Abstract
Androgen receptor (AR) is important for prostate cancer development and progression. Genome-wide mapping of AR binding sites in prostate cancer have found that the majority of AR binding sites are located within non-promoter regions. These distal AR binding regions regulate AR target genes (e.g. UBE2C) involved in prostate cancer growth through chromatin looping. In addition to long-distance gene regulation, looping has been shown to induce spatial proximity of two genes otherwise located far away along the genomic sequence and the formation of double strand DNA breaks, resulting in aberrant gene fusions (e.g. TMPRSS2-ERG) that also contribute to prostate tumorigenesis. Elucidating the mechanisms of AR-driven chromatin looping will increase our understanding of prostate carcinogenesis and may lead to the identification of new therapeutic targets.
Androgen receptor (AR) signaling in prostate cancer
Although significant strides have been made in prostate cancer research and treatment, prostate cancer remains one of the most commonly diagnosed cancers and the second leading cause of cancer deaths in American men1. The “male” hormones or androgens, exerting their biological effect through androgen receptor (AR), play important role in prostate cancer development and progression2, 3. Therefore androgen ablation therapy, including surgical and chemical castration, is the first-line therapeutic approach for advanced androgen-dependent prostate cancer (ADPC). Although this therapy is initially effective, ADPC ultimately progresses into an incurable, castration-resistant stage of the disease (CRPC), involving the reactivation of AR signaling. The mechanisms for AR reactivation after castration include AR amplification, increased androgen sensitivity, increased intracellular synthesis of androgen, a constitutively active AR lacking ligand binding domain (LBD), activation of growth factor pathways, and retinoblastoma (RB) loss-induced, E2F1-mediated AR overexpression3–6. Thus, aberrantly active AR signaling exists in both ADPC and CRPC.
AR is a ligand-dependent transcription factor belonging to the nuclear hormone receptor (NR) superfamily7. To understand how AR signaling contributes to ADPC and CRPC, recent studies have utilized genome-wide chromatin immunoprecipitation (ChIP) techniques to define AR binding sites across the entire human genome in prostate cancer cells8–13. In these studies, AR ChIP-enriched DNA was amplified and hybridized to tiling DNA microarrays (ChIP-on-chip), or subjected to massively parallel high throughput sequencing (ChIP-seq)14, 15. These studies have greatly advanced understanding of AR binding to chromatin, the functional interplay between AR and chromatin, and AR-mediated regulation of target genes involved in prostate tumorigenesis. In this review, we discuss how the majority of AR binding sites in the genome is located within non-promoter distal regions and the importance of this for transcriptional regulation. At these sites, several transcription factors act as positive or negative regulators of AR function and affect AR-mediated gene regulation. We then discuss two types of AR binding-driven chromatin looping in prostate cancer, the AR-driven chromatin looping leading to target gene expression (without genomic rearrangement) and the AR-driven chromatin looping that results in gene fusion (with genomic rearrangement). The elucidation of the mechanisms of AR signaling in prostate cancer has translational implications in the development of new therapies in prostate cancer.
Features of genome-wide AR binding atlas in prostate cancer
Distal AR binding sites: their location and importance
In the pre-genome-wide ChIP era, studies on the canonical AR target gene PSA (prostate specific antigen) found that AR primarily binds to the PSA enhancer rather than to the promoter region16. Consistent with the findings from the PSA gene, genome-wide mapping of AR binding sites in prostate cancer cells revealed that most AR binding sites are not within the promoter region of AR-regulated genes. Approximately 86%~–95% of AR binding sites identified in ADPC cell models (LNCaP12, 13 and VCaP13) and CRPC cell models (LNCaP-abl12 and C4-2B11) are located within non-promoter regions. Interestingly, similar distal AR binding patterns are also observed in an immortalized normal human prostate epithelium cell line (HPr-1)17, and non-prostate androgen-responsive cells (human primary skeletal muscle myoblasts18) or tissues (mouse epididymis19). These studies strongly indicate that distal binding may be a general rule for AR action.
Several lines of evidence suggest that at least part of these distal AR binding regions are important for transcriptional regulation. First, some AR binding regions, when cloned upstream of minimal promoters, function as enhancers in reporter gene assays8, 10, 11. Second, transcriptional coactivators (e.g. the histone acetyltransferases [HAT], BRM-containing chromatin-remodeling complex and the Mediator complex) and RNA polymerase II (Pol II) bind to some distal AR binding sites8, 10, 12, 13, 20, 21, indicating that these sites are transcriptionally active. Third, some distal AR binding sites are associated with active histone modifications including H3K4 mono- and di-methylation (H3K4me1 and H3K4me2) and H3 acetylation (H3Ac)10–13, which create a permissive chromatin environment that facilitates active transcription. Fourth, the nucleosomes present at some distal AR binding regions contain the H2A.Z variant22, which has been reported to correlate with gene activation23, 24. Collectively, these observations suggest that some distal AR binding sites act as transcriptional enhancers.
