Abstract
Objectives
To present an adapted Clinical Trial Processor (CTP) test set-up for receiving, anonymising and saving Digital Imaging and Communications in Medicine (DICOM) data using external input from the original database of an existing clinical study information system to guide the anonymisation process.
Methods
Two methods are presented for an adapted CTP test set-up. In the first method, images are pushed from the Picture Archiving and Communication System (PACS) using the DICOM protocol through a local network. In the second method, images are transferred through the internet using the HTTPS protocol.
Results
In total 25,000 images from 50 patients were moved from the PACS, anonymised and stored within roughly 2 h using the first method. In the second method, an average of 10 images per minute were transferred and processed over a residential connection. In both methods, no duplicated images were stored when previous images were retransferred. The anonymised images are stored in appropriate directories.
Conclusions
The CTP can transfer and process DICOM images correctly in a very easy set-up providing a fast, secure and stable environment. The adapted CTP allows easy integration into an environment in which patient data are already included in an existing information system.
Key Points
Store DICOM images correctly in a very easy set-up in a fast, secure and stable environment
Allows adaptation of the software to perform a certain task based on specific needs
Allows easy integration into an existing environment
Reduce the possibility of inappropriate anonymisation
Keywords: Anonymisation tool, Clinical trial processor, Privacy, Clinical trials, Software, Patient data
Introduction
Digital Imaging and Communications in Medicine (DICOM) [1] was developed to standardise medical image data and to easily share medical image data between computer systems. It is currently the global standard for handling, storing, printing and transmitting information in medical imaging. A DICOM image consists of a DICOM header and the viewable image. The DICOM header saves identifying information of patients and images which may include patient information, study information, institution information, etc. The DICOM format is now used by most of the medical imaging community, not only for clinical practice but also for clinical research raising the possibility of data sharing or exchange. However, sharing sensitive medical image data to a third party demands protection of the data itself to ensure data safety and patient privacy.
Gonzales et al. [2] stated that it is desirable and good clinical practice that patient data are rendered “anonymous” before transferral. The UK Medical Research Council (MRC) [3] described anonymised data as data prepared from personal information, but from which the person cannot be identified by the recipient of the information. This anonymity can contain coded information that could be used to identify people by using external information that is not generally known.
Data anonymisation is the simplest but most secure approach to providing privacy and integrity of DICOM data. This method is used to remove confidential entries from DICOM files and is generally irreversible. Confidential entries include tags in the standard DICOM Data Dictionary that could in itself or in combination with other entries be used to derive the patient’s real identity [1]. There are numerous tools for anonymising DICOM data, both commercially and open source, which employ various approaches to removing patient-related information in a more or less automated way [4–6].
However, anonymisation often is not done properly. The use of fully automated software may cause less awareness of fields being anonymised. One default scheme in the software may completely remove the inappropriate fields of the DICOM headers which might be needed by a specific task, patient’s age in months for example in paediatric studies. On the other hand, it is also possible for the software not to anonymise crucial or confidential information that may lead to the recovery of the patient’s identity. A non-guided anonymisation also will lead to duplication that may consume a lot of space in the storage.
The RSNA Clinical Trial Processor (CTP) [7] is a highly configurable and extensible stand-alone application that provides processing features such as import services, export services, storage service and processor services for clinical trials. The processor service also includes a DICOM anonymisation stage that can be configured via a script language. The CTP can anonymise a DICOM object based on the script mentioned in the configuration. The configuration can also refer to a look-up table so that the anonymisation process for certain tags will be done based on the predefined list.
Besides the image data, other information is also gathered for clinical research including reports and patient information. This information is usually entered into an information system separate from the image data system. Consequently, anonymisation of information has to be performed twice leading to possible mistakes leading to a mismatch between the image data and the other information.
In this paper, we present an adapted CTP test set-up for receiving, anonymising and saving DICOM data into storage through the local intranet and also through the internet for implementation in large, multi-centre, clinical trial studies using external input from the original database of an existing clinical study information system to guide the anonymisation process.
Materials and methods
The CTP is a stand-alone program that utilises the processing features of the RSNA Medical Imaging Resource Center (MIRC) [8] for clinical trials in a highly configurable and extensible application. It is developed to satisfy the requirements of trials that need complex processing that cannot be handled by MIRC. CTP has some key features such as support for configurable multiple pipelines, pre-defined implementation for key components, and web-based monitoring of the application’s status. It is open source software and can be downloaded for free from the RSNA website [9]. The software is written in Java and runs on both Linux and Microsoft Windows operating systems. It requires Java 1.6 (or higher) Java Runtime Environment (JRE). Some pipelines also need Java Advance Imaging ImageIO Tools [10] installed on the system used for the CTP software.
