Abstract
Several protein misfolding diseases are associated with the conversion of native proteins into ordered protein aggregates known as amyloid. Studies of amyloid assemblies have indicated that non-native proteins are responsible for initiating aggregation in vitro and in vivo. Despite the importance of these species for understanding amyloid disease, the structural and dynamic features of amyloidogenic intermediates and the molecular details of how they aggregate remain elusive. This review focuses on recent advances in developing a molecular description of the folding and aggregation mechanisms of the human amyloidogenic protein β2-microglobulin under physiologically relevant conditions. In particular, the structural and dynamic properties of the non-native folding intermediate IT and its role in the initiation of fibrillation and the development of dialysis-related amyloidosis are discussed.
Keywords: amyloid, conformational conversion, dialysis-related amyloidosis, dynamics, NMR, prion
The role of β2-microglobulin in amyloid disease
β2-microglobulin (β2m) is the non-covalently bound light chain of the major histocompatibility complex class I (MHC I), wherein the protein plays an essential role in chaperoning assembly of the complex for antigen presentation (Fig. 1A) [1–3]. Wild-type β2m contains 99 amino acids and has a classical β-sandwich fold comprising seven anti-parallel β-strands that is stabilized by its single inter-strand disulfide bridge between β-strands B and F (Fig. 1B) [4–6]. The high resolution structures of monomeric native β2m from humans and several of its variants have been solved by solution NMR [7–10] and X-ray crystallography [4,11–16]. β2m contains five peptidyl–prolyl bonds, one of which (His31-Pro32) adopts the thermodynamically unfavoured cis-isomer in the native state (Fig. 1B) [4,7,9]. Another interesting feature of monomeric native β2m is the conformational dynamics of the D-strand and the loop that connects the D- and E-strands (the DE-loop) (Fig. 1B). This region forms contacts with the MHC I heavy chain [17], but shows dynamics on a microsecond to millisecond timescale when a monomer in solution [7] and variability in different crystal structures (Fig. 1C,D) [13]. This rationalizes hydrogen–deuterium exchange studies on monomeric native β2m showing that the DE-loop region exhibits enhanced backbone dynamics compared with the non-covalently MHC I bound state [18]. Notably, a link between the dynamic properties of monomeric native β2m, particularly in the D-strand and the DE-loop region, and its potential to assemble into amyloid fibrils has been proposed [7,10,11,18–20].
Fig. 1.
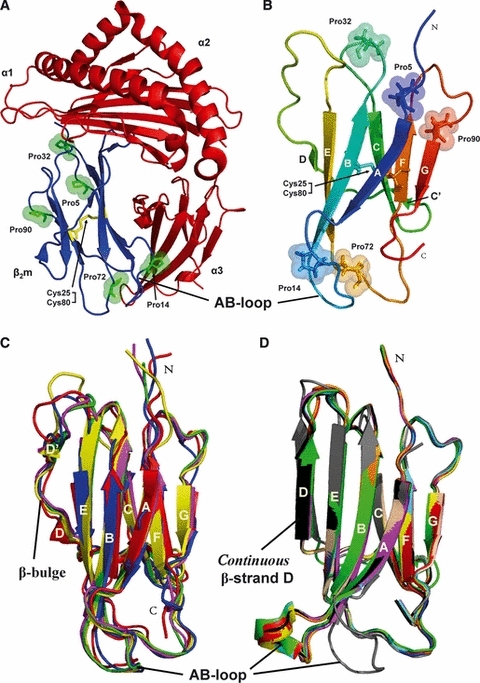
Monomeric β2m plays a key role in DRA. (A) Cartoon representation of human MHC I (PDB code 3MYJ [136]) showing the heavy chain (α1, α2, α3 in red) and the light chain (β2m in blue). Highlighted are the residues Pro5, Pro14, Pro32, Pro72 and Pro90 (in green sticks, spheres) and the disulfide bond between residues Cys25 and Cys80 (in yellow sticks). (B) Cartoon representation of the solution structure of monomeric native wild-type β2m (PDB code 2XKS [9]) showing β-strands A (6–11), B (21–28), C (36–41), C′ (44–45), D (50–51), E (64–70), F (79–83) and G (91–94). Highlighted are the residues Pro5, Pro14, Pro32, Pro72 and Pro90 (in sticks, spheres) and the disulfide bond between residues Cys25 and Cys80 (in sticks). N, N-terminus; C, C-terminus. (C) Structures displaying a β-bulge and an attached AB-loop: wild-type β2m (PDB code 1JNJ [7]) in red, H31Y (PDB code 1PY4 [15]) in green, W60G (PDB code 2VB5 [16]) in blue, H13F (PDB code 3CIQ [55]) in yellow and MHC I (PDB code 3MYJ [136]) in magenta. (D) Structures displaying a straight β-strand D: wild-type β2m (PDB code 1LDS [11]) in red, L39W/W60F/W95F (PDB code 2D4D [137]) in green, wild-type β2m (PDB code 2D4F [137]) in blue, wild-type β2m (PDB code 2YXF [12]) in yellow, W60G (PDB code 2Z9T [16]) in magenta, W60C (PDB code 3DHJ [14]) in cyan, D59P (PDB code 3DHM [14]) in orange, W60G (PDB code 3EKC [14]) in wheat, K58P/W60G (PDB code 3IB4 [121]) in black and P32A (PDB code 2F8O [58]) in grey.
Catabolism of β2m following its dissociation from the MHC I heavy chain occurs predominantly in the proximal tubules in the kidney [21,22]. As a consequence, the concentration of β2m circulating in the serum of patients suffering from renal dysfunction is enhanced up to 60-fold compared with healthy individuals. This causes the deposition of β2m as amyloid fibrils in osteoarticular tissues, leading to pathological bone destruction and the condition known as dialysis-related amyloidosis (DRA) (Fig. 2) [23]. However, a poor correlation between the β2m concentration in the serum and fibril load in osteoarticular tissues in long-term dialysis patients suggests that additional factors must be responsible for the initiation of β2m aggregation in vivo [24]. Consistent with these results, in vitro studies have shown that β2m is remarkably intransigent to assembly into amyloid fibrils at neutral pH, remaining predominantly monomeric for several months at pH 7.5, 37 °C, when incubated at protein concentrations more than 20-fold higher than those found in dialysis patients (∼ 3.2 μm [21]) [25,26]. As a consequence of these findings, factors have been sought that could facilitate protein aggregation of β2m in vivo, including the age of patients [27], the duration of kidney failure [28], the dialysis procedure itself [29–31], post-translational modifications of full-length β2m [32–40] and bimolecular collision between β2m and biological molecules abundant in osteoarticular tissues or encountered during dialysis [26,41–51]. As a result, a multitude of factors have been shown to enhance the aggregation of β2m in vitro and are implicated in vivo, including Cu2+ [47,52–59], glycosaminoglycans [26,41,60], lysophosphatidic acid [49], non-esterified fatty acids [48,50] and collagen [41,42,61].
Fig. 2.
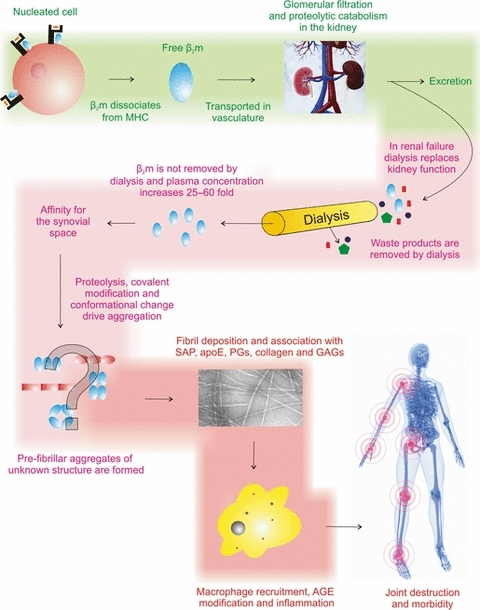
Schematic of the key processes which result in the pathological symptoms experienced in DRA (reproduced, with permission, from [138]).
Amyloid formation of β2m under physiological pH conditions (around pH 7.0) commences from the fully folded native protein state [62]. Analysis of the thermodynamic stability of native wild-type β2m and an array of variants, however, showed no correlation between the thermodynamic stability of β2m and its potential to assemble into amyloid-like fibrils in vitro [62]. Instead, the formation of one or more non-native precursors that are accessible by dynamic fluctuations from the native protein is required before aggregation can occur [9,18–20,63–69]. Such fluctuations may expose aggregation-prone sequences normally sequestered in the native structure [70], consistent with local and/or more global unfolding events being a common feature in the aggregation mechanisms of globular proteins [58,67,71–80].
Peptidyl–prolyl isomerization initiates β2m amyloid assembly at physiological pH
In pioneering work, Chiti et al. [81] used a series of spectroscopic probes to show that wild-type β2m folds via two structurally distinct intermediates, known as I1 and I2, en route to the globular native state. The first intermediate along the folding reaction coordinate, I1, is populated within 5 ms of dilution of the protein from denaturant. This species shows substantial elements of non-random structure and contains a disorganized hydrophobic core in which several hydrophobic residues remain exposed to solvent [81]. The second folding intermediate, I2, forms within milliseconds of the population of I1 and displays native-like secondary structure and ordered packing of side chains within the hydrophobic core. Further folding of I2 occurs on a timescale of seconds to minutes at 30 °C, suggesting substantial energetic barriers to the attainment of the globular native fold [62,81]. Although folding of wild-type β2m is a cooperative process as judged by equilibrium denaturation [81], I2 nonetheless accumulates, reaching a population of about 14 ± 8% at equilibrium at pH 7.4, 30 °C, as judged by capillary electrophoresis [82]. Importantly, the concentration of I2 was found to correlate with the rate of elongation using seeds formed from ex vivo amyloid fibrils at pH 7.4, 30 °C, consistent with this native-like folding intermediate being directly (or indirectly via further conformational changes) capable of amyloid elongation [82]. A slow folding intermediate, reminiscent of I2, has also been described by others [34,83].
