Abstract
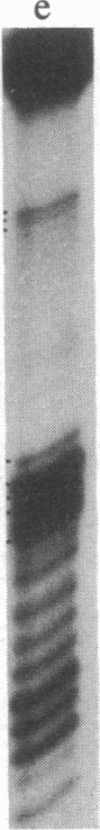
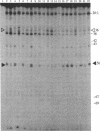
The crystal structure of yeast tRNAAsp enables visualization of an anticodon-anticodon interaction at the molecular level. Except for differences in the base stacking and twist, the overall conformation of the anticodon loop is quite similar to that of yeast tRNAPhe. The anticodon nucleotide triplets, GUC, of two symmetrically related molecules form a minihelix of the RNA type 11. The modified base m1G37 stacks on both sides of the triplets and enforces the continuity with the anticodon stems. Anticodon association induces long-range conformational changes in the region of the dihydrouracil and thymine loops. Experimental evidence includes the variation in the distribution of temperature factors between yeast tRNAPhe and tRNAAsp, the difference in the self-splitting patterns of tRNAAsp in crystal and solution, and the differential accessibility of cytidines to dimethyl sulfate in free and duplex tRNAAsp. These observations are linked to the fragility and disruption of the G.C Watson-Crick base pair at the corner of the molecule formed by the dihydrouracil and thymine loops.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Boutorin A. S., Clark B. F., Ebel J. P., Kruse T. A., Petersen H. U., Remy P., Vassilenko S. A study of the interaction of Escherichia coli elongation factor-Tu with aminoacyl-tRNAs by partial digestion with cobra venom ribonuclease. J Mol Biol. 1981 Nov 5;152(3):593–608. doi: 10.1016/0022-2836(81)90271-0. [DOI] [PubMed] [Google Scholar]
- Brown R. S., Hingerty B. E., Dewan J. C., Klug A. Pb(II)-catalysed cleavage of the sugar-phosphate backbone of yeast tRNAPhe--implications for lead toxicity and self-splicing RNA. Nature. 1983 Jun 9;303(5917):543–546. doi: 10.1038/303543a0. [DOI] [PubMed] [Google Scholar]
- Bruce A. G., Uhlenbeck O. C. Reactions at the termini of tRNA with T4 RNA ligase. Nucleic Acids Res. 1978 Oct;5(10):3665–3677. doi: 10.1093/nar/5.10.3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davanloo P., Sprinzl M., Cramer F. Proton nuclear magnetic resonance of minor nucleosides in yeast phenylalanine transfer ribonucleic acid. Conformational changes as a consequence of aminoacylation, removal of the Y base, and codon--anticodon interaction. Biochemistry. 1979 Jul 24;18(15):3189–3199. doi: 10.1021/bi00582a001. [DOI] [PubMed] [Google Scholar]
- Donis-Keller H., Maxam A. M., Gilbert W. Mapping adenines, guanines, and pyrimidines in RNA. Nucleic Acids Res. 1977 Aug;4(8):2527–2538. doi: 10.1093/nar/4.8.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisinger J. Complex formation between transfer RNA'S with complementary anticodons. Biochem Biophys Res Commun. 1971 May 21;43(4):854–861. doi: 10.1016/0006-291x(71)90695-4. [DOI] [PubMed] [Google Scholar]
- Farber N., Cantor C. R. Comparison of the structures of free and ribosome-bound tRNAPhe by using slow tritium exchange. Proc Natl Acad Sci U S A. 1980 Sep;77(9):5135–5139. doi: 10.1073/pnas.77.9.5135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller W., Hodgson A. Conformation of the anticodon loop intRNA. Nature. 1967 Aug 19;215(5103):817–821. doi: 10.1038/215817a0. [DOI] [PubMed] [Google Scholar]
