Abstract
Survivors of pediatric Hodgkin lymphoma (HL) are at significant risk for radiation therapy (RT)-induced second malignant neoplasms (SMNs). We identified two variants at chromosome 6q21 associated with SMNs in HL survivors treated with RT as children but not as adults. The variants comprise a risk locus associated with decreased basal PRDM1 expression and impaired induction of PRDM1 by radiation exposure. These data suggest a novel gene-exposure interaction that may implicate PRDM1 in the etiology of RT-induced SMNs.
Patients treated successfully for Hodgkin lymphoma (HL) in childhood are at significant risk for radiation therapy-induced second malignant neoplasms (SMNs), with a cumulative incidence of 18.4% by 30 years after treatment and an absolute excess risk of 6.9 per 1,000 person-years of follow-up1. This high prevalence makes SMNs the second leading cause of mortality in HL survivors. SMNs primarily affect organs in the involved mediastinal RT field, including the thyroid, skin, gastrointestinal tract, and female breast2,3. Risk is positively associated with cumulative radiation dose and inversely correlated with age at treatment4,5.
Despite the clinical significance of this devastating late consequence of RT exposure, little is known about predisposing risk factors. We performed a genome-wide association study (GWAS) to identify variants associated with RT-induced SMNs in HL survivors. In studies of sporadic cancers, non-genetic heterogeneity can obscure genetic associations6, but here, RT exposure is common to both HL patients who do and do not develop SMNs. Thus, we hypothesized that limiting our study to RT-treated survivors would improve our power to detect the genetic contribution to SMN risk.
The discovery set consisted of 100 SMN cases and 89 SMN-free controls (Supplementary Table 1a, Supplementary Table 2). All cases and controls were diagnosed with HL as children (median age: 15.6, range: 8–20) and treated with 25–44 Gy RT +/− alkylating chemotherapy7. Cases developed SMNs with a mean latency of 20.0 years (s.d. = 5.8 years, range: 6–34). Controls were followed for at least 27 years (median: 32 years, range: 27–38) to ensure that the maximal contamination of controls by future cases was < 2%. For a detailed description of the study populations and experimental protocols, see the Supplementary Methods.
