Abstract
The production of androgenetic embryos in large animals is a complex procedure. Androgenetic embryos have been produced so far only in cattle and sheep using pronuclear transfer (PT) between zygotes derived from in vitro fertilization (IVF) of previously enucleated oocytes. PT is required due to the poor developmental potential of androgenotes derived from IVF of enucleated oocytes. Here we compare the developemt to blastocyst of androgenetic embryos produced by the standard pronuclear transfer and by fertilization of oocytes enucleated in Ca2+/Mg2+-free medium, without pronuclear transfer. The enucleation in Ca2+/Mg2+-free medium abolished almost completely the manipulation-induced activation, significantly improving the development to blastocyst of the androgenetic embryos (IVF followed by PT; 18.6%: IVF only; 17.7%, respectively). Karyotype analysis of IVF revealed a similar proportion of diploid embryos in androgenetic and control blastocysts (35% and 36%, respectively), although mixoploid blastocysts were frequently observed in both groups (64%). Androgenotes had lower total cell numbers than control and parthenogenetic embryos, but more cells in ICM cells comparing to parthenogenotes (30.42 vs. 17.15%). Higher expression of the pluripotency-associated gene NANOG, and trophoblastic-specific gene CDX2, were also observed in androgenotes compared to parthenogenotes and controls. The global methytion profile of androgenetic embryos was comparable to controls, but was lower than parthenogenetic embryos. The cell composition and methylation pattern we have detected in monoparental sheep monoparental embryos are unprecedented, and differ considerably from the standard reference mouse embryos. Altogether, these finding indicate significant differences across species in the molecular mechanisms regulating early development of monoparental embryos, and highlights the need to study postimplantation development of androgenetic embryos in sheep.
Introduction
Monoparental embryos have a diploid chromosome complement that is derived from only the mother (parthenogenetic/gynogenetic) or from the father (androgenetic embryos). Parthenogenetic embryos are produced by interfering with the meiotic reduction of the chromosomes, leading to the development of a diploid embryos with maternal chromosomes only. Gynogenetic and androgenetic embryos are produced by reconstructing fertilized oocytes (zygotes) with maternal and paternal pronuclei, respectively. Monoparental embryos were originally produced to demonstrate the complementary parental contribution to development (McGrath and Solter, 1984; Surani et al., 1984). Since then, the mouse has become the reference model for genomic imprinting and epigenetic dynamics in normal and monoparental embryos (Dean et al., 2001). However, recent data challenge the universality of the mouse model (Haaf, 2006). Indeed, it appears that major epigenetic modifications, such as the genome-wide demethylation waves during preimplantation development, differ remarkably across mammals (Beaujean et al., 2004a; Fulka et al., 2004, Loi et al., 2008; Shi et al., 2004). Therefore, the recent interest in the production of monoparental embryos from alternative animal models to study genomic imprinting and the parental contribution to embryo development is justified.
Parthenogenetic embryos are easily obtained by artificial activation of oocytes in all species, whereas the production of androgenetic embryos is more complicated, particularly in large animals. We have recently produced androgenetic sheep embryos (Matsukawa et al., 2007) according to a protocol originally established in the mouse (Kono et al., 1993), and subsequently applied to the bovine (Lagutina et al., 2004). Briefly, androgenetic embryos were produced by pronuclear transfer between diploid and haploid “zygotes” that were produced by in vitro fertilization (IVF) of enucleated oocytes. The pronuclear transfer (PT) was required because on the contrary of the mice (Kono et al. 1993), the development of IVF enucleated oocytes was very poor (1.8% blastocyst rate) (Matsukawa et al., 2007). Although we succeeded in increasing the number of androgenetic blastocysts, the procedure is cumbersome and time consuming, thus limiting the number of androgenetic embryos that can be produced. These difficulties can explain the lack of information on androgenetic embryos in large animals.
In this work, we first set out to identify and remove the causes of the poor development of IVF-derived sheep androgenotes. We then investigated the cellular composition [i.e., the number of inner cell mass (ICM) and trophoblast (TR) cells], the global methylation profile, and chromosome composition of androgenetic, parthenogenetic, and control IVF blastocysts. Finally, we analyzed the expression levels of the totipotency genes OCT4 and NANOG and trophectoderm-specific gene CDX2 in androgenetic and parthenogenetic embryos.
We demonstrate that preventing the manipulation-induced oocyte activation dramatically improves the development of androgenetic embryos to blastocyst stage. We also show that androgenetic blastocysts, like IVF controls, are diploid with a significant proportion of mixoploidy. Unexpectedly, androgenetic embryos have a higher ICM/TR cell ratio, different global methylation profiles, and expression of totipotency-associated genes than parthenogenetic embryos.
Materials and Methods
All chemicals, unless otherwise indicated, were purchased from Sigma Chemicals Co. (St. Louis, MO).
In vitro maturation
Sheep ovaries were obtained at local abattoirs and transferred at 37°C to the laboratory within 2 h. Cumulus–oocyte complexes were recovered by aspirating the follicles and cultured in four-well Nunc plastic dishes, filled with 500 μL of TCM-199 medium without covering oil (Gibco Life Technologies, Rockville, MD) in humidified air with 5% CO2 at 38.5°C for 24 h. TCM-199 was supplemented with 2 mM glutamine (Gibco), 100 μM cysteamine, 0.3 mM sodium pyruvate, 10% fetal bovine serum (Gibco), 5 μg/mL FSH (Ovagen, ICP, Auckland, New Zealand), 5 μg/mL LH, and 1 μg/mL estradiol. Methods for in vitro embryo production were as previously described (Ptak et al., 1999, 2002).
