was supported by the following grants P01 CACA85878, R01CA129623, R21CA131859 and P50CA093990. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Schmidt-Weber CB, Blaser K. Regulation and role of transforming growth factor-beta in immune tolerance induction and inflammation. Curr Opin Immunol. 2004;16:709–716. doi: 10.1016/j.coi.2004.09.008. [DOI] [PubMed] [Google Scholar]
- 2.Massague J. TGFbeta in Cancer. Cell. 2008;134:215–230. doi: 10.1016/j.cell.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Meulmeester E, Ten Dijke P. The dynamic roles of TGF-beta in cancer. The Journal of pathology. 2011;223:205–218. doi: 10.1002/path.2785. [DOI] [PubMed] [Google Scholar]
- 4.Ross S, Hill CS. How the Smads regulate transcription. Int J Biochem Cell Biol. 2008;40:383–408. doi: 10.1016/j.biocel.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 5.Kang Y, He W, Tulley S, Gupta GP, Serganova I, Chen CR, et al. Breast cancer bone metastasis mediated by the Smad tumor suppressor pathway. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:13909–13914. doi: 10.1073/pnas.0506517102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Giampieri S, Manning C, Hooper S, Jones L, Hill CS, Sahai E. Localized and reversible TGFbeta signalling switches breast cancer cells from cohesive to single cell motility. Nat Cell Biol. 2009;11:1287–1296. doi: 10.1038/ncb1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hida N, Awais M, Takeuchi M, Ueno N, Tashiro M, Takagi C, et al. High-sensitivity real-time imaging of dual protein-protein interactions in living subjects using multicolor luciferases. PLoS One. 2009;4:e5868. doi: 10.1371/journal.pone.0005868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Korpal M, Yan J, Lu X, Xu S, Lerit DA, Kang Y. Imaging transforming growth factor-beta signaling dynamics and therapeutic response in breast cancer bone metastasis. Nature medicine. 2009;15:960–966. doi: 10.1038/nm.1943. [DOI] [PubMed] [Google Scholar]
- 9.Serganova I, Moroz E, Vider J, Gogiberidze G, Moroz M, Pillarsetty N, et al. Multimodality imaging of TGFbeta signaling in breast cancer metastases. Faseb J. 2009;23:2662–2672. doi: 10.1096/fj.08-126920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chong AK, Satterwhite T, Pham HM, Costa MA, Luo J, Longaker MT, et al. Live imaging of Smad2/3 signaling in mouse skin wound healing. Wound Repair Regen. 2007;15:762–766. doi: 10.1111/j.1524-475X.2007.00299.x. [DOI] [PubMed] [Google Scholar]
- 11.Luo J, Lin AH, Masliah E, Wyss-Coray T. Bioluminescence imaging of Smad signaling in living mice shows correlation with excitotoxic neurodegeneration. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:18326–18331. doi: 10.1073/pnas.0605077103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lin AH, Luo J, Mondshein LH, ten Dijke P, Vivien D, Contag CH, et al. Global analysis of Smad2/3-dependent TGF-beta signaling in living mice reveals prominent tissue-specific responses to injury. J Immunol. 2005;175:547–554. doi: 10.4049/jimmunol.175.1.547. [DOI] [PubMed] [Google Scholar]
- 13.Macias-Silva M, Abdollah S, Hoodless PA, Pirone R, Attisano L, Wrana JL. MADR2 is a substrate of the TGFbeta receptor and its phosphorylation is required for nuclear accumulation and signaling. Cell. 1996;87:1215–1224. doi: 10.1016/s0092-8674(00)81817-6. [DOI] [PubMed] [Google Scholar]