Following genotype quality control, 665,313 single nucleotide polymorphisms (SNPs) were successfully genotyped in 96 cases and 82 controls. We compared allele frequencies between cases and controls using a Chi-square test of homogeneity. A quantile-quantile (Q-Q) plot of the expected and observed distribution of P values revealed no evidence for systematic genotype calling error or hidden population substructure (genomic control λ = 1.007) (Supplementary Fig. 1)8. Principal component analysis using Eigenstrat indicated cases and controls were of European descent (Supplementary Fig. 2)9.
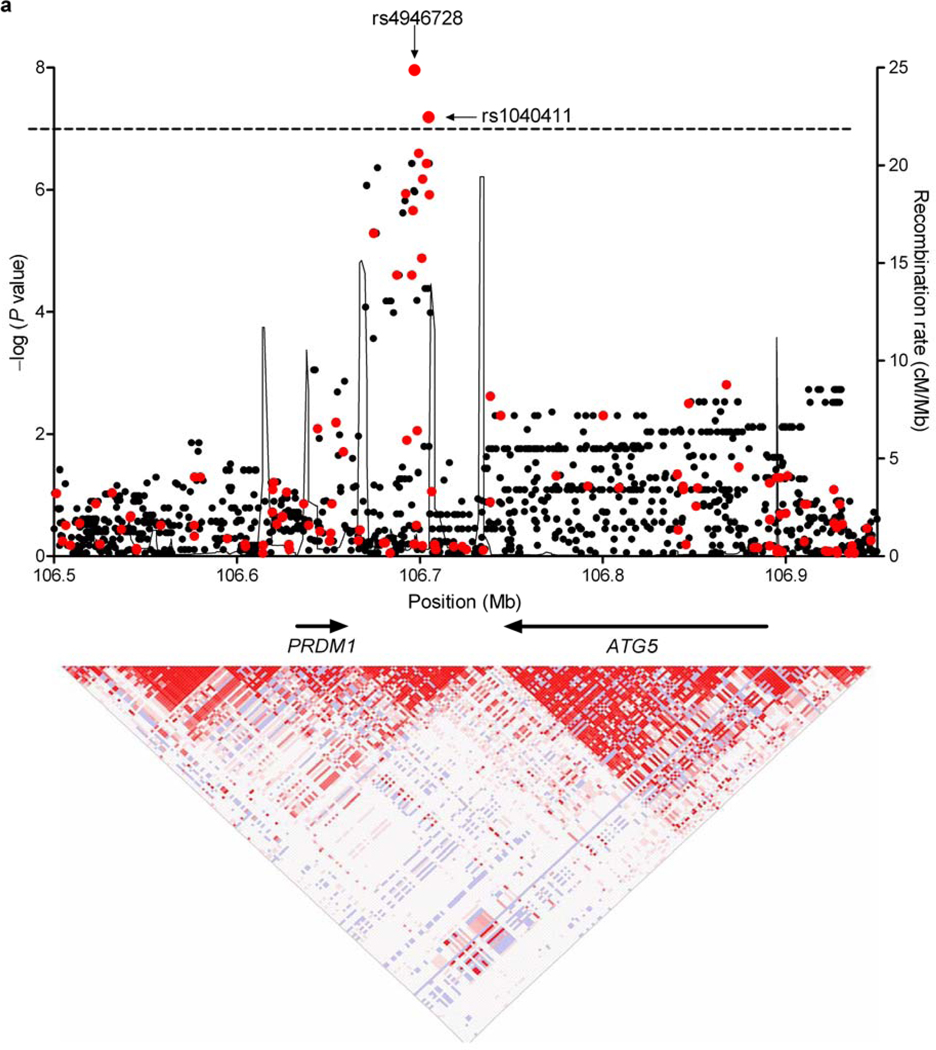
We empirically determined the threshold for a genome-wide type I error rate of 0.05 by permutation (P < 1.0×10−7). At this threshold, our study had 80% power to detect a SNP with a frequency of 35% and an odds ratio of 3.5 (Supplementary Fig. 3). Three SNPs (rs4946728, rs1040411, rs8083533) achieved genome-wide significance (Supplementary Fig. 4 and Table 1). rs4946728 and rs1040411 mapped to chromosome 6q21, intergenic between ATG5 and PRDM1. The strongest evidence for association in this region was for rs4946728 (P = 1.09×10−8, ORallelic = 4.22 [95% CI = 2.53–7.05]). rs8083533 mapped to 18q11.2, intronic to TAF4B (P = 4.98×10−8, ORallelic = 3.78 [95% CI = 2.31–6.18]). Logistic regression, adjusting for gender, age at diagnosis, year of HL diagnosis, gonadal radiation (in females), and alkylating chemotherapy exposure, demonstrated that these risk variables had no effect on the observed associations (Supplementary Table 3).
We sought to replicate these findings in an independent set of 62 cases with SMNs and 71 SMN-free controls, all treated for HL in childhood with 25–44 Gy mediastinal RT (Supplementary Table 1b). We observed significant associations with SMNs for both SNPs on chromosome 6q21, rs4946728 (P = 0.002) and rs1040411 (P = 0.03), but not for rs8083533 (P = 0.82) (Table 1). In the combined set, odds of an SMN were increased over 3-fold per copy of the major allele for rs4946728 (ORallelic = 3.32 [95% CI = 2.25–4.90], combined P = 5.99×10−10) and over 2-fold for rs1040411 (ORallelic = 2.39 [95% CI = 1.73–3.30], combined P = 1.18×10−7).
Table 1.
Association of SNPs with SMNs following HL treatment.