The flexibility and configurability in the program’s approach to de-identification of selected patient data can handle the variation of pertinent rules and regulations, which can vary from one facility to another [11]. It can protect and maintain the security of health-related records and fulfil the need in a clinical trial or research study to de-identify patient information.
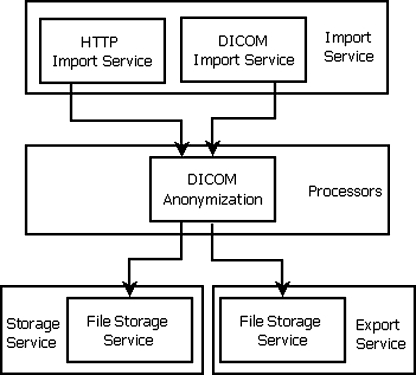
The processing stages are divided into four types, which are import service, processor, storage service and export service. The import service receives objects and queues them for processing by subsequent stages. A processor performs some kind of processing on an object and passes the result on to the next stage in the pipeline. An example of such a processer is an anonymiser. A storage service stores an object in a file system. An export service provides queued transmission to an external system via a defined protocol (e.g. HTTP, HTTPs or DICOM protocol). Pipelines contain sequences of processing stages and must at least have one import service. The pipelines and its stages can each be configured either through a configuration file which is located in the same directory as the program or through the application monitoring web page. In this study, one pipeline was defined with three main stages (import service, anonymiser and storage service). The illustration of the pipeline is shown in Fig. 1.
Fig. 1.
Illustration of the four main stages in the pipeline used for this experiment
The DICOM anonymiser provided by CTP has a simple scripting language in which each of the DICOM elements can have its own replacement script. The unnecessary patient’s protected healthcare information (PHI) will be removed before being stored. It minimises the amount of PHI of the objects as much as possible depending on the study requirements. The anonymiser provides many functions to perform the anonymisation task such as ‘function 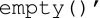 which will be used to return a zero-length string for the chosen tags and ‘function
which will be used to return a zero-length string for the chosen tags and ‘function 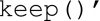 which forces the element to be preserved as an anonymised DICOM object. It can be extended to meet specialised requirements by editing the script file. A look-up function in the anonymiser maps values through a local look-up table, which is intended to perform the anonymisation based on the table to meet the pre-defined requirements of the anonymised DICOM object. The look-up table itself is a property file that should be referenced in the anonymisation stage configuration when needed. A Storage Service stores an object in a file system. It is not queued, and therefore it must be complete before subsequent stages can proceed. When storing files, the storage service automatically defines subdirectories beneath its root directory and populates them accordingly.
which forces the element to be preserved as an anonymised DICOM object. It can be extended to meet specialised requirements by editing the script file. A look-up function in the anonymiser maps values through a local look-up table, which is intended to perform the anonymisation based on the table to meet the pre-defined requirements of the anonymised DICOM object. The look-up table itself is a property file that should be referenced in the anonymisation stage configuration when needed. A Storage Service stores an object in a file system. It is not queued, and therefore it must be complete before subsequent stages can proceed. When storing files, the storage service automatically defines subdirectories beneath its root directory and populates them accordingly.
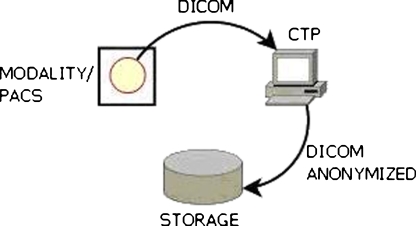
In this study, two methods were tested. In the first method, a total of 25,000 images from 50 patients are pushed from the picture archiving and communication systems (PACS) using the DICOM protocol into a machine with adapted CTP installed. The adapted CTP receives the images, anonymises them and saves the anonymised images into local storage (Fig. 2). This set-up can typically be used for research studies within one institution. Data from 50 patients consisting of a total of 25,000 images were transferred using this method.
Fig. 2.