Building on the observations made by Chiti and colleagues [82], more detailed studies of the folding and unfolding mechanism of wild-type β2m, combined with mutagenesis of the sequence, demonstrated that the transition between the slow folding intermediate I2 and the native fold is rate limited by trans to cis isomerization of the His31-Pro32 peptide bond, which led to the kinetically trapped intermediate being termed IT [67–69]. Consistent with these findings, folding studies of a variant of β2m in which Pro32 is replaced with Val using manual mixing experiments at low temperature (2.8–4.0 °C) monitored by CD and NMR revealed that the slow folding step is abolished, trapping β2m in a non-native species presumably with a trans His31-Val32 peptide bond [68]. Pro32 is highly conserved in β2m in different organisms [84] and trans to cis peptidyl–prolyl isomerization at this site has been shown previously to be responsible for the slow refolding commonly found in other immunoglobulin domains [85–91]. Interestingly, however, P32V β2m is not able to elongate amyloid fibrillar seeds in vitro or to nucleate fibril formation, suggesting that a trans His31-Xaa peptide bond is necessary, but not sufficient, to endow β2m with its amyloidogenic properties [68].
To gain a more detailed understanding of the kinetic folding mechanism of β2m and the role of different partially folded species in linking the folding and aggregation energy landscapes, Jahn and co-workers [67] analysed the folding and unfolding kinetics of β2m under an array of conditions, including analysis of the folding mechanism of the variant P32G. Using global analysis of the resulting kinetic data, the authors proposed a five-state model for the folding mechanism of wild-type β2m involving parallel folding pathways initiated from cis or trans His31-Pro32 in the unfolded state [67]. The five-state model has been challenged by Sakata and co-workers [69] who proposed that a simpler four-state model satisfies their obtained microscopic and macroscopic rates of β2m unfolding and refolding using chevron analysis. In particular, using their approach Sakata et al. were unable to detect spectroscopically the accumulation of the folding intermediate containing a native cis-His31-Pro32 peptide bond (IC), suggesting that this species is non-existent or populated to levels below the detection limit. Despite these differences, both folding models suggest that IT is low but significantly populated under physiological conditions at equilibrium, consistent with the poor ability of wild-type β2m to elongate fibrillar seeds at neutral pH in vitro [26,67]. Replacement of Pro32 with glycine (P32G) resulted in a simple three-state folding mechanism in which an intermediate, presumably with a trans His31-Gly32 peptide bond akin to IT, accumulates during folding, reaching an equilibrium concentration of approximately 30% [67]. Importantly, by titrating the population of IT populated at equilibrium for the wild-type protein and P32G by varying the solution conditions, Jahn et al. [67] showed that the population of IT correlates with the rate of fibril elongation in vitro, suggesting that IT is a key link between the folding and aggregation energy landscapes for this protein. This could occur directly by this species showing an ability to elongate amyloid seeds, or indirectly via further conformational excursions to other species accessible from this folding intermediate [9,20,66,67]. Interrogation of the conformational properties of P32G using NMR suggested large conformational changes involving residues in the BC- and FG-loops, the D-strand and the N-terminal region of the protein that presumably arise from the isomerization of Pro32 and subsequent partial unfolding of the protein [67]. These regions map precisely to the regions reported previously to be perturbed in the kinetic folding intermediate IT, suggesting a close structural relationship of the two species [67].
The intransigence of wild-type β2m to form amyloid fibrils when incubated for extended periods of time at neutral pH at concentrations substantially higher than those found in vivo [21,25,26] can be rationalized in light of the finding that the amyloidogenic precursor, IT, is both transiently sampled and maintained at low concentrations at equilibrium in the wild-type protein under ambient conditions [25,67,82]. In order to explore the thermodynamics and kinetics of amyloid assembly from β2m at physiological pH in vitro, therefore, a plethora of conditions have been used to increase the population of species akin (but not necessarily identical) to IT at equilibrium. These include the addition of Cu2+ ions and urea [46,47,53,92], organic solvents [60,83], collagen [41,42], glycosaminoglycans or other biologically relevant factors [26,60,93], SDS or lysophospholipids [48–51,94]. Changes in the physicochemical environment, including ultrasonication [95], heat treatment [96], high salt and stirring/agitation [97], have also been employed. These apparently very different conditions have in common the principle that they perturb the equilibrium position of the cis/trans His31-Pro32 peptide bond and hence enhance the amyloidogenic potential of the wild-type protein [25]. Mutations in the N- and/or C-terminal regions of the sequence have also been shown to enhance amyloid formation of β2m at physiological pH [8,9,25,26,32,98,99], whilst other mutations that focus on the DE-loop region demonstrated variable effects on the thermodynamic stability of the protein depending on the amount of strain introduced [14,16,20,100,101]. DE-loop mutations such as D59P that introduce loop strain show a decreased folding free energy compared with the wild-type protein and an enhanced potential to aggregate, whereas a release of loop strain such as in W60G leads to super-stable variants which have reduced amyloidogenic features [13,14,16]. However, DE-loop cleavage variants such as ΔK58 or cK58 (which contain a specific cleavage at Lys58 with or without removal of Lys58, respectively) have been demonstrated to be highly aggregation-prone [34,102–104]. Together these studies are indicative of a fragile and delicate amino acid network required for the stabilization of the cis isomer at His31-Pro32 that is required both for binding to the MHC I heavy chain [16] and to maintain a soluble native structure for the monomeric protein.
β2m assembly mechanisms at atomic resolution
Clinical studies have shown that dialysis patients treated with Cu2+-free filter membranes have a > 50% reduced incidence of DRA compared with patients who were exposed to traditional Cu2+-containing dialysis membranes [27,105]. These studies suggest that Cu2+ ions may play a role in initiating or enhancing aggregation of wild-type β2m in DRA. Indeed, Cu2+ has been shown to bind to native human β2m with moderate affinity (Kapp = 2.7 μm) and specificity (Cu2+ > Zn2+ >> Ni2+) [46,106]. Binding involves coordination to the imidizole ring of His31 [7,107]. Non-native states of wild-type β2m also bind Cu2+ ions; in this case the three other histidines in the sequence (His13, His51, His84) coordinate Cu2+ with a Kapp ∼ 41 μm [107]. As a consequence, binding of Cu2+ ions increases the concentration of non-native (so-called ‘activated’) forms of monomeric β2m, named by Miranker and co-authors as M*, which triggers the formation of dimeric, tetrameric and hexameric species (< 1 h) believed to be on-pathway to amyloid-like fibrils [47,106]. Cu2+ binding is required for the conformational changes leading to the formation of M* and to the generation of early oligomeric species. However, once these oligomeric species and subsequent fibrillar aggregates are formed, Cu2+ is not essential for their stability [52,54,56,57,108]. By creating two variants, P32A and H13F, Miranker and colleagues [55,58] were able to crystallize dimeric and hexameric forms of β2m (the latter after Cu2+-induced oligomerization). These studies revealed that dimeric P32A and hexameric H13F contain a trans His31-Ala32 and a trans His31-Pro32 peptide bond, respectively. Each oligomer is composed of monomers that retain a native-like fold, yet display significant alterations in the organization of aromatic side chains within the hydrophobic core, most notably Phe30, Phe62 and Trp60 (Fig. 3A,B, in blue), which the authors speculate could be important determinants of amyloid assembly [53,55,58]. How these static structures relate to the transient intermediates formed during folding or populated during aggregation, however, remain unclear. Importantly in this regard, P32A and H13F lack an enhanced ability to assemble into amyloid fibrils compared with wild-type β2m [55,58], reminiscent of the behaviour of P32V [68,69]. Despite containing a trans His31-Xaa32 peptide bond, these species lack structural and/or dynamical properties critical for amyloid formation.
Fig. 3.
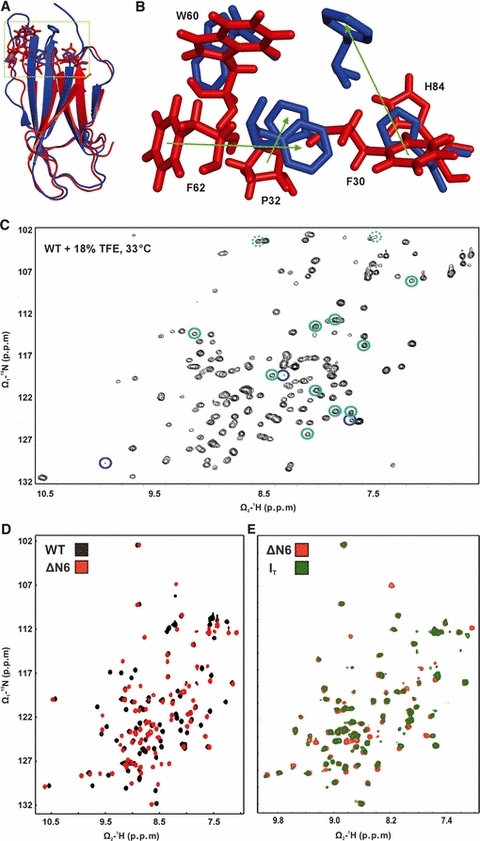
Molecular description of the IT state using X-ray crystallography and high resolution solution NMR. (A) The ribbon overlay shows one monomer of the hexameric crystal structure of H13F (PDB code 3CIQ [55], in blue) and the lowest energy structure of ΔN6 (PDB code 2XKU) [9] (in red). The residues Phe30, Pro32, Trp60, Phe62 and His84 are highlighted in sticks. The dashed green box indicates a zoom-in for this region shown in (B). (C) 1H–15N HSQC of wild-type β2m in 18% (v/v) TFE at pH 6.6 and 33 °C (reproduced, with permission, from [20]). Green circles are assigned resonances for IT, while blue circles indicate the TFE induced, structurally disordered D state that is thought to be precursor for fibril elongation under these conditions. (D) 1H–15N HSQC overlay of wild-type β2m (black) and ΔN6 (red) recorded in 25 mm sodium phosphate buffer pH 7.5, 25 °C. (E) 1H–15N SOFAST HMQC overlay of ΔN6 (red) and the kinetic intermediate IT (green) recorded approximately 2 min after refolding was initiated (25 mm sodium phosphate buffer pH 7.5, 0.8 m residual urea, 25 °C). Reproduced with permission from [9].