| Genotype counta | OR [95% CI]b | |||||||
|---|---|---|---|---|---|---|---|---|
| SNP | Risk Allele |
Cases | Controls | Stage | Per Allele | Heterozygotec | Homozygoted | P valuee |
| rs4946728f | C | 2/23/71 | 12/43/27 | Discoveryg | 4.22 [2.53–7.05] | 3.21 [0.66–15.59] | 15.78 [3.31–75.18] | 1.09×10−8 |
| 1/17/43 | 6/32/33 | Replicationh | 2.43 [1.33–4.46] | 3.19 [0.35–28.69] | 7.82 [0.90–68.14] | 0.002 | ||
| 3/40/114 | 18/75/60 | Combinedi | 3.32 [2.25–4.90] | 3.20 [0.89–11.52] | 11.4 [3.23–40.25] | 5.99×10− 10 | ||
| rs1040411f | T | 7/47/42 | 27/45/10 | Discovery | 3.27 [2.11–5.06] | 4.03 [1.60–10.17] | 16.2 [5.50–47.71] | 6.43×10−8 |
| 9/30/22 | 19/33/18 | Replication | 1.59 [0.97–2.59] | 1.92 [0.75–4.89] | 2.58 [0.94–7.07] | .03 | ||
| 16/77/64 | 46/78/28 | Combined | 2.39 [1.73–3.30] | 2.84 [1.48–5.44] | 6.57 [3.19–13.52] | 1.18×10−7 | ||
| rs8083533 | T | 21/44/31 | 4/21/57 | Discovery | 3.78 [2.31–6.18] | 2.51 [0.76–8.23] | 9.65 [3.04–30.65] | 4.98×10−8 |
| 2/31/26 | 8/33/29 | Replication | 0.89 [0.52–1.53] | 0.27 [0.05–1.35] | 0.28 [0.05–1.43] | .82 | ||
| 23/75/57 | 12/54/86 | Combined | 1.86 [1.32–2.62] | 1.38 [0.63–3.01] | 2.89 [1.33–6.27] | 4.00×10−4 | ||
Number of individuals genotyped as homozygous for the protective allele/heterozygous/homozygous for the risk allele
Odds ratio [95% confidence interval]
OR for heterozygous carriage of risk allele compared to homozygous carriage of protective allele
OR for homozygous carriage of risk allele compared to homozygous carriage of protective allele
Two-sided Chi-squared P value for discovery and combined sets; one-sided for replication
rs4946728 and rs1040411 are in linkage disequilibrium (r2 = 0.4).
Discovery set: 96 cases and 82 controls
Replication set: 62 cases and 71 controls
Combined set: 158 cases and 153 control
We found no evidence that the association of rs4946728 and rs1040411 differed between breast cancer and other SMNs (Phet = 0.41 for rs4946728 and Phet = 0.58 for rs1040411) or between males and females (Phet = 0.83 for rs4846728 and Phet = 0.29 for rs1040411) (Supplementary Tables 4a and 4b). To determine whether rs4946728 or rs1040411 were associated with SMNs after adult HL, we genotyped both SNPs in 57 SMN cases and 37 controls who were treated with RT as adults (median age: 24.0, range: 21–43) (Supplementary Table 1b). We did not observe an association for either rs4946728 (P = 0.87) or rs1040411 (P = 0.65) (Supplementary Table 5) suggesting that age of RT-exposure modifies the association between these variants and SMN risk. These results should be interpreted with caution, however, given the small number of individuals genotyped.
Both rs4946728 and rs1040411 (r2 = 0.4) are noncoding SNPs located between PRDM1 and ATG5 on chromosome 6q21 (Fig. 1a). Imputation of the locus with the 1000 Genomes reference panel10 did not reveal any variant with a stronger association than either genotyped SNP (Supplementary Table 6). Logistic regression conditioning on rs4946728 revealed a modest residual association for rs1040411 (P = 0.05) (Supplementary Table 7), suggesting that an unobserved causal variant may be correlated with a haplotype harboring both SNPs.
Figure 1. Variants at 6q21 are associated with both RT-induced SMNs and PRDM1 levels before and after radiation exposure.