Transcription factors that positively or negatively affect AR mediated gene expression
De novo motif searching (i.e. searching for common sequence patterns) or known motif scanning within the AR binding regions across the genome has identified that, in addition to AR binding motifs, many other transcription factor motifs (e.g. Forkhead, GATA, OCT, ETS, and NKX3-1) are significantly enriched within the AR binding regions, compared with genomic background. These observations suggest that transcription factors recognizing these motifs may be recruited to AR binding regions and play important roles in AR-mediated gene expression in prostate cancer, either by enhancing or repressing AR action. Consistent with this hypothesis, the pioneer factors FoxA1 and GATA2 were reported to be recruited to a significant portion of AR binding sites in ADPC and CRPC cells, facilitating AR binding and enhancing AR-mediated transcription8, 11, 13, 22. In addition, OCT1, a member of the POU domain family of transcription factors, may facilitate Pol II binding to AR-bound regions after short-term and prolonged androgen stimulation8, 22. By contrast, E26 transformation-specific (ETS) family member ERG binding sites that significantly overlap with AR binding sites in VCaP cells, repress AR action and AR target gene expression13. These studies suggest that AR-mediated gene expression in prostate cancer involves combinatorial transcriptional regulation.
AR-driven chromatin looping leads to target gene expression
Distal AR binding regions communicate with target gene promoters through chromatin looping
As the majority of AR binding sites are located at distal regions, how these distal binding sites interact with their target gene promoters and regulate target gene expression remains an open question. The looping model proposes that distal transcription factor binding sites interact with the proximal promoter regions with the intervening DNA looped out25, 26. Chromosome conformation capture (3C) technology, developed almost a decade ago27, allows for testing this proposed model. In this assay, the distal and proximal genomic regions, which have been cross-linked, are digested by the same restriction enzyme and re-ligated under extensively diluted DNA concentrations. The re-ligation of distal/proximal regions should be detected by PCR, if the distal region is physically associated with the proximal region27, 28. Using this assay and its derived ChIP-3C (ChIP combined with 3C) assay, recent studies have demonstrated that distal AR binding sites regulate well-known AR target genes such as PSA16 and TMPRSS28, 21 through chromatin looping. Chromatin looping also drives the expression of critical AR target genes directly involved in prostate cancer growth. For example, the CRPC-specific AR target gene UBE2C (ubiquitin-conjugating enzyme E2C) has an essential role in promoting CRPC cell growth12, 21 by inactivation of the M-phase checkpoint29 or increasing the pool of active anaphase promoting complex/cyclosome (APC/C)30. It was recently demonstrated that two CRPC cell-specific AR-bound enhancers, located −32.8 kb and +41.6 kb from the transcription start site (TSS) of UBE2C gene, interact with the UBE2C promoters through chromatin looping in CRPC but not in ADPC cells, which is required for UBE2C gene expression and CRPC growth12, 21.
Although the 3C assay is a powerful approach for studying enhancer/promoter interaction, it is not feasible to perform standard 3C for each AR binding site to identify or validate its target gene owing to more than 10,000 AR binding sites in the genome. The development of high-throughput adaptation of 3C assays, 4C (circular 3C or 3C-on-chip)31, 5C (3C-carbon copy)32, Hi-C33 and CHIA-PET (chromatin interaction analysis by paired-end sequencing)34, should allow for genome-wide assessment of AR-bound enhancer/promoter interactions in the near future.
Mechanisms for AR-mediated chromatin looping in CRPC
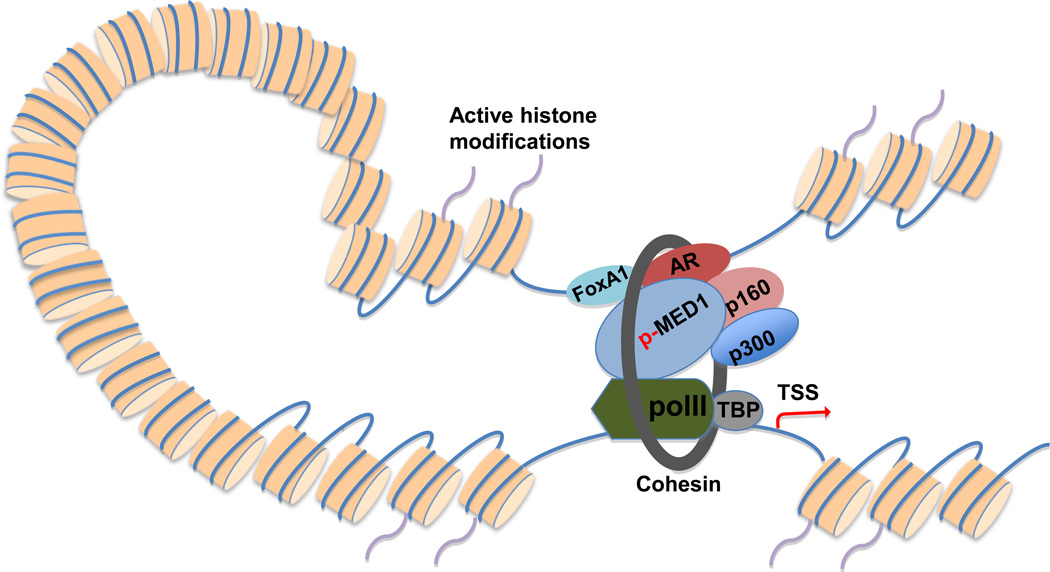
Chromatin looping is established and/or maintained by protein-protein interaction between enhancer-bound transcription factors and promoter-bound proteins25, 26, 35. While the distally bound transcription factors may directly interact with promoter-bound proteins, in many cases transcriptional coregulators or Pol II facilitate and/or mediate such interactions26, 36. Indeed, recent studies demonstrated that both transcription factors (e.g. AR, estrogen receptor [ER], FoxA1 and GATA-1)8, 16, 21, 34, 37 and coactivators (e.g. Med12, SRC-1, p300/CBP and BRG1)38–41 play essential roles in looping formation and/or maintenance. Among these coactivators, Mediator, an evolutionally conserved multiprotein complex including ~30 subunits42, is of particular interest. While previous