System setting for method 1. Images from the investigation or PACS server sent through a DICOM protocol to a clinical trial processor (CTP) machine that anonymises and stores them in proper storage
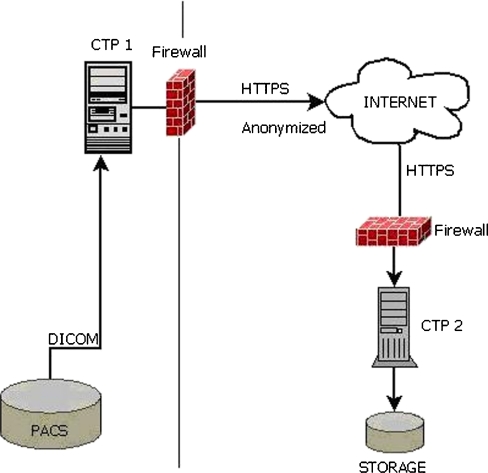
The other method is designed to test the CTP data transfer performance using HTTPS networking through the internet. There are two sites both running a server with the adapted CTP installed. One site acts as sender and the other as receiver. Both servers are geographically separated machines where the sender is a server located in The Netherlands and the receiver is a server located in the United States. The anonymisation is performed on the server at the sender site before it is transferred to the receiver site.
The CTP machine at site 1 is configured to import images from the local research PACS, to anonymise the images and to export the resulting images to the CTP machine at site 2 using the secure HTTPS networking protocol. The CTP machine at site 2 receives the anonymised images and saves them into local storage (Fig. 3). In both methods, the system only accepts DICOM images. Files that do not conform to the DICOM standard requirements will be transferred to a quarantine folder.
Fig. 3.
System setting for method 2. A CTP machine from one site exports images received from local PACS into another CTP machine at the other site over secure HTTP networking. The images received are saved in storage at the receiver site
Experiment and results
As the anonymisation process will be integrated into an ongoing study set-up, the anonymisation properties will be inherited from this study. There are 40 tags defined to be replaced or made blank to omit relevant information from the object. These include the patient’s personal data, studies, and other crucial information that can, in itself or in combination, refer directly to the patient. In Table 1 the modified fields of the DICOM header in our anonymisation are shown. The fields were all blanked except the Patient ID and Patient Name, which must be filled in based on the look-up table constructed from the external study information system. This look-up table was automatically updated for every image sent from the PACS based on identity mapping with the sql-database server running the database of our clinical study information system. A function was added to monitor if there were any images sent from the PACS. This function will query the database to receive the correct pair of the original ID of the DICOM images and the anonymisation value and will subsequently write it into the lookup file. The pairs of values will be removed automatically after the whole set of images is successfully anonymised and stored in the appropriate file system.
Table 1.
Fields in the DICOM header defined to be modified (M) or made blank
| Tag ID | Tag Name |
|---|---|
| 0008,0020 | StudyDate |
| 0008,0021 | SeriesDate |
| 0008,0022 | AcquisitionDate |
| 0008,0023 | ContentDate |
| 0008,0024 | OverlayDate |
| 0008,0025 | CurveDate |
| 0008,002A | AcquisitionDatetime |
| 0008,0030 | StudyTime |
| 0008,0031 | SeriesTime |
| 0008,0032 | AcquisitionTime |
| 0008,0033 | ContentTime |
| 0008,0034 | OverlayTime |
| 0008,0035 | CurveTime |
| 0008,0050 | AccessionNumber |
| 0008,0080 | InstitutionName |
| 0008,0081 | InstitutionAddress |
| 0008,0090 | ReferringPhysiciansName |
| 0008,0092 | ReferringPhysiciansAddress |
| 0008,0094 | ReferringPhysiciansTelephoneNumber |
| 0008,0096 | ReferringPhysicianIDSequence |
| 0008,1040 | InstitutionalDepartmentName |
| 0008,1048 | PhysicianOfRecord |
| 0008,1049 | PhysicianOfRecordIDSequence |
| 0008,1050 | PerformingPhysiciansName |
| 0008,1052 | PerformingPhysicianIDSequence |
| 0008,1060 | NameOfPhysicianReadingStudy |
| 0008,1062 | PhysicianReadingStudyIDSequence |
| 0008,1070 | OperatorsName |
| 0010,0010 | PatientsName (M) |
| 0010,0020 | PatientID (M) |
| 0010,0021 | IssuerOfPatientID |
| 0010,0030 | PatientsBirthDate |
| 0010,0032 | PatientsBirthTime |
| 0010,0040 | PatientsSex |
| 0010,1000 | OtherPatientIDs |
| 0010,1001 | OtherPatientNames |
| 0010,1005 | PatientsBirthName |
| 0010,1010 | PatientsAge |
| 0010,1040 | PatientsAddress |
| 0010,1060 | PatientsMothersBirthName |
| 0010,2150 | CountryOfResidence |
| 0010,2152 | RegionOfResidence |
| 0010,2154 | PatientsTelephoneNumbers |
| 0020,0010 | StudyID |
| 0038,0300 | CurrentPatientLocation |
| 0038,0400 | PatientsInstitutionResidence |
| 0040,A120 | DateTime |
| 0040,A121 | Date |
| 0040,A122 | Time |
| 0040,A123 | PersonName |
The anonymised images are saved in local storage under the file storage service. This service will save fully processed objects in a file system. It creates the directory stated in the root element in the service’s configuration. It also creates subfolders and groups the images based on the element set in the configuration. These subfolders can also be defined using an element from the DICOM Header. Default settings for the file storage allow the service to create more than one copy of an image. This duplication may occur due to double transfers, intentionally or not, from the PACS server. Similar to the import service, the storage service can be set to accept certain objects. Rejected objects will be moved into the quarantine folder.
