Abstract
5-Aza-2′-deoxycytidine (AzC), trichostatin A (TSA), and its natural mimetic, sodium butyrate (NaB), are antineoplastic drugs that can modify the epigenetic status of donor cells prior to somatic cell nuclear transfer (SCNT). In this study, we used fibroblast cells treated with these drugs to investigate the direct and indirect effects of induced changes in DNA methylation and acetylation of the lysine 9 residue of histone H3 (H3K9). Additionally, we assayed cellular characteristics (cell growth, cell proliferation, cell cycle progression, and apoptosis) and SCNT efficiency in response to these drugs as well as monitoring these effects 24 h after removing the drugs. We observed the following: (1) AzC, TSA, and NaB all showed dose-dependent effects on different cellular characteristics; (2) TSA and NaB induced H3K9 hyperacetylation accompanied by DNA hypermethylation, whereas AzC induced DNA hypomethylation with no effect on H3K9 hyperacetylation; (3) TSA and NaB improved cloning efficiency, whereas AzC reduced it; and (4) unlike AzC, the effects of TSA and NaB on cellular characteristics and SCNT efficiency were reversed following drug removal. Our results indicate that somatic cells treated with TSA and NaB show better survival and recovery rates following the removal of these drugs. Moreover, H3K9 hyperacetylation (induced with TSA and NaB), but not DNA hypomethylation (induced with AzC), favors cloning efficiency.
Introduction
The fifth base of mammalian DNA, 5-methyl cytosine (5-mC), and the acetylation status of lysine residues on histones H3 and H4 are two central elements that regulate gene expression (Jones et al., 1998; Tse et al., 1998). In 1997, the birth of the first cloned mammal (Dolly, the sheep) through somatic cell nuclear transfer (SCNT) technology proved that differentiated cells can be reprogrammed to revert to the embryonic state (Wilmut et al., 1997). However, almost 15 years later, the efficiency of SCNT is still very low. Among the different factors involved in this process, aberrant epigenetic reprogramming of the nuclei donor cell has been considered to be the most important for determining cloning efficiency (Dean et al., 2001; Deshmukh et al., 2011; Lan et al., 2010; Santos et al., 2003; Sawai et al., 2010).
5-Aza-2′-deoxycytidine (AzC) and trichostatin A (TSA) are two synthetic antineoplastic drugs that inhibit DNA methyltransferase (DNMTase) and histone deacetylase (HDAC) enzymes, respectively (Kharroubi et al., 2001; Yoshida et al., 1990). Sodium butyrate (NaB), another antineoplastic drug, is a natural mimetic of TSA and is normally present in the large intestine, where it inhibits excessive cell proliferation (Candido et al., 1978). AzC induces DNA hypomethylation through the inactivation of DNMTase by acting as a substrate analog and covalently binding to the enzyme in CpG islands of DNA (Christman, 2002). In addition, AzC indirectly causes hyperacetylation by disrupting HDAC recruitment by methyl-binding proteins, whose binding sites have been lost due to AzC incorporation (Jones et al., 1998).
Considerable efforts have been made to treat somatic donor cells with some epigenetic drugs prior to SCNT. Enright et al. (2003b), Ding et al. (2008) and Li et al. (2008) have shown that pre-SCNT treatment of donor cells with TSA could improve in vitro development of cloned embryos. However, reports about the impact of AzC on SCNT are disappointing (Enright et al., 2003b, 2005; Jones et al., 2001). It is still unclear, however, which of these epigenetic changes (DNA methylation or histone acetylation) is more important for reprogramming and somatic cell cloning. Although it has been reported that induced DNA hypomethylation by AzC stimulates histone hyperacetylation, it is not known whether induced histone hyperacetylation by TSA and NaB can also result in DNA hypomethylation. Given that TSA, NaB, and AzC are potentially toxic, in addition to understanding their role in epigenetics, it is also important to systematically investigate their effects on different cellular characteristics (cell growth, proliferation, cell cycle progression, and apoptosis) of somatic cells that are candidates for epigenetic modification prior to SCNT, which is investigated in the first part of this study. Moreover, it is unknown whether the effects of these drugs are reversible. The second part of this study aims to determine the extent to which cellular characteristics and cloning efficiencies may be affected following drug removal and cell refreshment.
Materials and Methods
Unless otherwise specified, chemicals and media were obtained from Sigma Aldrich Chemicals (St. Louis, MO, USA) and Gibco (Invitrogen Corporation, Grand Island, NY, USA), respectively. This study received the approval of the Ethical Committee of Royan Institute (www.royaninstitute.org).
Adult somatic cell collection, culture, and characterization
Adult bovine ear fibroblast (BEF) cells were prepared from a 9-month-old bull as described by Enright et al. (2003b) and Hosseini et al. (2008). Briefly, a skin biopsy was taken from the ear, cut into small pieces (2–3 mm2), and the explants were cultured in Dulbecco's modified Eagle medium F-12 (DMEM/F-12) containing 10% fetal calf serum (FCS) and 1% penicillin-streptomycin at 37°C in a humidified atmosphere of 5% CO2 until confluence. Fibroblast lineage was confirmed by immunocytochemical staining as described by Kubota et al. (2000). Briefly, confluent cultured cells were prepared over coverslips, washed with phosphate buffer saline (PBS) containing 0.05% Tween-20 (PBS-T), and then fixed with fresh 4% paraformaldehyde (PF) in PBS (pH=7.2–7.4) for 45 min. After thorough washing with PBS-T (to avoid PF-autofluorescence), the samples were permeabilized with 0.2% Triton X-100 for 30 min at room temperature (RT). Samples were incubated with blocking solution [3% bovine serum albumin (BSA) in PBS] for 1 h at RT and then incubated with either 100 μL of primary antibody against antivimentin clone 11 (Sigma, A6630 at 1:40 dilution for detection of intermediate filaments vimentin of fibroblasts) or antipan cytokeratin clone 9 (Sigma, A2913 at 1:400 dilution for detection of epithelial cells) for 1 h at 37°C. After washing three times with PBS-T, samples were incubated with fluorescent-conjugated secondary antibody (Chemicon®, AP124F at 1:50 dilution in PBS plus 1% BSA). Finally, samples were washed and counterstained with 1 μg/mL 4′,6- diamidino-2-phenylindol (DAPI)] for 5 min to detect DNA using an epifluorescence microscope (Olympus, BX51) (excitation: 494 nm, emission: 518 nm).