Assessment of manipulation-induced activation
Matured oocytes were stripped of granulosa cells by a combined treatment with 300 U/mL hyaluronidase and gentle pipetting. For the determination of manipulation-induced oocyte activation, 270 oocytes were divided in four groups. Group A (n=85) was handled in normal manipulation medium [i.e., TCM 199 with antibiotics, 4 mg/mL bovine serum albumin (BSA), 7.5 μg/mL Cytochalasin B] and group B (n=60) manipulated in Ca2+/Mg2+-free medium [i.e., Ca2+/Mg2+-free phosphate-buffered saline (PBS) with antibiotics, 4 mg/mL BSA, and 7.5 μg/mL Cytochalasin B]. For both groups manipulation consisted in the removal of a small piece of cytoplasm (opposite to the metaphase plate), as normally carried on during enucleation. In groups A1 and B1, oocytes were exposed to normal manipulation medium (group A1, n=83) or to Ca2+/Mg2+-free medium (group B1, n=42), but without manipulation. Oocytes were then cultured in standard conditions for 10–15 h, then fixed in 3:1 acetic acid:methanol overnight, and stained with 2% aceto-orcein for pronuclear identification.
Androgenetic embryo production
Androgenetic embryo production by IVF followed by pronuclear transfer (IVF-PT): Enucleation of oocytes
At 22 h of maturation, oocytes were denuded of granulosa cells in the presence of hyaluronidase. Oocytes with extruded first polar bodies were incubated in 10 mg/mL Hoechst 33342 in TCM 199 for 10 min, and then enucleated by aspirating the metaphase II plate in TCM 199 medium with 4 mg/mL BSA, antibiotics and 7.5 μg/mL cytochalasin B, under UV light with a Narishighe Micromanipulator fitted to an inverted Nikon microscope (Melville, NY).
IVF
The IVF procedure used for enucleated oocytes is as described below but with higher concentration of spermatozoa (25×106 mL).
Pronuclear Transfer IVF/PT (enucleated, in vitro fertilized, then pronuclear transfer)
Ten hours after IVF, embryos (n=118) were incubated in HEPES-buffered TCM 199 with 7.5 μg/mL cytochalasin B for 10 min, then centrifuged at 12,000×g for 5 min in order to visualize the pronuclei. Single pronuclei surrounded by a small volume of cytoplasm were aspirated with a beveled pipette from diploid androgenotes and transferred in the perivitelline space of haploid androgenotes. The reconstructed embryos were manually aligned under the micromanipulator and electro-fused using a BTX ECM 830 apparatus in 0.27 M mannitol solution, containing 50 μM CaCl2 and 100 μM MgCl2, by a single direct-current pulse (80 μsec) of 1.2 kV/cm.
Androgenetic embryo production by IVF only (enucleated and in vitro fertilized)
Enucleation of oocytes (n=190) was carried out as above, but using Ca2+/Mg2+-free manipulation medium. IVF was as described below but with higher concentration of spermatozoa (25×106), as above. A subset of presumptive androgenetic embryos (n=61) was collected at 10 h post-IVF, incubated in TCM 199 with BSA and antibiotics and 7.5 μg/mL Cytochalasin B for 10 min and centrifuged at 12,000×g for 5 min to stratify the cytoplasm. The number of pronuclei in these embryos was scored under a microscope. The others were cultured to blastocyst stage.
Parthenogenetic embryo production
In vitro matured metaphase II oocytes (n=190) were activated with a combined treatment of ionomycin and 6-dimethylaminopurine, in SOF medium, as previously described (Loi et al., 1998).
IVF of control embryos
Matured oocytes (n=443) were partially stripped of cumulus cells by repeated pipetting. Frozen semen was rapidly thawed at 37°C and washed twice by centrifugation at 500×g for 5 min with bicarbonate-buffered SOF with 4 mg/mL BSA. IVF was carried out in 50 μL drops, using 5×106 sperm/mL and a maximum of 15 oocytes per drop, at 38.5°C in 5 % CO2 for 20 h. The IVF medium was bicarbonate-buffered SOF enriched with 20% (v/v) heat-inactivated oestrous sheep serum, 2.9 mM Ca2+ lactate, and 16 μM isoproterenol.
In vitro culture
All classes of embryos were transferred into 20 μL drops of SOF enriched with 1% (v:v) Basal Medium Eagle (BME), essential amino acids, 1% (v:v) Minimum Essential Medium (MEM), nonessential amino acids (Gibco), 1 mM glutamine, and 8 mg/mL fatty acid-free BSA (SOFaa-BSA). Zygotes were cultured in a humidified atmosphere of 5% CO2, 7% O2, 88% N2 at 38.5°C, and the medium changed at day 3 (supplemented with amino acids) and day 5 (supplemented with 10% FBS). Cleavage was assessed at day 2 and blastocyst formation was recorded at days 7 and 8.
Differential cell staining of parthenogenetic, androgenetic, and control blastocysts
Embryos were differentially stained as described by Thouas et al. (2001), with minor modification. Briefly, blastocysts were incubated in 500 mL of solution 1 (PBS with 1% Triton X-100 and 100 mg/mL propidium iodide) for 20 sec. Blastocyst were then directly transferred to 500 mL of solution 2 (100% ethanol with 25 mg/mL bisbensimide Hoechst 33258) and stored at 4°C overnight. Blastocysts were then mounted onto a microscope slide in a drop of glycerol and flattened with a cover slide. Cell counting was performed directly on an inverted microscope fitted with an ultraviolet lamp and excitation filter (460 nm for blue and red fluorescence).