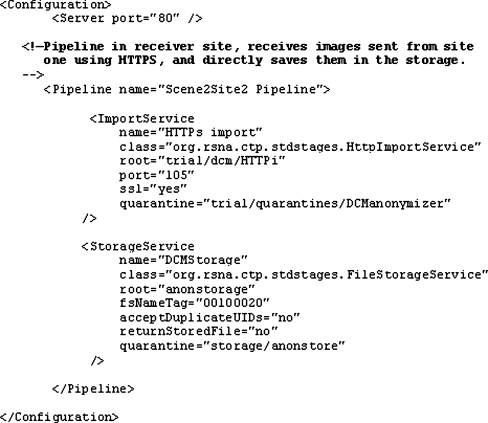
In method 1, transfers are initiated by pushing images from PACS into the CTP machine using the DICOM protocol. The DICOM images are received by the CTP Import Service through the defined listener port. The images are directly stored in a defined file system stated in the system configuration after being anonymised. These three services are configured together in one machine between the source (PACS) and the storage. The configuration file for the experiment using the local network is shown in Fig. 4.
Fig. 4.
Configuration for the experiment using method 1
There are two additional stages in the experiment using the second method, which are the HTTP export service with secure transfer configured at site 1, and the receiver at site 2. The HTTP export service queues objects and transmits them using the standard HTTP protocol. Considering the security of the transfer process, a secure socket layer (ssl) is used to initiate the connection. As the receiver has to receive images using the same protocol, the HTTP import service with an ssl is configured at the receiver site. Both HTTP export and import can determine which object can be accepted or rejected. There is no need for the receiver site to anonymise the images again, therefore at site 2 there are only two main stages to import and then directly save the objects into the file system. Configuration from the CTP machines at site 1 and site 2 can be seen in Figs. 5 and 6 respectively.
Fig. 5.
Sender configuration for the experiment using method 2
Fig. 6.
Receiver configuration for experiment using method 2
Using the first method, adapted CTP can successfully receive patient image data sent from a PACS server, anonymise and then store them in local storage. The total time needed to transfer all images is roughly 2 h, which means every second there are on average four images moved from the PACS, anonymised, and then saved in storage. This time was calculated based on the difference between the first file being received by the DICOM Import Service and the time logged from the last file stored in the file system. The adapted CTP correctly anonymised all images based on the lookup table and stored them in an appropriate directory. The CTP machine ran stable during the tests. Additionally, several transfers were made with the same original patient ID, none of them resulting in duplication of the data.
The second method also correctly de-identified and stored the anonymised image data in the correct file system. The average time needed to transfer the images is 10 images per minute or one image every 6 s over a home internet connection with upstream network transfer speed of approximately 0.48 Mbps. The sender is configured using a Microsoft Windows XP environment and the receiver using a CentOS Linux environment. Data are anonymised and transferred through normal HTTP using secure socket layer. The resulting anonymised images were all saved without any duplication occurring. The adapted CTP was running stable throughout all tests.
Discussion
The needs of data trackback to its origins raised the consideration of using pseudonymisation instead of anonymisation in some research [12–14]. While anonymisation removes or blanks the PHI from the DICOM header, pseudonymisation only replaces the person-related data with unique identifiers. This will allow both follow-up of the studies and the high level maintenance of patient data. CTP offers the possibility of pseudonymisation through some of the available functions at its anonymisation stage by using simple data modification or the utilisation of a hash of an element’s value.
In our system, the anonymisation process is done by emptying most of the PHI-related fields and using the previously registered pairs of original and anonymised values for patient name and ID from the study information system database. Therefore, the anonymisation will cover the security of patient-related data while data trackback is still possible by querying the data using the anonymisation ID. The access to the study information system database is limited to authorised personnel and can be obtained through our internal network only, thus securing the access to the trackback information.
The proposed set-up can be easily integrated into existing research set-ups because of the use of the anonymisation database from the existing system thus facilitating easy inclusion of digital image data and decreasing or eliminating the need for data transfer onto physical media (CD, DVD, etc.).