- 14.Luker KE, Smith MC, Luker GD, Gammon ST, Piwnica-Worms H, Piwnica-Worms D. Kinetics of regulated protein-protein interactions revealed with firefly luciferase complementation imaging in cells and living animals. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:12288–12293. doi: 10.1073/pnas.0404041101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nyati S, Ranga R, Ross BD, Rehemtulla AMSB. Molecular Imaging of GSK3β and CK1α kinases. Analytical Biochemistry. 2010 doi: 10.1016/j.ab.2010.06.020. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kang YB, Siegel PM, Shu WP, Drobnjak M, Kakonen SM, Cordon-Cardo C et al. A multigenic program mediating breast cancer metastasis to bone. Cancer cell. 2003;3:537–549. doi: 10.1016/s1535-6108(03)00132-6. [DOI] [PubMed] [Google Scholar]
- 17.Zawel L, Dai JL, Buckhaults P, Zhou S, Kinzler KW, Vogelstein B, et al. Human Smad3 and Smad4 are sequence-specific transcription activators. Mol Cell. 1998;1:611–617. doi: 10.1016/s1097-2765(00)80061-1. [DOI] [PubMed] [Google Scholar]
- 18.Abramoff MD, Magalhaes PJ, Ram SJ. Image processing with ImageJ. Biophotonics International. 2004;11:36–42. [Google Scholar]
- 19.Chen Y, Kam CS, Liu FQ, Liu Y, Lui VC, Lamb JR, et al. LPS-induced up-regulation of TGF-beta receptor 1 is associated with TNF-alpha expression in human monocyte-derived macrophages. J Leukoc Biol. 2008;83:1165–1173. doi: 10.1189/jlb.0807521. [DOI] [PubMed] [Google Scholar]
- 20.Segawa S, Goto D, Yoshiga Y, Sugihara M, Hayashi T, Chino Y, et al. Inhibition of transforming growth factor-beta signalling attenuates interleukin (IL)-18 plus IL-2-induced interstitial lung disease in mice. Clin Exp Immunol. 160:394–402. doi: 10.1111/j.1365-2249.2010.04094.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Callahan JF, Burgess JL, Fornwald JA, Gaster LM, Harling JD, Harrington FP, et al. Identification of novel inhibitors of the transforming growth factor beta1 (TGF-beta1) type 1 receptor (ALK5) J Med Chem. 2002;45:999–1001. doi: 10.1021/jm010493y. [DOI] [PubMed] [Google Scholar]
- 22.Laping NJ, Grygielko E, Mathur A, Butter S, Bomberger J, Tweed C, et al. Inhibition of transforming growth factor (TGF)-beta1-induced extracellular matrix with a novel inhibitor of the TGF-beta type I receptor kinase activity: SB-431542. Mol Pharmacol. 2002;62:58–64. doi: 10.1124/mol.62.1.58. [DOI] [PubMed] [Google Scholar]
- 23.Inman GJ, Nicolas FJ, Hill CS. Nucleocytoplasmic shuttling of Smads 2, 3, and 4 permits sensing of TGF-beta receptor activity. Mol Cell. 2002;10:283–294. doi: 10.1016/s1097-2765(02)00585-3. [DOI] [PubMed] [Google Scholar]
- 24.Matsuyama S, Iwadate M, Kondo M, Saitoh M, Hanyu A, Shimizu K, et al. SB-431542 and Gleevec inhibit transforming growth factor-beta-induced proliferation of human osteosarcoma cells. Cancer research. 2003;63:7791–7798. [PubMed] [Google Scholar]
- 25.Zhang X, Arnott JA, Rehman S, Delong WG, Jr, Sanjay A, Safadi FF, et al. Src is a major signaling component for CTGF induction by TGF-beta1 in osteoblasts. J Cell Physiol. 2010;224:691–701. doi: 10.1002/jcp.22173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hanke JH, Gardner JP, Dow RL, Changelian PS, Brissette WH, Weringer EJ, et al. Discovery of a novel, potent, and Src family-selective tyrosine kinase inhibitor. Study of Lck-and FynT-dependent T cell activation. J Biol Chem. 1996;271:695–701. doi: 10.1074/jbc.271.2.695. [DOI] [PubMed] [Google Scholar]