Increased conformational dynamics has emerged as a common feature of the assembly of β2m monomers into amyloid fibrils at neutral pH from a wealth of studies under varied solution conditions [9,10,18–20,32,65–67,92,103,109], akin to the findings on other proteins that also assemble into amyloid fibrils commencing from folded monomeric states [64,71,73,76,77,80,110–116]. Accordingly, ΔN6 (in which β2m is cleaved at Lys6) [32], cK58 and ΔK58 [34,102,103,117,118] and wild-type β2m in the presence of SDS/2,2,2-trifluoroethanol (TFE)/other additives [20,41,42,50,51,66,119] all exhibit decreased solubility, increased local and global unfolding events and enhanced amyloidogenicity at pH values close to physiological. Of particular interest is the variant ΔN6, since this species is found as a significant component (∼ 26%) in ex vivo amyloid deposits and exhibits an increased affinity for collagen compared with the wild-type protein, suggesting a role for this protein in the development of DRA [61,120]. Pioneering work by Esposito and colleagues showed that ΔN6 experiences a global decrease in conformational stability compared with wild-type β2m and, using molecular dynamics simulations, the authors proposed that the D-strand facilitates intermolecular interactions to form oligomeric assemblies prior to the development of long straight amyloid fibrils at pH 6.5, 37 °C [32]. Similarly, the variants cK58 and ΔK58 were found to be highly aggregation-prone, presumably due to enhanced conformational dynamics, especially for strand D, and a concomitant increase in concentration of the amyloidogenic folding intermediates at equilibrium [34,103]. In contrast, the mutation W60G which also lies in the DE-loop diminishes the potential of this variant to extend fibrillar seeds of the human wild-type protein at pH 7.4 in the presence of 20% (v/v) TFE [16], consistent with the dynamics within this region of the protein playing a crucial role in β2m assembly at neutral pH [13,14,19,20,66,121]. These studies therefore reinforce the importance of interrogating the conformational dynamics of β2m and its truncation variants in more detail in order to understand the aggregation properties of this species and, more generally, how other non-native species that retain a globular fold aggregate in vitro and in vivo [116].
Major breakthroughs in understanding the properties that endow non-native states of β2m with their amyloidogenic properties have arisen from NMR studies of wild-type β2m and several variants of the protein by exploiting the capabilities of modern NMR methods for rapid and sensitive data acquisition [7,9,11,20,32,55,58,66–68,103,109]. Accordingly, recent studies of the folding kinetics of wild-type β2m using real-time NMR combined with amino acid selective labelling of Phe, Val and Leu provided the first glimpses of the amyloid precursor of β2m under conditions close to physiological [109]. However, extensive peak broadening caused by conformational dynamics on a microsecond to millisecond timescale ruled out detailed assignment and structure elucidation of IT. Following on from this work, studies of the folding kinetics of wild-type β2m in different concentrations of TFE using real-time NMR revealed that the native protein is generated with double exponential kinetics from IT for all resonances studied, indicative of an energy landscape that is more complex than the single barrier suspected hitherto [66,67,69]. By contrast with the behaviour of the wild-type protein, W60G folds to the native state from IT with mono-exponential kinetics, indicative of a more simple folding energy landscape for this less amyloidogenic variant [66]. Based on these results, the authors propose that a species that is more disordered than IT (named a ‘native-unlike’ or D state), formed maximally in 20% (v/v) TFE, is responsible for elongating wild-type β2m seeds [20]. The wild-type protein under those conditions has also been simulated using molecular dynamics [122]. Exploiting the sensitivity of β2m conformations to the concentration of TFE, the authors were able to find conditions wherein IT is maximally populated from W60G, reaching 30–40% population in 18% (v/v) TFE (at pH 6.6, 33 °C), and were able to assign 63 backbone amide resonances (out of 93 amide bonds) unambiguously for this species (BMRB code 16587) (Fig. 3C) [20]. Incomplete assignment of the IT state in W60G and considerable peak overlap by native state resonances, however, hampered the assignment of the backbone conformation of the peptidyl–prolyl bond at Pro32 and a more detailed structural and dynamic characterization of this intermediate [20].
Most recently, the difficulties in determining the conformational properties of IT have been overcome by using the β2m truncation variant ΔN6 as a structural mimic of this species (Fig. 3A,B, in red) [9,25]. High resolution NMR studies directly comparing the 1H–15N HSQC spectra of ΔN6 and IT revealed that the major species populated by ΔN6 in solution at pH 7.5, 25 °C, closely resembles the transient folding intermediate IT (Fig. 3D,E). Using ΔN6 as a structural model for IT, full resonance assignment and structural elucidation were possible, revealing the structural and dynamical properties of this non-native conformer of β2m. The results showed that under the conditions employed ΔN6 retains a native fold but undergoes a major re-packing of several side chains within the hydrophobic core to accommodate the non-native trans-conformation of the His31-Pro32 peptide bond (Fig. 3A,B, in red). Intriguingly, the side chains involved map predominantly to the same residues that undergo structural reorganization in the presence of Cu2+ ions, although the precise packing of residues remains different in many cases (Fig. 3A,B) [9,55,58]. Despite adopting a thermodynamically stable [9,25] native-like topology, ΔN6 is a highly dynamic entity, possessing only limited protection from hydrogen exchange together with pH- and concentration-dependent sensitivity of its backbone dynamics on a microsecond to millisecond timescale. These data suggest that increased conformational dynamics of ΔN6 correlate with an increase in its amyloidogenic properties presumably by enabling the formation of one or more rarely populated conformers that have an enhanced potential to assemble into amyloid fibrils [9,32,123]. One of the key events in this amyloid switch is protonation of His84, which experiences a large pKa shift from ∼ 4 to ∼ 7 upon peptidyl–prolyl isomerization of the His31-Pro32 peptide bond (Fig. 4A) [9]. The involvement of His84 in the initiation of β2m amyloid fibril formation has been proposed previously using computational methods [61]. Oligomeric structures which become available after peptidyl–prolyl isomerization and exploration of conformational space upon His84 protonation have been proposed previously in association with Cu2+ binding [55,58], in the presence of dithiothreitol [124] or by the binding of nanobodies [125]. Interestingly, the last two conditions result in the formation of oligomers that are domain swapped, as proposed hitherto for β2m assembly under native conditions using computational methods [126] or Cu2+ treatment [106]. Whether domain swapping occurs in DRA, however, remains to be elucidated. Another open question is the structural and dynamic similarities and differences between trans intermediates formed under different conditions (such as alterations of pH and temperature, Cu2+ treatment, mutagenesis (ΔN6) or addition of organic solvent (TFE)) and how these map to the structure determined for ΔN6 at neutral pH [9] or that of the more ephemeral amyloid precursors that form from this protein or from the folding intermediate IT. Nonetheless, these data are suggestive of a mechanism of assembly under different solution conditions that contains many features in common.
Fig. 4.
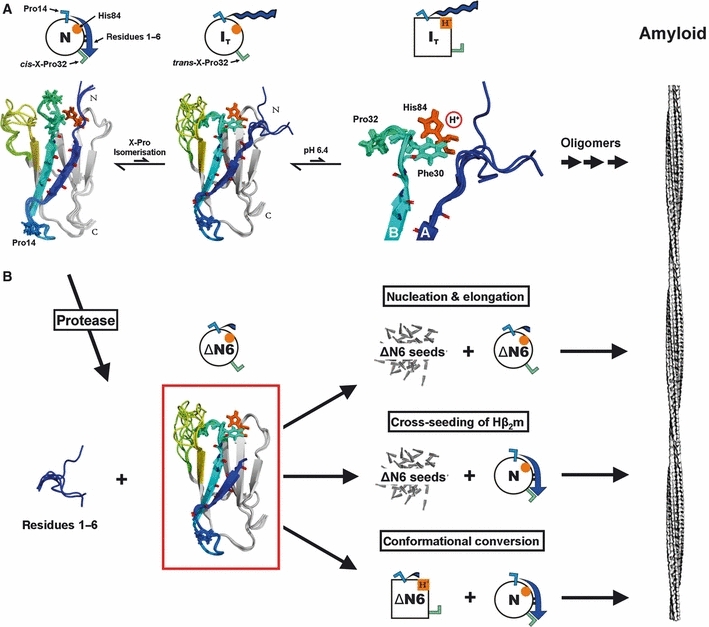
Prion-like conversion during amyloid formation. (A) Summary showing the structures of wild-type β2m (PDB code 2XKS) and a model of IT. Above, keys for these conformational states. Native wild-type β2m (leftmost), shown above as a circle with cis His31-Pro32 (green Γ), trans His13-Pro14 (blue Γ), His84 (orange circle) and the N-terminal region (residues 1–6, blue arrow). Backbone atoms of residues which establish strong hydrogen bonding between β-strands A and B in the native state are shown in sticks. Upon dissociation of the N-terminal region, the His31-Pro32 peptide bond is free to relax into the trans-conformation, causing further conformational changes that lead to the formation of the non-native IT conformer (shown as a circle above a model of its structure). Protonation of His84 under mildly acidic conditions (shown in red ball and stick and as an orange square in the model above), which lies adjacent to Pro32, enhances the amyloid potential of IT further. Oligomerization of these aggregation-prone species then leads to the formation of β2m amyloid fibrils. Assuming that the fibrils formed at neutral pH are structurally similar to those formed at acidic pH, as suggested by FTIR [135] and solid state NMR [133,134], large conformational changes are required in order to transform the anti-parallel β-sheet arrangement of ΔN6 into the parallel in-register arrangement of β-strands characteristic of β2m amyloid fibrils, as reported recently [132] (reproduced, with permission, from [9]). (B) Summary showing the consequences of β2m cleavage of the N-terminal hexapeptide that generates ΔN6 as a persistent IT state (PDB code 2XKU). Once formed ΔN6 is able to nucleate and elongate its own fibrils and also to cross-seed elongation of its fibrillar seeds with the wild-type protein, leading to the development of long straight amyloid-like fibrils (the image of the fibrils was redrawn from the cryo-EM structure of β2m amyloid fibrils from [139]). Furthermore, ΔN6 can transform the innocuous native state of β2m via bimolecular collision. The formation of catalytic amounts of ΔN6 thus has been proposed to be a cataclysmic event during the development of DRA.