Karyotype analysis of blastocyst stage embryos
Blastocyst stage monoparental and control embryos were incubated with 0.05 μg/mL Colcemid (Sigma) for 3 h, then transferred to hypotonic solution, 0.8% sodium citrate (37°C) for 10 min and then into 0.56% KCl (37°C) for another 10 min. Blastocysts were fixed in 3:1 methanol:acetic acid for 2 h. Spreading was done by dropping a 1:1 methanol:acetic acid solution under an inverted phase contrast microscope. Slides were stained with 4% Giemsa for 5 min. Photographs were taken using a 100×objective and immersion oil.
Immunodetection of global DNA methylation
Blastocyst stage embryos were washed five times in 0.4% PBS/PVP, fixed in 4% paraformaldehyde for 15 min, and then permeabilized in 0.1% Triton X-100 for 30 min. Blastocysts were washed again and hydrolyzed in 4 N HCL for 10 min, neutralized in 100 mM Tris/HCl (pH=8.5) for 15 min, washed in PBS/0.4% PVP (5 min×3), then blocked in PBS plus 1% BSA, 0.05% Tween 20 at 4°C overnight. Embryos were incubated with a mouse anti-5-Methyl Cytidine antibody (Santa Cruz Biotechnology sc-56615, Santa Cruz, CA) at room temperature for 2 h, washed in blocking medium three times and incubated with goat antimouse IgG FITC conjugate antibody (Sigma F9137) at room temperature for 1 h. Mounted specimens were analyzed with an epifluorescence microscope (Nikon). The quantification of the fluorescence intensity for 5-MethyCytidine determination was carried out with the software ImageJ (Image processing and Analysis in JAVA).
Gene expression analysis
Isolation of mRNA and RT-PCR
Poly(A)+ RNA was isolated from single frozen (PBS and PVP 0.4%) blastocysts using the Dynabeads mRNA DIRECT Kit (Invitrogen Dynal AS, Oslo, Norway). The procedure was carried out according to the manufacturer's instructions, with minor modifications. Briefly, single embryos (stored at −80°C) were lysed in 150 μL lysis/binding buffer (100 mM Tris–HCl pH=8.0, 500 mM LiCl, 10 mM EDTA, 1% LiDS, 5 mM DDT). Subsequently, 10 μL of prewashed Dynabeads oligo (dT)25 was added to each tube containing single embryos, and after a short incubation at room temperature the beads were separated using a magnetic separator (Dynal MPC-P-12 magnet; Invitrogen Dynal). Beads/mRNA complexes were washed once with 100 μL washing buffer A (10 mM Tris–HCl pH=8.0, 0.15 M LiCl, 1 mM EDTA, 0.1% LiDS) and twice with 100 μL washing buffer B (10 mM Tris–HCl pH=8.0, 0.15 M LiCl, 1.0 mM EDTA). Poly(A)+ RNA was then eluted from the beads by incubation in 10 μL of 10 mM Tris–HCl at 70°C for 3 min. RT was carried out using 80% of the eluted Poly(A)+ RNA in a total volume of 20 μL using the QuantiTect Reverse Transcription Kit (Qiagen, Chatsworth, CA). Briefly, samples were first incubated in gDNA Wipeout Buffer at 42°C for 2 min to remove contaminating genomic DNA. Then, according to the manufacturer's recommended protocol, the RT reaction was carried out at 42°C for 15 min followed by a final step at 95°C for 3 min to inactivate the transcriptase
Real-time PCR
The obtained cDNA was diluted (1:2) and then used for real-time PCR amplification to quantify the expression of NANOG, OCT4, and CDX2 using the following primer pairs: NANOG: (Ovis Aries, FJ 970651) forward: aaaccattgtccccatctgc; reverse: tagctgaggttcaagatgttgg; OCT4: (Bos Taurus, NM_174580) forward: aagctcctaaagcagaagagg; reverse: ttctcgttgttgtcagcttcc; CDX2: Bos Taurus (XM_871005.3) forward: aagacaaataccgggtcgtg; reverse: ctctgcggttctgaaaccaa.
The reaction was performed using Platinum SYBR Green qPCR SuperMix UDG with ROX (Invitrogen) and ABI PRISM 7900 Real-Time PCR System (Applied Biosystems, Bedford, MA) according to the manufacturer's instructions. Amplification conditions were: 2 min at 50°C, 2 min at 95°C, followed by 45 cycles at 95°C for 15 sec and 56°C for 60 sec. To avoid false-positive signals, dissociation-curve analyses were performed at the end of each run; the conditions of the dissociation step were 15 sec at 95°C, 15 sec at 60°C, and 15 sec at 95°C. Relative gene expression data were calculated using the comparative threshold cycle (Ct) method with β-Actin as endogenous control.
Statistical analysis
Embryo development
Differences between the experimental groups were verified using the chi-square test. A value of p<0.05 was considered significant.
Pronuclear stage and cell numbers
Differences between the experimental groups were verified with the one-way analysis of variance (ANOVA) test (GraphPad Prism). A value of p<0.05 was considered significant.
Image analysis
The quantification of the fluorescence intensity for 5-MethyCytidine determination was carried out with the software ImageJ (Image processing and Analysis in JAVA). Differences between the experimental groups were verified with the one-way ANOVA test (GraphPad Prism). A value of p<0.05 was considered significant.
Gene expression analysis
Data reported are the mean (±SEM) of at least five independent determinations, each in triplicate. Statistical analysis was performed with the nonparametric Mann-Whitney t-test (GraphPAD Prism). Differences were considered significant when p<0.05.