Experimental design
At passage three, fibroblast cells were added in equal densities to 6-mm culture dishes containing DMEM/F-12 plus 10% FCS to be randomly subjected to one of the following treatments: TSA [(0.0, 0.03, 0.08, and 1.0 μM) for 24 h], NaB [(0.0, 0.5, 1.0, and 2.0 mM) for 96 h], or AzC [(0.0, 0.01, 0.08, and 0.3 μM) for 72 h]. Concentrations and durations of TSA, NaB, and AzC were selected based on previous reports (Enright et al., 2003b, 2005; Shi et al., 2003). For each treatment we seeded a specific cell density in 60-mm culture dishes, avoiding confluency (REF 353004, BD Falcon, Bedford, MA, USA): for drug treatment, 4×105, 5×104, and 1×105 cells were seeded for TSA, NaB, and AzC treatment, respectively, and for drug removal, 2×105, 25×103 and 5×104 cells were seeded for TSA, NaB, and AzC, respectively. The 22-mm2 coverslips were placed in 60-mm culture dishes for immunofluorescence staining. At the end of each treatment, treated cells along with their corresponding untreated groups were used for analysis of different cellular characteristics (cell growth, cell proliferation, cell cycle progression, apoptosis, and epigenetic markers) or SCNT as described below. TSA, NaB, and AzC were added to the culture medium for up to 96 h to assess their effects on cell growth. To investigate the effect of drug removal on cellular characteristics and cloning efficiency, for each of the above treatments, treated and nontreated (control) cells were refreshed (in the absence of drugs) for 24 h before somatic cell analyses and SCNT. Cells were refreshed by thoroughly washing them with PBS and further cultured in the presence of fresh preincubated DMEM/F12 plus 10% FCS for 24 h, then assessed as the treated cells were. This experiment was performed in triplicate. As AzC is relatively unstable and has a short half-life, the culture media was refreshed after 48 h.
Somatic cell assessments
Six experiments were conducted to assess the effects of varying concentrations of TSA, NaB, and AzC on different characteristics of cultured somatic donor cells. In addition, the capacity of treated cells to reverse these changes was investigated after drug removal. Each treatment was repeated three times, and for each treatment and replicate group, a corresponding control group was assayed.
Cell growth and morphology
To evaluate the effects of varying concentrations of each epigenetic drug at 24, 48, 72, and 96 h posttreatment on growth and morphology, fibroblast cells at an initial density of 2×104 were added to 12 60-mm culture dishes (BD, Falcon) containing preincubated DEMEM/F12 plus 10% FCS. The cultured cells were rested for 4 h to allow them to attach to the dish before treatment with the different concentrations of the epigenetic drugs (four dishes per concentration). The culture dishes were regularly observed under an inverted phase-contrast microscope (Olympus, CKX 41, Japan) and the digital images of cultured cells were taken at 200× magnification using a camera (Olympus, DP-72, Japan) operated on DP2-BSW software. Changes in cell morphology were interpreted based on the related studies of Mohana Kumar et al. (2007) and Enright et al. (2005). At 24, 48, 72, and 96 h postdrug treatment, the culture dishes were trypsinized (0.05% Trypsin–EDTA) and the cells were harvested to assess cell growth using a hemocytometer. Trypan blue was used to obtain the number of viable cell (Mohana Kumar et al., 2007).
Cell cycle analysis
Flowcytometric analysis of cell cycle was conducted as described by Nasr-Esfahani et al. (2011). Briefly, cultured cells in different treatment groups were trypsinized, washed with PBS by centrifugation (700×g for 10 min), and the resulting pellet was resuspended in ice-cold 75% ethanol for 1 h. The fixed cells were washed with PBS before being resuspended in staining solution [propidium iodide (50 μg/mL), RNase A (100 μg/mL), and Triton X-100 (0.5%) in PBS] for 20 min in the dark at RT. The cells were then filtered through 40-μm nylon mesh (BD Falcon, REF 352340) to remove cell aggregations. A total of 20,000 cells were collected on a fluorescence-activated cell sorter (FACS) Caliber (Becton Dickinson, San Jose, CA) and were analyzed using Modfit® software (Boquest et al., 1999).
Cell proliferation
Assessment of cell proliferation was conducted as described previously (Shi et al., 2003). Briefly, treated cells were cultured over 22-mm2 glass in the presence of 5-bromo-2′-deoxyuridine (BrdU: 10 μM, Sigma 858811) for 24 h before being washed with PBS-T and fixed in PF. The cells were then immunostained with mouse monoclonal anti-BrdU (diluted 1:750) as described for analysis of cell-specific markers except for an additional treatment with 4 N HCl before blocking with 3% BSA. The prepared samples were observed by epifluorescent microscopy (Olympus, BX51) to detect BrdU-incorporated cells (Emission 518 nm and excitation 494 nm).
DNA fragmentation
Assessment of DNA fragmentation was conducted using terminal deoxynucleotidyl transferase (TdT)-mediated dUTP-digoxigenin nick end labeling (TUNEL) with an in situ cell-death detection kit (Promega® Diagnostic Corporation, Madison, WI, USA) according to the manufacturer's recommendation. Briefly, cells grown over 22-mm2 glass coverslips in 12-well plates were washed with PBS-T and fixed with PF for 25 min at 4°C. After washing twice with PBS-T (5 min each), cells were permeabilized with 0.2% Triton X-100 in PBS for 5 min at RT and incubated in 100 μL of equilibration buffer for 10 min at RT. Subsequently, 50 μL TdT reaction mix (45 μL reaction buffer+5 μL TdT enzyme+1 μL nucleotide) was added to the cells and they were further incubated for 60 min at 37°C in a humidified chamber in the absence of light. The reaction was stopped with 2× SSC for 15 min. After washing with PBS-T, nuclei were counterstained with 2 μg/mL PI for 2 min. Finally, localized yellow fluorescence of apoptotic cells was detected using fluorescence microscopy.
DNA methylation and H3K9 acetylation
The effects of epigenetic drugs on DNA methylation and histone H3K9 acetylation levels of the treated cells were assessed using two techniques, epifluorescence microscopy and flow cytometry, as described below.
Epifluorescence microscopy
Fibroblasts were cultured on 22-mm2 glass coverslips in 12-well plates and treated with TSA, NaB, and AzC as described above. DNA methylation was detected using immunofluorescence staining with mouse monoclonal anti-5-methyl-cytosine (Eurogentec P/N. BI-MEC1Y-0100), as described for cell proliferation. Immunofluorescence staining for detection of H3K9 acetylation with mouse monoclonal anti-H3K9 (Sigma, H0913) was as described for characterization of fibroblast cells.
Flowcytometry
Quantitative assessment of DNA methylation and histone H3K9 acetylation (Enright et al., 2003a) was conducted by incubating the treated cells with 1:400 and 1:200 dilutions of mouse anti-5-methyl cytosine and anti-H3K9 monoclonal antibodies, respectively. Fluorescein isothiocyanate conjugated goat antimouse immunoglobin was used as the secondary antibody at a 1:50 dilution. Corresponding controls for each experiment were included. Cells were filtered through 40-μm nylon mesh in order to exclude aggregation. Ten thousand cells were collected with the FACS-Caliber and were analyzed using CELL QUEST® 3.1 software (Becton Dickinson). Three replicates were conducted for each treatment with appropriate controls to eliminate the possible effects of autofluorescence and nonspecific binding by the secondary antibody.
