Abstract
When screening a brain cDNA library, we found that the N-methyl-d-aspartate receptor subunit NR3A binds to microtubule-associated protein (MAP) 1S/chromosome 19 open reading frame 5 (C19ORF5). The interaction was confirmed in vitro and in vivo, and binding of MAP1S was localized to the membrane-proximal part of the NR3A C-terminus. MAP1S belongs to the same family as MAP1A and MAP1B, and was found to be abundant in both postnatal and adult rat brain. In hippocampal neurons the distribution-pattern of MAP1S resembled that of β-tubulin III, but a fraction of the protein colocalized with synaptic markers synapsin and postsynaptic density protein 95 (PSD95), in β-tubulin III-negative filopodia-like protrusions. There was coexistance between MAP1S and NR3A immunoreactivity in neurite shafts and occasionally in filopodia-like processes. MAP1S potentially links NR3A to the cytoskeleton, and may stabilize NR3A-containing receptors at the synapse and regulate their movement between synaptic and extrasynaptic sites.
Keywords: N-Methyl-d-aspartate receptor, NR3A, Microtubule associated-protein, MAP1S, C19ORF5
The N-methyl-D-aspartate receptor (NMDAR) is most likely a tetrameric assembly of different combinations of the subunits NR1, NR2A-D and NR3A-B, and is ubiquitously expressed in neurons of the mammalian brain (reviewed in [1]). The human NR3A subunit has been cloned [2,3] and shown to binds glycine with a similar affinity as NR1 [4]. When NR3A assembles with NR1 and NR2 they form receptors with lower unitary conductance, reduced Ca2+ permeability and altered open time, compared to receptors composed only of NR1 and NR2 [5,6]. In agreement with NR3A’s attenuating effect, NR3A knockout mice show increased NMDAR currents [5]. In addition, the mice have an increased density of dendritic spines, a phenotype that is most pronounced during the postnatal period when NR3A expression levels are normally high [7]. In contrast to the other NMDAR subunits, NR3A is to a large extent expressed at perisynaptic and extrasynaptic sites and appears to be more loosely associated with the postsynaptic density [8]. Recently, PACSIN1/syndapin1 was reported to associate with NR3A and mediate endocytosis of NR3A-containing NMDARs in an activity-dependent manner [8]. This suggests that the expression of NR3A at the cell surface is regulated differently from the other NMDAR subunits. Since PAC-SIN1 does not bind to the other NMDAR subunits this interaction might facilitate the removal of NR3A-containing receptors from the synapse as the brain matures [8]. Moreover, protein phosphatase 2A (PP2A) has been reported to interact with NR3A in the rat brain and this association can regulate the phosphorylation state of NR1 [9].
The NMDAR is transported along microtubules and NR2B-containing vesicles are linked to the motor-protein kinesin superfamily protein 17 (KIF17), which transports cargo through the dendrites [10]. Depolymerization of microtubules, or knock-down of KIF17 expression, inhibits NMDAR-mediated currents in cortical neurons [11]. These data suggest that an intact microtubular cytoskeleton is essential for NMDAR transport and function. NMDARs are also indirectly linked to microtubules through MAP1A, which interacts with the NMDAR-binding protein postsynaptic density protein 93 (PSD93)/Chapsyn-110 [12]. PSD93 belongs to the membrane-associated guanylate kinase (MAGUK) family, which also comprises PSD95 and synapse associated proteins 102 and 97 (SAP102 and SAP97). The MAGUKs are scaffolding proteins and have been shown to be important for regulating clustering and stabilization of glutamate receptors at synaptic sites (reviewed in [13]). Moreover, SAP102, which is the major MAGUK in developing neurons, is involved in NMDAR trafficking through its interactions with the exo-cyst complex [14].
In this study, we show a novel interaction between NR3A and MAP1S [15] in the mammalian brain. MAP1S was originally identified by Liu and McKeehan who termed the protein C19ORF5 and suggested it to be a member of the MAP family [16]. We show that MAP1S is widely expressed in the central nervous system (CNS), predominately in neurons, where its expression pattern partly overlaps with that of NR3A.
Materials and methods
Yeast two-hybrid screen
Matchmaker Gal4 Two-Hybrid System 3 (Clontech) was used according to the manufacturer’s instructions, to screen a pretransformed fetal, human brain cDNA library, for proteins interacting with the intracellular tail of human NR3A (amino acids 952–1115). Truncations of the NR3A bait were generated with the Quick-Change Site-directed mutagenesis Kit from Stratagene, and direct mating used to determine which part of the NR3A C-terminus is involved in the MAP1S interaction.