Results
Assessment of manipulation-induced oocyte activation
To evaluate whether and to what extent the manipulation required to enucleate the oocytes activates them, we compared oocytes that were sham manipulated (aspiration of a portion of cytoplasm near the first polar body, group A) with oocytes that were simply maintained in the same medium with Ca2+/Mg2+, but unmanipulated (group A1). More than half of the group A oocytes (56%) showed well organized pronuclei, indicative of activation induced by the manipulation, whereas activation was only observed occasionally in the control group A1 (0.6%) (Table 1). Conversely, when Ca2+/Mg2+-free manipulation medium was used, activation was strongly reduced (3.3%) in sham manipulated oocytes (group B), and completely abolished in the control group (group B1, Table 1).
Table 1.
Activation Frequencies (Assessed on the Pronuclear Organization) in Sham- Manipulated and Control Oocytes That Were Fixed in 3:1 Acetic Acid:Methanol and Stained with Aceto-orcein 2%
| Group | Treatment | No. oocytes | Activated (pronuclei) (%) |
|---|---|---|---|
| A | TCM 199, sham manipulated | 85 | 48 (56)a |
| A1 | TCM199, control | 83 | 4 (0,6)b |
| B | Ca2+/Mg2+ free PBS—sham manipulated | 60 | 2 (3,3)b |
| B1 | Ca2+/Mg2+ free PBS—control | 42 | 0 (0) |
Different superscripts indicate p<0.0001.
Number of pronuclei in zygotes obtained by IVF of oocytes enucleated in Ca2+/Mg2+-free PBS
Based on these results, we next evaluated the fertilization rate of oocytes that were enucleated in Ca2+/Mg2+-free medium, by counting the number of pronuclei 10 h after IVF. Most of the oocytes were fertilized (82%), with 26.2% haploid androgenotes cone pronucleus (PN), 52.42% diploid (two PN), and only a minority triploid (three PN, 3.2%; Table 2).
Table 2.
Number of Pronuclei in Living Androgenetic Embryos Produced by IVF of Oocytes Enucleated in Ca2+ /Mg 2+-Free Medium (Observed after Centrifugation at 12,000×g, 10 Hours after IVF)
| No. of pronuclei | No. zygotes (%) |
|---|---|
| 0 PN | 10/61 (16.4%) |
| 1 PN | 16/61 (26.2%) |
| 2 PN | 32/61 (52.4%) |
| 3 PN | 2/61 (3.37%) |
Development to blastocyst stage of IVF only (enucleated and in vitro fertilized) and IVF/PT (enucleated, in vitro fertilized, plus pronuclear transfer) androgenotes at days 7–8
We next compared the development potential to blastocyst stage of androgenetic embryos that were produced either by IVF of oocytes enucleated in normal manipulation medium followed by Pronuclear Transfer (IVF-PT) (Matsukawa et al., 2007), or by IVF of oocytes enucleated in Ca2+/Mg2+-free medium (IVF only). Biparental IVF (control) and parthenogenetic embryos were produced in every replicate (n=6) as controls. No differences were found between IVF only (Fig. 1) and IVF-PT androgenotes in their capacity to reach blastocyst stage (18.60 and 17.70%, respectively); the development to blastocyst stage of both groups of androgenotes was slightly lower than control and parthenogenetic embryos (20.7 and 18.9%, respectively; Table 3).
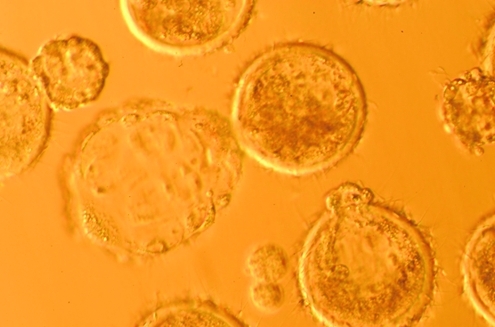
FIG. 1.
Hatching androgenetic blastocysts at day 7 (40×).
Table 3.
In Vitro Development to Blastocyst Stage of Control and Monoparental Embryos
| Group | No. oocytes | Fully expanded blastocyst |
|---|---|---|
| Control IVF | 443 | 92 (20.7%)a |
| Andro IVF Only | 129 | 24 (18.6%)a |
| Andro IVF/PT | 118 | 21 (17.7%)a |
| Parthenogenetic | 190 | 36 (18.9%)a |
Different superscripts indicate p<0.0001.
Assessment of cell composition and chromosome constitution of the IVF only androgenetic embryos
Staining with propidium iodide and Hoechst 33342 to differentiate between ICM and TR cells showed that the total cell number was comparable in control and parthenogenetic blastocysts (n=127±28 and n=129±117, respectively), but was significantly reduced in androgenotes (n=100±51) (Table 5). Moreover, androgenetic embryos showed a higher ICM/TR cell ratio than parthenogenetic embryos (Table 5).
Table 5.
Differential Staining of Monoparental (AN, PA) and Biparental (IVF-Control) Blastocysts on Day 8 of Development
| |
|
Cell number |
|
||
|---|---|---|---|---|---|
| Blastocysts | No. embryos | Total | ICM (%) | TR (%) | Ratio ICM/TR |
| IVF | 6 | 127±28 | 38±9 (29.88±0.09) | 89±24 (70.66±0.09) | 0.44±0.13a |
| AN | 10 | 100±51 | 27±14 (30.42±0.18) | 73±42 (69.86±0.17) | 0.59±0.7b |
| PA | 10 | 129±117 | 25±15 (17.15±0.13) | 105±95 (82.05±0.14) | 0.44±0.2a |
Different superscripts indicate p<0.0031.
We have analyzed the chromosome composition of IVF only androgenetic blastocysts (n=56) and control embryos (n=23). The analysis of the metaphase spreads revealed that 35.7% of control and 30.43% and androgenetic embryos were euploid (2n=54) (Fig. 2) and that in both groups mixoploidy was frequent (64.3% control and 69.57% androgenetic embryos; Table 4).