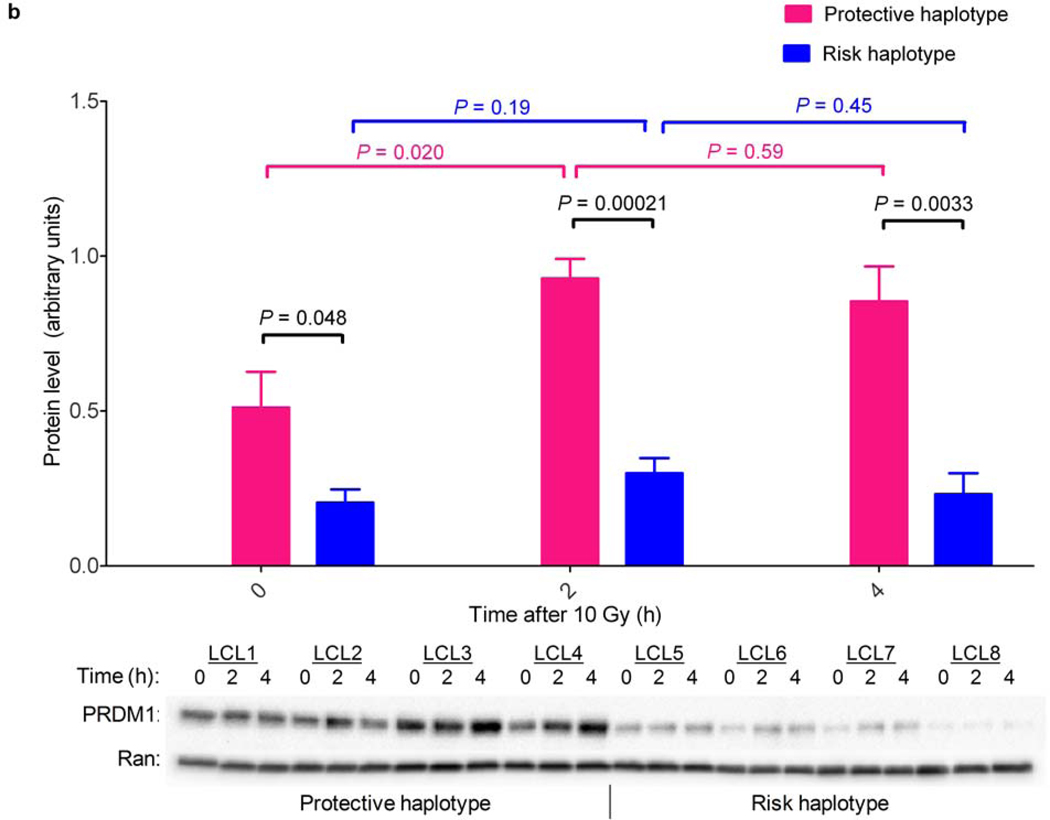
(a) Shown is the regional association plot of the 6q21 locus. The −log (P value) for SNPs in this region are shown with respect to genomic position. Genotyped SNPs are in red; imputed SNPs are in black. The line at −log (P) = 7 denotes the threshold for genome-wide significance. Recombination rates (taken from HapMap) and genes within this region are also shown (top panel). LD structure based on D’ values for the GWAS data are shown (bottom panel). NCBI build 36 was used for all map locations. (b) Shown is the western blot analysis over time for PRDM1 in eight lymphoblastoid cell lines, four homozygous for the chromosome 6q21 protective haplotype and four homozygous for the risk haplotype, treated with 10 Gy gamma irradiation. The protective haplotype was associated with both higher baseline PRDM1 protein levels than the risk haplotype and a marked induction of PRDM1 following irradiation. In contrast, PRDM1 was not induced by irradiation in cells homozygous for the risk haplotype.
rs4946728 and rs1040411 form three common haplotypes in Caucasians that represent 99.9% of the haplotypes at this locus (Supplementary Table 8). As noncoding risk variants frequently regulate gene expression11, we performed expression quantitative trait locus (eQTL) analysis to determine whether these haplotypes were associated with expression of PRDM1, ATG5 or other genes within five megabases. We found that increasing dosage of the risk haplotype (comprised of the risk alleles for both rs4946728 and rs1040411) was significantly associated with lower PRDM1 mRNA expression (P = 0.03) (Supplementary Fig. 5). In contrast, no association with expression was observed for any other gene, including ATG5 (P = 0.39).
Because SMNs after HL are caused by radiation exposure, we investigated the relationship between ionizing radiation (IR) exposure and PRDM1 protein levels in cell lines homozygous for either the risk haplotype (n = 4) or the protective haplotype (comprised of the protective alleles for both rs4946728 and rs1040411, n = 4). In untreated cells, PRDM1 was more abundant in cells homozygous for the protective haplotype than in cells homozygous for the risk haplotype (P = 0.048) and was significantly induced within 2 hours of IR-exposure (P = 0.020) (Fig. 1b). Strikingly, PRDM1 was not induced by IR in cells homozygous for the risk haplotype (P = 0.19).