As all DICOM data transferred were CT images that have a file size per image of 0.5 MB, a transfer speed of 2 MB per second or 16 Mb per second was achieved during the first method. Based on our measurements, the transfer of 25,000 images over the second method’s connection speed would take approximately 41 h to complete. Although this could be acceptable in clinical research studies, it is definitely too time-consuming in clinical practice. However, faster connections that are in place between enterprises will partly solve this problem.
While no significant problems occurred during our tests while the adapted CTP was receiving, anonymising, exporting and storing images, there are some limitations to this application. For example, Gonzales et al. [2] mentioned that CTP still does not have a standard DICOM anonymisation mechanism and also has limitations in adapting to new anonymisation methods. Furthermore, it is stated on the official CTP website [7] that this application is still under development and some possible improvements are scheduled. The main issue raised to improve the performance of the CTP is the use of the DCM4CHE2 library, instead of the currently used DCM4CHE library, which is claimed to provide faster transfer and system processing.
Conclusion
The experimental results show that CTP can transfer, receive, anonymise and store DICOM images correctly in a very easy set-up in a fast, secure and stable environment. CTP’s configurability will enable the anonymisation of various tasks with different schemes. This will reduce the possibility of inappropriate anonymisation.
Its open source availability allows adaptation of the software to perform a certain task based on specific needs. Our adaptations to the original CTP allow easy integration into environments in which patient data are already included in an information system by using the existing database from this system to guide the anonymisation process. Resulting from this, the mismatch in data that can occur when using two separate databases, is eliminated. Furthermore, the possibility of duplicate data entry is also prohibited.
Acknowledgments
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- 1.Pianykh OS. Digital Imaging and Communications in Medicine (DICOM)—A practical introduction and survival guide. Heidelberg: Springer; 2008. [Google Scholar]
- 2.Gonzales DR, Carpenter T, van Hemert JI, Wardlaw J. An open source toolkit for medical imaging de-identification. Eur Radiol. 2010;20(8):1896–1904. doi: 10.1007/s00330-010-1745-3. [DOI] [PubMed] [Google Scholar]
- 3.Medical Research Council (2000) Personal Information in Medical Research, Swindon. Available via http://www.mrc.ac.uk/Utilities/Documentrecord/index.htm?d=MRC002452. Accessed July 2010
- 4.Santesof, Sante DICOM Editor Integrated Anonymizer, Athens. Available via http://www.santesoft.com/howto/anonymize.html. Accessed July 2010
- 5.IBM Haifa Labs, Universal De-Identification Platform, Haifa. Available via https://www.research.ibm.com/haifa/projects/software/udip/. Accessed July 2010
- 6.Grassroots DICOM library, Available via http://gdcm.sourceforge.net. Accessed July 2010
- 7.Radiological Society of North America, Inc. CTP-The RSNA Clinical Trial Processor, Oak Brook. Available via http://mircwiki.rsna.org/index.php?title=CTP-The_RSNA_Clinical_Trial_Processor. Accessed March 2010
- 8.Radiological Society of North America, Inc., MIRCwiki, Oak Brook. Available via http://mircwiki.rsna.org. Accessed February 2010
- 9.Radiological Society of North America, Inc., Oak Brook. Available via http://www.rsna.org. Accessed January 2010
- 10.Java Advanced Imaging Image I/O Tools, http://download.java.net/media/jai-imageio/builds/release/1.1. Accessed February 2010
- 11.Mendelson DS, Bak PRG, Menschik E, Siegel E. Informatics in radiology: image exchange: IHE and the Evolution of Image Sharing. RadioGraphics. 2008;28:1817–1833. doi: 10.1148/rg.287085174. [DOI] [PubMed] [Google Scholar]
- 12.Rajala T, Savio S, Penttinen J, Dastidar P, and Kähönen M, et al. (2010) Development of a Research Dedicated Archival System (TARAS) in a University Hospital. J Digit Imaging. [Epub ahead of print]. doi:10.1007/s10278-010-9350-1 [DOI] [PMC free article] [PubMed]
- 13.Neubauer T, Heurix J. A methodology for the pseudonymization of medical data. Int J Med Inform. 2011;80(3):190–204. doi: 10.1016/j.ijmedinf.2010.10.016. [DOI] [PubMed] [Google Scholar]
- 14.Onken M, Riesmeier J, Engel M, Yabanci A, Zabel B, et al. Reversible anonymization of DICOM images using automatically generated policies. Stud Health Technol Inform. 2009;150:861–865. [PubMed] [Google Scholar]