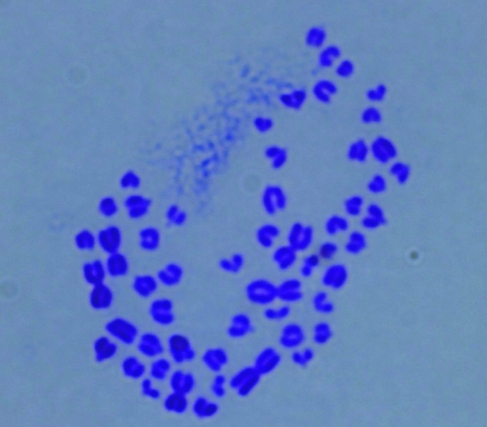
FIG. 2.
Normal (n=54) karyotype in sheep androgenetic blastocyst (×100).
Table 4.
Chromosome Composition of Control and IVF-Only Androgenetic Blastocysts
| Group | Diploid (%) | Mixoploid (%) |
|---|---|---|
| IVF blastocysts | 20/56 (35.7%)a | 36/56 (64.3%)a |
| Androgenetic blastocysts | 7/23 (30.43%)a | 16/23 (69.5%)a |
Different superscripts indicate p<0.0001.
DNA methylation of monoparental and control embryos
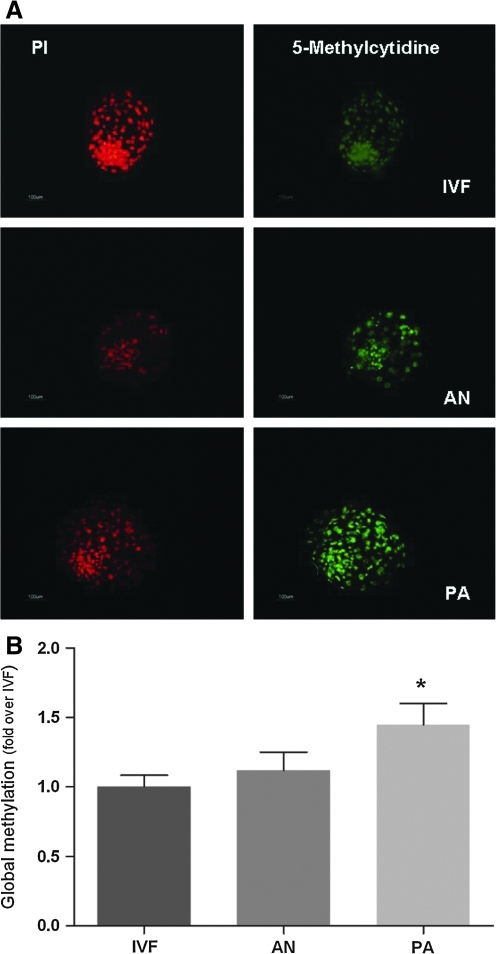
The global methylation profile (detected using a mouse anti-5-Methyl Cytidine antibody) of androgenotes overlapped with that of control embryos, although it was significantly higher in parthenogenetic embryos (Fig. 3B). ICM and trophoblast cells were equally methylated in all classes of embryos (Fig. 3A).
FIG. 3.
Global DNA methylation of monoparental (AN, PA) and biparental (IVF-Control) sheep blastocysts. (A) Immunostaining anti-5-methyl.(B) Semiquantitative analysis of fluorescence intensity. Different superscripts indicate p<0.001.
Analysis of OCT4, NANOG, and CDX2 expression in individual blastocysts
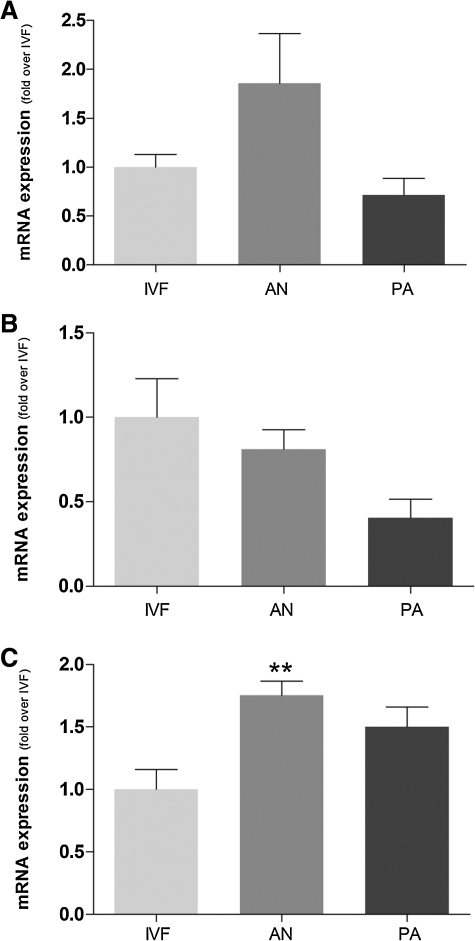
The totipotency-associated genes OCT4 and NANOG were expressed at higher level in androgenetic embryos, particularly NANOG, comparing to parthenogenetic ones (Fig. 4A–B). CDX2, a master gene regulating early differentiation of trophoblastic cells, was also expressed at higher level in androgenotes, comparing to parthenogenetic embryos (Fig. 4C).
FIG. 4.
Expression level of OCT 4 (A), NANOG (B), and CDX2 (C) in blastocyst stage monoparental (AN, PA) and biparental (IVF-control) embryos. Different superscripts indicate p<0.008.