Somatic cell nuclear transfer
In vitro maturation and zona-free enucleation
Abattoir-derived ovaries were used as the source of oocyte. The process of oocyte in vitro maturation was as described previously (Moulavi et al., 2006). In brief, cumulus–oocyte complexes were aspirated of antral follicles (2–8 mm) and cultured in tissue culture medium 199 (TCM199) containing 10% FCS, FSH (10 μg/mL), LH (10 μg/mL), estradiol-17β (1 μg/mL), cysteamine (0.1 mM) at 39°C, 5% CO2 and humidified air for 18–20 h. Matured oocytes were denuded by vortexing for 3–5 min in HEPES-buffered TCM199 (HTCM199) supplemented with 300 IU/mL hyalorunidase. For SCNT, the high throughput zona-free SCNT method of Oback et al. (2003) was used with minor modifications. In brief, denuded oocytes were treated with pronase (5 mg/mL, 1 min) to remove zona. Oocyte were then incubated with 5 μg/mL H33342 for 5 min, transferred on the microscope stage (Olympus; IX71) equipped with Narishige micromanipulators (Olympus). At the 100× magnification, zona-free oocytes were enucleated using a blunt (10–15 μm inner diameter) micropipettes and a blind separation micropipettes oocytes under UV exposure.
Nuclear transfer, artificial activation, and embryo development
For renucleation, oocytes were adhered to individual fibroblasts in medium containing 10 mg/mL phytohemagglutinin. The couplets were then electrofused [a sinusoidal electric current (7 V/cm) for 10 sec followed by two direct currents (1.75 kV/cm for 30 μsec and 1 sec delay)] in 290 mOsm fusion buffer free of Ca2+ and Mg2+ and the reconstructed oocytes were activated using ionomycin (5 μM, 5 min) followed by incubation with 2 mM 6-DMAP for 4 h (Hosseini et al., 2006). Accordingly, embryos in groups of five to seven were cultured in wells drained in 20 μL of modified synthetic oviductal fluid (mSOF) (Tervit et al., 1972) under mineral oil at 39.0°C, 5% CO2, 5% O2, and humidified air (Vajta et al., 2000).
Blastocyst analysis
At day 8, blastocysts in each group were assigned to differential staining to detect total cell number (TCN), number of cells in the inner cell mass (ICM) and the ratio of ICM/TCN (Hosseini et al., 2008). Briefly, blastocysts were incubated in 500 μL of 1% Triton X-100 and 100 μg/mL propidium iodide for up to 30 sec, depending on the size of the embryos, and then immediately transferred into a 500 μL solution of 100% ethanol plus 25 μg/mL H33342 and stored at 4°C overnight. Fixed and stained embryos were subsequently mounted on glass microscope slides in one drop of glycerol, gently flattened with a coverslip, and visualized for cell counting on a fluorescence microscope (Olympus) using 460-nm excitation filter for blue and 560 nm for red. TE cells were visualized as blue and ICM as pink to red. TCN was calculated by counting the numbers of both ICM and TE.
Statistical analysis
Data reflecting the percentages of cloned embryo development were modeled using the binomial model of parameters by ArcSin transformation. The transformed data, along with the crude data of the cellular characteristics, were analyzed by the one-way analysis of variance (ANOVA) model of SPSS 17. Differences were compared by Tukey's multiple comparison post hoc test. All data for drug treatment and drug recovery were compared and analyzed by the paired sample t-test. All data were presented as means±SEM and differences were considered to be significant at a cutoff value of p<0.05.
Results
Effect of drug treatment and postdrug recovery on cellular characteristics
Morphology
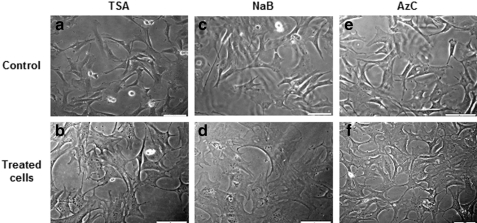
The majority (over 90%) of untreated cells showed compact cytoplasm with intact plasmalemma and clear borders. In contrast, treated cells showed alterations in morphology that included cell size increase, flattening, and occurrence of cell processes—a phenotype commonly observed in senescent cells (Shi et al., 2003) (Fig. 1B). These changes in cellular morphology were observable in the presence of all three drugs, but most evident in NaB-treated cells, and more prominent at the highest drug concentrations. Overall, following drug removal, the refreshed cells exhibited restored, normal morphology at all but the highest concentrations of the three drugs.
FIG. 1.
Morphology of untreated and treated cells with the highest concentration of TSA, NaB, and AzC. Morphology of donor cells treated with (b) 1 μM TSA or with (d) 2 mM NaB or with (f ) 0.3 μM 5-aza-dC in comparison with (a, c, and e) untreated cells. Original magnification ×200. The bars represent 50 μm.
Growth and proliferation
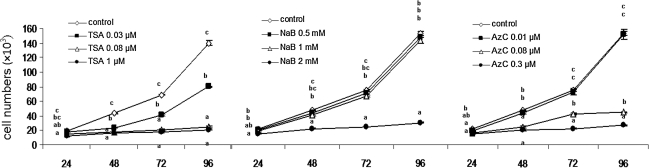
Figure 2 represents growth curves of cells treated with TSA, NaB, and AzC for 24, 48, 72, and 96 h. As shown, irrespective of the type of drug used, dose-dependent growth inhibition was observed that became more evident as the duration of drug treatment increased. Two concentrations of TSA (0.08 and 1.0 μM) and AzC (0.08 and 0.3 μM), and the highest concentration of NaB (2.0 mM) significantly reduced cell growth at all durations when compared with the untreated cells (p<0.05).
FIG. 2.
Effect of various concentrations of trichostatin A (TSA), sodium butyric (NaB) and 5-aza-2′-deoxycytidine on (AzC) on cell growth treated for 24, 48, 72, and 96 h, respectively. Different subscripts within one time point is significantly different at p<0.05.
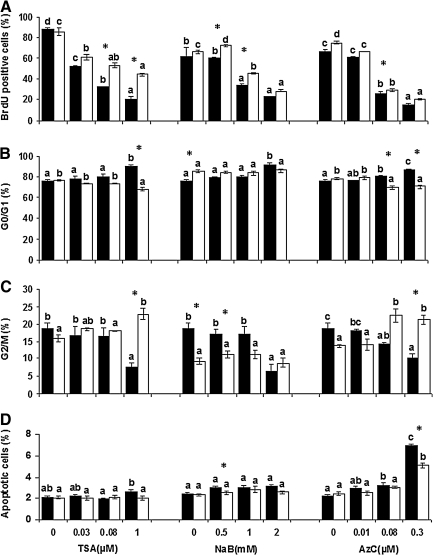
BrdU assessment of cell proliferation (Fig. 3A) indicated that the percentages of cells that were capable of BrdU incorporation decreased as the concentrations of TSA, NaB, and AzC increased. Percentages of BrdU-positive cells treated with all concentrations of TSA (0.03, 0.08, 1.0 μM), the two highest concentrations of NaB (1.0 and 2.0 mM), and AzC (0.08 and 0.3 μM) were lower than untreated cells. After drug removal, a higher capacity for BrdU incorporation was observed in the refreshed cells for all treatments compared with unrefreshed cells. These increases reached a significant level at 0.08 μM (53.4±2.2%), 1.0 μM (44.4±1.6%)] TSA, 0.5 mM (72.7±1.0), and 1 mM (45.4±0.7) NaB, and 0.08 μM (29.3±1.0) AzC (Fig. 3.A).
FIG. 3.
