Abstract
Tropomyosin-related kinase (TRK) receptor B (TRKB) mediates the supportive actions of neurotrophin 4/5 and brain-derived neurotrophic factor on early ovarian follicle development. Absence of TRKB receptors reduces granulosa cell (GC) proliferation and delays follicle growth. In the present study, we offer mechanistic insights into this phenomenon. DNA array and quantitative PCR analysis of ovaries from TrkB-null mice revealed that by the end of the first week of postnatal life, Jagged1, Hes1, and Hey2 mRNA abundance is reduced in the absence of TRKB receptors. Although Jagged1 encodes a NOTCH receptor ligand, Hes1 and Hey2 are downstream targets of the JAGGED1-NOTCH2 signaling system. Jagged1 is predominantly expressed in oocytes, and the abundance of JAGGED1 is decreased in TrkB−/− oocytes. Lack of TRKB receptors also resulted in reduced expression of c-Myc, a NOTCH target gene that promotes entry into the cell cycle, but did not alter the expression of genes encoding core regulators of cell-cycle progression. Selective restoration of JAGGED1 synthesis in oocytes of TrkB−/− ovaries via lentiviral-mediated transfer of the Jagged1 gene under the control of the growth differentiation factor 9 (Gdf9) promoter rescued c-Myc expression, GC proliferation, and follicle growth. These results suggest that neurotrophins acting via TRKB receptors facilitate early follicle growth by supporting a JAGGED1-NOTCH2 oocyte-to-GC communication pathway, which promotes GC proliferation via a c-MYC-dependent mechanism.
Ovarian follicle development is tightly controlled by various endocrine, paracrine, and autocrine factors that act in a coordinated manner to regulate growth of the oocyte and its surrounding granulosa and theca cell layers (1–4). In rodents, initial follicle recruitment and the transition from primary to secondary follicles are mainly regulated by intraovarian factors, several of which have been identified (2). One of these regulatory systems uses neurotrophins (NT) as ligands and tropomyosin-related kinase (TRK) receptors, in addition to a common p75NTR receptor, for signaling (5–7). The NT were originally described as a family of polypeptide growth factors essential for the survival and differentiation of various neuronal populations in the central and peripheral nervous system (8, 9). It is now clear that they are also required for the development and function of organs as diverse as those comprising the cardiovascular, immune, endocrine, and reproductive systems (reviewed in Ref. 10). The four known mammalian NT include nerve growth factor, brain-derived neurotrophic factor (BDNF), NT3, and NT4/5. They are recognized by different TRK receptors: TRKA binds nerve growth factor, TRKB recognizes BDNF and NT4/5, and TRKC binds NT3 (6). The four NT are recognized by the pan-p75NTR (5). All of these molecules are expressed in feto-neonatal rodent ovaries and fetal human ovaries before the initiation of follicle assembly (11–18). BDNF and NT4/5 have been shown to promote oocyte maturation (19) and in vitro follicular assembly (20), strongly suggesting a role for TRKB signaling in these processes.
Using TrkB-null mice, we (21) and others (12) demonstrated that TRKB signaling is required for oocyte survival and preantral follicular development. Because the ovary expresses both full-length, kinase domain-containing TRKB receptors and a truncated TRKB isoform lacking the tyrosine kinase domain of the receptor, we studied mice lacking both TRKB isoforms. Follicle assembly is reduced in these mutants (22), which in addition suffer a stage-selective deficiency in early follicular development that compromises the ability of follicles to grow beyond the primary stage. Proliferation of granulosa cells (GC), required for this transition, and expression of FSH receptors, which reflects the degree of biochemical differentiation of growing follicles, are also reduced. Although these observations and those of Spears et al. (12) demonstrate the importance of TRKB receptors in early ovarian development, they also raise the question as to the downstream molecules and cellular mechanisms underlying these novel functions of TRKB signaling in the ovary.
Earlier studies demonstrated that the JAGGED1-NOTCH2 complex contributes to maintaining oocyte-GC communication (23–27), with oocytes expressing JAGGED1 (the ligand) and GC expressing NOTCH2 (the receptor) (28). More recent studies have shown that NOTCH2 signaling is required for both follicle formation (29) and GC cell proliferation during early follicle development (27). Using DNA arrays as a high throughput strategy for gene discovery and a combination of molecular, morphologic, and gene transfer approaches, we now report that deficits in follicle development and GC proliferation observed in TrkB-null mice are due, to a significant extent, to perturbation of the JAGGED1-NOTCH2 cell-cell communication pathway. Our results also indicate that this perturbation results in reduced expression of c-Myc, a direct target of NOTCH signaling (30, 31) that drives cell cycle progression by promoting entry into the S (DNA synthesis) phase of the cell cycle (32, 33). Finally, our study indicates that oocytes play a critical role in sustaining TRKB-dependent early follicle growth, as evidenced by the effectiveness of oocyte-specific restoration of JAGGED1 synthesis to rescue the defects in c-Myc expression, follicle growth, and GC proliferation caused by the absence of TRKB receptors. A preliminary report of these findings has been presented (34).