GST pull-down
The MAP1S-encoding cDNA, isolated in the yeast screen, was subcloned into pGEX-6P-2 (GE Healthcare) and expressed as a GST fusion protein in E. Coli BL-21 Gold (Stratagene). At log-phase, 100 µM isopropyl β-d-thiogalactoside (IPTG) was added and the culture incubated at 30 °C for 1 h. Cells were resuspended in PBS with Complete™ protease inhibitor cocktail (Roche), sonicated on ice and the supernatant used for the pull-down assay. After confirming protein expression by Western blot, equal amounts of GST-MAP1S or GST were bound to 100 µl Glutathione–Sepharose 4B beads (GE Healthcare). The beads were rinsed 2× in PBS-0.5% Triton X-100 and full-length human NR3A, from transfected HEK293 cells, added and mixed at 4 °C overnight. Finally, the beads were washed 5× in PBS-0.5% Triton X-100, resuspended in sample buffer, boiled and proteins analyzed by Western blot.
Immunoprecipitation
Eight-day-old Sprague–Dawley rats were decapitated and forebrains homogenized in ice-cold buffer (0.32 M sucrose, 4 mM Hepes, pH 7.4, and protease inhibitors). The homogenate was centrifuged at 950g, the pellet resuspended in homogenization buffer, the centrifugation repeated and the supernatants pooled. The pooled supernatants were centrifuged at 25,400g, the pellet resuspended in 50 mM Tris–HCl, pH 9.0, 1% DOC, and solubilized for 1 h at 37 °C under continuous shaking, and the mixture was centrifuged at 100,000g. The supernatant was dialyzed in 50 mM Tris–HCl, pH 7.5, 100 mM NaCl, 1 mM EDTA with protease inhibitors. Solubilized homogenate was diluted 10× in PBS and precleared for 1 h at room temperature (RT) with 10 µl of goat-anti-mouse IgG Dynabeads (Dynal). Three micrograms of MAP1S or NR3A antibody was added to the precleared homogenate, the mixture incubated overnight at 4 °C, transferred to RT and incubated with 40 µl Dynabeads for 2 h. Finally, the beads were washed in RIPA buffer and 3× in PBS, 0.5% Triton X-100, resuspended in sample buffer, boiled and proteins analyzed by Western blot.
Western blot
Rat and human tissue were homogenized in cold buffer (130 µM Tris–HCl, pH 6.8, 2% SDS, and 10% glycerol) and protein concentrations were determined using the BCA Protein Assay Kit (Pierce). Samples were separated by SDS–PAGE on 7% Tris–glycine gels, transferred to nitrocellulose membranes, incubated with primary antibody overnight and with secondary antibody for 1 h, both at RT. CCD camera LAS-3000 (Fuji-film) was used for detection and band intensities quantified using Multi Gauge v3.0 (Fuji film). Rat tissue was obtained from decapitated adult and eight-day-old Sprague–Dawley rats and adult human brain tissue was acquired from Netherlands Brain Bank. Embryonic human tissue was collected during routine, first trimester abortions and written consent had been given by the pregnant women. The use of human tissue was performed in compliance with Dutch and Swedish law and procedures involving human and animal tissue were approved by the Regional Ethical Committee in Stockholm and by the Ethical Committee on Animal Research.
Cell cultures and immunocytochemistry
Hippocampus and cerebral cortex were dissected from 18-day-old rat fetuses and from nine week old human embryos, 26,000 cells/well plated on poly-d-lysine-coated cover slips and grown in Neurobasal media with B27 supplement and 0.5 mM l-glutamine. At 10 days in vitro cells were transfected with 2 µg of human MAP1S-EGFP cDNA in pEGFP-C3 (Clontech) using Lipofectamine 2000 prepared in DMEM media without serum. Cerebrocortical cells were cultured for two weeks in either Neurobasal media as above, to select for neuronal cells, or in DMEM/F-12 with Glutamax®, N2 supplement and 10% fetal calf serum, to allow glial cell expansion. HEK293 cells were grown in RPMI 1640 media with Glutamax® and 10% fetal calf serum and transfected using Fugene 6 (Roche). All other media and reagents were purchased from Gibco Invitrogen Corporation. At 14 days in vitro, hippocampal cells were fixed with 4% paraformaldehyde, 10 min at 4 °C, washed with PBS and permeabilized with PBS-0.2% Triton X-100 at RT for 10 min. Blocking was done with 1.5% normal goat serum at RT for 30 min and cells incubated with primary antibody overnight at 4 °C. The next morning, cells were washed in PBS, incubated 1 h at RT with secondary antibody and washed again. The antibodies were diluted in PBS-0.3% Triton X-100.