Different cellular characteristics including (A) BrdU labeling, cell in (B) G0/G1, and (C) G2/M stage, and (D) apoptotic cells in control and bovine fibroblast cells treated with various concentrations of trichostatin A (TSA) (0. 0.03, 0.08, and 1 μM), sodium butyric (NaB) (0, 0.5, 1, and 2 mM) and 5-aza-2′-deoxycytidine (AzC) (0, 0.01, 0.08, and 0.3 μM) for 24, 96, and 72 h, respectively (black columns), and after drug removal for 24 h (white columns). Comparisons were carried out at three levels: (1) within the drug-treated groups, (2) within the drug removal groups, and (3) between drug-treated and drug-removal groups. Accordingly, values with at least one common letter on black or white columns indicate nonsignificant difference within the drug treated and drug removal groups, whereas asterisks indicate significant difference between drug treatment and drug removal for each drug treatment at each concentration at p<0.05.
Cell cycle
Figure 3B and C compare the distribution of cells in two cell-cycle stages (G0/G1 and G2/M) immediately after drug treatment and 24 h after drug recovery.
After treatment with 1.0 μM TSA for 24 h, a significant increase in the G0/G1 fraction (90.2±1.3%) along with a significant decrease in the G2/M ratio (7.7±1.2%) was observed compared with cells treated with 0.03 and 0.08 μM TSA. Upon drug removal, a reduction in G0/G1 ratios and an increase in G2/M ratios were observed which were significant at 1.0 μM TSA (67.5±2.0% and 22.9±1.7%, respectively; p<0.05).
For NaB, the trend of cell-cycle changes was similar to that observed in TSA-treated cells. When cells were treated with 2.0 mM NaB for 96 h, the percentages of cells in G0/G1 (91.4±2.2%) and G2/M (6.4±2.0%) were both significantly different when compared with cells treated at lower concentrations of NaB (0, 0.5, and 1.0 mM) (p<0.05). Drug removal did not dramatically change these ratios, except for the percentages of cells located in G2/M stage (11.2±1.0%) for 0.5 mM NaB.
After 72-h exposure to 0.08 μM and 0.3 μM AzC, the respective percentages of cells in G0/G1 were 80.9±0.5% and 86.5±1.4%, which were significantly different compared with the same rates of untreated cells (75.6±1.6%) (p<0.05). However, for the G2/M stage, the only significant difference was observed between 0.01 μM (18.2±0.2%) and 0.3 μM (10.2±1.2%) (p<0.05) AzC-treated cells. After drug removal, a significant reduction in the percentage of cells in G0/G1 was observed for both the 0.08 μM and 0.3 μM AzC-treated cells (69.5±2.0% and 70.7±1.5%, respectively), but a significant increase in the G2/M fraction was only seen for 0.3 μM AzC-treated cells (21.3±1.4%) compared with unrefreshed cells (p<0.05).
Apoptosis
As depicted in Figure 3D, a negligible amount of untreated cells (less than 2.4±0.1%) showed signs of apoptosis. For TSA- and NaB-treated cells, there was a slight, insignificant increase in apoptotic rates compared with untreated cells. However, with an increase in AzC concentration, there was also an increase in the apoptosis rate, which was significant at 0.08 μM and 0.3 μM AzC concentration (3.2±0.2% and 6.9±0.2%, respectively) when compared with untreated cells. After drug removal, the mean percentages of apoptotic cells were not significantly different than unrefreshed cells, with the exception of 0.5 mM NaB and 0.3 μM AzC (2.5±0.2% and 5.2±0.3%, respectively) (p<0.05).
DNA methylation and histone acetylation
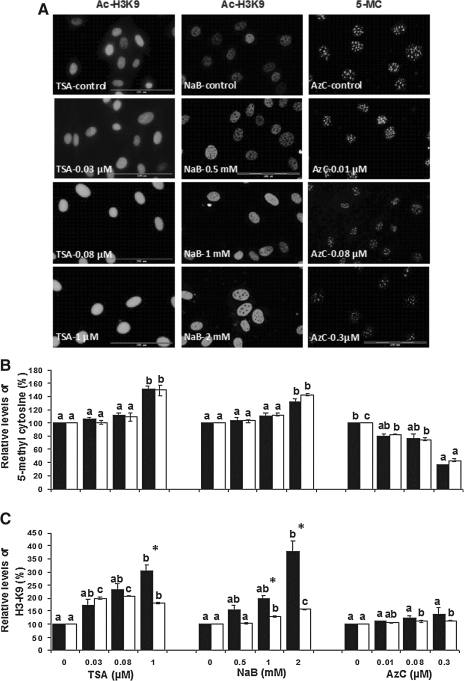
Figure 4A shows the immunostaining of BEF cells treated with TSA and NaB for Ac-H3K9 and cells treated with AzC for 5-mC. Figure 4B and C shows the effects of drug treatment and postdrug recovery on the levels of DNA methylation and histone acetylation as detected by FACS.
FIG. 4.
(A) Immunostaining of BEF cells treated with TSA and NaB for Ac-H3K9 and cells treated with AzC for 5-MC. Cells were immunostained with anti-H3K9 or anti-5MC (grey). In nontreated cells intensity of Ac-H3K9 was weak, whereas in TSA- and NaB-treated cells intensity was strong. On the other hand, 5-MC intensity is also strong in nontreated cells compared with AzC-treated cells. Scale bar represents 200 μm. Relative levels of (B) DNA methylation and (C) histone H3K9 acetylation in control and treated bovine fibroblast cells treated with various concentrations of trichostatin A (TSA), sodium butyric (NaB), and 5-aza-2′-deoxycytidine (AzC) for 24, 96, and 72 h, respectively (black column) and after drug removal for 24 h (white column). Comparison was carried out between drug groups or the drug removal groups. Different subscript indicate significantly difference between the groups, whereas asterisks indicate significant difference between drug treatment and drug removal for each drug treatment at each concentration at p<0.05.
As depicted in Figure 4B, there was a significant increase in DNA methylation level of cells that was treated with 1 μM TSA in comparison to cells treated with 0.03 and 0.08 μM TSA and untreated cells. H3K9 acetylation levels in cells treated with 1.0 μM TSA were significantly higher than untreated cells. After drug removal there were no significant changes in methylation, but a significant reduction in acetylation was observed at 1.0 μM TSA compared with unrefreshed cells. However, this lowered level of acetylation after drug removal was still significantly higher than untreated cells (p<0.05).
The methylation level in 2.0 mM NaB-treated cells was significantly higher than 0, 0.5, and 1 mM NaB-treated cells, and a dose-dependent increase in acetylation was observed. At 2.0 mM NaB, the acetylation value was significantly higher than untreated cells. After drug removal, no group showed changes in methylation levels. However, there was a clear reduction in acetylation levels of refreshed versus unrefreshed cells that attained a significant value at 1.0 and 2.0 mM NaB. These reduced levels of acetylation after drug removal were significantly higher than untreated cells (p<0.05).
As concentrations of AzC increased, the levels of DNA methylation decreased, which was significant at the highest concentration compared with untreated cells (p<0.05). However, unlike the indirect effect of TSA and NaB on induced hypermethylation, AzC-related hypomethylation did not indirectly result in an elevation of acetylation in treated cells. Moreover, drug removal did not have a specific effect on acetylation or methylation.