Prion-like conversion during β2m amyloid assembly
Despite the finding that ΔN6 comprises ∼ 26% of β2m in amyloid deposits in patients with DRA, this species is not found in the serum of people with renal dysfunction [127]. As a consequence of these findings, formation of ΔN6 has been proposed to occur as a post-assembly event [123]. Most recently, however, it has been demonstrated that ΔN6 is not only able to nucleate fibrillogenesis efficiently in vitro at physiological pH as discussed above (Fig. 4B) [9,25,26] but, as a persistent trans-Pro32 state, ΔN6 is also able to convert wild-type β2m into an aggregation-competent conformer by bimolecular collision between the two monomers (Fig. 4B) [9]. Accordingly, only catalytic amounts (1%) of ΔN6 are sufficient to convert significant quantities of the wild-type protein into amyloid fibrils (Fig. 4B). Detailed interrogation of bimolecular collision between native wild-type β2m and ΔN6 using NMR revealed the molecular mechanism by which this prion-like templating might occur [9]. First, ΔN6 binds specifically, but transiently, to native wild-type β2m, possibly involving residues of β-strands A, B and D and the DE-loop. This interaction changes the native configuration of Pro14 within the AB-loop which is highly dynamic as indicated by molecular dynamics simulations [63,122] and X-ray crystallography (Fig. 1C,D). Pro14 dynamics have been shown hitherto to be responsible for an alternative β2m conformation in which the hydrogen bonding between β-strands A and B is severely impaired [15]. Inter-strand hydrogen bonding between those two strands, together with the correct attachment of the N-terminal hexapeptide, has been demonstrated to be crucial in maintaining a low concentration of IT at equilibrium [25]. Binding of ΔN6 to wild-type β2m, therefore, leads to the disruption of important interactions between the N-terminal hexapeptide and the BC-loop, leading to accelerated relaxation kinetics towards the amyloidogenic trans His31-Pro32 isomeric state. The truncation variant ΔN6 is thus capable of driving the innocuous native wild-type protein into aggregation-competent entities, reminiscent of the action of prions. Such an observation rationalizes the lack of circulating ΔN6 in the serum and, given the natural affinity of this species for collagen (which is enhanced relative to wild-type β2m [61]), explains why assembly of fibrils occurs most readily in collagen-rich joints. Rather than being an innocuous post-assembly event, therefore, proteolytic cleavage of β2m to create one or more species truncated at the N-terminus could be a key initiating event in DRA, enabling the formation of a species that is not only able to assemble de novo into amyloid fibrils but can enhance fibrillogenesis of wild-type β2m. The latter is accomplished by initiating the ability of the wild-type protein to nucleate its own assembly, or by cross-seeding fibril elongation of ΔN6 seeds with wild-type monomers (Fig. 4). Identifying the proteases responsible for the production of ΔN6 or using the high resolution structure of ΔN6 as a target for the design of small molecules able to intervene in assembly may provide new approaches for therapeutic intervention in DRA.
Outlook: towards a complete molecular description of β2m amyloidosis
In this review we have highlighted the importance of conformational dynamics for the initiation and development of β2m amyloid formation commencing from the natively folded state. Detailed analysis of the folding, stability and amyloidogenicity of a number of different proteins has revealed that a polypeptide chain can adopt a diversity of structures within a multidimensional energy landscape, the thermodynamics and kinetics of which are dependent on the protein sequence and solution conditions employed [128]. One key feature that appears to identify amyloidogenic proteins from their non-amyloidogenic counterparts is a lack of structural cooperativity that is revealed by enhanced conformational dynamics on a microsecond to millisecond timescale, often portrayed by increased rates of proteolysis, hydrogen exchange and R2 NMR relaxation rates [115]. Such motions may expose sequences with high amyloid potential that are usually hidden within the native structure [70] or may endow surface properties that enable new protein–protein interactions to form. Studies of β2m have contributed substantially to this view, resulting most recently in a high resolution structure for the amyloid-initiating folding intermediate IT and the beginnings of a molecular understanding of why increased conformational dynamics make this species highly aggregation-prone [9]. Rather than an innocuous post-assembly event, the work suggests proteolytic cleavage as a cataclysmic event that releases a species that is not only able to spawn further aggregation-prone species but is also able to convert the wild-type protein into an amyloidogenic state via conformational conversion akin to the activity famously associated with prions [129–131]. Finally, many studies of β2m amyloid assembly under a wide range of conditions, some close to physiological and others utilizing metal ions or solvent additives to drive fibrillogenesis at neutral pH, have together revealed common principles of β2m self-assembly which are related by the formation of non-native species initiated by a cis to trans His31-Pro32 switch despite the wide range of conditions employed. Further work is now needed to define the origins of molecular recognition between monomers and oligomers that form as assembly progresses into amyloid fibrils at neutral pH and to define the extent of further conformational changes required to form the cross-β structure of amyloid [132–135]. This will entail greater structural knowledge about the multitude of protein states populated on the folding and aggregation energy landscapes and how these species are formed and interconnected.
Acknowledgments
We thank David Brockwell and members of the Radford and Homans research groups for helpful discussions. We acknowledge, with thanks, the Wellcome Trust (062164 and GR075675MA) and the University of Leeds for funding.
Glossary
Abbreviations
- β2m
β2-microglobulin
- DRA
dialysis-related amyloidosis
- MHC I
major histocompatibility complex class I
- TFE
2,2,2-trifluoroethanol
References
- 1.Zijlstra M, Bix M, Simister NE, Loring JM, Raulet DH, Jaenisch R. Beta2-microglobulin deficient mice lack CD4-8+ cytolytic T cells. Nature. 1990;344:742–746. doi: 10.1038/344742a0. [DOI] [PubMed] [Google Scholar]
- 2.Hughes EA, Hammond C, Cresswell P. Misfolded major histocompatibility complex class I heavy chains are translocated into the cytoplasm and degraded by the proteasome. Proc Natl Acad Sci USA. 1997;94:1896–1901. doi: 10.1073/pnas.94.5.1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Paulsson KM, Wang P, Anderson PO, Chen S, Pettersson RF, Li S. Distinct differences in association of MHC class I with endoplasmic reticulum proteins in wild-type, and beta2-microglobulin- and TAP-deficient cell lines. Int Immunol. 2001;13:1063–1073. doi: 10.1093/intimm/13.8.1063. [DOI] [PubMed] [Google Scholar]
- 4.Becker JW, Reeke GN., Jr Three-dimensional structure of beta2-microglobulin. Proc Natl Acad Sci USA. 1985;82:4225–4229. doi: 10.1073/pnas.82.12.4225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Smith DP, Radford SE. Role of the single disulphide bond of beta2-microglobulin in amyloidosis in vitro. Protein Sci. 2001;10:1775–1784. doi: 10.1110/ps.4901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Katou H, Kanno T, Hoshino M, Hagihara Y, Tanaka H, Kawai T, Hasegawa K, Naiki H, Goto Y. The role of disulfide bond in the amyloidogenic state of beta2-microglobulin studied by heteronuclear NMR. Protein Sci. 2002;11:2218–2229. doi: 10.1110/ps.0213202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Verdone G, Corazza A, Viglino P, Pettirossi F, Giorgetti S, Mangione P, Andreola A, Stoppini M, Bellotti V, Esposito G. The solution structure of human beta2-microglobulin reveals the prodromes of its amyloid transition. Protein Sci. 2002;11:487–499. doi: 10.1110/ps.29002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Corazza A, Pettirossi F, Viglino P, Verdone G, Garcia J, Dumy P, Giorgetti S, Mangione P, Raimondi S, Stoppini M, et al. Properties of some variants of human beta2-microglobulin and amyloidogenesis. J Biol Chem. 2004;279:9176–9189. doi: 10.1074/jbc.M310779200. [DOI] [PubMed] [Google Scholar]