Antibodies
Primary antibodies used for Western blot (WB) and immunocytochemistry (ICC): GST 1:10,000 (GE Healthcare); NR3A 1:4000 for WB, 1:500 for ICC (custom made by Zymed Invitrogen Corporation, raised against the 15C-terminal amino acids of human NR3A); NR1 1:500 for WB, 1:300 for ICC (54.1, BD Pharmingen); GluR2 1:500 (MAB 397, Chemicon); MAP1S 1:4000 (mAb4G1[21]); tubulin 1:2000 (YL1/2, Abcam); β-tubulin neuronal class III 1:500 (Biosite); PSD-95 1:2000 (K28/43, Upstate Cell Signaling Solutions); synapsin 1:300 (Molecular Probes); GFAP 1:1000 (Promega); actin 1:1000 (AC-40, Abcam). Secondary antibodies used for WB and ICC: Horseradish peroxidase linked IgG (GE Healthcare), Cy™3-conjugated IgG 1:2000 (Jackson Immuno Research) and AlexaFluor®488-conjugated IgG 1:1400 (Molecular Probes). As negative control for Immunoprecipitation, 3 µg of normal rabbit IgG (R&D Systems) was used.
Results and discussion
NR3A binds to MAP1S in vitro and in rat brain
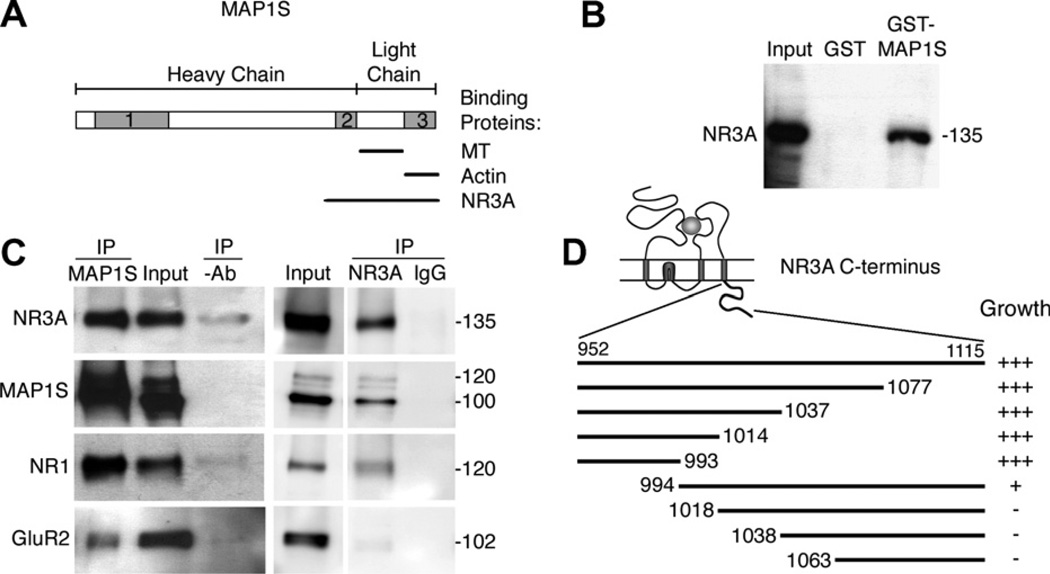
We used the yeast two-hybrid system to screen a fetal, human brain cDNA library for proteins binding to the intracellular C-terminus of human NR3A. One of the isolated clones was shown to encode MAP1S [15]/C19ORF5 [16], a member of the MAP1 family, previously also termed variable charge Y chromosome 2 interacting protein 1 (VCY2IP-1) [17] and RAS-associated domain family protein 1A-binding protein 1 (RAB1) [18]. The MAP1S protein, is considerably smaller but shows high over-all sequence homology to MAP1A (42%) and MAP1B (59%), and contains three short regions termed MAP1 homology domains [15], where the sequence is more highly conserved, (Fig. 1A) (reviewed in [19]). The sequence we isolated from the cDNA library encodes the microtubule and actin binding light chain, and the C-terminal part of the heavy chain of MAP1S. To confirm the interaction between NR3A and MAP1S we performed in vitro GST pull-down experiments. Full-length NR3A protein was precipitated by GST-MAP1S but not by the GST-tag alone (Fig. 1B). This shows that NR3A and MAP1S specifically bind to each other in vitro.
Fig. 1.
NR3A binds to MAP1S both in vitro and in vivo. (A) Schematic picture of MAP1S. The full-length protein is post-translationally cleaved into heavy and light chains. Shaded boxes 1, 2, and 3 indicate the MAP1 homology domains [15]. The microtubule (MT), actin and NR3A-binding regions are indicated. (B) NR3A was coprecipitated with GST-MAP1S but not with the GST-tag alone. (C) NR3A, NR1 and small amounts of GluR2 were coimmunoprecipitated with MAP1S. MAP1S and NR1, but not GluR2 were coimmunoprecipitated with NR3A. (D) Schematic picture of NR3A showing the different parts of NR3A tested for interaction with MAP1S. Abundant, sparse or no growth of colonies is indicated by +++, +, and −. Input in B and C corresponds to 5% of the protein used in the experiments.