In vitro cloned embryo development in association with fibroblast cell epigenetic treatments
Table 1 represents the results of in vitro development and quality of cloned embryos derived from drug-treated cells. In this experiment, the best quality oocytes (with intact, round, and evenly granulated cytoplasm and first polar-body extruded/extruding) were selected to reduce interexperiment variation. As described in the experimental design, a corresponding control group was included along with each drug. However, because the developmental data of the control groups were very similar, these data were pooled and presented as a single control group. As shown, there was no significant difference between the cleavage rates of different groups. However, as the concentrations of TSA and NaB increased, the capabilities of treated fibroblast cells to develop to the blastocyst stage were enhanced. Accordingly, the percentages of blastocyst production for fibroblast cells treated with the highest concentrations of TSA (1.0 μM) and NaB (2.0 mM) were 45.2±0.8% and 46.5±0.8%, which were insignificantly different when compared with the related rates of fibroblast cells treated with 0.08 μM TSA (37.9±1.8%) and 1.0 mM NaB (39.7±2.1%). However, they were significantly higher than the related rates of cells treated with the lowest concentrations of TSA (0.03 μM: 29.6±1.1%), NaB (0.5 mM: 27.4±1.3%), and the untreated group (28.7±1.8%). Contrary to the findings with TSA and NaB, treatment with AzC negatively affected the potency of treated fibroblast cells to develop to the blastocyst stage. Importantly, this negative effect of AzC became more evident as AzC concentrations were increased; at the highest AzC concentration (0.3 μM), the proportion of cleaved embryos that developed to blastocyst was the lowest (1.5±1.5%) and significantly different from the other treatments. Analysis of TCN, ICM and ICM/TCN ratios of the SCNT-derived blastocysts indicated that TSA and NaB, in contrast with AzC, positively and dose-dependently improved the quality of the SCNT-derived blastocysts. After treatment with 1.0 μM TSA and 2.0 mM NaB, the mean number of ICM and TCN and the ICM/TCN ratio (%) were significantly greater than the related rates of blastocysts that developed from other groups.
Table 1.
Effect of Donor Cell Treatment with Different Concentrations of TSA, NaB, and AzC on Developmental Competence and Quality of the SCNT-Derived Embryo
| |
|
|
|
Embryo development (%) |
Blastocyst quality |
||||
|---|---|---|---|---|---|---|---|---|---|
| No. replicates | Activated oocytes (n) | Cleavage | Blastocyst | Blastocyst (n) | ICM | TCN | ICM/TCN (%) | ||
| Untreated | 9 | 403 | 356 (94.7±1.4) ab | 102 (28.7±1.8) a | 15 | 31.1±1.9 a | 121.1±3.9a | 25.7±1.5 a | |
| 0.03 | 4 | 116 | 98 (83.9±5.6) ab | 29 (29.6±1.2) ab | 10 | 37.3±2.9 ab | 131.4±6.3 ab | 28.2±1.6 ab | |
| TSA (μM) | 0.08 | 4 | 141 | 124 (88.0±3.1) ab | 47 (37.9±1.8) bc | 9 | 44.9±3.3 b | 134.8±6.4 ab | 33.1±1.4 bc |
| 1.0 | 3 | 112 | 93 (89.4±4.7) ab | 42 (45.2±0.8) c | 12 | 57.1±2.9 c | 146.1±3.9 b | 38.9±1.4 c | |
| 0.5 | 3 | 124 | 102 (89.5±1.8) ab | 28 (27.4±1.3) a | 9 | 37.2±3.7 ab | 118.8±5.7 a | 30.9±2.0 a | |
| NaB (mM) | 1.0 | 5 | 161 | 146 (93.6±2.2) ab | 58 (39.7±2.1) b | 10 | 43.3±2.7 b | 140.1±5.7 b | 30.8±1.3 a |
| 2.0 | 3 | 110 | 95 (95.9±2.1) ab | 46 (46.5±0.8) b | 12 | 54.3±2.8 c | 146.6±5.1 b | 36.9±1.0 b | |
| 0.01 | 5 | 175 | 154 (97.5±0.7) b | 36 (23.3±1.3) a | 8 | 37.8±3.3 a | 121.5±7.0 a | 30.7±1.1 a | |
| AzC (μM) | 0.08 | 3 | 104 | 93 (97.9±2.1) b | 19 (20.4±3.9) a | 7 | 24.4±2.2 ab | 105.6±6.9 ab | 22.9±0.8 ab |
| 0.3 | 4 | 157 | 133 (89.3±0.9) a | 2 (1.5±1.5) b | 2 | 16.5±1.5 b | 83.0±6.0 b | 19.9±0.4 b | |
Within each column, values with at least one different letter are significantly different at p<0.05.
SCNT, somatic cell nuclear transfer; TSA, trichostatin A; AzC, 5-Aza-2′ -deoxycytidine; NaB, sodium butyrate.
Table 2 represents the effect of postdrug recovery on yield and quality of the cloned embryos. As shown, there was no significant difference between the cleavage rates of the different groups. Also, the competency of fibroblast cells after drug removal was the same between untreated cells, those treated with different concentrations of TSA and NaB, and cells treated with the lowest concentration of AzC. However, no blastocysts were produced from refreshed cells previously treated with 0.3 μM AzC. No conclusive difference in the quality of blastocysts was confirmed.
Table 2.
Effect of 24-h Refreshment After Donor Cell Treatment with Different Concentrations of TSA, NaB, and AzC on Developmental Competence and Quality of the SCNT-Derived Embryos
| |
|
|
|
Embryo development (%) |
Blastocyst quality |
||||
|---|---|---|---|---|---|---|---|---|---|
| Activated oocytes (n) | Cleavage | Blastocyst | Blastocyst (n) | ICM | TCN | ICM/TCN (%) | |||
| Untreated | 5 | 340 | 302 (93.6±2.2) ab | 78 (25.8±2.1) b | 10 | 34.3±2.9 a | 126.1±6.1 a | 26.8±1.1 a | |
| 3 | 0.03 | 120 | 101 (91.8±1.5) ab | 31 (30.7±0.6) b | 10 | 37.6±3.3 a | 129.2±7.7 a | 28.7±1.3 a | |
| TSA (μM) | 3 | 0.08 | 135 | 121 (95.3±0.7) ab | 38 (31.4±1.0 ) b | 8 | 41.3±4.1 ab | 135.8±6.1 a | 29.9±1.8 a |
| 3 | 1.0 | 105 | 88 (94.7±1.5 ) ab | 30 (34.1±1.1) b | 10 | 53.6±3.8 b | 142.2±6.0 a | 37.3±1.4 b | |
| 3 | 0.5 | 100 | 83 (89.2±2.1 ) ab | 25 (30.1±1.5) b | 9 | 36.7±4.2 ab | 120.1±5.4 a | 29.8±2.3 ab | |
| NaB (mM) | 3 | 1.0 | 125 | 102 (92.7±0.7 ) ab | 36 (35.3±1.7 ) b | 10 | 43.4±3.6 ab | 135.2±6.9 a | 31.6±1.2 ab |
| 3 | 2.0 | 110 | 96 (96.0±2.1 ) ab | 34(35.4±0.8 ) b | 10 | 48.7±3.9 b | 138.5±6.6 a | 34.7±1.3 b | |
| 3 | 0.01 | 170 | 150 (96.2±1.3 ) b | 42 (28±0.8 ) b | 7 | 42.4±4.2 b | 121.7±8.0 a | 34.4±1.4 b | |
| AzC (μM) | 3 | 0.08 | 145 | 125 (94.7±2.6 ) ab | 25 (20±0.9) b | 7 | 29.3±2.5 a | 118.4±6.0 a | 24.4±1.0 a |
| 3 | 0.3 | 132 | 104 (86.7±1.5 ) a | 0.0 (0.0±0.0) a | — | — | — | — | |
Within each column, values with at least one different letter are significantly different at p<0.05 for each drug in compare with untreated group.