Discussion
In this work we have demonstrated that the production of sheep androgenetic embryos is improved by circumventing oocyte activation during the enucleation. Originally reconstructed by PT between zygotes (Surani et al., 1984), the production of mice androgenetic embryos has been further simplified, first by fertilizing enucleated oocytes (Kono et al.,1993), and more recently by the direct injection of two round spermatids into enucleated oocytes (Miki et al., 2009; Zhao Q et al., 2010). Thus, although making androgenetic embryos is a robust procedure in mice, the situation is more complicated in other animals, particularly in sheep, where IVF of enucleated oocytes gave poor results (Matsukawa et al., 2007).
Here we suggest that the low developmental potential of sheep androgenotes obtained by IVF (Matsukawa et al., 2007) might be due to the oocyte activation caused by the enucleation procedure, which inhibits the penetration of the second spermatozoa, essential for diploidization. Moreover, we have demonstrated that the enucleation is carried out in Ca2+/Mg2+-free medium abolishes almost completely the activation of the oocytes (Table 1).
It is known that a mechanical stress triggers oocyte activation in amphibian oocytes (Yang and Sachs, 1988). Because stretch-activated ion channels are sensitive to pressure as well as suction (Yang and Sachs, 1990), it is possible that the suction applied to the oolemma during enucleation might open the stretch-activated channels, thus triggering the Ca2+ dependent activation. Stretch-activated ion channels are present at the oocyte membrane of Xenopus Levis (Yang and Sachs, 1988) and in a wide range of cells (Sachs, 2010), but no information is available concerning mammalian oocytes. If we assume that stretch-activated ion channels are present in sheep oocyte, it is plausible to find them on the mouse oocyte. How can we then explain the difference in the developmental capacity of sheep and mice IVF androgenotes (1.8 vs. 43%, respectively), given that enucleation of mice oocytes was conducted in M2, a Ca2+ containing medium (Kono et al., 1993)? A possible explanation might be the differences in the polyspermic block between species. (Gardner and Evans, 2006; Gardner et al., 2007). The polyspermic block occurs rapidly in sheep (Gardner and Evans, 2006), whereas mice oocytes can be penetrated by a second spermatozoa even 2 h after the first one (Gardner et al., 2007). The promptness of the polyspermic block in sheep oocytes might therefore explain the low frequency of development of IVF androgenetic embryos, given that only diploid androgenotes (dispermic) develop to blastocyst stage.
The simple measure of performing the enucleation in Ca2+/Mg 2+-free manipulation medium significantly improved the development of IVF-only androgenotes, which reached blastocyst stage at rates comparable to those produced by IVF followed by Pronuclear Transfer (IVF-PT) (18.60 and 17.70%, respectively; Table 3).
The development of both classes of androgenetic embryos to blastocyst stage was lower comparing to control and parthenogenetic embryos (Table 5), consistent with the presence of the lethal YY combination in about one quarter of embryos (Latham et al.,. 2000). All the IVF-only androgenetic blastocysts were diploid, although a significant proportion of mixoploid embryos were also observed. The chromosome mixoploidy were found in androgenetic and control biparental embryos (69 and 64%, respectively). These data are in agreement with previous work conducted in the bovine (Alexander et al., 2006; Van De Velde et al., 1999). The real significance of the chromosomal mixoploidy is under intense investigation at the moment, but still remains unclear (Alfarawati et al., 2010; Curlej et al., 2010; Gomez et al., 2009; Hornak et al., 2009). The fact that chromosomal abnormalities are present in control IVF (this study) and in normal human embryos (Vanneste et al., 2009) suggests that a certain degree of aneuploidy/mixoploidy should be considered as a default in early development, although further studies will be required to dissect the aneuploidy/mixoploidy between ICM and trophectoderm.
The androgenetic embryos had a reduced number of cells and higher ICM/TR cells ratio than parthenogenetic embryos (0.59 and 0.28, respectively). A lower total cell number was also described in bovine androgenotes, but the proportion of ICM/TR cells was not evaluated (Lagutina et al., 2004). The higher ICM “cellularity” of the androgenotes might be due to their higher expression of the totipotency-associated genes OCT4 and NANOG in comparison to the parthenogenetic embryos (Fig. 4A and B).
In their original article, Surani and coworkers (1984) reported that mouse androgenotes showed a preferential proliferation of trophectoderm-derived cells and defective development of ICM-derived structures. In preimplantation sheep androgenetic embryos, we have observed an opposite trend, with a higher number of ICM cells and a very poor trophectoderm. This finding suggests that major phenotypic differences between species in preimplantation development of monoparental embryos might exist, although further studies on postimplantation development of androgenetic embryos are required. Alternatively, the increased ICM/TR ratio in sheep androgenotes might represent a cell dosage compensation, resulting from a counting mechanism that operates in preimplantation embryos and finalized to counterbalance the absence of the maternal contribution. The reduced proliferation of trophoblastic cells in sheep androgenotes occurs despite the unregulated expression of CDX2 (Fig. 4), the primary gene regulating trophectoderm differentiation and proliferation (Strumpf et al., 2005). The increased expression of CDX2 observed in androgenotes might result from either a compensatory mechanism, or from the genetic noise caused by the aneuploidy (Birchler et al., 2005) frequently observed in embryos. Whatever induced, the increased transcription of CDX2 might be itself responsible for the defective proliferation of trophoblastic cells (Xie et al., 2010).