To investigate if the interaction between NR3A and MAP1S occurs in vivo, we solubilized brain membranes and their associated protein complexes from eight-day old rats, and performed coimmunoprecipitation experiments. Both NR1 and NR3A coprecipitated with MAP1S, indicating that NR3A and MAP1S interact in vivo, and that NR1 is assembled with NR3A when the interaction with MAP1S occurs (Fig. 1C). We were equally able to coprecipitate MAP1S together with NR3A, further confirming the interaction between the two proteins. The α-amino-3-hydroxy-5-methyl-4-isoxazole propionic acid (AMPA) receptor subunit GluR2 did not coprecipitate with NR3A, although small amounts were coprecipitated with MAP1S.
To determine which part of the NR3A C-terminus that interacts with MAP1S we constructed different N- and C-terminal truncations of the NR3A intracellular tail (Fig. 1D). We found the membrane proximal part of the C-terminus to be essential for MAP1S-binding. Amino acids 952–993 bound MAP1S approximately as well as the full-length C-terminus, while amino acids 994–1115 gave rise only to a week interaction. The interaction between NR3A and PP2A involves the membrane-proximal 37 amino acids of the NR3A intracellular tail [20]. This region overlaps with the sequence that binds to MAP1S, suggesting that MAP1S and PP2A compete for the same binding region. In contrast, the full-length intracellular tail is required to bind PACSIN1 [8].
Expression of MAP1S in cells and tissue
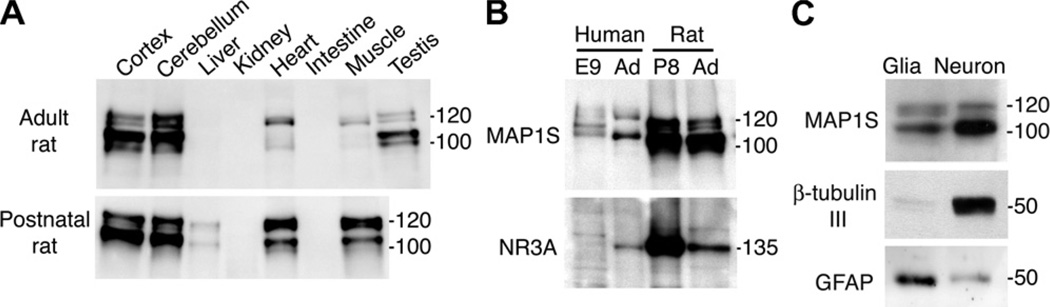
To date there is only one published study on the expression of MAP1S protein in different tissues, and this was performed in the adult mouse [15]. Considering that the highest levels of NR3A in the rodent brain are seen during the second postnatal week, we examined the levels of MAP1S protein in postnatal rat and compared them to those of the adult. In both the postnatal and adult rat the highest levels of MAP1S protein was found in cerebral cortex and cerebellum (Fig. 2A). MAP1S protein was also detected in heart and skeletal muscle at both ages, and in testis of the adult animal. Low expression could also be seen in postnatal, but not adult, liver. However, neither kidney nor intestines expressed detectable amounts of MAP1S protein. In brain and testis the levels of proteolytically cleaved heavy chain (100 kDa) predominated over the full-length precursor protein (120 kDa). In heart and skeletal muscle the relationship was the reverse, and in liver equal amounts of the two forms were present. Equal amounts of protein were loaded in all wells and actin used as a loading control (data not shown).
Fig. 2.
Expression of MAP1S protein. (A) MAP1S-expression in organs of adult and eight-day-old rat. (B) Expression of MAP1S and NR3A at embryonic week nine (E9) and in adult (Ad) human forebrain and in postnatal day eight (P8) and adult rat forebrain. (C) Expression of MAP1S in glia and neurons. The neuronal marker β-tubulin III and the astrocyte marker glial fibrillarly acidic protein (GFAP), were used to check the purity of the cell cultures.