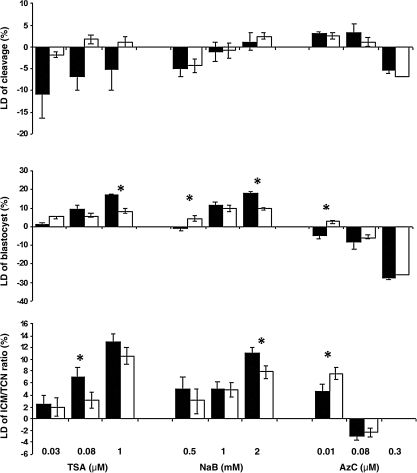
Figure 5 represents a statistical comparison of the level of difference (LD) between the developmental rate and blastocyst analysis of cloned embryos derived from drug-treated versus drug-refreshed cells. LD was calculated based on the difference between the treated and the corresponding untreated group for each replicate. Overall, although there was no specific effect of drug treatment and removal on cleavage rate, refreshed cells previously treated with TSA and NaB had a reduced capability to develop into cloned blastocysts, as evidenced by the significant difference between the LD values of unrefreshed versus refreshed cells treated with 1.0 μM TSA and 0.5 and 2.0 mM NaB. Despite nonspecific differences between LD values of blastocysts produced with 0.08 and 0.3 μM AzC, the blastocyst LD value of 0.01 μM AzC was significantly in favor of refreshed cells.
FIG. 5.
Show the level of difference (LD) for each treatment relative to the control at cleavage, blastocyst stage, and the ratio of inner cell mass (ICM) over the total cell number (TCN). The level of difference was obtained by subtracting the value of each treatment group from its related control. Asterisks indicate significant difference between drug treatment (black column) and drug removal (white column) for each drug at p<0.05. Three replicates were carried out for this experiment.
Discussion
Epigenetic modification of donor cells with anticancer drugs for SCNT is becoming a focal point of research in the cloning field. Our observations of cells treated with TSA, NaB, and AzC indicate that histone acetylation and DNA methylation are not independent phenomena, and any alteration in methylation is accompanied by an alteration in acetylation, and vice versa. We showed that treatment with TSA and NaB led to a significant increase in acetylation of histone H3K9, which was accompanied by a significant increase in the levels of methylation of CpG islands at the highest concentrations of these drugs. In contrast, AzC induced DNA hypomethylation that was not associated with a significant increase in acetylation levels. In concordance with these findings, Enright et al. (2003b) reported that AzC induced global hypomethylation that indirectly resulted in global hyperacetylation.
A comparison between TSA and NaB indicated that both drugs caused similar alterations in acetylation and methylation levels in somatic cells. Contrary to this notion, Shi et al. (2003) found that TSA treatment, but not NaB treatment, of bovine somatic cells increased acetylation of H3 and H4 up to sixfold compared with untreated cells. However, another study performed by the same group (Yang et al., 2007) found that treatment of rabbit fetal fibroblast cells with NaB significantly increased the level of H3K9 acetylation. The exact reason for this discrepancy is unclear, although it may be due to species-specific differences.
Szyf et al. (1985) reported an indirect effect of HDAC inhibitors (including NaB) that resulted in DNA hypomethylation, which is in contrast to our observation. Furthermore Enright et al. (2003b) showed that HDAC inhibitors induced hypermethylation rather than hypomethylation. Cervoni and Szyf (2001) have proposed that these changes might be replication independent; however, this suggestion has not been proven (Cervoni et al., 2002; Detich et al., 2003; Milutinovic et al., 2006).
Comparisons between developmental competences of embryos derived from TSA- and NaB-treated cells with cells treated with AzC suggested that induced hyperacetylation by TSA and NaB favored cloned blastocyst production (Enright et al., 2003b; Shi et al., 2003). These contrasting effects of TSA and NaB with AzC were clearly evident when the percentages of cloned blastocysts produced with the highest drug concentrations were compared with each other [TSA (45.2±0.8%), NaB (46.5±0.8%), and AzC (1.5±1.5%)]. Quality assessment of cloned blastocysts (assessed from TCN and ICM ratios) further validated this observation. However, to make a final conclusion on the impact of each drug on in vitro SCNT efficiency, molecular studies investigating gene expression as well as DNA methylation and histone acetylation are required. In addition, the present study indicated that treatment of fibroblast cells with the natural TSA mimetic, NaB, similarly resulted in a significant increase in histone acetylation and accompanying DNA hypermethylation, as well as improved developmental rates and quality of the resultant embryos. The beneficial HDAC-inhibitory effects of TSA and NaB on SCNT that were observed in this study are in agreement with other reports (Iager et al., 2008; Wee et al., 2006, 2007). Moreover, Iager et al. (2008) observed that TSA increased DNMT3b expression in treated cells, which may agree with the findings in this experiment.
To determine if the positive effects of TSA and NaB and the deleterious effect of AzC might originate from different mechanisms, we systematically investigated the effects of different concentrations of TSA, NaB, and AzC on the cell cycle, apoptosis and epigenesis. We also simultaneously assessed the reversibility of drug treatment on somatic cell characteristics and cloning efficiency. Our results indicated that although the cell cycle was unchanged between different drugs, higher AzC concentrations could induce apoptosis at significant rates. Thus, the AzC-induced apoptosis may partially explain the reduced cloning efficiency observed in cells treated with AzC. However, to address the AzC-induced epigenetic alterations on cloning efficiency, a safer mimetic form of AzC with lower apoptotic activity should be used. Considering the potential hazards of epigenetic drugs, the assessment of drug reversibility can provide a platform to select the drug of choice for subsequent studies.