Finally, the global methylation profile of androgenotes overlapped with the IVF controls, whereas the parthenogenetic embryos presented higher level of methylation (Fig. 3). Both ICM and trophoblast were methylated, a feature that is conserved in other large animals (Haaf, 2006; Loi et al., 2008; Young and Beaujeau, 2004), but not in mice, where DNA methylation is confined to the ICM (Reik et al., 2003). Species-specific differences in the implantation timing and dynamics might account for the variations in methylation pattern at blastocyst stage. The higher methylation of parthenogenetic embryos was surprising. Because the paternal pronucleus is not actively demethylated in sheep zygotes (Beaujean et al., 2004b), we would have expected higher methylation in androgenetic embryos, as they derive from two spermatozoa, normally more methylated than the oocyte (Reik et al., 2003). Again, this might reflect differences in the epigenetic asset of preimplantation development of monoparental embryos across species.
In conclusion, we have demonstrated that it is possible to significantly improve the developmental potential of diploid androgenetic sheep embryos by simply preventing the manipulation-induced oocyte activation, a protocol that might be easily transferred to other species, including human. This procedural simplification empowered us to dispose of suitable numbers of androgenotes that allowed us to characterize the difference in cell composition and methylation profile between androgenetic, parthenogenetic, and control, fertilized embryos in sheep. The overall picture we have observed diverge significantly from the data published in monoparental mouse embryos, a finding that highlights the importance of extending the investigation on postimplantation development of androgenetic conceptuses in sheep.
Acknowledgments
The authors acknowledge Antonella Fidanza, for the help given in gene expression analysis. The research leading to these results has received funding from the European Research Council under the European Community Seventh Framework Programme (FP7/2007-2013)/ERC grant agreement No. 210103; GP: PRIN 2007, No. 2007MY2M92 to G.P. P.L. acknowledges the support of the EU FP7-KBBE-2009-3 Programme, project No. 244356, NextGene.
Author Disclosure Statement
The authors declare that no conflicting financial interests exist.
References
- Alexander B. Coppola G. Di Berardino D., et al. The effect of 6-dimethylaminopurine (6-DMAP) and cycloheximide (CHX) on the development and chromosomal complement of sheep parthenogenetic and nuclear transfer embryos. Mol. Reprod. Dev. 2006;73:20–30. doi: 10.1002/mrd.20372. [DOI] [PubMed] [Google Scholar]
- Alfarawati S. Fragouli E. Colls P., et al. The relationship between blastocyst morphology, chromosomal abnormality, and embryo gender. Fertil. Steril. 2010;95:520–524. doi: 10.1016/j.fertnstert.2010.04.003. [DOI] [PubMed] [Google Scholar]
- Beaujean N. Taylor J. Gardner J., et al. Effect of limited DNA methylation reprogramming in the normal sheep embryo on somatic cell nuclear transfer. Biol. Reprod. 2004a;71:185–193. doi: 10.1095/biolreprod.103.026559. [DOI] [PubMed] [Google Scholar]
- Beaujean N. Taylor J.E. McGarry M., et al. The effect of interspecific oocytes on demethylation of sperm DNA. Proc. Natl. Acad. Sci. USA. 2004b;101:7636–7640. doi: 10.1073/pnas.0400730101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birchler J.A. Riddle N.C. Auger D.L., et al. Dosage balance in gene regulation: biological implications. Trends Genet. 2005;21:219–226. doi: 10.1016/j.tig.2005.02.010. [DOI] [PubMed] [Google Scholar]
- Curlej J. Bulla J. Chrenek P. Occurrence of chromosomal aneuploidy in rabbit oocytes and embryos at different developmental stages. Zygote. 2010;18:203–207. doi: 10.1017/S0967199409990207. [DOI] [PubMed] [Google Scholar]
- Dean W.L. Kelsey G. Reik W. Generation of monoparental embryos for investigation into genomic imprinting. Methods Mol. Biol. 2001;181:1–19. doi: 10.1385/1-59259-211-2:1. [DOI] [PubMed] [Google Scholar]
- Fulka H. Mrazek M. Tepla O., et al. DNA methylation pattern in human zygotes and developing embryos. Reproduction. 2004;128:703–708. doi: 10.1530/rep.1.00217. [DOI] [PubMed] [Google Scholar]
- Gardner A.J. Evans J.P. Mammalian membrane block to polyspermy: new insights into how mammalian eggs prevent fertilisation by multiple sperm. Reprod. Fertil. Dev. 2006;18:53–61. doi: 10.1071/rd05122. [DOI] [PubMed] [Google Scholar]
- Gardner A.J. Williams C.J. Evans J.P. Establishment of the mammalian membrane block to polyspermy: evidence for calcium-dependent and -independent regulation. Reproduction. 2007;133:383–393. doi: 10.1530/REP-06-0304. [DOI] [PubMed] [Google Scholar]
- Gómez E. Gutiérrez-Adán A. Díez C., et al. Biological differences between in vitro produced bovine embryos and parthenotes. Reproduction. 2009;137:285–295. doi: 10.1530/REP-08-0220. [DOI] [PubMed] [Google Scholar]
- Haaf T. Methylation dynamics in the early mammalian embryo: implications of genome reprogramming defects for development. Curr. Top. Microbiol. Immunol. 2006;310:13–22. doi: 10.1007/3-540-31181-5_2. [DOI] [PubMed] [Google Scholar]