studies imply that Mediator is important for chromatin looping in several loci41, 43, a recent study found that Mediator may mediate genome-wide enhancer/promoter interactions. The global Mediator-mediated looping is stabilized by cohensin, a protein which embraces the loop39. These studies suggest that Mediator is an essential chromatin architectural factor. Consistent with this notion, a recent study demonstrated that silencing of the Mediator subunit MED1, an AR coactivator16, 44, significantly decreased the interaction between the AR-bound UBE2C enhancers and the UBE2C promoter in the CRPC cell model LNCaP-abl, leading to decreased UBE2C gene expression21. Importantly, phosphorylation of MED1 at threonine (T) 1032 by phosphatidylinositol 3-kinase (PI3K)/AKT was proposed as a mechanism for MED1-mediated looping in CRPC cells by enhancing interactions between enhancer-bound FoxA1/AR and promoter-bound Pol II and TATA binding protein21. Based on the findings from these studies, we propose a looping model for AR-mediated gene regulation in CRPC (Figure 1). Furthermore, it seems reasonable to hypothesize that other looping-related coactivators such as SRC-1, p300/CBP38, and cohesin39 may also be involved in AR-mediated looping in CRPC (Figure 1). Future studies are needed to test these hypotheses.
Figure 1.
A proposed looping model for AR-regulated gene expression in CRPC. Active histone modifications (e.g. H3K4me1 and H3K4me2) facilitate the binding of AR and its collaborating transcription factors (e.g. FoxA1) to distal nucleosome-depleted regions12, 22. p-MED1 mediates chromatin looping by facilitating interactions between distal transcription factors and the basal transcriptional apparatus21, 39. Other coactivators (e.g. p160 coactivators and p30038) may also enhance chromatin looping. Cohension may stabilize chromatin looping by embracing the loop39. AR, androgen receptor; H3K4me1 and H3K4me2, H3K4 mono- and di-methylation; p-MED1, PI3K/AKT phosphorylated MED1.
AR-driven chromatin looping results in gene fusion
Overview of AR-regulated fusion genes in prostate cancer
Although genomic rearrangements are important for physiological processes such as VDJ recombination and class switch recombination (CSR) in lymphocytes, aberrant genomic rearrangements may lead to gene fusions in prostate cells contributing to prostate cancer initiation and progression45, 46. For example, the TMPRSS2-ERG fusion gene, which is expressed and functional in ~50% of primary prostate cancer and ~30% of CRPC patients45, 47, 48, is generated by the juxtaposition of the 5’ untranslated region of the AR target gene TMPRSS2 (21q22. 3)8 and the 5’ end of the oncogene ERG (21q22.2)45, 49. The juxtaposition of TMPRSS2 and ERG occurs either through balanced translocations with a “closed chain” pattern (i.e. without loss of genetic material) or through interstitial deletions (Edel)50–52.
The TMPRSS2-ERG fusion gene was first identified by Chinnaiyan’s group using an integrative computational and experimental approach49. An algorithm, termed COPA (cancer outlier profile analysis), was used to identify ERG as an outlier gene (i.e., a gene overexpressed in a subset of patients) from gene expression datasets in the Oncomine database53. Exon-walk PCR and 5’-RNA-ligase-mediated rapid amplification of complementary DNA ends (5’-RLM-RACE) assays were then performed to identify the fusion between TMPRSS2 and ERG45, 49. Using the same approach, many other androgen-regulated genes fused with members of the ETS gene family (e.g. TMPRSS2-ETV149, SLC45A3-ETV154, and CANT1-ETV455, 56), have been identified. Recently, more androgen-regulated gene fusions including non-ETS fusions (e.g. NDRG1-ERG57, SLC45A3-BRAF58 and TMPRSS2-FKBP5-ERG59) have been identified in prostate cancer by using the newly developed paired-end RNA sequencing (PE RNA-seq)60. In this approach, the total RNA is fragmented and converted into double-stranded cDNA. The cDNA fragments go through adapter ligation and PCR amplification processes, and the final cDNA library is used for paired-end high throughput sequencing. By integrating publically available AR ChIP-seq data13 and standard AR ChIP data8, 61, 62 with gene expression and FISH (fluorescence in situ hybridization) analysis data, we have summarized the published AR-regulated fusion genes (Table S1).
In addition to AR-regulated fusion genes, many non-AR regulated fusion genes have been reported. For example, ETV1 was found to be fused with the housekeeping gene HNRPA2B154. In addition, none of the most recently identified 5’ gene fusion partners (e.g. ALG5, PIGU, and TNPO1) by PE RNA-seq are androgen-regulated59. These findings suggest that multiple mechanisms exist for regulation of fusion gene expression in prostate cancer.