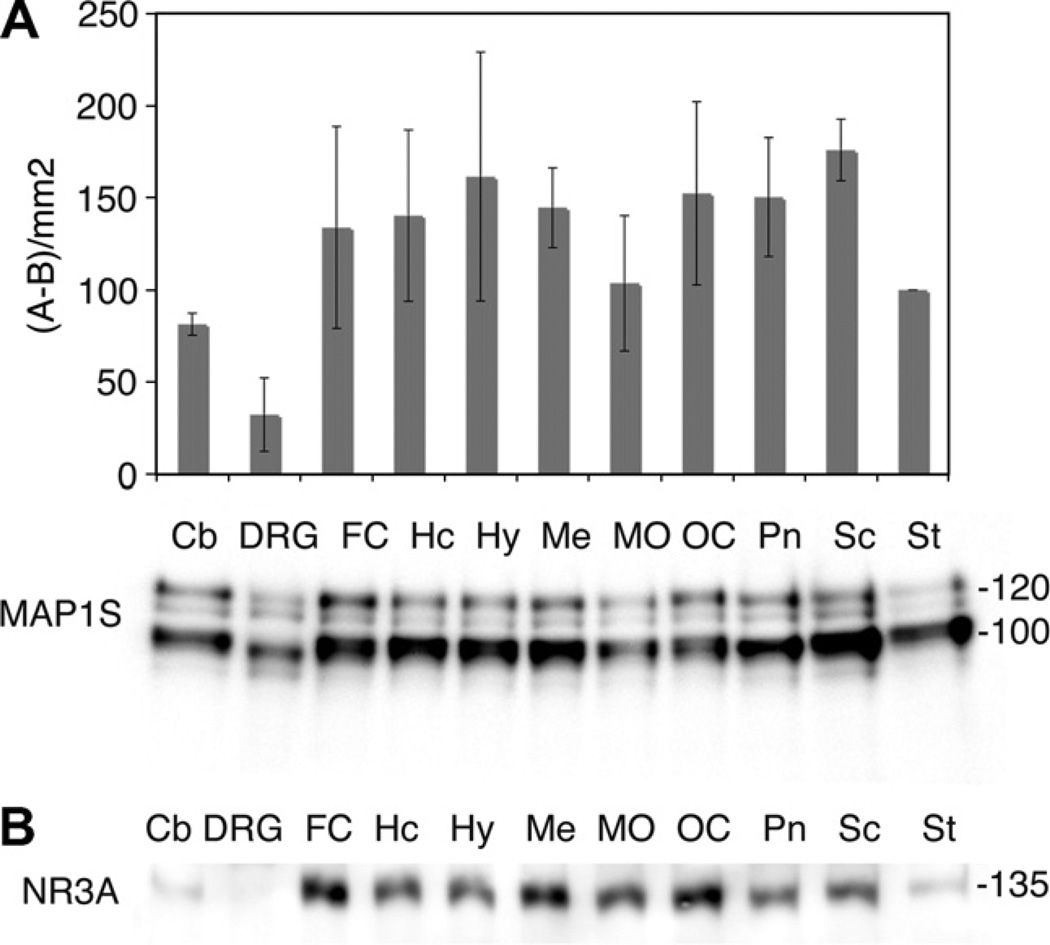
Human MAP1S contains an additional 86 amino acids compared to the mouse ortholog. The slower migration of the human MAP1S in Fig. 2B suggests that there is a similar difference in molecular weight between the human and rat species. Compared to the rat, MAP1S immunoreactive bands were weaker in the human cerebral cortex. In the eight-day old rat cortex MAP1S protein is highly expressed, the levels are slightly lower in the adult cortex, but most striking is that the amount of full-length protein is considerably lower in the adult animal. The expression pattern of NR3A follows that of MAP1S, being highly expressed in the young rat brain and with more moderate protein levels in the adult cortex. The nine-week human embryonic brain showed very week NR3A expression. We also investigated the relative expression of MAP1S in glial and neuronal cells and found MAP1S to be preferentially expressed in neurons, but lower levels were also present in astrocytes (Fig. 2C). Further, we examined the expression of MAP1S in 11 different regions of the postnatal rat CNS, and found the protein to be expressed in all of them (Fig. 3A). The highest MAP1S protein levels were found in hippocampus, hypothalamus, mesencephalon, spinal cord, and cortical regions. Lower levels were detected in cerebellum, dorsal root ganglia, medulla oblongata, and striatum. The expression pattern of MAP1S to some extent followed that of NR3A, which also is most abundant in the cortical regions, mesencephalon and hippocampus (Fig. 3B).
Fig. 3.
MAP1S and NR3A are widely expressed in the postnatal rat CNS. (A) MAP1S expression. The Y-axis shows the relative optical density. Values are normalized to striatum and n = 3–4. (B) NR3A expression. (Cb, cerebellum; DRG, dorsal root ganglia; FC, frontal cortex; Hc, hippocampus; Hy, hypothalamus; Me, mesencephalon; MO, medulla oblongata; OC, occipital cortex; Pn, pons; Sc, spinal cord; St, striatum).
In non-neuronal cell-lines MAP1S binds to hyper-stabilized microtubules and high MAP1S levels cause mitochondrial aggregation and cell death [21]. Considering the high levels of MAP1S we find in the adult mammalian brain, it is reasonable to believe that MAP1S serves other functions in the brain than those involving regulation of the cell cycle, especially since we find MAP1S predominantly in postmitotic neurons. In neurons, MAP1S more likely has functions resembling those of its neuronal homologues MAP1A and MAP1B.