- Hornak M. Hulinska P. Musilova P., et al. Investigation of chromosome aneuploidies in early porcine embryos using comparative genomic hybridization. Cytogenet. Genome Res. 2009;126:210–216. doi: 10.1159/000245922. [DOI] [PubMed] [Google Scholar]
- Kono T. Sotomaru Y. Sato Y., et al. Development of androgenetic mouse embryos produced by in vitro fertilization of enucleated oocytes. Mol. Reprod. Dev. 1993;34:43–46. doi: 10.1002/mrd.1080340107. [DOI] [PubMed] [Google Scholar]
- Lagutina I. Lazzari G. Duchi R., et al. Developmental potential of bovine androgenetic an parthenogenetic embryos: a comparative study. Biol. Reprod. 2004;70:400–405. doi: 10.1095/biolreprod.103.021972. [DOI] [PubMed] [Google Scholar]
- Latham K.E. Patel B. Bautista F.D., et al. Effects of X chromosome number and parental origin on X-linked gene expression in preimplantation mouse embryos. Biol. Reprod. 2000;63:64–73. doi: 10.1095/biolreprod63.1.64. [DOI] [PubMed] [Google Scholar]
- Loi P. Ledda S. Fulka J., Jr, et al. Development of parthenogenetic and cloned ovine embryos: effect of activation protocols. Biol. Reprod. 1998;58:1177–1187. doi: 10.1095/biolreprod58.5.1177. [DOI] [PubMed] [Google Scholar]
- Loi P. Beaujean N. Khochbin S., et al. Asymmetric nuclear reprogramming in somatic cell nuclear transfer? Bioessays. 2008;30:66–74. doi: 10.1002/bies.20684. [DOI] [PubMed] [Google Scholar]
- Matsukawa K. Turco M.Y. Scapolo P.A., et al. Development of sheep androgenetic embryos is boosted following transfer of male pronuclei into androgenetic hemizygotes. Cloning Stem Cells. 2007;9:374–381. doi: 10.1089/clo.2006.0016. [DOI] [PubMed] [Google Scholar]
- McGrath J. Solter D. Completion of mouse embryogenesis requires both the maternal and paternal genomes. Cell. 1984;37:179–183. doi: 10.1016/0092-8674(84)90313-1. [DOI] [PubMed] [Google Scholar]
- Miki H. Hirose M. Ogonuki N., et al. Efficient production of androgenetic embryos by round spermatid injection. Genesis. 2009;47:155–160. doi: 10.1002/dvg.20451. [DOI] [PubMed] [Google Scholar]
- Ptak G. Loi P. Dattena M., et al. Offspring from one month old lambs: studies on the developmental capability of prepubertal oocytes. Biol. Reprod. 1999;61:1568–1574. doi: 10.1095/biolreprod61.6.1568. [DOI] [PubMed] [Google Scholar]
- Ptak G. Clinton M. Tischner M., et al. Improving delivery and offspring viability of in vitro-produced and cloned sheep embryos. Biol. Reprod. 2002;67:1719–1725. doi: 10.1095/biolreprod.102.006171. [DOI] [PubMed] [Google Scholar]
- Reik W. Santos F. Mitsuya K., et al. Epigenetic asymmetry in the mammalian zygote and early embryo: relationship to lineage commitment? Philos. Trans. R. Soc. Lond. B Biol. Sci. 2003;358:1403–1409. doi: 10.1098/rstb.2003.1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs F. Stretch-activated ion channels: what are they? Physiology (Bethesda) 2010;25:50–56. doi: 10.1152/physiol.00042.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi W. Dirim F. Wolf E., et al. Methylation reprogramming and chromosomal aneuploidy in in vivo fertilized and cloned rabbit preimplantation embryos. Biol. Reprod. 2004;71:340–347. doi: 10.1095/biolreprod.103.024554. [DOI] [PubMed] [Google Scholar]
- Strumpf D. Mao C.A. Yamanaka Y., et al. Cdx2 is required for correct cell fate specification and differentiation of trophectoderm in the mouse blastocyst. Development. 2005;132:2093–2102. doi: 10.1242/dev.01801. [DOI] [PubMed] [Google Scholar]
- Surani M.A.H. Barton S.C. Norris M.L. Development of reconstituted mouse eggs suggested imprinting of the genome during gametogenesis. Nature. 1984;308:548–550. doi: 10.1038/308548a0. [DOI] [PubMed] [Google Scholar]
- Thouas G.A. Korfiatis N.A. French A.J., et al. Simplified technique for differential staining of inner cell mass and trophectoderm cells of mouse and bovine blastocysts. Reprod. Biomed. Online. 2001;3:25–29. doi: 10.1016/s1472-6483(10)61960-8. [DOI] [PubMed] [Google Scholar]
- Van De Velde A. Liu L. Bols P.E., et al. Cell allocation and chromosomal complement of parthenogenetic and IVF bovine embryos. Mol. Reprod. Dev. 1999;54:57–62. doi: 10.1002/(SICI)1098-2795(199909)54:1<57::AID-MRD8>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- Vanneste E. Voet T. Le Caignec C., et al. Chromosome instability is common in human cleavage-stage embryos. Nat. Med. 2009;15:577–583. doi: 10.1038/nm.1924. [DOI] [PubMed] [Google Scholar]
- Xie Y. Li L. Wang X., et al. Overexpression of Cdx2 inhibits progression of gastric cancer in vitro. Int. J. Oncol. 2010;36:509–516. [PubMed] [Google Scholar]
- Yang X.C. Sachs F. Characterization of stretch-activated ion channels in Xenopus oocytes. J. Physiol. 1990;431:103–122. doi: 10.1113/jphysiol.1990.sp018322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X.C. Sachs F. Stretch-activated (SA) ion channel in Xenopus oocytes: permeation and block. Biophys. J. 1988;53:412a. [Google Scholar]
- Young L.E. Beaujean N. DNA methylation in the preimplantation embryo: the differing stories of the mouse and sheep. Anim. Reprod. Sci. 2004;83:61–78. doi: 10.1016/j.anireprosci.2004.05.020. [DOI] [PubMed] [Google Scholar]
- Zhao Q. Wang J. Zhang Y., et al. Generation of histocompatible androgenetic embryonic stem cells using spermatogenic cells. Stem Cells. 2010;28:229–239. doi: 10.1002/stem.283. [DOI] [PubMed] [Google Scholar]