AzC inhibits DNA methyltransferases (Christman et al., 1983; Creusot et al., 1982; Taylor and Jones, 1982), and thus it may be loss of DNMTase 1s (which is present in somatic cells) that leads to a decrease in DNA methylation levels of the AzC-treated cells, which in turn may favor development of cloned embryos. However, AzC-inhibitory effects on DNMTase 3a and DNMTase 3b, the two DNMT enzymes that are crucially required during later stages of embryo development and especially during postgenomic activation for de novo methylation (Schneider-Stock et al., 2005), may compromise further embryo development. Interestingly, we observed that the majority of cloned embryos reconstituted from AzC-treated cells failed to develop beyond the 8–16 cell stage. Enright et al. (2005) observed a significant reduction in DNA methylation levels in the cloned blastocysts that developed from AzC-treated cells. Taken together, this indicates that AzC-induced global hypomethylation does not favor reprogramming and development of cloned embryos. Drug-treated cells cultured in the absence of drugs for 24 h showed a trend of reversion in characteristics of cells treated with TSA and NaB. However, refreshed cells that had been treated with the highest concentrations of drugs still showed increases in acetylation levels, developmental rates, and numbers of cells in the ICM and TCN. This notion was further confirmed when the LD values for yield and quality of blastocysts in control group were compared with treated and treated/refreshed groups (Fig. 5). Despite similar changes in different cellular characteristics of TSA- and NaB-treated cells, refreshed cells in the NaB group did not recover as well as the cells in the TSA group, based on the G2/M ratios. Interestingly, after NaB-treated cells were refreshed, the reduction we observed in the G2/M ratio was accompanied by a significant increase in the BrdU-incorporation rate. It is unclear what the mechanism for this relationship may be; however, it may be due to the fact that the refreshed cells in NaB groups have a higher capacity to progress from the G2/M stage toward G0/G1 and S phase of the next cell cycle. Importantly, AzC-related changes detected in treated cells were not significantly reversed, a finding that could be attributed to the irreversible binding of DNMTase to AzC residues in DNA (Christman et al., 1983; Taylor and Jones, 1982). Another explanation for this fact may be that methylation (hypo or hyper) is a more stable epigenetic mark than histone acetylation and therefore less likely to rapidly turnover.
In conclusion, this study is the first report of a reversible effect specifically in bovine SCNT. Our findings indicate that induced hyperacetylation by TSA and NaB is a reversible event and interrelated with hypermethylation. However, AzC-related hypomethylation may be irreversible and has no specific effect on hyperacetylation. In addition, we demonstrated that induced hyperacetylation combined with moderate hypermethylation enhanced the developmental competence of the reconstructed embryos. Conversely, AzC treatment led to hypomethylation and negatively affected cloning efficiency. However, it has been shown that AzC can increase apoptosis, and therefore it is not clear whether the reduced cloning efficiency we observed in AzC-treated cells was due to its apoptotic effect or to hypomethylation. Further studies on AzC are required to answer this question.
Acknowledgments
The authors express their gratitude to Royan Institute for their full support. This work was supported by a Royan Institute grant (IR-GNO: 12486).
Author Disclosure Statement
There are no conflicts of interest for this article.
References
- Boquest A. C. Day B. N. Prather R. S. Flow cytometric analysis of cultured porcine fetal fibroblast cells. Biol. Reprod. 1999;60:1013–1019. doi: 10.1095/biolreprod60.4.1013. [DOI] [PubMed] [Google Scholar]
- Candido E. P. M. Reeves R. Davie J. R. Sodium butyrate inhibits histone deacetylation in cultured cells. Cell. 1978;14:105–113. doi: 10.1016/0092-8674(78)90305-7. [DOI] [PubMed] [Google Scholar]
- Cervoni N. Szyf M. Demethylase activity is directed by histone acetylation. J. Biol. Chem. 2001;276:40778–40787. doi: 10.1074/jbc.M103921200. [DOI] [PubMed] [Google Scholar]
- Cervoni N. Detich N. Seo S. B., et al. The oncoprotein Set/TAF-1beta, an inhibitor of histone acetyltransferase, inhibits active demethylation of DNA, integrating DNA methylation and transcriptional silencing. J. Biol. Chem. 2002;277:25026–25031. doi: 10.1074/jbc.M202256200. [DOI] [PubMed] [Google Scholar]
- Christman J. K. 5-Azacytidine and 5-aza-2′-deoxycytidine as inhibitors of DNA methylation: mechanistic studies and their implications for cancer therapy. Oncogene. 2002;21:5483–5495. doi: 10.1038/sj.onc.1205699. [DOI] [PubMed] [Google Scholar]
- Christman J. K. Mendelsohn N. Herzog D., et al. Effect of 5-azacytidine on differentiation and DNA methylation in human promyelocytic leukemia cells (HL-60) Cancer Res. 1983;43:763–769. [PubMed] [Google Scholar]
- Creusot F. Acs G. Christman J. K. Inhibition of DNA methyltransferase and induction of Friend erythroleukemia cell differentiation by 5-azacytidine and 5-aza-2′-deoxycytidine. J. Biol. Chem. 1982;257:2041–2048. [PubMed] [Google Scholar]
- Dean W. Santos F. Stojkovic M., et al. Conservation of methylation reprogramming in mammalian development: aberrant reprogramming in cloned embryos. Proc. Natl. Acad. Sci. USA. 2001;98:13734–13738. doi: 10.1073/pnas.241522698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshmukh R. S. Ostrup O. Ostrup E., et al. DNA methylation in porcine preimplantation embryos developed in vivo and produced by in vitro fertilization, parthenogenetic activation and somatic cell nuclear transfer. Epigenetics. 2011;6:177–187. doi: 10.4161/epi.6.2.13519. [DOI] [PubMed] [Google Scholar]
- Detich N. Bovenzi V. Szyf M. Valproate induces replication independent active DNA demethylation. J. Biol. Chem. 2003;278:27586–27592. doi: 10.1074/jbc.M303740200. [DOI] [PubMed] [Google Scholar]
- Ding X. Wang Y. Zhang D., et al. Increased pre-implantation development of cloned bovine embryos treated with 5-aza-2′-deoxycytidine and trichostatin A. Theriogenology. 2008;70:622–630. doi: 10.1016/j.theriogenology.2008.04.042. [DOI] [PubMed] [Google Scholar]
- Enright B. P. Jeong B. S. Yang X., et al. Epigenetic characteristics of bovine donor cells for nuclear transfer: levels of histone acetylation. Biol. Reprod. 2003a;69:1525–1530. doi: 10.1095/biolreprod.103.019950. [DOI] [PubMed] [Google Scholar]
- Enright B. P. Kubota C. Yang X., et al. Epigenetic characteristics and development of embryos cloned from donor cells treated by trichostatin A or 5-aza-2′-deoxycytidine. Biol. Reprod. 2003b;69:896–901. doi: 10.1095/biolreprod.103.017954. [DOI] [PubMed] [Google Scholar]