NR3A and MAP1S colocalize in neurons
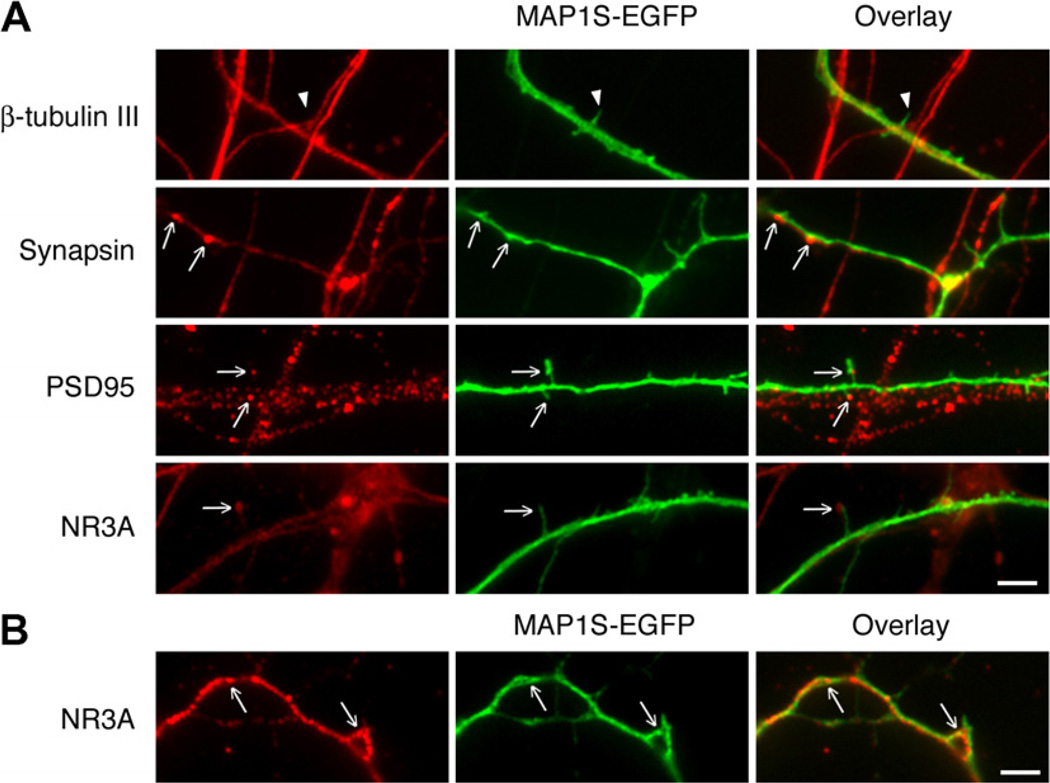
To investigate the subcellular distribution of MAP1S, hippocampal cells were transfected with EGFP-tagged MAP1S and then stained with antibodies recognizing different cellular structures. There was considerable overlap between MAP1S and β-tubulin III immunoreactivity in neurites, confirming that MAP1S is associated with microtubules in neurons (Fig. 4A). However, MAP1S was also abundant in filopodia-like protrusions lacking β-tubulin III, indicating that a pool of MAP1S does not interact with microtubules. A partial overlap between MAP1S and staining for the pre-and postsynaptic markers, synapsin and PSD95 could be seen, suggesting that a small proportion of MAP1S is localized to synapses. Similar to MAP1S, only a fraction of the NR3A pool has an obvious synaptic localization, the rest being distributed throughout soma and neurites. There was an overlap between MAP1S and NR3A immunoreactivity in neurites, and occasionally in filopodia-like protrusions. The colocalization between MAP1S and NR3A was most clearly seen in the human cell cultures (Fig. 4B), most likely because the NR3A antibody is raised against the human NR3A sequence. Our data suggest that MAP1S is expressed throughout neurite shafts, in filopodia-like protrusions and in certain synapses, and that the interaction with NR3A predominately takes place in neurite shafts.
Fig. 4.
Distribution of MAP1S-EGFP in hippocampal cell cultures. (A) Rat cells transfected with MAP1S-EGFP and immunostained with β-tubulin III, synapsin, PSD95 and NR3A antibodies. Arrowhead in upper panel indicates a β-tubulin III negative filopodia-like protrusion. Arrows show colocalization of MAP1S-EGFP with synapsin, PSD95 and NR3A respectively. (B) Human cells transfected with MAP1S-EGFP and immunostained with NR3A antibody. Arrows indicate colocalization of MAP1S-EGFP and NR3A immunoreactivity. Scale bar, 2.5 µm.