- 9.Eichner T, Kalverda AP, Thompson GS, Homans SW, Radford SE. Conformational conversion during amyloid formation at atomic resolution. Mol Cell. 2011;41:161–172. doi: 10.1016/j.molcel.2010.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Esposito G, Corazza A, Viglino P, Verdone G, Pettirossi F, Fogolari F, Makek A, Giorgetti S, Mangione P, Stoppini M, et al. Solution structure of beta2-microglobulin and insights into fibrillogenesis. Biochim Biophys Acta. 2005;1753:76–84. doi: 10.1016/j.bbapap.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 11.Trinh CH, Smith DP, Kalverda AP, Phillips SE, Radford SE. Crystal structure of monomeric human beta2-microglobulin reveals clues to its amyloidogenic properties. Proc Natl Acad Sci USA. 2002;99:9771–9776. doi: 10.1073/pnas.152337399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Iwata K, Matsuura T, Sakurai K, Nakagawa A, Goto Y. High-resolution crystal structure of beta2-microglobulin formed at pH 7.0. J Biochem. 2007;142:413–419. doi: 10.1093/jb/mvm148. [DOI] [PubMed] [Google Scholar]
- 13.Ricagno S, Raimondi S, Giorgetti S, Bellotti V, Bolognesi M. Human beta2-microglobulin W60V mutant structure: implications for stability and amyloid aggregation. Biochem Biophys Res Commun. 2009;380:543–547. doi: 10.1016/j.bbrc.2009.01.116. [DOI] [PubMed] [Google Scholar]
- 14.Ricagno S, Colombo M, de Rosa M, Sangiovanni E, Giorgetti S, Raimondi S, Bellotti V, Bolognesi M. DE loop mutations affect beta2-microglobulin stability and amyloid aggregation. Biochem Biophys Res Commun. 2008;377:146–150. doi: 10.1016/j.bbrc.2008.09.108. [DOI] [PubMed] [Google Scholar]
- 15.Rosano C, Zuccotti S, Mangione P, Giorgetti S, Bellotti V, Pettirossi F, Corazza A, Viglino P, Esposito G, Bolognesi M. Beta2-microglobulin H31Y variant 3D structure highlights the protein natural propensity towards intermolecular aggregation. J Mol Biol. 2004;335:1051–1064. doi: 10.1016/j.jmb.2003.11.040. [DOI] [PubMed] [Google Scholar]
- 16.Esposito G, Ricagno S, Corazza A, Rennella E, Gumral D, Mimmi MC, Betto E, Pucillo CE, Fogolari F, Viglino P, et al. The controlling roles of Trp60 and Trp95 in beta2-microglobulin function, folding and amyloid aggregation properties. J Mol Biol. 2008;378:887–897. doi: 10.1016/j.jmb.2008.03.002. [DOI] [PubMed] [Google Scholar]
- 17.Khan AR, Baker BM, Ghosh P, Biddison WE, Wiley DC. The structure and stability of an HLA-A*0201/octameric tax peptide complex with an empty conserved peptide-N-terminal binding site. J Immunol. 2000;164:6398–6405. doi: 10.4049/jimmunol.164.12.6398. [DOI] [PubMed] [Google Scholar]
- 18.Hodkinson JP, Jahn TR, Radford SE, Ashcroft AE. HDX-ESI-MS reveals enhanced conformational dynamics of the amyloidogenic protein beta2-microglobulin upon release from the MHC-1. J Am Soc Mass Spectrom. 2009;20:278–286. doi: 10.1016/j.jasms.2008.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rennella E, Corazza A, Fogolari F, Viglino P, Giorgetti S, Stoppini M, Bellotti V, Esposito G. Equilibrium unfolding thermodynamics of beta2-microglobulin analyzed through native-state H/D exchange. Biophys J. 2009;96:169–179. doi: 10.1529/biophysj.108.142448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rennella E, Corazza A, Giorgetti S, Fogolari F, Viglino P, Porcari R, Verga L, Stoppini M, Bellotti V, Esposito G. Folding and fibrillogenesis: clues from beta2-microglobulin. J Mol Biol. 2010;401:286–297. doi: 10.1016/j.jmb.2010.06.016. [DOI] [PubMed] [Google Scholar]
- 21.Floege J, Bartsch A, Schulze M, Shaldon S, Koch KM, Smeby LC. Clearance and synthesis rates of beta2-microglobulin in patients undergoing hemodialysis and in normal subjects. J Lab Clin Med. 1991;118:153–165. [PubMed] [Google Scholar]
- 22.Floege J, Ketteler M. Beta2-microglobulin-derived amyloidosis: an update. Kidney Int Suppl. 2001;78:S164–S171. doi: 10.1046/j.1523-1755.2001.59780164.x. [DOI] [PubMed] [Google Scholar]
- 23.Otsubo S, Kimata N, Okutsu I, Oshikawa K, Ueda S, Sugimoto H, Mitobe M, Uchida K, Otsubo K, Nitta K, et al. Characteristics of dialysis-related amyloidosis in patients on haemodialysis therapy for more than 30 years. Nephrol Dial Transplant. 2009;24:1593–1598. doi: 10.1093/ndt/gfn706. [DOI] [PubMed] [Google Scholar]
- 24.Gejyo F, Odani S, Yamada T, Honma N, Saito H, Suzuki Y, Nakagawa Y, Kobayashi H, Maruyama Y, Hirasawa Y, et al. Beta2-microglobulin: a new form of amyloid protein associated with chronic hemodialysis. Kidney Int. 1986;30:385–390. doi: 10.1038/ki.1986.196. [DOI] [PubMed] [Google Scholar]
- 25.Eichner T, Radford SE. A generic mechanism of beta2-microglobulin amyloid assembly at neutral pH involving a specific proline switch. J Mol Biol. 2009;386:1312–1326. doi: 10.1016/j.jmb.2009.01.013. [DOI] [PubMed] [Google Scholar]
- 26.Myers SL, Jones S, Jahn TR, Morten IJ, Tennent GA, Hewitt EW, Radford SE. A systematic study of the effect of physiological factors on beta2-microglobulin amyloid formation at neutral pH. Biochemistry. 2006;45:2311–2321. doi: 10.1021/bi052434i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van Ypersele de Strihou C, Jadoul M, Malghem J, Maldague B, Jamart J. Effect of dialysis membrane and patient's age on signs of dialysis-related amyloidosis. The Working Party on Dialysis Amyloidosis. Kidney Int. 1991;39:1012–1019. doi: 10.1038/ki.1991.128. [DOI] [PubMed] [Google Scholar]
- 28.Davison AM. Beta2-microglobulin and amyloidosis: who is at risk? Nephrol Dial Transplant. 1995;10(Suppl. 10):48–51. [PubMed] [Google Scholar]
- 29.Zingraff JJ, Noel LH, Bardin T, Atienza C, Zins B, Drueke TB, Kuntz D. Beta2-microglobulin amyloidosis in chronic renal failure. N Engl J Med. 1990;323:1070–1071. doi: 10.1056/NEJM199010113231514. [DOI] [PubMed] [Google Scholar]
- 30.Moriniere P, Marie A, el Esper N, Fardellone P, Deramond H, Remond A, Sebert JL, Fournier A. Destructive spondyloarthropathy with beta2-microglobulin amyloid deposits in a uremic patient before chronic hemodialysis. Nephron. 1991;59:654–657. doi: 10.1159/000186661. [DOI] [PubMed] [Google Scholar]
- 31.Fuchs D, Norkrans G, Wejstal R, Reibnegger G, Weiss G, Weiland O, Schvarcz R, Fryden A, Wachter H. Changes of serum neopterin, beta2-microglobulin and interferon-gamma in patients with chronic hepatitis C treated with interferon-alpha 2b. Eur J Med. 1992;1:196–200. [PubMed] [Google Scholar]
- 32.Esposito G, Michelutti R, Verdone G, Viglino P, Hernandez H, Robinson CV, Amoresano A, Dal Piaz F, Monti M, Pucci P, et al. Removal of the N-terminal hexapeptide from human beta2-microglobulin facilitates protein aggregation and fibril formation. Protein Sci. 2000;9:831–845. doi: 10.1110/ps.9.5.831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bellotti V, Gallieni M, Giorgetti S, Brancaccio D. Dynamic of beta2-microglobulin fibril formation and reabsorption: the role of proteolysis. Semin Dial. 2001;14:117–122. doi: 10.1046/j.1525-139x.2001.00030.x. [DOI] [PubMed] [Google Scholar]
- 34.Heegaard NH, Roepstorff P, Melberg SG, Nissen MH. Cleaved beta2-microglobulin partially attains a conformation that has amyloidogenic features. J Biol Chem. 2002;277:11184–11189. doi: 10.1074/jbc.M108837200. [DOI] [PubMed] [Google Scholar]
- 35.Miyata T, Taneda S, Kawai R, Ueda Y, Horiuchi S, Hara M, Maeda K, Monnier VM. Identification of pentosidine as a native structure for advanced glycation end products in beta2-microglobulin-containing amyloid fibrils in patients with dialysis-related amyloidosis. Proc Natl Acad Sci USA. 1996;93:2353–2358. doi: 10.1073/pnas.93.6.2353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miyata T, Inagi R, Iida Y, Sato M, Yamada N, Oda O, Maeda K, Seo H. Involvement of beta2-microglobulin modified with advanced glycation end products in the pathogenesis of hemodialysis-associated amyloidosis. Induction of human monocyte chemotaxis and macrophage secretion of tumor necrosis factor-alpha and interleukin-1. J Clin Invest. 1994;93:521–528. doi: 10.1172/JCI117002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Niwa T, Katsuzaki T, Miyazaki S, Momoi T, Akiba T, Miyazaki T, Nokura K, Hayase F, Tatemichi N, Takei Y. Amyloid beta2-microglobulin is modified with imidazolone, a novel advanced glycation end product, in dialysis-related amyloidosis. Kidney Int. 1997;51:187–194. doi: 10.1038/ki.1997.23. [DOI] [PubMed] [Google Scholar]
- 38.Niwa T. Dialysis-related amyloidosis: pathogenesis focusing on AGE modification. Semin Dial. 2001;14:123–126. doi: 10.1046/j.1525-139x.2001.00031.x. [DOI] [PubMed] [Google Scholar]
- 39.Capeillere-Blandin C, Delaveau T, Descamps-Latscha B. Structural modifications of human beta2-microglobulin treated with oxygen-derived radicals. Biochem J. 1991;277(Pt 1):175–182. doi: 10.1042/bj2770175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Odani H, Oyama R, Titani K, Ogawa H, Saito A. Purification and complete amino acid sequence of novel beta2-microglobulin. Biochem Biophys Res Commun. 1990;168:1223–1229. doi: 10.1016/0006-291x(90)91159-p. [DOI] [PubMed] [Google Scholar]