- Enright B. P. Sung L. Y. Chang C. C., et al. Methylation and acetylation characteristics of cloned bovine embryos from donor cells treated with 5-aza-2′-deoxycytidine. Biol. Reprod. 2005;72:944–948. doi: 10.1095/biolreprod.104.033225. [DOI] [PubMed] [Google Scholar]
- Hosseini S. M. Hajian M. Moulavi F., et al. Optimized combined electrical and chemical activation of in vitro matured bovine oocytes. Anim. Reprod. Sci. 2006;108:122–133. doi: 10.1016/j.anireprosci.2007.07.011. [DOI] [PubMed] [Google Scholar]
- Hosseini S. M. Moulavi F. Foruzanfar M., et al. Effect of donor cell type and gender on the efficiency of in vitro sheep somatic cell cloning. Small Rum. Res. 2008;78:162–168. [Google Scholar]
- Iager A. E. Ragina N. P. Ross P. J., et al. Trichostatin A improves histone acetylation in bovine somatic cell nuclear transfer early embryos. Cloning Stem Cells. 2008;10:371–379. doi: 10.1089/clo.2007.0002. [DOI] [PubMed] [Google Scholar]
- Jones K. L. Hill J. Shin T. Y., et al. DNA hypomethylation of karyoplasts for bovine nuclear transplantation. Mol. Reprod. Dev. 2001;60:208–213. doi: 10.1002/mrd.1079. [DOI] [PubMed] [Google Scholar]
- Jones P. L. Veenstra G. J. Wade P. A., et al. Methylated DNA and MECP2 recruit histone deacetylase to repress transcription. Nat. Genet. 1998;19:187–191. doi: 10.1038/561. [DOI] [PubMed] [Google Scholar]
- Kharroubi A. E. Piras G. Stewart C. L. DNA demethylation reactivates a subset of imprinted genes in uniparental mouse embryonic fibroblasts. J. Biol. Chem. 2001;276:8674–8680. doi: 10.1074/jbc.M009392200. [DOI] [PubMed] [Google Scholar]
- Kubota C. Yamakuchi H. Todoroki J., et al. Six cloned calves produced from adult fibroblast cells after long-term culture. Proc. Natl. Acad. Sci. USA. 2000;97:990–995. doi: 10.1073/pnas.97.3.990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan J. Hua S. Zhang H., et al. Methylation patterns in 5′ terminal regions of pluripotency-related genes in bovine in vitro fertilized and cloned embryos. J. Genet. Genomics. 2010;37:297–304. doi: 10.1016/S1673-8527(09)60047-3. [DOI] [PubMed] [Google Scholar]
- Li J. Svarcova O. Villemoes K., et al. High in vitro development after somatic cell nuclear transfer and trichostatin A treatment of reconstructed porcine embryos. Theriogenology. 2008;70:800–808. doi: 10.1016/j.theriogenology.2008.05.046. [DOI] [PubMed] [Google Scholar]
- Milutinovic S. D'Alessio A. C. Detich N., et al. Valproate induces widespread epigenetic reprogramming which involves demethylation of specific genes. Carcinogenesis. 2006;2:560–571. doi: 10.1093/carcin/bgl167. [DOI] [PubMed] [Google Scholar]
- Mohana Kumar B. Song H. J. Cho S. K., et al. Effect of histone acetylation modification with sodium butyrate, a histone deacetylase inhibitor, on cell cycle, apoptosis, ploidy and gene expression in porcine fetal fibroblasts. J. Reprod. Dev. 2007;53:903–913. doi: 10.1262/jrd.18180. [DOI] [PubMed] [Google Scholar]
- Moulavi F. Hosseini S. M. Ashtiani S. K., et al. Can Vero cell co-culture improve in-vitro maturation of bovine oocytes? Reprod. Biomed. Online. 2006;13:404–411. doi: 10.1016/s1472-6483(10)61446-0. [DOI] [PubMed] [Google Scholar]
- Nasr-Esfahani M. H. Hosseini S. M. Hajian M., et al. Development of an optimized zona-free method of somatic cell nuclear transfer in the goat. Cell. Reprogram. 2011;13:157–170. doi: 10.1089/cell.2010.0083. [DOI] [PubMed] [Google Scholar]
- Oback B. Wiersema A. T. Gaynor P., et al. Cloned cattle derived from a novel zona-free embryo reconstruction system. Cloning Stem Cells. 2003;5:3–12. doi: 10.1089/153623003321512111. [DOI] [PubMed] [Google Scholar]
- Santos F. Zakhartchenko V. Stojkovic M., et al. Epigenetic marking correlates with developmental potential in cloned bovine preimplantation embryos. Curr. Biol. 2003;13:1116–1121. doi: 10.1016/s0960-9822(03)00419-6. [DOI] [PubMed] [Google Scholar]
- Sawai K. Takahashi M. Fujii T., et al. DNA methylation status of bovine blastocyst embryos obtained from various procedures. J. Reprod. Dev. 2010;57:236–241. doi: 10.1262/jrd.10-035a. [DOI] [PubMed] [Google Scholar]
- Schneider-Stock R. Diab-Assef M. Rohrbeck A., et al. 5-Aza-cytidine is a potent inhibitor of DNA methyltransferase 3a and induces apoptosis in HCT-116 colon cancer cells via Gadd45- and p53-dependent mechanisms. J. Pharm. Exp. 2005;312:525–536. doi: 10.1124/jpet.104.074195. [DOI] [PubMed] [Google Scholar]
- Shi W. Hoeflich A. Flaswinkel H., et al. Induction of a senescent-like phenotype does not confer the ability of bovine immortal cells to support the development of nuclear transfer embryos. Biol. Reprod. 2003;69:301–309. doi: 10.1095/biolreprod.102.012112. [DOI] [PubMed] [Google Scholar]
- Szyf M. Eliasson L. Mann V., et al. Cellular and viral DNA hypomethylation associated with induction of Epstein-Barr virus lytic cycle. Proc. Natl. Acad. Sci. USA. 1985;82:8090–8094. doi: 10.1073/pnas.82.23.8090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor S. M. Jones P. A. Mechanism of action of eukaryotic DNA methyltransferase. Use of 5-azacytosine-containing DNA. J. Mol. Biol. 1982;162:679–692. doi: 10.1016/0022-2836(82)90395-3. [DOI] [PubMed] [Google Scholar]
- Tervit H. R. Whittingham D. G. Rowson L. E. A. Successful culture in vitro of sheep and cattle ova. J. Reprod. Fertil. 1972;30:493–497. doi: 10.1530/jrf.0.0300493. [DOI] [PubMed] [Google Scholar]
- Tse C. Sera T. Wolffe A. P., et al. Disruption of higher-order folding by core histone acetylation dramatically enhances transcription of nucleosomal arrays by RNA polymerase III. Mol. Cell. Biol. 1998;18:4629–4638. doi: 10.1128/mcb.18.8.4629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vajta G. Peura T. T. Holm P., et al. New method for culture of zona-included or zona-free embryos: the Well of the Well (WOW) system. Mol. Reprod. Dev. 2000;55:256–264. doi: 10.1002/(SICI)1098-2795(200003)55:3<256::AID-MRD3>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- Wee G. Koo D. B. Song B. S., et al. Inheritable histone H4 acetylation of somatic chromatins in cloned embryos. J. Biol. Chem. 2006;281:6048–6057. doi: 10.1074/jbc.M511340200. [DOI] [PubMed] [Google Scholar]
- Wee G. Shim J. J. Koo D. B., et al. Epigenetic alteration of the donor cells does not recapitulate the reprogramming of DNA methylation in cloned embryos. Reproduction. 2007;134:781–787. doi: 10.1530/REP-07-0338. [DOI] [PubMed] [Google Scholar]
- Wilmut I. Schnieke A. E. McWhir J., et al. Viable offspring derived from fetal and adult mammalian cells. Nature. 1997;385:810–813. doi: 10.1038/385810a0. [DOI] [PubMed] [Google Scholar]
- Yang F. Hao R. Kessler B., et al. Rabbit somatic cell cloning: effects of donor cell type, histone acetylation status and chimeric embryo complementation. Reproduction. 2007;133:219–230. doi: 10.1530/rep.1.01206. [DOI] [PubMed] [Google Scholar]
- Yoshida M. Kijima M. Akita M., et al. Potent and specific inhibition of mammalian histone deacetylase both in vivo and in vitro by Trichostatin A. J. Biol. Chem. 1990;265:17174–17179. [PubMed] [Google Scholar]