The presence of MAP1S in dendritic spines opens for novel functions of the protein, distinct from those involving binding to microtubules. Instead, MAP1S may interact with the actin fibrils abundant in synapses. Even though the function of the interaction between NR3A and MAP1S is not yet clear, parallels can be drawn to the interaction between MAP1B and the γ-aminobutyric acid (GABA)C receptor in the retina. Hanley and coworkers [22] showed that MAP1B specifically binds to the ρ1 subunit of the GABAC receptor and that the proteins colocalize at postsynaptic sites. Furthermore, Wang et al. [23] reported that a GABAA-receptor-associated protein (GABARAP) links the GABAA receptor to the cytoskeleton. GABARAP has sequence-similarities to light chain 3 of MAP1A and MAP1B and binds to tubulin. These studies demonstrate that MAPs are able to link ligand-gated receptors to the cytoskeleton and to influence their distribution at the plasma membrane.
Another possible function of the interaction between MAP1S and NR3A could be to mediate the transport of NR3A-containing receptors through the dendritic shaft. NMDARs have been shown to be transported along microtubules in transport packages and these packages are highly mobile during synaptogenesis [24] In addition, NR2B is linked to the kinesin motor KIF17 through a protein complex including mLin7, mLin-2 and mLin-10 [10]. Depolymerization of the microtubular cytoskeleton has been shown to inhibit NMDAR-mediated currents in neurons in a Ca2+-independent but GTP-dependent manner [11]. These studies point to the central role of microtubules in facilitating transport of NMDAR and thereby indirectly regulating NMDAR responses. MAP1S may thus control the transport of NR3A-containing NMDAR and regulate the localization of NR3A subunits to synaptic and extrasynaptic sites.
Acknowledgments
We thank the Netherlands Brain Bank for providing us with adult human post-mortem brain tissue. This work was supported by the Swedish Research Council, Marianne and Marcus Wallenberg Foundation and the research funds at Karolinska Institutet.
References
- 1.Dingledine R, Borges K, Bowie D, Traynelis SF. The glutamate receptor ion channels. Pharmacol. Rev. 1999;51:7–62. [PubMed] [Google Scholar]
- 2.Eriksson M, Nilsson A, Froelich-Fabre S, Akesson E, Dunker J, Seiger A, Folkesson R, Benedikz E, Sundstrom E. Cloning and expression of the human N-methyl-d-aspartate receptor subunit NR3A. Neurosci. Lett. 2002;321:177–181. doi: 10.1016/s0304-3940(01)02524-1. [DOI] [PubMed] [Google Scholar]
- 3.Andersson O, Stenqvist A, Attersand A, von Euler G. Nucleotide sequence, genomic organization, and chromosomal localization of genes encoding the human NMDA receptor subunits NR3A and NR3B. Genomics. 2001;78:178–184. doi: 10.1006/geno.2001.6666. [DOI] [PubMed] [Google Scholar]
- 4.Nilsson A, Duan J, Mo-Boquist LL, Benedikz E, Sundstrom E. Characterisation of the human NMDA receptor subunit NR3A glycine binding site. Neuropharmacology. 2007;52:1151–1159. doi: 10.1016/j.neuropharm.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 5.Das S, Sasaki YF, Rothe T, Premkumar LS, Takasu M, Crandall JE, Dikkes P, Conner DA, Rayudu PV, Cheung W, Chen HS, Lipton SA, Nakanishi N. Increased NMDA current and spine density in mice lacking the NMDA receptor subunit NR3A. Nature. 1998;393:377–381. doi: 10.1038/30748. [DOI] [PubMed] [Google Scholar]
- 6.Perez-Otano I, Schulteis CT, Contractor A, Lipton SA, Trimmer JS, Sucher NJ, Heinemann SF. Assembly with the NR1 subunit is required for surface expression of NR3A-containing NMDA receptors. J. Neurosci. 2001;21:1228–1237. doi: 10.1523/JNEUROSCI.21-04-01228.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wong HK, Liu XB, Matos MF, Chan SF, Perez-Otano I, Boysen M, Cui J, Nakanishi N, Trimmer JS, Jones EG, Lipton SA, Sucher NJ. Temporal and regional expression of NMDA receptor subunit NR3A in the mammalian brain. J. Comp. Neurol. 2002;450:303–317. doi: 10.1002/cne.10314. [DOI] [PubMed] [Google Scholar]