- 41.Relini A, Canale C, De Stefano S, Rolandi R, Giorgetti S, Stoppini M, Rossi A, Fogolari F, Corazza A, Esposito G, et al. Collagen plays an active role in the aggregation of beta2-microglobulin under physiopathological conditions of dialysis-related amyloidosis. J Biol Chem. 2006;281:16521–16529. doi: 10.1074/jbc.M513827200. [DOI] [PubMed] [Google Scholar]
- 42.Relini A, De Stefano S, Torrassa S, Cavalleri O, Rolandi R, Gliozzi A, Giorgetti S, Raimondi S, Marchese L, Verga L, et al. Heparin strongly enhances the formation of beta2-microglobulin amyloid fibrils in the presence of type I collagen. J Biol Chem. 2008;283:4912–4920. doi: 10.1074/jbc.M702712200. [DOI] [PubMed] [Google Scholar]
- 43.Athanasou NA, Puddle B, Sallie B. Highly sulphated glycosaminoglycans in articular cartilage and other tissues containing beta2-microglobulin dialysis amyloid deposits. Nephrol Dial Transplant. 1995;10:1672–1678. [PubMed] [Google Scholar]
- 44.Yamamoto S, Hasegawa K, Yamaguchi I, Goto Y, Gejyo F, Naiki H. Kinetic analysis of the polymerization and depolymerization of beta2-microglobulin-related amyloid fibrils in vitro. Biochim Biophys Acta. 2005;1753:34–43. doi: 10.1016/j.bbapap.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 45.Pepys MB. Amyloidosis. Annu Rev Med. 2006;57:223–241. doi: 10.1146/annurev.med.57.121304.131243. [DOI] [PubMed] [Google Scholar]
- 46.Morgan CJ, Gelfand M, Atreya C, Miranker AD. Kidney dialysis-associated amyloidosis: a molecular role for copper in fiber formation. J Mol Biol. 2001;309:339–345. doi: 10.1006/jmbi.2001.4661. [DOI] [PubMed] [Google Scholar]
- 47.Eakin CM, Miranker AD. From chance to frequent encounters: origins of beta2-microglobulin fibrillogenesis. Biochim Biophys Acta. 2005;1753:92–99. doi: 10.1016/j.bbapap.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 48.Ookoshi T, Hasegawa K, Ohhashi Y, Kimura H, Takahashi N, Yoshida H, Miyazaki R, Goto Y, Naiki H. Lysophospholipids induce the nucleation and extension of beta2-microglobulin-related amyloid fibrils at a neutral pH. Nephrol Dial Transplant. 2008;23:3247–3255. doi: 10.1093/ndt/gfn231. [DOI] [PubMed] [Google Scholar]
- 49.Pal-Gabor H, Gombos L, Micsonai A, Kovacs E, Petrik E, Kovacs J, Graf L, Fidy J, Naiki H, Goto Y, et al. Mechanism of lysophosphatidic acid-induced amyloid fibril formation of beta2-microglobulin in vitro under physiological conditions. Biochemistry. 2009;48:5689–5699. doi: 10.1021/bi900356r. [DOI] [PubMed] [Google Scholar]
- 50.Hasegawa K, Tsutsumi-Yasuhara S, Ookoshi T, Ohhashi Y, Kimura H, Takahashi N, Yoshida H, Miyazaki R, Goto Y, Naiki H. Growth of beta2-microglobulin-related amyloid fibrils by non-esterified fatty acids at a neutral pH. Biochem J. 2008;416:307–315. doi: 10.1042/BJ20080543. [DOI] [PubMed] [Google Scholar]
- 51.Yamamoto S, Hasegawa K, Yamaguchi I, Tsutsumi S, Kardos J, Goto Y, Gejyo F, Naiki H. Low concentrations of sodium dodecyl sulfate induce the extension of beta2-microglobulin-related amyloid fibrils at a neutral pH. Biochemistry. 2004;43:11075–11082. doi: 10.1021/bi049262u. [DOI] [PubMed] [Google Scholar]
- 52.Srikanth R, Mendoza VL, Bridgewater JD, Zhang G, Vachet RW. Copper binding to beta2-microglobulin and its pre-amyloid oligomers. Biochemistry. 2009;48:9871–9881. doi: 10.1021/bi901172y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Calabrese MF, Miranker AD. Metal binding sheds light on mechanisms of amyloid assembly. Prion. 2009;3:1–4. doi: 10.4161/pri.3.1.8601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Blaho DV, Miranker AD. Delineating the conformational elements responsible for Cu(2+)-induced oligomerization of beta2-microglobulin. Biochemistry. 2009;48:6610–6617. doi: 10.1021/bi900540j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Calabrese MF, Eakin CM, Wang JM, Miranker AD. A regulatable switch mediates self-association in an immunoglobulin fold. Nat Struct Mol Biol. 2008;15:965–971. doi: 10.1038/nsmb.1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Antwi K, Mahar M, Srikanth R, Olbris MR, Tyson JF, Vachet RW. Cu(II) organizes beta2-microglobulin oligomers but is released upon amyloid formation. Protein Sci. 2008;17:748–759. doi: 10.1110/ps.073249008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Calabrese MF, Miranker AD. Formation of a stable oligomer of beta2-microglobulin requires only transient encounter with Cu(II) J Mol Biol. 2007;367:1–7. doi: 10.1016/j.jmb.2006.12.034. [DOI] [PubMed] [Google Scholar]
- 58.Eakin CM, Berman AJ, Miranker AD. A native to amyloidogenic transition regulated by a backbone trigger. Nat Struct Mol Biol. 2006;13:202–208. doi: 10.1038/nsmb1068. [DOI] [PubMed] [Google Scholar]
- 59.Deng NJ, Yan L, Singh D, Cieplak P. Molecular basis for the Cu2+ binding-induced destabilization of beta2-microglobulin revealed by molecular dynamics simulation. Biophys J. 2006;90:3865–3879. doi: 10.1529/biophysj.105.064444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yamamoto S, Yamaguchi I, Hasegawa K, Tsutsumi S, Goto Y, Gejyo F, Naiki H. Glycosaminoglycans enhance the trifluoroethanol-induced extension of beta2-microglobulin-related amyloid fibrils at a neutral pH. J Am Soc Nephrol. 2004;15:126–133. doi: 10.1097/01.asn.0000103228.81623.c7. [DOI] [PubMed] [Google Scholar]
- 61.Giorgetti S, Rossi A, Mangione P, Raimondi S, Marini S, Stoppini M, Corazza A, Viglino P, Esposito G, Cetta G, et al. Beta2-microglobulin isoforms display an heterogeneous affinity for type I collagen. Protein Sci. 2005;14:696–702. doi: 10.1110/ps.041194005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Smith DP, Jones S, Serpell LC, Sunde M, Radford SE. A systematic investigation into the effect of protein destabilisation on beta2-microglobulin amyloid formation. J Mol Biol. 2003;330:943–954. doi: 10.1016/s0022-2836(03)00687-9. [DOI] [PubMed] [Google Scholar]
- 63.Armen RS, Daggett V. Characterization of two distinct beta2-microglobulin unfolding intermediates that may lead to amyloid fibrils of different morphology. Biochemistry. 2005;44:16098–16107. doi: 10.1021/bi050731h. [DOI] [PubMed] [Google Scholar]
- 64.Armen RS, DeMarco ML, Alonso DO, Daggett V. Pauling and Corey's alpha-pleated sheet structure may define the prefibrillar amyloidogenic intermediate in amyloid disease. Proc Natl Acad Sci USA. 2004;101:11622–11627. doi: 10.1073/pnas.0401781101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fogolari F, Corazza A, Viglino P, Zuccato P, Pieri L, Faccioli P, Bellotti V, Esposito G. Molecular dynamics simulation suggests possible interaction patterns at early steps of beta2-microglobulin aggregation. Biophys J. 2007;92:1673–1681. doi: 10.1529/biophysj.106.098483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Corazza A, Rennella E, Schanda P, Mimmi MC, Cutuil T, Raimondi S, Giorgetti S, Fogolari F, Viglino P, Frydman L, et al. Native-unlike long-lived intermediates along the folding pathway of the amyloidogenic protein beta2-microglobulin revealed by real-time two-dimensional NMR. J Biol Chem. 2010;285:5827–5835. doi: 10.1074/jbc.M109.061168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jahn TR, Parker MJ, Homans SW, Radford SE. Amyloid formation under physiological conditions proceeds via a native-like folding intermediate. Nat Struct Mol Biol. 2006;13:195–201. doi: 10.1038/nsmb1058. [DOI] [PubMed] [Google Scholar]
- 68.Kameda A, Hoshino M, Higurashi T, Takahashi S, Naiki H, Goto Y. Nuclear magnetic resonance characterization of the refolding intermediate of beta2-microglobulin trapped by non-native prolyl peptide bond. J Mol Biol. 2005;348:383–397. doi: 10.1016/j.jmb.2005.02.050. [DOI] [PubMed] [Google Scholar]
- 69.Sakata M, Chatani E, Kameda A, Sakurai K, Naiki H, Goto Y. Kinetic coupling of folding and prolyl isomerization of beta2-microglobulin studied by mutational analysis. J Mol Biol. 2008;382:1242–1255. doi: 10.1016/j.jmb.2008.08.003. [DOI] [PubMed] [Google Scholar]
- 70.Pawar AP, Dubay KF, Zurdo J, Chiti F, Vendruscolo M, Dobson CM. Prediction of ‘aggregation-prone’ and ‘aggregation-susceptible’ regions in proteins associated with neurodegenerative diseases. J Mol Biol. 2005;350:379–392. doi: 10.1016/j.jmb.2005.04.016. [DOI] [PubMed] [Google Scholar]
- 71.Canet D, Last AM, Tito P, Sunde M, Spencer A, Archer DB, Redfield C, Robinson CV, Dobson CM. Local cooperativity in the unfolding of an amyloidogenic variant of human lysozyme. Nat Struct Biol. 2002;9:308–315. doi: 10.1038/nsb768. [DOI] [PubMed] [Google Scholar]
- 72.Dumoulin M, Canet D, Last AM, Pardon E, Archer DB, Muyldermans S, Wyns L, Matagne A, Robinson CV, Redfield C, et al. Reduced global cooperativity is a common feature underlying the amyloidogenicity of pathogenic lysozyme mutations. J Mol Biol. 2005;346:773–788. doi: 10.1016/j.jmb.2004.11.020. [DOI] [PubMed] [Google Scholar]
- 73.Plakoutsi G, Taddei N, Stefani M, Chiti F. Aggregation of the acylphosphatase from Sulfolobus solfataricus: the folded and partially unfolded states can both be precursors for amyloid formation. J Biol Chem. 2004;279:14111–14119. doi: 10.1074/jbc.M312961200. [DOI] [PubMed] [Google Scholar]