Function of AR-regulated fusion genes
The binding of liganded AR to the regulatory elements of the 5’ partner of a fusion gene drives the overexpression of the 3’ partner, which is often an oncogene (e.g. ERG), thus contributing to prostate carcinogenesis. For example, it has been reported that AR-driven expression of the ETS family members ETV1, ETV5 and ERG promotes invasion of benign prostate cells (e.g. RWPE, PrEC and BPH-1) and ADPC cells (e.g. LNCaP and VCaP) by activating an invasion-associated transcriptional program and thus inducing the plasminogen activator pathway13, 54, 63–65. While overexpression of ETV1 and ETV5 showed no effect on cell proliferation54, 63, recent studies found that ERG overexpression also suppresses the AR-mediated differentiation program to promote LNCaP and VCaP cell growth13, 65. Although ERG expression is necessary for prostate cancer cell invasion and growth, transgenic overexpression of ERG in mouse prostate is insufficient for inducing prostatic intraepithelial neoplasia (PIN) and prostate cancer64, 65, suggesting that ERG overexpression cooperates with other genetic alterations in prostate tumorigenesis. Consistent with this hypothesis, recent studies using mouse models indicated that loss of the tumor suppressor gene PTEN collaborates with ERG overexpression to develop PIN and prostate cancer66, 67. In addition, assessment of human prostate samples identified an association of TMPRSS2-ERG with deletion of chromosome 3p14, which includes the potential tumor suppressor genes FoxP1, RYBP and SHQ1, suggesting a potential new cooperation in prostate tumorigenesis68.
In addition to AR-regulated ETS fusion genes, AR-driven non-ETS fusion genes also have a critical role in prostate cancer growth and invasion. For example, the AR target gene SLC45A3 was reported to be fused with BRAF, a gene encoding a serine/threonine-specific protein kinase involved in mitogen-activated protein kinase (MAPK) signaling pathway, in less than 2% of prostate cancer patients. Overexpression of SLC45A3-BRAF in RWPE cells increases cell proliferation and invasion, and formation of small tumors in immunodeficient mice. While ETS fusion genes are considered as poor therapeutic targets, the function of the SLC45A3-BRAF fusion gene can be readily inhibited by RAF and mitogen-activated protein kinase kinase (MAP2K1) inhibitors, indicating that this fusion gene is a druggable target58.
Mechanisms for AR-mediated gene fusion
In general, gene fusions require the spatial proximity of two genomic regions otherwise located far away on the genome, the formation of double strand DNA breaks (DSB), and error-prone DNA repair, which together might lead to illegitimate DNA recombination46, 69. In addition to the ability of AR to directly regulate expression of fusion genes, as described above, recent studies focusing on the TMPRSS2-ERG fusion have found that androgen and/or AR might contribute to the processes that drive gene fusion. For example, androgen stimulation increases spatial association of TMPRSS2 and ERG. Inspired by the finding that estrogen is able to induce chromatin looping38, two independent studies61,70 found that dihydrotestosterone (DHT) treatment of LNCaP cells induces colocalization of TMPRSS2 and ERG. Further studies found that this androgen-induced chromatin looping is mediated by AR, which is recruited to the TMPRSS2 and ERG breakpoints upon DHT treatment61, 62 (Table S1). In agreement with the notion that breakpoints are associated with active histone modifications46, these AR binding regions at the TMPRSS2 and ERG breakpoints are enriched in histone H3K79 methylation and H4K16 acetylation61. A recent study that combined prostate cancer whole genome sequencing data with VCaP ChIP-seq data13 further revealed a genome-wide association of AR binding and active histone modifications (H3K4me3, H3K36me3 and H3Ac) near breakpoints in TMPRSS2-ERG positive patients50. These active histone modifications may facilitate AR binding and gene fusions.
In addition, DHT-bound AR recruits enzymes capable of inducing DSB. DHT stimulation has been found to induce AR-mediated recruitment of topoimerase II beta (TOP2B) to breakpoints62, an enzyme producing transient DSB that are required for ER-71, 72 and AR-regulated transcription62. Of note is also the observation that in the presence of exogenous genotoxic stress (e.g. irradiation), DHT treatment markedly increases the mRNA and protein expression of activation-induced cytidine deaminase (AID)61, a lymphoid-specific enzyme that changes cytosine to uracil and may ultimately lead to DSB73. AR then recruits AID to chromatin via its coactivator Gadd4561. Along these lines, genotoxic stress also increases the expression of long interspersed nucleotide element-1 (LINE-1) retroelement encoded open reading frame 2(ORF2) endonuclease. While ORF2 is recruited to translocation regions in a DHT-dependent manner, no physical interaction between AR and ORF2 has been detected61.