Materials and Methods
TrkB-null mice
The TrkB-null mice used in this study, as well as additional details concerning their postnatal phenotype, were previously described (21, 35). TrkB−/+ mice were bred to wild-type (WT) animals of the same genetic background, and the F1 progeny was used to produce TrkB-null and WT controls. The animals were maintained on a 12-h light, 12-h dark cycle (lights off at 1900 h), with food and water available ad libitum.
The breeders were fed with LabDiet 5001 (PMI Nutrition International Brentwood, St. Louis, MO). Animal usage was duly approved by the Institutional Animal Care and Use Committee of the Oregon National Primate Research Center, in accordance to the guidelines provided by the National Institutes of Health Guide and Use of Laboratory Animals.
Collection of ovarian tissue and genotyping
Ovaries from entire litters were collected at 0 (day of birth), 2, 3, 4, 6, 7, and 12 d after birth and used for different procedures (organ and cell culture, RNA extraction, immunohistochemistry, in vitro hybridization, and morphometric analysis) before establishing each genotype. Once the genotype was known, the ovaries were assigned to either a WT (TrkB+/+) or a knockout (KO) (TrkB−/−) group. No heterozygotes were included in the analyses. For identification of TrkB alleles, we used DNA isolated from 2-mm tail biopsies. Tail samples were lysed in a DirectPCR buffer containing proteinase K (Viagen Biotech, Inc., Los Angeles, CA), and 1 μl of the crude lysates was used for PCR. The TrkB WT allele and a DNA segment comprising both the targeting vector and a gene-specific sequence were detected using a set of three primers: TrkB-C8 (5′-ACTGACATCCGTAAGCCAGT-3′), TrkB-N2 (5′-ATGTCGCCCTGGCTGAAGTG-3′), and PGK-3.1 (5′-GGTTCTAAGTACTGTGGTTTCC-3′). The size of the PCR product was 400 bp for the WT allele and 200 bp for the deleted allele (21, 22).
DNA microarrays
To identify mRNA that are differentially expressed in ovaries from TrkB−/− mice as compared with WT animals during the initiation of early follicular growth, we employed 7-d-old mice and a two-dye spotted cDNA microarray containing 8400 gene probes generated from a mouse NIA 15K gene set, printed in duplicate on glass slides by the Gene Microarray Shared Resource Facility of the Oregon Health and Science University (http://www.ohsu.edu/xd/research/research-cores/gmsr/). The procedure employed for RNA extraction, synthesis of cDNA from total RNA, cDNA labeling, and hybridization to DNA microarrays has been previously described (36). The signal intensities were analyzed using print tip group lowess, as recommended by Yang et al. (37), and implemented by Sandrine Dutoit in BioConductor (http://www.bioconductor.org/). The array results have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) and are accessible through Gene Expression Omnibus Series accession no. GSE8528.
Culture of ovaries with BDNF
To determine the effect of TRKB activation on Jagged1, c-Myc, and ornithine decarboxylase 1 (Odc1) mRNA levels, ovaries from 4-d-old WT mice were dissected under a stereomicroscope using aseptic conditions, placed on sterile lens paper, and cultured on metal grids in a 24-well plate at the interface of air/culture medium, under an atmosphere of 60% O2-35% N2-5% CO2, as described (21, 38). One ovary from each animal was cultured in the presence of BDNF (100 ng/ml); the contralateral ovary served as an untreated control. After 8 h of incubation, the tissues were frozen in dry ice and stored at −85 C until RNA extraction.
Culture of ovaries for lentivirus (LV)-mediated gene transfer
To increase Jagged1 expression in oocytes of TrkB−/− mice, a cDNA containing the rat Jagged1-coding region fused to a sequence encoding a human influenza hemagglutinin (HA) epitope (kindly provided by Gerry Weinmaster; Department of Biological Chemistry, University of California, Los Angeles, CA) was inserted into the multiple cloning site of a LV vector (39). To restrict Jagged1 expression to oocytes, the cytomegalovirus promoter of this vector was replaced by the Gdf9 promoter (kindly provided by Austin Cooney; Baylor College of Medicine, Houston, TX). Infectious viral particles were produced and quantified as reported (40). The ovaries from 3-d-old TrkB−/− mice were dissected and incubated as described above. One ovary from each animal was treated for 4 d with this LV construct (termed LV-Jagged1-HA) and the contralateral ovary with a vector lacking Jagged1 (LV-no Jagged1), each at 7 × 106 transducer units per milliliter per well. At the end of the culture period, the ovaries were either fixed for immunohistochemistry and morphometric analysis or frozen on dry ice and stored at −85 C until RNA extraction (see below).