- 74.Plakoutsi G, Bemporad F, Calamai M, Taddei N, Dobson CM, Chiti F. Evidence for a mechanism of amyloid formation involving molecular reorganisation within native-like precursor aggregates. J Mol Biol. 2005;351:910–922. doi: 10.1016/j.jmb.2005.06.043. [DOI] [PubMed] [Google Scholar]
- 75.Colon W, Kelly JW. Partial denaturation of transthyretin is sufficient for amyloid fibril formation in vitro. Biochemistry. 1992;31:8654–8660. doi: 10.1021/bi00151a036. [DOI] [PubMed] [Google Scholar]
- 76.Lai Z, Colon W, Kelly JW. The acid-mediated denaturation pathway of transthyretin yields a conformational intermediate that can self-assemble into amyloid. Biochemistry. 1996;35:6470–6482. doi: 10.1021/bi952501g. [DOI] [PubMed] [Google Scholar]
- 77.Liu K, Cho HS, Lashuel HA, Kelly JW, Wemmer DE. A glimpse of a possible amyloidogenic intermediate of transthyretin. Nat Struct Biol. 2000;7:754–757. doi: 10.1038/78980. [DOI] [PubMed] [Google Scholar]
- 78.Elam JS, Taylor AB, Strange R, Antonyuk S, Doucette PA, Rodriguez JA, Hasnain SS, Hayward LJ, Valentine JS, Yeates TO, et al. Amyloid-like filaments and water-filled nanotubes formed by SOD1 mutant proteins linked to familial ALS. Nat Struct Biol. 2003;10:461–467. doi: 10.1038/nsb935. [DOI] [PubMed] [Google Scholar]
- 79.Banci L, Bertini I, D'Amelio N, Gaggelli E, Libralesso E, Matecko I, Turano P, Valentine JS. Fully metallated S134N Cu,Zn-superoxide dismutase displays abnormal mobility and intermolecular contacts in solution. J Biol Chem. 2005;280:35815–35821. doi: 10.1074/jbc.M506637200. [DOI] [PubMed] [Google Scholar]
- 80.Nordlund A, Oliveberg M. Folding of Cu/Zn superoxide dismutase suggests structural hotspots for gain of neurotoxic function in ALS: parallels to precursors in amyloid disease. Proc Natl Acad Sci USA. 2006;103:10218–10223. doi: 10.1073/pnas.0601696103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chiti F, Mangione P, Andreola A, Giorgetti S, Stefani M, Dobson CM, Bellotti V, Taddei N. Detection of two partially structured species in the folding process of the amyloidogenic protein beta2-microglobulin. J Mol Biol. 2001;307:379–391. doi: 10.1006/jmbi.2000.4478. [DOI] [PubMed] [Google Scholar]
- 82.Chiti F, De Lorenzi E, Grossi S, Mangione P, Giorgetti S, Caccialanza G, Dobson CM, Merlini G, Ramponi G, Bellotti V. A partially structured species of beta2-microglobulin is significantly populated under physiological conditions and involved in fibrillogenesis. J Biol Chem. 2001;276:46714–46721. doi: 10.1074/jbc.M107040200. [DOI] [PubMed] [Google Scholar]
- 83.Heegaard NH, Sen JW, Kaarsholm NC, Nissen MH. Conformational intermediate of the amyloidogenic protein beta2-microglobulin at neutral pH. J Biol Chem. 2001;276:32657–32662. doi: 10.1074/jbc.M104452200. [DOI] [PubMed] [Google Scholar]
- 84.Benyamini H, Gunasekaran K, Wolfson H, Nussinov R. Beta2-microglobulin amyloidosis: insights from conservation analysis and fibril modelling by protein docking techniques. J Mol Biol. 2003;330:159–174. doi: 10.1016/s0022-2836(03)00557-6. [DOI] [PubMed] [Google Scholar]
- 85.Goto Y, Azuma T, Hamaguchi K. Refolding of the immunoglobulin light chain. J Biochem. 1979;85:1427–1438. doi: 10.1093/oxfordjournals.jbchem.a132470. [DOI] [PubMed] [Google Scholar]
- 86.Goto Y, Hamaguchi K. Unfolding and refolding of the reduced constant fragment of the immunoglobulin light chain. Kinetic role of the intrachain disulfide bond. J Mol Biol. 1982;156:911–926. doi: 10.1016/0022-2836(82)90147-4. [DOI] [PubMed] [Google Scholar]
- 87.Goto Y, Hamaguchi K. Unfolding and refolding of the constant fragment of the immunoglobulin light chain. J Mol Biol. 1982;156:891–910. doi: 10.1016/0022-2836(82)90146-2. [DOI] [PubMed] [Google Scholar]
- 88.Thies MJ, Mayer J, Augustine JG, Frederick CA, Lilie H, Buchner J. Folding and association of the antibody domain CH3: prolyl isomerization precedes dimerization. J Mol Biol. 1999;293:67–79. doi: 10.1006/jmbi.1999.3128. [DOI] [PubMed] [Google Scholar]
- 89.Feige MJ, Walter S, Buchner J. Folding mechanism of the CH2 antibody domain. J Mol Biol. 2004;344:107–118. doi: 10.1016/j.jmb.2004.09.033. [DOI] [PubMed] [Google Scholar]
- 90.Feige MJ, Groscurth S, Marcinowski M, Yew ZT, Truffault V, Paci E, Kessler H, Buchner J. The structure of a folding intermediate provides insight into differences in immunoglobulin amyloidogenicity. Proc Natl Acad Sci USA. 2008;105:13373–13378. doi: 10.1073/pnas.0802809105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Feige MJ, Groscurth S, Marcinowski M, Shimizu Y, Kessler H, Hendershot LM, Buchner J. An unfolded CH1 domain controls the assembly and secretion of IgG antibodies. Mol Cell. 2009;34:569–579. doi: 10.1016/j.molcel.2009.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Villanueva J, Hoshino M, Katou H, Kardos J, Hasegawa K, Naiki H, Goto Y. Increase in the conformational flexibility of beta2-microglobulin upon copper binding: a possible role for copper in dialysis-related amyloidosis. Protein Sci. 2004;13:797–809. doi: 10.1110/ps.03445704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Borysik AJ, Morten IJ, Radford SE, Hewitt EW. Specific glycosaminoglycans promote unseeded amyloid formation from beta2-microglobulin under physiological conditions. Kidney Int. 2007;72:174–181. doi: 10.1038/sj.ki.5002270. [DOI] [PubMed] [Google Scholar]
- 94.Kihara M, Chatani E, Sakai M, Hasegawa K, Naiki H, Goto Y. Seeding-dependent maturation of beta2-microglobulin amyloid fibrils at neutral pH. J Biol Chem. 2005;280:12012–12018. doi: 10.1074/jbc.M411949200. [DOI] [PubMed] [Google Scholar]
- 95.Ohhashi Y, Kihara M, Naiki H, Goto Y. Ultrasonication-induced amyloid fibril formation of beta2-microglobulin. J Biol Chem. 2005;280:32843–32848. doi: 10.1074/jbc.M506501200. [DOI] [PubMed] [Google Scholar]
- 96.Sasahara K, Yagi H, Naiki H, Goto Y. Heat-induced conversion of beta2-microglobulin and hen egg-white lysozyme into amyloid fibrils. J Mol Biol. 2007;372:981–991. doi: 10.1016/j.jmb.2007.06.088. [DOI] [PubMed] [Google Scholar]
- 97.Sasahara K, Yagi H, Sakai M, Naiki H, Goto Y. Amyloid nucleation triggered by agitation of beta2-microglobulin under acidic and neutral pH conditions. Biochemistry. 2008;47:2650–2660. doi: 10.1021/bi701968g. [DOI] [PubMed] [Google Scholar]
- 98.Jones S, Smith DP, Radford SE. Role of the N and C-terminal strands of beta2-microglobulin in amyloid formation at neutral pH. J Mol Biol. 2003;330:935–941. doi: 10.1016/s0022-2836(03)00688-0. [DOI] [PubMed] [Google Scholar]
- 99.Piazza R, Pierno M, Iacopini S, Mangione P, Esposito G, Bellotti V. Micro-heterogeneity and aggregation in beta2-microglobulin solutions: effects of temperature, pH, and conformational variant addition. Eur Biophys J. 2006;35:439–445. doi: 10.1007/s00249-006-0051-0. [DOI] [PubMed] [Google Scholar]
- 100.Giorgetti S, Stoppini M, Tennent GA, Relini A, Marchese L, Raimondi S, Monti M, Marini S, Ostergaard O, Heegaard NH, et al. Lysine 58-cleaved beta2-microglobulin is not detectable by 2D electrophoresis in ex vivo amyloid fibrils of two patients affected by dialysis-related amyloidosis. Protein Sci. 2007;16:343–349. doi: 10.1110/ps.062563507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Colombo M, Ricagno S, Barbiroli A, Santambrogio C, Giorgetti S, Raimondi S, Bonomi F, Grandori R, Bellotti V, Bolognesi M. The effects of an ideal beta-turn on beta2-microglobulin fold stability. J Biochem. 2011 doi: 10.1093/jb/mvr034. in press. doi: 10.1093/jb/mvr034. [DOI] [PubMed] [Google Scholar]
- 102.Heegaard NH, Jorgensen TJ, Rozlosnik N, Corlin DB, Pedersen JS, Tempesta AG, Roepstorff P, Bauer R, Nissen MH. Unfolding, aggregation, and seeded amyloid formation of lysine-58-cleaved beta2-microglobulin. Biochemistry. 2005;44:4397–4407. doi: 10.1021/bi047594t. [DOI] [PubMed] [Google Scholar]
- 103.Mimmi MC, Jorgensen TJ, Pettirossi F, Corazza A, Viglino P, Esposito G, De Lorenzi E, Giorgetti S, Pries M, Corlin DB, et al. Variants of beta2-microglobulin cleaved at lysine-58 retain the main conformational features of the native protein but are more conformationally heterogeneous and unstable at physiological temperature. FEBS J. 2006;273:2461–2474. doi: 10.1111/j.1742-4658.2006.05254.x. [DOI] [PubMed] [Google Scholar]
- 104.Corlin DB, Johnsen CK, Nissen MH, Heegaard NH. A beta2-microglobulin cleavage variant fibrillates at near-physiological pH. Biochem Biophys Res Commun. 2009;381:187–191. doi: 10.1016/j.bbrc.2009.02.041. [DOI] [PubMed] [Google Scholar]
- 105.Miura Y, Ishiyama T, Inomata A, Takeda T, Senma S, Okuyama K, Suzuki Y. Radiolucent bone cysts and the type of dialysis membrane used in patients undergoing long-term hemodialysis. Nephron. 1992;60:268–273. doi: 10.1159/000186764. [DOI] [PubMed] [Google Scholar]