PRDM1 (PR domain containing 1, with ZNF domain (also known as BLIMP1)) (OMIM# 603423), encodes a zinc finger transcriptional repressor involved in a variety of cellular processes including proliferation, differentiation, and apoptosis12. It was recently shown to be a tumor suppressor in activated B cell-like diffuse large B cell lymphoma13,14, and is frequently lost in many cancer types, including solid tumors15. Of note, loss of heterozygosity at chromosome 6q was found to be significantly more common in breast cancers following RT for HL than in sporadic breast cancers (42% vs 10%), suggesting this region may be targeted for loss in these RT-induced cancers16.
PRDM1 negatively regulates pro-proliferative genes, such as MYC17. Therefore, we investigated whether the 6q21 variants were associated with repression of MYC by radiation concomitant with PRDM1 induction. Though basal MYC levels did not correlate with carriage of the 6q21 risk haplotype (P = 0.19), MYC was significantly more repressed following IR exposure in cells homozygous for the protective haplotype than in cells homozygous for the risk haplotype (P = 0.02) (Supplementary Fig. 6).
In summary, these data demonstrate that variants at 6q21 are strongly associated with risk for SMNs following RT treatment for HL in childhood and suggest that common variants can have large effect sizes in the context of specific exposures. The SNPs we identified are associated with basal and radiation-induced PRDM1 expression, as well as radiation-induced MYC repression. Taken together, our findings support a novel role for PRDM1 as a radiation-responsive tumor suppressor. We cannot rule out, however, either long-range effects of these variants on other genes or tissue-specific differences in PRDM1 function. Additionally, the observation that SNPs intergenic between PRDM1 and ATG5 are associated with autoimmune disease18,19 raises the intriguing possibility that altered immune function or inflammation may be associated with SMN risk. Although the RT doses used currently to treat HL are considerably lower than the RT doses used to treat the children analyzed in this study, recent data indicate that children treated with lower-dose RT for HL remain at significant risk for SMNs20. Thus, our findings may be important for understanding the etiology of SMNs in these pediatric HL survivors, as well as in other cancer patients treated with RT.
Supplementary Material
Acknowledgements
This work was supported by grants from the National Institutes of Health (HD0433871, CA129045 and CA40046 to KOn, CA55727 to LLR, GM089941 and CA139278 to RSH, CA58839 to TMM, and CA110836 to WC); NIH/NIGMS Pharmacogenomics of Anticancer Agents grant U01GM61393 (RSH); Contract No N01-PC-35139 (WC); the USAMRMC (Department of Defense PR054600 to WC, and Department of Defense DAMD 17-097-1-7147 to KOf); the American Cancer Society – Illinois Division (KOn); the American Lebanese Syrian Associated Charities (LLR); Leukemia Lymphoma Society (TR 6137-07 to WC, and TR 6202-09 to TK); the Breast Cancer Research Foundation (SMD, KLN, and KOf); the Lymphoma Foundation (KOf); the Robert and Kate Niehaus Research Fund (KOf); University of Chicago Cancer Center Support Grant (#P30 CA14599) (RSH); Breast Cancer SPORE Career Development Award (RSH); and the Cancer Research Foundation (KOn).
The authors thank the University of Chicago PAAR cell core for providing lymphoblastoid cell lines. The collection of some HL patients used in this publication was supported by the California Department of Health Services as part of the statewide cancer reporting program mandated by California Health and Safety Code Section 103885. The ideas and opinions expressed herein are those of the authors, and no endorsement by the State of California, Department of Health Services is intended or should be inferred.
We thank NA Ellis for many productive discussions and thoughtful feedback. We are especially grateful for the contributions of the many participating patients and their parents, without whom this work would not have been possible.
Footnotes
Author Contributions
TB and KOn designed the study and wrote the manuscript with substantial contributions from ADS, YY, SB, LCS, RSH, TMM, DVC, KOff, WC, and LLR; TB performed the experiments and undertook the analysis; DL, ADS, TK, and YY performed data analysis; SAJ, SMD, KLN, OIO, WC, and LLR provided clinical samples and performed analysis of patient data; KOn directed the project. All authors contributed to the final manuscript.