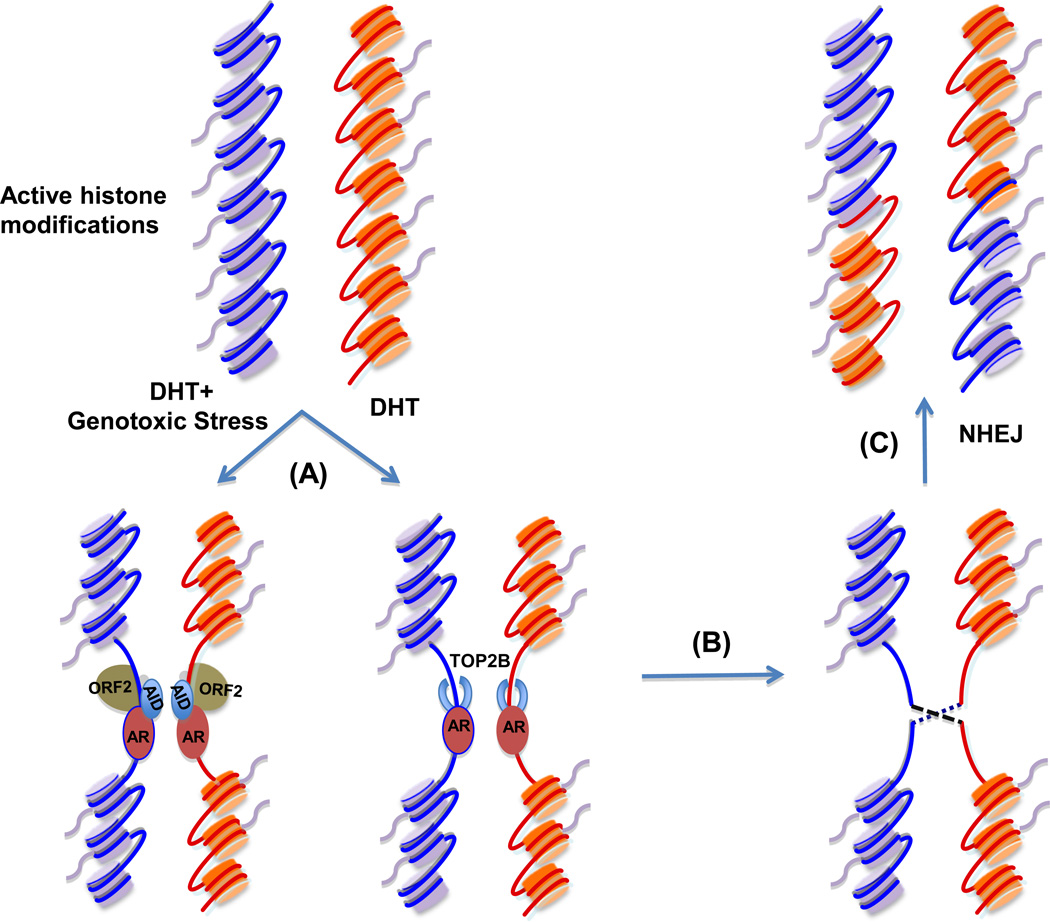
Finally, androgen stimulation increases recruitment of proteins involved in non-homologous end joining (NHEJ). There are two main pathways for DSB repair, namely the homologous recombination (HR) pathway and the NHEJ pathway. The error-prone NHEJ is the major pathway for the repair of DSB in eukaryotes74. It has been reported that upon DHT stimulation and increased DSB, several proteins involved in NHEJ including Ku70–Ku80 and ataxia telangiectasia-mutated protein (ATM), are recruited to the breakpoints, resulting in TMPRSS2-ERG fusion61, 62. Significantly, pharmacologic inhibitors and small interfering RNAs (siRNAs) targeting NHEJ pathway components have been shown to reduce the formation of de novo TMPRSS2-ERG gene fusion62. Based on these findings, a model for AR-mediated TMPRSS2-ERG fusion in prostate cancer is proposed (Figure 2). Evidence suggests that this model is generalizable to other AR-regulated fusion genes, such as SLC45A3-ETV161.
Figure 2.
AR-mediated TMPRSS2-ERG gene fusion in prostate cancer cells. (A) In the presence of genotoxic stress, AR binds to TMPRSS2 and ERG breakpoints and recruits AID upon DHT treatment. Exposure of prostate cancer cells under genotoxic stress to DHT also increases ORF2 recruitment to breakpoints61. DHT stimulation only leads to AR-TOP2B complex loading on breakpoints62. (B) The recruitment of AID, ORF2 and TOP2B to breakpoints causes DSB61, 62. (C) NHEJ machinery is recruited to the breakpoints, leading to error-prone DSB repair and gene fusions such as TMPRSS2-ERG gene fusion61, 62. AID, activation-induced cytidine deaminase; DHT, dihydrotestosterone; ORF2, open reading frame 2; TOP2B, topoimerase II beta; NHEJ, non-homologous end joining.
Concluding remarks and future directions
AR expression and functionality have been well documented in both ADPC and CRPC2, 3. Recent studies have found that the distal binding AR transcription complex, including AR and associated transcription factors and coactivators, regulates expression of several AR target genes involved in prostate cancer growth through chromatin looping. By using a global 3C assay, future studies should address whether such a long-range combinatorial regulation can be generalized to include other AR-dependent genes in the genome. This would allow for the identification of all AR direct target genes involved in prostate tumorigenesis and might lead to the development of new therapies for the disease. In addition to the looping mechanism, future studies should also examine whether other mechanisms for enhancer function (e.g. spreading and non-coding RNA26) participate in AR-mediated long-range gene regulation.
With regard to AR-mediated gene fusion, significant progress has been made in our understanding of the mechanisms of AR-driven chromatin looping that leads to gene fusions. The finding that inhibition of the NHEJ pathway reduces TMPRSS2-ERG gene fusion62 suggest that future clinical trials may consider combining agents targeting both the AR and NHEJ pathway proteins. Despite progress on gene fusion mechanisms, it is still unclear on why gene fusions only occur in a limited number of genomic regions. It has been proposed that histone modifications, coactivators or noncoding RNAs may assist in the selection of these regions75. Future work should test these possibilities. Finally, as gene fusion at the RNA level without genomic rearrangement has recently been reported59, 76, 77, future investigations should include whether AR has a role in driving the formation of such chimeric RNAs that also promote prostate carcinogenesis.