- 27.Kim JH, Jang YS, Eom KS, Hwang YI, Kang HR, Jang SH, et al. Transforming growth factor beta1 induces epithelial-to-mesenchymal transition of A549 cells. J Korean Med Sci. 2007;22:898–904. doi: 10.3346/jkms.2007.22.5.898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kasai H, Allen JT, Mason RM, Kamimura T, Zhang Z. TGF-beta 1 induces human alveolar epithelial to mesenchymal cell transition (EMT) Resp Res. 2005;6 doi: 10.1186/1465-9921-6-56. - [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang Y, Hutter D, Liu Y, Wang X, Sheikh MS, Chan AM, et al. Transforming growth factor-beta 1 suppresses serum deprivation-induced death of A549 cells through differential effects on c-Jun and JNK activities. J Biol Chem. 2000;275:18234–18242. doi: 10.1074/jbc.M909431199. [DOI] [PubMed] [Google Scholar]
- 30.Yagi K, Furuhashi M, Aoki H, Goto D, Kuwano H, Sugamura K, et al. c-myc is a downstream target of the Smad pathway. J Biol Chem. 2002;277:854–861. doi: 10.1074/jbc.M104170200. [DOI] [PubMed] [Google Scholar]
- 31.Osherov N, Gazit A, Gilon C, Levitzki A. Selective inhibition of the epidermal growth factor and HER2/neu receptors by tyrphostins. J Biol Chem. 1993;268:11134–11142. [PubMed] [Google Scholar]
- 32.Solovyan VT, Keski-Oja J. Proteolytic activation of latent TGF-beta precedes caspase-3 activation and enhances apoptotic death of lung epithelial cells. J Cell Physiol. 2006;207:445–453. doi: 10.1002/jcp.20607. [DOI] [PubMed] [Google Scholar]
- 33.Jeong JH, An JY, Kwon YT, Li LY, Lee YJ. Quercetin-induced ubiquitination and downregulation of Her-2/neu. Journal of cellular biochemistry. 2008;105:585–595. doi: 10.1002/jcb.21859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Phan TT, Lim IJ, Chan SY, Tan EK, Lee ST, Longaker MT. Suppression of transforming growth factor beta/smad signaling in keloid-derived fibroblasts by quercetin: implications for the treatment of excessive scars. J Trauma. 2004;57:1032–1037. doi: 10.1097/01.ta.0000114087.46566.eb. [DOI] [PubMed] [Google Scholar]
- 35.Wendt MK, Allington TM, Schiemann WP. Mechanisms of the epithelial-mesenchymal transition by TGF-beta. Future Oncol. 2009;5:1145–1168. doi: 10.2217/fon.09.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Luo J, Wyss-Coray T. Bioluminescence analysis of Smad-dependent TGF-beta signaling in live mice. Methods in molecular biology. (Clifton, NJ. 2009;574:193–202. doi: 10.1007/978-1-60327-321-3_16. [DOI] [PubMed] [Google Scholar]
- 37.de Caestecker MP, Parks WT, Frank CJ, Castagnino P, Bottaro DP, Roberts AB, et al. Smad2 transduces common signals from receptor serine-threonine and tyrosine kinases. Genes Dev. 1998;12:1587–1592. doi: 10.1101/gad.12.11.1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Derynck R, Zhang YE. Smad-dependent and Smad-independent pathways in TGF-beta family signalling. Nature. 2003;425:577–584. doi: 10.1038/nature02006. [DOI] [PubMed] [Google Scholar]
- 39.Park SH. Fine tuning and cross-talking of TGF-beta signal by inhibitory Smads. J Biochem Mol Biol. 2005;38:9–16. doi: 10.5483/bmbrep.2005.38.1.009. [DOI] [PubMed] [Google Scholar]