- Gassen H. G. Ligand-induced conformational changes in ribonucleic acids. Prog Nucleic Acid Res Mol Biol. 1980;24:57–86. doi: 10.1016/s0079-6603(08)60671-6. [DOI] [PubMed] [Google Scholar]
- Geerdes H. A., Van Boom J. H., Hilbers C. W. Nuclear magnetic resonance studies of codon-anticodon interaction in tRNAPhe. I. Effect of binding complementary tetra and pentanucleotides to the anticodon. J Mol Biol. 1980 Sep 15;142(2):195–217. doi: 10.1016/0022-2836(80)90045-5. [DOI] [PubMed] [Google Scholar]
- Grosjean H. J., de Henau S., Crothers D. M. On the physical basis for ambiguity in genetic coding interactions. Proc Natl Acad Sci U S A. 1978 Feb;75(2):610–614. doi: 10.1073/pnas.75.2.610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosjean H., Chantrenne H. On codon- anticodon interactions. Mol Biol Biochem Biophys. 1980;32:347–367. doi: 10.1007/978-3-642-81503-4_27. [DOI] [PubMed] [Google Scholar]
- Helk B., Sprinzl M. Interaction of unfolded tRNA with the 3'-terminal region of E. coli 16S ribosomal RNA. Nucleic Acids Res. 1985 Sep 11;13(17):6283–6298. doi: 10.1093/nar/13.17.6283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huong P. V., Audry E., Giege R., Moras D., Thierry J. C., Comarmond M. B. Conformational change in yeast tRNAAsp. Biopolymers. 1984 Jan;23(1):71–81. doi: 10.1002/bip.360230107. [DOI] [PubMed] [Google Scholar]
- Moras D., Comarmond M. B., Fischer J., Weiss R., Thierry J. C., Ebel J. P., Giegé R. Crystal structure of yeast tRNAAsp. Nature. 1980 Dec 25;288(5792):669–674. doi: 10.1038/288669a0. [DOI] [PubMed] [Google Scholar]
- Nilsson L., Rigler R., Laggner P. Structural variability of tRNA: small-angle x-ray scattering of the yeast tRNAphe-Escherichia coli tRNAGlu2 complex. Proc Natl Acad Sci U S A. 1982 Oct;79(19):5891–5895. doi: 10.1073/pnas.79.19.5891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pace B., Matthews E. A., Johnson K. D., Cantor C. R., Pace N. R. Conserved 5S rRNA complement to tRNA is not required for protein synthesis. Proc Natl Acad Sci U S A. 1982 Jan;79(1):36–40. doi: 10.1073/pnas.79.1.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peattie D. A., Gilbert W. Chemical probes for higher-order structure in RNA. Proc Natl Acad Sci U S A. 1980 Aug;77(8):4679–4682. doi: 10.1073/pnas.77.8.4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quigley G. J., Teeter M. M., Rich A. Structural analysis of spermine and magnesium ion binding to yeast phenylalanine transfer RNA. Proc Natl Acad Sci U S A. 1978 Jan;75(1):64–68. doi: 10.1073/pnas.75.1.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romby P., Giegé R., Houssier C., Grosjean H. Anticodon-anticodon interactions in solution. Studies of the self-association of yeast or Escherichia coli tRNAAsp and of their interactions with Escherichia coli tRNAVal. J Mol Biol. 1985 Jul 5;184(1):107–118. doi: 10.1016/0022-2836(85)90047-6. [DOI] [PubMed] [Google Scholar]
- Romby P., Moras D., Bergdoll M., Dumas P., Vlassov V. V., Westhof E., Ebel J. P., Giegé R. Yeast tRNAAsp tertiary structure in solution and areas of interaction of the tRNA with aspartyl-tRNA synthetase. A comparative study of the yeast phenylalanine system by phosphate alkylation experiments with ethylnitrosourea. J Mol Biol. 1985 Aug 5;184(3):455–471. doi: 10.1016/0022-2836(85)90294-3. [DOI] [PubMed] [Google Scholar]