Supplementary Material
Acknowledgement
The studies performed in the laboratory of Q.W. were supported by grants from National Institute of Health (R00 CA126160, U54 CA113001), and The Ohio State University Comprehensive Cancer Center. We also thank Drs. Jindan Yu, Wei Li and Zhong Chen for critically reviewing of the manuscript. We apologize to the colleagues whose relevant work was not cited due to space limitation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Jemal A, et al. Cancer statistics, 2010. CA Cancer J Clin. 2010;60:277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 2.Heinlein CA, Chang C. Androgen receptor in prostate cancer. Endocr Rev. 2004;25:276–308. doi: 10.1210/er.2002-0032. [DOI] [PubMed] [Google Scholar]
- 3.Knudsen KE, Penning TM. Partners in crime: deregulation of AR activity and androgen synthesis in prostate cancer. Trends Endocrinol Metab. 2010;21:315–324. doi: 10.1016/j.tem.2010.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Feldman BJ, Feldman D. The development of androgen-independent prostate cancer. Nat Rev Cancer. 2001;1:34–45. doi: 10.1038/35094009. [DOI] [PubMed] [Google Scholar]
- 5.Attard G, et al. Steroid hormone receptors in prostate cancer: a hard habit to break? Cancer Cell. 2009;16:458–462. doi: 10.1016/j.ccr.2009.11.006. [DOI] [PubMed] [Google Scholar]
- 6.Sharma A, et al. The retinoblastoma tumor suppressor controls androgen signaling and human prostate cancer progression. J Clin Invest. 2010;120:4478–4492. doi: 10.1172/JCI44239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mangelsdorf DJ, et al. The nuclear receptor superfamily: the second decade. Cell. 1995;83:835–839. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang Q, et al. A hierarchical network of transcription factors governs androgen receptor-dependent prostate cancer growth. Mol Cell. 2007;27:380–392. doi: 10.1016/j.molcel.2007.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Massie CE, et al. New androgen receptor genomic targets show an interaction with the ETS1 transcription factor. EMBO Rep. 2007;8:871–878. doi: 10.1038/sj.embor.7401046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Takayama K, et al. Identification of novel androgen response genes in prostate cancer cells by coupling chromatin immunoprecipitation and genomic microarray analysis. Oncogene. 2007;26:4453–4463. doi: 10.1038/sj.onc.1210229. [DOI] [PubMed] [Google Scholar]
- 11.Jia L, et al. Genomic androgen receptor-occupied regions with different functions, defined by histone acetylation, coregulators and transcriptional capacity. PLoS ONE. 2008;3:e3645. doi: 10.1371/journal.pone.0003645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang Q, et al. Androgen receptor regulates a distinct transcription program in androgen-independent prostate cancer. Cell. 2009;138:245–256. doi: 10.1016/j.cell.2009.04.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yu J, et al. An integrated network of androgen receptor, polycomb, and TMPRSS2-ERG gene fusions in prostate cancer progression. Cancer Cell. 2010;17:443–454. doi: 10.1016/j.ccr.2010.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang Q, Brown M. Mapping the androgen receptor cistrome. In: Tindall D, a.M.J, editors. Androgen action in prostate cancer. Springer Science + Business Media; 2009. pp. 663–680. [Google Scholar]
- 15.Chen Z, et al. Histone modifications and chromatin organization in prostate cancer. Epigenomics. 2010;2:551–560. doi: 10.2217/epi.10.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang Q, et al. Spatial and temporal recruitment of androgen receptor and its coactivators involves chromosomal looping and polymerase tracking. Mol Cell. 2005;19:631–642. doi: 10.1016/j.molcel.2005.07.018. [DOI] [PubMed] [Google Scholar]
- 17.Bolton EC, et al. Cell- and gene-specific regulation of primary target genes by the androgen receptor. Genes Dev. 2007;21:2005–2017. doi: 10.1101/gad.1564207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wyce A, et al. Research Resource: The androgen receptor modulates expression of genes with critical roles in muscle development and function. Mol Endocrinol. 2010;24:1665–1674. doi: 10.1210/me.2010-0138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hu S, et al. Research resource: Genome-wide mapping of in vivo androgen receptor binding sites in mouse epididymis. Mol Endocrinol. 2010;24:2392–2405. doi: 10.1210/me.2010-0226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Makkonen H, et al. Long-range activation of FKBP51 transcription by the androgen receptor via distal intronic enhancers. Nucleic Acids Res. 2009;37:4135–4148. doi: 10.1093/nar/gkp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen Z, et al. Phospho-MED1-enhanced UBE2C locus looping drives castration-resistant prostate cancer growth. EMBO J. 2011;30:2405–2419. doi: 10.1038/emboj.2011.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.He HH, et al. Nucleosome dynamics define transcriptional enhancers. Nat Genet. 2010;42:343–347. doi: 10.1038/ng.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barski A, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 24.Wang Z, et al. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet. 2008;40:897–903. doi: 10.1038/ng.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dekker J. Gene regulation in the third dimension. Science. 2008;319:1793–1794. doi: 10.1126/science.1152850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bulger M, Groudine M. Functional and mechanistic diversity of distal transcription enhancers. Cell. 2011;144:327–339. doi: 10.1016/j.cell.2011.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dekker J, et al. Capturing chromosome conformation. Science. 2002;295:1306–1311. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- 28.Hagege H, et al. Quantitative analysis of chromosome conformation capture assays (3C-qPCR) Nat Protoc. 2007;2:1722–1733. doi: 10.1038/nprot.2007.243. [DOI] [PubMed] [Google Scholar]