- 40.Moustakas A, Souchelnytskyi S, Heldin CH. Smad regulation in TGF-beta signal transduction. J Cell Sci. 2001;114:4359–4369. doi: 10.1242/jcs.114.24.4359. [DOI] [PubMed] [Google Scholar]
- 41.Martin MM, Buckenberger JA, Jiang J, Malana GE, Knoell DL, Feldman DS, et al. TGF-beta1 stimulates human AT1 receptor expression in lung fibroblasts by cross talk between the Smad, p38 MAPK, JNK, and PI3K signaling pathways. Am J Physiol Lung Cell Mol Physiol. 2007;293:L790–L799. doi: 10.1152/ajplung.00099.2007. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 42.Jiang W, Zhang Y, Wu H, Zhang X, Gan H, Sun J, et al. Role of cross-talk between the Smad2 and MAPK pathways in TGF-beta1-induced collagen IV expression in mesangial cells. Int J Mol Med. 2010;26:571–576. doi: 10.3892/ijmm_00000501. [DOI] [PubMed] [Google Scholar]
- 43.Dziembowska M, Danilkiewicz M, Wesolowska A, Zupanska A, Chouaib S, Kaminska B. Cross-talk between Smad and p38 MAPK signalling in transforming growth factor beta signal transduction in human glioblastoma cells. Biochem Biophys Res Commun. 2007;354:1101–1106. doi: 10.1016/j.bbrc.2007.01.113. [DOI] [PubMed] [Google Scholar]
- 44.Yu L, Hebert MC, Zhang YE. TGF-beta receptor-activated p38 MAP kinase mediates Smad-independent TGF-beta responses. The EMBO journal. 2002;21:3749–3759. doi: 10.1093/emboj/cdf366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu S, Kasisomayajula K, Peng J, Bancalari E. Inhibition of JNK enhances TGF-beta1-activated Smad2 signaling in mouse embryonic lung. Pediatr Res. 2009;65:381–386. doi: 10.1203/PDR.0b013e3181991c67. [DOI] [PubMed] [Google Scholar]
- 46.Engel ME, McDonnell MA, Law BK, Moses HL. Interdependent SMAD and JNK signaling in transforming growth factor-beta-mediated transcription. J Biol Chem. 1999;274:37413–37420. doi: 10.1074/jbc.274.52.37413. [DOI] [PubMed] [Google Scholar]
- 47.Bakin AV, Tomlinson AK, Bhowmick NA, Moses HL, Arteaga CL. Phosphatidylinositol 3-kinase function is required for transforming growth factor beta-mediated epithelial to mesenchymal transition and cell migration. J Biol Chem. 2000;275:36803–36810. doi: 10.1074/jbc.M005912200. [DOI] [PubMed] [Google Scholar]
- 48.Abdel-Wahab N, Wicks SJ, Mason RM, Chantry A. Decorin suppresses transforming growth factor-beta-induced expression of plasminogen activator inhibitor-1 in human mesangial cells through a mechanism that involves Ca2+-dependent phosphorylation of Smad2 at serine-240. Biochem J. 2002;362:643–649. doi: 10.1042/0264-6021:3620643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Meyer-ter-Vehn T, Sieprath S, Katzenberger B, Gebhardt S, Grehn F, Schlunck G. Contractility as a prerequisite for TGF-beta-induced myofibroblast transdifferentiation in human tenon fibroblasts. Invest Ophthalmol Vis Sci. 2006;47:4895–4904. doi: 10.1167/iovs.06-0118. [DOI] [PubMed] [Google Scholar]
- 50.Inman GJ, Nicolas FJ, Callahan JF, Harling JD, Gaster LM, Reith AD, et al. SB-431542 is a potent and specific inhibitor of transforming growth factor-beta superfamily type I activin receptor-like kinase (ALK) receptors ALK4, ALK5, and ALK7. Mol Pharmacol. 2002;62:65–74. doi: 10.1124/mol.62.1.65. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.