- Rubin J. R., Sundaralingam M. Lead ion binding and RNA chain hydrolysis in phenylalanine tRNA. J Biomol Struct Dyn. 1983 Dec;1(3):639–646. doi: 10.1080/07391102.1983.10507471. [DOI] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R. The use of thin acrylamide gels for DNA sequencing. FEBS Lett. 1978 Mar 1;87(1):107–110. doi: 10.1016/0014-5793(78)80145-8. [DOI] [PubMed] [Google Scholar]
- Schevitz R. W., Podjarny A. D., Krishnamachari N., Hughes J. J., Sigler P. B., Sussman J. L. Crystal structure of a eukaryotic initiator tRNA. Nature. 1979 Mar 8;278(5700):188–190. doi: 10.1038/278188a0. [DOI] [PubMed] [Google Scholar]
- Schwarz U., Menzel H. M., Gassen H. G. Codon-dependent rearrangement of the three-dimensional structure of phenylalanine tRNA, exposing the T-psi-C-G sequence for binding to the 50S ribosomal subunit. Biochemistry. 1976 Jun 1;15(11):2484–2490. doi: 10.1021/bi00656a035. [DOI] [PubMed] [Google Scholar]
- Sussman J. L., Holbrook S. R., Warrant R. W., Church G. M., Kim S. H. Crystal structure of yeast phenylalanine transfer RNA. I. Crystallographic refinement. J Mol Biol. 1978 Aug 25;123(4):607–630. doi: 10.1016/0022-2836(78)90209-7. [DOI] [PubMed] [Google Scholar]
- Vlassov V. V., Giegé R., Ebel J. P. Tertiary structure of tRNAs in solution monitored by phosphodiester modification with ethylnitrosourea. Eur J Biochem. 1981 Sep;119(1):51–59. doi: 10.1111/j.1432-1033.1981.tb05575.x. [DOI] [PubMed] [Google Scholar]
- Wagner R., Garrett R. A. Chemical evidence for a codon-induced allosteric change in tRNALys involving the 7-methylguanosine residue 46. Eur J Biochem. 1979 Jul;97(2):615–621. doi: 10.1111/j.1432-1033.1979.tb13151.x. [DOI] [PubMed] [Google Scholar]
- Werner C., Krebs B., Keith G., Dirheimer G. Specific cleavages of pure tRNAs by plumbous ions. Biochim Biophys Acta. 1976 May 3;432(2):161–175. doi: 10.1016/0005-2787(76)90158-1. [DOI] [PubMed] [Google Scholar]
- Westhof E., Dumas P., Moras D. Crystallographic refinement of yeast aspartic acid transfer RNA. J Mol Biol. 1985 Jul 5;184(1):119–145. doi: 10.1016/0022-2836(85)90048-8. [DOI] [PubMed] [Google Scholar]
- Westhof E., Dumas P., Moras D. Loop stereochemistry and dynamics in transfer RNA. J Biomol Struct Dyn. 1983 Oct;1(2):337–355. doi: 10.1080/07391102.1983.10507446. [DOI] [PubMed] [Google Scholar]
- Wikman F. P., Siboska G. E., Petersen H. U., Clark B. F. The site of interaction of aminoacyl-tRNA with elongation factor Tu. EMBO J. 1982;1(9):1095–1100. doi: 10.1002/j.1460-2075.1982.tb01302.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese C. Molecular mechanics of translation: a reciprocating ratchet mechanism. Nature. 1970 May 30;226(5248):817–820. doi: 10.1038/226817a0. [DOI] [PubMed] [Google Scholar]
- Woo N. H., Roe B. A., Rich A. Three-dimensional structure of Escherichia coli initiator tRNAfMet. Nature. 1980 Jul 24;286(5771):346–351. doi: 10.1038/286346a0. [DOI] [PubMed] [Google Scholar]
- Yarus M. Translational efficiency of transfer RNA's: uses of an extended anticodon. Science. 1982 Nov 12;218(4573):646–652. doi: 10.1126/science.6753149. [DOI] [PubMed] [Google Scholar]
- Yokoyama S., Miyazawa T., Iitaka Y., Yamaizumi Z., Kasai H., Nishimura S. Three-dimensional structure of hyper-modified nucleoside Q located in the wobbling position of tRNA. Nature. 1979 Nov 1;282(5734):107–109. doi: 10.1038/282107a0. [DOI] [PubMed] [Google Scholar]