- 29.Reddy SK, et al. Ubiquitination by the anaphase-promoting complex drives spindle checkpoint inactivation. Nature. 2007;446:921–925. doi: 10.1038/nature05734. [DOI] [PubMed] [Google Scholar]
- 30.van Ree JH, et al. Overexpression of the E2 ubiquitin-conjugating enzyme UbcH10 causes chromosome missegregation and tumor formation. J Cell Biol. 2010;188:83–100. doi: 10.1083/jcb.200906147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao Z, et al. Circular chromosome conformation capture (4C) uncovers extensive networks of epigenetically regulated intra- and interchromosomal interactions. Nat Genet. 2006;38:1341–1347. doi: 10.1038/ng1891. [DOI] [PubMed] [Google Scholar]
- 32.Dostie J, et al. Chromosome Conformation Capture Carbon Copy (5C): a massively parallel solution for mapping interactions between genomic elements. Genome Res. 2006;16:1299–1309. doi: 10.1101/gr.5571506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lieberman-Aiden E, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fullwood MJ, et al. An oestrogen-receptor-alpha-bound human chromatin interactome. Nature. 2009;462:58–64. doi: 10.1038/nature08497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miele A, Dekker J. Long-range chromosomal interactions and gene regulation. Mol Biosyst. 2008;4:1046–1057. doi: 10.1039/b803580f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nolis IK, et al. Transcription factors mediate long-range enhancer-promoter interactions. Proc Natl Acad Sci U S A. 2009;106:20222–20227. doi: 10.1073/pnas.0902454106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vakoc CR, et al. Proximity among distant regulatory elements at the beta-globin locus requires GATA-1 and FOG-1. Mol Cell. 2005;17:453–462. doi: 10.1016/j.molcel.2004.12.028. [DOI] [PubMed] [Google Scholar]
- 38.Hu Q, et al. Enhancing nuclear receptor-induced transcription requires nuclear motor and LSD1-dependent gene networking in interchromatin granules. Proc Natl Acad Sci U S A. 2008;105:19199–19204. doi: 10.1073/pnas.0810634105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kagey MH, et al. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010;467:430–435. doi: 10.1038/nature09380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim SI, et al. BRG1 requirement for long-range interaction of a locus control region with a downstream promoter. Proc Natl Acad Sci U S A. 2009;106:2259–2264. doi: 10.1073/pnas.0806420106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Park SW, et al. Thyroid hormone-induced juxtaposition of regulatory elements/factors and chromatin remodeling of Crabp1 dependent on MED1/TRAP220. Mol Cell. 2005;19:643–653. doi: 10.1016/j.molcel.2005.08.008. [DOI] [PubMed] [Google Scholar]
- 42.Malik S, Roeder RG. The metazoan Mediator co-activator complex as an integrative hub for transcriptional regulation. Nat Rev Genet. 2010;11:761–772. doi: 10.1038/nrg2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Degenhardt T, et al. Population-level transcription cycles derive from stochastic timing of single-cell transcription. Cell. 2009;138:489–501. doi: 10.1016/j.cell.2009.05.029. [DOI] [PubMed] [Google Scholar]
- 44.Wang Q, et al. A coregulatory role for the TRAP-mediator complex in androgen receptor-mediated gene expression. J Biol Chem. 2002;277:42852–42858. doi: 10.1074/jbc.M206061200. [DOI] [PubMed] [Google Scholar]
- 45.Kumar-Sinha C, et al. Recurrent gene fusions in prostate cancer. Nat Rev Cancer. 2008;8:497–511. doi: 10.1038/nrc2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mani RS, Chinnaiyan AM. Triggers for genomic rearrangements: insights into genomic, cellular and environmental influences. Nat Rev Genet. 2010;11:819–829. doi: 10.1038/nrg2883. [DOI] [PubMed] [Google Scholar]
- 47.Cai C, et al. Reactivation of androgen receptor-regulated TMPRSS2:ERG gene expression in castration-resistant prostate cancer. Cancer Res. 2009;69:6027–6032. doi: 10.1158/0008-5472.CAN-09-0395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Attard G, et al. Characterization of ERG, AR and PTEN gene status in circulating tumor cells from patients with castration-resistant prostate cancer. Cancer Res. 2009;69:2912–2918. doi: 10.1158/0008-5472.CAN-08-3667. [DOI] [PubMed] [Google Scholar]
- 49.Tomlins SA, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–648. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 50.Berger MF, et al. The genomic complexity of primary human prostate cancer. Nature. 2011;470:214–220. doi: 10.1038/nature09744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mehra R, et al. Characterization of TMPRSS2-ETS gene aberrations in androgen-independent metastatic prostate cancer. Cancer Res. 2008;68:3584–3590. doi: 10.1158/0008-5472.CAN-07-6154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mehra R, et al. Heterogeneity of TMPRSS2 gene rearrangements in multifocal prostate adenocarcinoma: molecular evidence for an independent group of diseases. Cancer Res. 2007;67:7991–7995. doi: 10.1158/0008-5472.CAN-07-2043. [DOI] [PubMed] [Google Scholar]