- 106.Eakin CM, Attenello FJ, Morgan CJ, Miranker AD. Oligomeric assembly of native-like precursors precedes amyloid formation by beta2-microglobulin. Biochemistry. 2004;43:7808–7815. doi: 10.1021/bi049792q. [DOI] [PubMed] [Google Scholar]
- 107.Eakin CM, Knight JD, Morgan CJ, Gelfand MA, Miranker AD. Formation of a copper specific binding site in non-native states of beta2-microglobulin. Biochemistry. 2002;41:10646–10656. doi: 10.1021/bi025944a. [DOI] [PubMed] [Google Scholar]
- 108.Mendoza VL, Antwi K, Baron-Rodriguez MA, Blanco C, Vachet RW. Structure of the preamyloid dimer of beta2-microglobulin from covalent labeling and mass spectrometry. Biochemistry. 2010;49:1522–1532. doi: 10.1021/bi901748h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kameda A, Morita EH, Sakurai K, Naiki H, Goto Y. NMR-based characterization of a refolding intermediate of beta2-microglobulin labeled using a wheat germ cell-free system. Protein Sci. 2009;18:1592–1601. doi: 10.1002/pro.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Booth DR, Sunde M, Bellotti V, Robinson CV, Hutchinson WL, Fraser PE, Hawkins PN, Dobson CM, Radford SE, Blake CC, et al. Instability, unfolding and aggregation of human lysozyme variants underlying amyloid fibrillogenesis. Nature. 1997;385:787–793. doi: 10.1038/385787a0. [DOI] [PubMed] [Google Scholar]
- 111.Moraitakis G, Goodfellow JM. Simulations of human lysozyme: probing the conformations triggering amyloidosis. Biophys J. 2003;84:2149–2158. doi: 10.1016/S0006-3495(03)75021-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kelly JW. The alternative conformations of amyloidogenic proteins and their multi-step assembly pathways. Curr Opin Struct Biol. 1998;8:101–106. doi: 10.1016/s0959-440x(98)80016-x. [DOI] [PubMed] [Google Scholar]
- 113.Marcon G, Plakoutsi G, Canale C, Relini A, Taddei N, Dobson CM, Ramponi G, Chiti F. Amyloid formation from HypF-N under conditions in which the protein is initially in its native state. J Mol Biol. 2005;347:323–335. doi: 10.1016/j.jmb.2005.01.034. [DOI] [PubMed] [Google Scholar]
- 114.Khare SD, Dokholyan NV. Common dynamical signatures of familial amyotrophic lateral sclerosis-associated structurally diverse Cu, Zn superoxide dismutase mutants. Proc Natl Acad Sci USA. 2006;103:3147–3152. doi: 10.1073/pnas.0511266103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Chiti F, Dobson CM. Protein misfolding, functional amyloid, and human disease. Annu Rev Biochem. 2006;75:333–366. doi: 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- 116.Chiti F, Dobson CM. Amyloid formation by globular proteins under native conditions. Nat Chem Biol. 2009;5:15–22. doi: 10.1038/nchembio.131. [DOI] [PubMed] [Google Scholar]
- 117.Corlin DB, Sen JW, Ladefoged S, Lund GB, Nissen MH, Heegaard NH. Quantification of cleaved beta2-microglobulin in serum from patients undergoing chronic hemodialysis. Clin Chem. 2005;51:1177–1184. doi: 10.1373/clinchem.2005.049544. [DOI] [PubMed] [Google Scholar]
- 118.Heegaard NH, Jorgensen TJ, Cheng L, Schou C, Nissen MH, Trapp O. Interconverting conformations of variants of the human amyloidogenic protein beta2-microglobulin quantitatively characterized by dynamic capillary electrophoresis and computer simulation. Anal Chem. 2006;78:3667–3673. doi: 10.1021/ac060194m. [DOI] [PubMed] [Google Scholar]
- 119.Yamaguchi K, Naiki H, Goto Y. Mechanism by which the amyloid-like fibrils of a beta2-microglobulin fragment are induced by fluorine-substituted alcohols. J Mol Biol. 2006;363:279–288. doi: 10.1016/j.jmb.2006.08.030. [DOI] [PubMed] [Google Scholar]
- 120.Bellotti V, Stoppini M, Mangione P, Sunde M, Robinson C, Asti L, Brancaccio D, Ferri G. Beta2-microglobulin can be refolded into a native state from ex vivo amyloid fibrils. Eur J Biochem. 1998;258:61–67. doi: 10.1046/j.1432-1327.1998.2580061.x. [DOI] [PubMed] [Google Scholar]
- 121.Santambrogio C, Ricagno S, Colombo M, Barbiroli A, Bonomi F, Bellotti V, Bolognesi M, Grandori R. DE-loop mutations affect beta2-microglobulin stability, oligomerization, and the low-pH unfolded form. Protein Sci. 2010;19:1386–1394. doi: 10.1002/pro.419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Fogolari F, Corazza A, Varini N, Rotter M, Gumral D, Codutti L, Rennella E, Viglino P, Bellotti V, Esposito G. Molecular dynamics simulation of beta2-microglobulin in denaturing and stabilizing conditions. Proteins. 2011;79:986–1001. doi: 10.1002/prot.22940. [DOI] [PubMed] [Google Scholar]
- 123.Monti M, Amoresano A, Giorgetti S, Bellotti V, Pucci P. Limited proteolysis in the investigation of beta2-microglobulin amyloidogenic and fibrillar states. Biochim Biophys Acta. 2005;1753:44–50. doi: 10.1016/j.bbapap.2005.09.004. [DOI] [PubMed] [Google Scholar]
- 124.Liu C, Sawaya MR, Eisenberg D. Beta-microglobulin forms three-dimensional domain-swapped amyloid fibrils with disulfide linkages. Nat Struct Mol Biol. 2010;18:49–55. doi: 10.1038/nsmb.1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Domanska K, Vanderhaegen S, Srinivasan V, Pardon E, Dupeux F, Marquez JA, Giorgetti S, Stoppini M, Wyns L, Bellotti V, et al. Atomic structure of a nanobody-trapped domain-swapped dimer of an amyloidogenic beta2-microglobulin variant. Proc Natl Acad Sci USA. 2011;108:1314–1319. doi: 10.1073/pnas.1008560108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Chen Y, Dokholyan NV. A single disulfide bond differentiates aggregation pathways of beta2-microglobulin. J Mol Biol. 2005;354:473–482. doi: 10.1016/j.jmb.2005.09.075. [DOI] [PubMed] [Google Scholar]
- 127.Stoppini M, Mangione P, Monti M, Giorgetti S, Marchese L, Arcidiaco P, Verga L, Segagni S, Pucci P, Merlini G, et al. Proteomics of beta2-microglobulin amyloid fibrils. Biochim Biophys Acta. 2005;1753:23–33. doi: 10.1016/j.bbapap.2005.07.019. [DOI] [PubMed] [Google Scholar]
- 128.Eichner T, Radford SE. A diversity of assembly mechanisms for a generic amyloid fold. Mol Cell. 2011 doi: 10.1016/j.molcel.2011.05.012. in press. [DOI] [PubMed] [Google Scholar]
- 129.Brundin P, Melki R, Kopito R. Prion-like transmission of protein aggregates in neurodegenerative diseases. Nat Rev Mol Cell Biol. 2010;11:301–307. doi: 10.1038/nrm2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Sindi SS, Serio TR. Prion dynamics and the quest for the genetic determinant in protein-only inheritance. Curr Opin Microbiol. 2009;12:623–630. doi: 10.1016/j.mib.2009.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Miller G. Neurodegeneration. Could they all be prion diseases? Science. 2009;326:1337–1339. doi: 10.1126/science.326.5958.1337. [DOI] [PubMed] [Google Scholar]
- 132.Ladner CL, Chen M, Smith DP, Platt GW, Radford SE, Langen R. Stacked sets of parallel, in-register beta-strands of beta2-microglobulin in amyloid fibrils revealed by site-directed spin labeling and chemical labeling. J Biol Chem. 2010;285:17137–17147. doi: 10.1074/jbc.M110.117234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Debelouchina GT, Platt GW, Bayro MJ, Radford SE, Griffin RG. Magic angle spinning NMR analysis of beta2-microglobulin amyloid fibrils in two distinct morphologies. J Am Chem Soc. 2010;132:10414–10423. doi: 10.1021/ja102775u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Debelouchina GT, Platt GW, Bayro MJ, Radford SE, Griffin RG. Intermolecular alignment in beta2-microglobulin amyloid fibrils. J Am Chem Soc. 2010;132:17077–17079. doi: 10.1021/ja107987f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Jahn TR, Tennent GA, Radford SE. A common beta-sheet architecture underlies in vitro and in vivo beta2-microglobulin amyloid fibrils. J Biol Chem. 2008;283:17279–17286. doi: 10.1074/jbc.M710351200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Borbulevych OY, Do P, Baker BM. Structures of native and affinity-enhanced WT1 epitopes bound to HLA-A*0201: implications for WT1-based cancer therapeutics. Mol Immunol. 2010;47:2519–2524. doi: 10.1016/j.molimm.2010.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Kihara M, Chatani E, Iwata K, Yamamoto K, Matsuura T, Nakagawa A, Naiki H, Goto Y. Conformation of amyloid fibrils of beta2-microglobulin probed by tryptophan mutagenesis. J Biol Chem. 2006;281:31061–31069. doi: 10.1074/jbc.M605358200. [DOI] [PubMed] [Google Scholar]
- 138.Hodkinson JP, Ashcroft AE, Radford SE. Springer; 2011. Protein misfolding and toxicity in dialysis related amyloidosis. Prefibrillar amyloidogenic protein assemblies – common cytotoxins underlying degenerative diseases. in press. [Google Scholar]
- 139.White HE, Hodgkinson JL, Jahn TR, Cohen-Krausz S, Gosal WS, Muller S, Orlova EV, Radford SE, Saibil HR. Globular tetramers of beta2-microglobulin assemble into elaborate amyloid fibrils. J Mol Biol. 2009;389:48–57. doi: 10.1016/j.jmb.2009.03.066. [DOI] [PMC free article] [PubMed] [Google Scholar]