- 8.Perez-Otano I, Lujan R, Tavalin SJ, Plomann M, Modregger J, Liu XB, Jones EG, Heinemann SF, Lo DC, Ehlers MD. Endocytosis and synaptic removal of NR3A-containing NMDA receptors by PACSIN1/syndapin1. Nat. Neurosci. 2006;9:611–621. doi: 10.1038/nn1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chan SF, Sucher NJ. An NMDA receptor signaling complex with protein phosphatase 2A. J. Neurosci. 2001;21:7985–7992. doi: 10.1523/JNEUROSCI.21-20-07985.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Setou M, Nakagawa T, Seog DH, Hirokawa N. Kinesin super-family motor protein KIF17 and mLin-10 in NMDA receptor-containing vesicle transport. Science. 2000;288:1796–1802. doi: 10.1126/science.288.5472.1796. [DOI] [PubMed] [Google Scholar]
- 11.Yuen EY, Jiang Q, Feng J, Yan Z. Microtubule regulation of N-methyl-d-aspartate receptor channels in neurons. J. Biol. Chem. 2005;280:29420–29427. doi: 10.1074/jbc.M504499200. [DOI] [PubMed] [Google Scholar]
- 12.Brenman JE, Topinka JR, Cooper EC, McGee AW, Rosen J, Milroy T, Ralston HJ, Bredt DS. Localization of Postsynaptic Density-93 to dendritic microtubules and interaction with Microtubule- Associated Protein 1A. J. Neurosci. 1998;18:8805–8813. doi: 10.1523/JNEUROSCI.18-21-08805.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boeckers TM. The postsynaptic density. Cell Tissue Res. 2006;326:409–422. doi: 10.1007/s00441-006-0274-5. [DOI] [PubMed] [Google Scholar]
- 14.Sans N, Prybylowski K, Petralia RS, Chang K, Wang YX, Racca C, Vicini S, Wenthold RJ. NMDA receptor trafficking through an interaction between PDZ proteins and the exocyst complex. Nat. Cell Biol. 2003;5:520–530. doi: 10.1038/ncb990. [DOI] [PubMed] [Google Scholar]
- 15.Orban-Nemeth Z, Simader H, Badurek S, Trancikova A, Propst F. Microtubule-associated protein 1S, a short and ubiquitously expressed member of the microtubule-associated protein 1 family. J. Biol. Chem. 2005;280:2257–2265. doi: 10.1074/jbc.M408984200. [DOI] [PubMed] [Google Scholar]
- 16.Liu L, McKeehan WL. Sequence analysis of LRPPRC and its SEC1 domain interaction partners suggests roles in cytoskeletal organization, vesicular trafficking, nucleocytosolic shuttling, and chromosome activity. Genomics. 2002;79:124–136. doi: 10.1006/geno.2001.6679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wong EY, Tse JY, Yao KM, Lui VC, Tam PC, Yeung WS. Identification and characterization of human VCY2-interacting protein: VCY2IP-1, a microtubule-associated protein-like protein. Biol. Reprod. 2004;70:775–784. doi: 10.1095/biolreprod.103.018531. [DOI] [PubMed] [Google Scholar]
- 18.Song MS, Chang JS, Song SJ, Yang TH, Lee H, Lim DS. The centrosomal protein RAS association domain family protein 1A (RASSF1A)-binding protein 1 regulates mitotic progression by recruiting RASSF1A to spindle poles. J. Biol. Chem. 2005;280:3920–3927. doi: 10.1074/jbc.M409115200. [DOI] [PubMed] [Google Scholar]
- 19.Halpain S, Dehmelt L. The MAP1 family of microtubule-associated proteins. Genome Biol. 2006;7:224.1–224.7. doi: 10.1186/gb-2006-7-6-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ma OK, Sucher NJ. Molecular interaction of NMDA receptor subunit NR3A with protein phosphatase 2A. Neuroreport. 2004;15:1447–1450. doi: 10.1097/01.wnr.0000132773.41720.2d. [DOI] [PubMed] [Google Scholar]
- 21.Liu L, Vo A, Liu G, McKeehan WL. Distinct structural domains within C19ORF5 support association with stabilized microtubules and mitochondrial aggregation and genome destruction. Cancer Res. 2005;65:4191–4201. doi: 10.1158/0008-5472.CAN-04-3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hanley JG, Koulen P, Bedford F, Gordon-Weeks PR, Moss SJ. The protein MAP-1B links GABA(C) receptors to the cytoskeleton at retinal synapses. Nature. 1999;397:66–69. doi: 10.1038/16258. [DOI] [PubMed] [Google Scholar]
- 23.Wang H, Bedford FK, Brandon NJ, Moss SJ, Olsen RW. GABA(A)-receptor-associated protein links GABA(A) receptors and the cytoskeleton. Nature. 1999;397:69–72. doi: 10.1038/16264. [DOI] [PubMed] [Google Scholar]
- 24.Washbourne P, Bennett JE, McAllister AK. Rapid recruitment of NMDA receptor transport packets to nascent synapses. Nat. Neurosci. 2002;5:751–759. doi: 10.1038/nn883. [DOI] [PubMed] [Google Scholar]