All authors declare no competing financial interests.
References
- 1.Friedman DL, et al. Subsequent neoplasms in 5-year survivors of childhood cancer: the Childhood Cancer Survivor Study. J Natl Cancer Inst. 2010;102:1083–1095. doi: 10.1093/jnci/djq238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Meadows AT, et al. Second neoplasms in survivors of childhood cancer: findings from the Childhood Cancer Survivor Study cohort. J Clin Oncol. 2009;27:2356–2362. doi: 10.1200/JCO.2008.21.1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Travis LB, et al. Breast cancer following radiotherapy and chemotherapy among young women with Hodgkin disease. Jama. 2003;290:465–475. doi: 10.1001/jama.290.4.465. [DOI] [PubMed] [Google Scholar]
- 4.Constine LS, et al. Subsequent malignancies in children treated for Hodgkin's disease: associations with gender and radiation dose. Int J Radiat Oncol Biol Phys. 2008;72:24–33. doi: 10.1016/j.ijrobp.2008.04.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Neglia JP, et al. Second malignant neoplasms in five-year survivors of childhood cancer: childhood cancer survivor study. J Natl Cancer Inst. 2001;93:618–629. doi: 10.1093/jnci/93.8.618. [DOI] [PubMed] [Google Scholar]
- 6.Thomas D. Gene-environment-wide association studies: emerging approaches. Nat Rev Genet. 2010;11:259–272. doi: 10.1038/nrg2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Robison LL, et al. Long-term outcomes of adult survivors of childhood cancer. Cancer. 2005;104:2557–2564. doi: 10.1002/cncr.21249. [DOI] [PubMed] [Google Scholar]
- 8.Devlin B, Roeder K. Genomic control for association studies. Biometrics. 1999;55:997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. [DOI] [PubMed] [Google Scholar]
- 9.Price AL, et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 10.Durbin RM, et al. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nicolae DL, et al. Trait-associated SNPs are more likely to be eQTLs: annotation to enhance discovery from GWAS. PLoS Genet. 2010;6:e1000888. doi: 10.1371/journal.pgen.1000888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Calame K. Blimp-1's maiden flight. J Immunol. 2010;185:3–4. doi: 10.4049/jimmunol.1090044. [DOI] [PubMed] [Google Scholar]
- 13.Calado DP, et al. Constitutive canonical NF-kappaB activation cooperates with disruption of BLIMP1 in the pathogenesis of activated B cell-like diffuse large cell lymphoma. Cancer Cell. 2010;18:580–589. doi: 10.1016/j.ccr.2010.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mandelbaum J, et al. BLIMP1 is a tumor suppressor gene frequently disrupted in activated B cell-like diffuse large B cell lymphoma. Cancer Cell. 2010;18:568–579. doi: 10.1016/j.ccr.2010.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beroukhim R, et al. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463:899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Behrens C, et al. Molecular changes in second primary lung and breast cancers after therapy for Hodgkin's disease. Cancer Epidemiol Biomarkers Prev. 2000;9:1027–1035. [PubMed] [Google Scholar]
- 17.Lin Y, Wong K, Calame K. Repression of c-myc transcription by Blimp-1, an inducer of terminal B cell differentiation. Science. 1997;276:596–599. doi: 10.1126/science.276.5312.596. [DOI] [PubMed] [Google Scholar]
- 18.Raychaudhuri S, et al. Genetic variants at CD28, PRDM1 and CD2/CD58 are associated with rheumatoid arthritis risk. Nat Genet. 2009;41:1313–1318. doi: 10.1038/ng.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gateva V, et al. A large-scale replication study identifies TNIP1, PRDM1, JAZF1, UHRF1BP1 and IL10 as risk loci for systemic lupus erythematosus. Nat Genet. 2009;41:1228–1333. doi: 10.1038/ng.468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.O'Brien MM, Donaldson SS, Balise RR, Whittemore AS, Link MP. Second malignant neoplasms in survivors of pediatric Hodgkin's lymphoma treated with low-dose radiation and chemotherapy. J Clin Oncol. 2010;28:1232–1239. doi: 10.1200/JCO.2009.24.8062. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.