- 53.Rhodes DR, et al. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6:1–6. doi: 10.1016/s1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tomlins SA, et al. Distinct classes of chromosomal rearrangements create oncogenic ETS gene fusions in prostate cancer. Nature. 2007;448:595–599. doi: 10.1038/nature06024. [DOI] [PubMed] [Google Scholar]
- 55.Han B, et al. A fluorescence in situ hybridization screen for E26 transformation-specific aberrations: identification of DDX5-ETV4 fusion protein in prostate cancer. Cancer Res. 2008;68:7629–7637. doi: 10.1158/0008-5472.CAN-08-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hermans KG, et al. Two unique novel prostate-specific and androgen-regulated fusion partners of ETV4 in prostate cancer. Cancer Res. 2008;68:3094–3098. doi: 10.1158/0008-5472.CAN-08-0198. [DOI] [PubMed] [Google Scholar]
- 57.Pflueger D, et al. N-myc downstream regulated gene 1 (NDRG1) is fused to ERG in prostate cancer. Neoplasia. 2009;11:804–811. doi: 10.1593/neo.09572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Palanisamy N, et al. Rearrangements of the RAF kinase pathway in prostate cancer, gastric cancer and melanoma. Nat Med. 2010;16:793–798. doi: 10.1038/nm.2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pflueger D, et al. Discovery of non-ETS gene fusions in human prostate cancer using next-generation RNA sequencing. Genome Res. 2011;21:56–67. doi: 10.1101/gr.110684.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Maher CA, et al. Chimeric transcript discovery by paired-end transcriptome sequencing. Proc Natl Acad Sci U S A. 2009;106:12353–12358. doi: 10.1073/pnas.0904720106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lin C, et al. Nuclear receptor-induced chromosomal proximity and DNA breaks underlie specific translocations in cancer. Cell. 2009;139:1069–1083. doi: 10.1016/j.cell.2009.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Haffner MC, et al. Androgen-induced TOP2B-mediated double-strand breaks and prostate cancer gene rearrangements. Nat Genet. 2010;42:668–675. doi: 10.1038/ng.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Helgeson BE, et al. Characterization of TMPRSS2:ETV5 and SLC45A3:ETV5 gene fusions in prostate cancer. Cancer Res. 2008;68:73–80. doi: 10.1158/0008-5472.CAN-07-5352. [DOI] [PubMed] [Google Scholar]
- 64.Klezovitch O, et al. A causal role for ERG in neoplastic transformation of prostate epithelium. Proc Natl Acad Sci U S A. 2008;105:2105–2110. doi: 10.1073/pnas.0711711105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tomlins SA, et al. Role of the TMPRSS2-ERG gene fusion in prostate cancer. Neoplasia. 2008;10:177–188. doi: 10.1593/neo.07822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Carver BS, et al. Aberrant ERG expression cooperates with loss of PTEN to promote cancer progression in the prostate. Nat Genet. 2009;41:619–624. doi: 10.1038/ng.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.King JC, et al. Cooperativity of TMPRSS2-ERG with PI3-kinase pathway activation in prostate oncogenesis. Nat Genet. 2009;41:524–526. doi: 10.1038/ng.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Taylor BS, et al. Integrative genomic profiling of human prostate cancer. Cancer Cell. 2010;18:11–22. doi: 10.1016/j.ccr.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nussenzweig A, Nussenzweig MC. Origin of chromosomal translocations in lymphoid cancer. Cell. 2010;141:27–38. doi: 10.1016/j.cell.2010.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mani RS, et al. Induced chromosomal proximity and gene fusions in prostate cancer. Science. 2009;326:1230. doi: 10.1126/science.1178124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ju BG, et al. A topoisomerase IIbeta-mediated dsDNA break required for regulated transcription. Science. 2006;312:1798–1802. doi: 10.1126/science.1127196. [DOI] [PubMed] [Google Scholar]
- 72.Perillo B, et al. DNA oxidation as triggered by H3K9me2 demethylation drives estrogen-induced gene expression. Science. 2008;319:202–206. doi: 10.1126/science.1147674. [DOI] [PubMed] [Google Scholar]
- 73.Nambiar M, Raghavan SC. How does DNA break during chromosomal translocations? Nucleic Acids Res. 2011 doi: 10.1093/nar/gkr223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lieber MR. The mechanism of human nonhomologous DNA end joining. J Biol Chem. 2008;283:1–5. doi: 10.1074/jbc.R700039200. [DOI] [PubMed] [Google Scholar]
- 75.Mathas S, Misteli T. The dangers of transcription. Cell. 2009;139:1047–1049. doi: 10.1016/j.cell.2009.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Maher CA, et al. Transcriptome sequencing to detect gene fusions in cancer. Nature. 2009;458:97–101. doi: 10.1038/nature07638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kannan K, et al. Recurrent chimeric RNAs enriched in human prostate cancer identified by deep sequencing. Proc Natl Acad Sci U S A. 2011;108:9172–9177. doi: 10.1073/pnas.1100489108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.