RNA extraction, semiquantitative PCR (qPCR), and real-time PCR
A Micro RNeasy kit (QIAGEN, Germantown, MD) was used to isolate total RNA, and 250 ng of total RNA were reverse transcribed using Omniscript reverse transcriptase, as previously described (36, 40). In one set of experiments, in which we treated ovaries in culture with BDNF or NT4, we used the semi-qPCR procedure of Ambion (Austin, TX) to detect Jagged1, c-Myc, and Odc1 mRNA. Before carrying out the amplification procedure, the optimal gene-specific primer concentrations, linear range of the PCR, and optimal primer concentration for the amplification of Ppia mRNA (used as an internal standard) were determined. Ppia mRNA encodes peptidylprolyl isomerase A, also known as cyclophilin A. The PCR were carried out in a 25-μl volume containing 0.5 μl of RT reaction and 25 pmol (Jagged1, c-Myc, or Odc) and 25 pmol (Ppia) of primers at a primer/competimer ratio of 1:1. The PCR amplification protocol consisted of 30 cycles of denaturing at 94 C (30 sec), annealing at 55 C (30 sec), and extension at 72 C (1 min). Equal volumes of the PCR were electrophoresed on 2% agarose gels stained with ethidium bromide, the gels were imaged in a Gel Doc 2000 (Bio-Rad Laboratories, Hercules, CA), and the images were quantitated using the image analysis Quantity One software (Bio-Rad Laboratories).
For all other studies, we measured mRNA levels by real-time PCR using SYBR Green PCR technology and reagents purchased from Applied Biosystems (Foster City, CA). Standard curves (threshold cycle number vs. log [RNA]) were constructed by using serial dilutions (1:10) of cDNA, assuming that the amount of cDNA is equal to the initial amount of mRNA. The threshold cycle number from each sample was referred to this curve to estimate the corresponding mRNA content, and each mRNA value was then normalized for procedural losses using the 18s rRNA values estimated from the relative standard curve.
All PCR primers were designed using the software Primer Select 6.0 (DNASTAR, Inc., Madison, WI) (Table 1).
Table 1.
RT-PCR primer list
| Gene | Forward primer | Reverse primer | GenBank |
|---|---|---|---|
| Jagged1a | GCACGCCGACAAAACACCCGAACT | ATTAGGACCGCTGGCAGATGTGGA | NM_013822 |
| Jagged1 | CCGAGGACTATGAGGGCAAGAA | GGGGACCACAGACGTTAGAAGAG | NM_013822 |
| Notch2 | GTGGACGGCATCAATCGCTACA | GGGGCATATACACCGGAAACCAT | NM_010928 |
| Hes1 | GCCGCCGCCGCTTGTGC | GGGATGACCGGGCCGCTGTGAG | NM_008235 |
| Hey2 | ATTTTGAAGATGCTCCAGGCTACAG | CACTCTCGGAATCCAATGCTCA | NM_013904 |
| Ppiaa | GGCAAATGCTGGACCAAACACAA | GGTAAAATGCCCGCAAGTCAAAAG | NM_008907 |
| Cdk2 | GGGGGATGACCGCAGTGT | GGGTCCCCAGAGTCCGAAAGAT | NM_183417 |
| Cdk4 | TGTACGGCTGATGGATGTCTGTGC | GCCCGGTGGAGGTGCTTTGTC | NM_009870 |
| CyclinD2 | GCCGCAGTCACCCCTCACGA | TGCTCCCACGCTTCCAGTTGC | NM_009829 |
| CyclinE1 | TGTCCTCGCTGCTTCTGCTTTGTA | CGGATAACCATGGCGAACGGAACC | NM_007633 |
| p15INK4b | AGGGGCGCGGCTGGATGT | CCTAGATGGGGCTGGGGAGAAAGA | NM_007670 |
| p16INK4a | CAAGAGCGGGGACATCAAGACATC | ACGTTCCCAGCGGTACACAAAGAC | NM_009877 |
| p18INK4c | GGGGCATCGGAACCATAAG | AACCCCATTTGCCTCCATCA | NM_007671 |
| p19INK4d | GAAGAAGGGAGTGGGAGGAGCAGT | CCAAAAGGGGTGAGAAAAACAAAT | NM_009878 |
| p21Cip1 | TGGGCCCGGAACATCTCAGG | CGTGGGCACTTCAGGGTTTTCTCT | NM_007669 |
| p27Kip1 | GGTGGACCAAATGCCTGACTCGT | TCTGTTCTGTTGGCCCTTTTGTTT | NM_009875 |
| p57Kip2 | GGGTGCTGAGCCGGGTGATGA | CTCCGGCTCCTCGTCCTTCTCCTC | NM_009876 |
| c-Myc | GCCCACCACCAGCAGCGACTCT | GGGGTTTGCCTCTTCTCCACAG | NM_001177352 |
| Odc1a | GCCCGGCTCTGACGATGAA | CCGCTCTCCTGGGCACAAG | NM_013614 |
Semiquantitative RT-PCR.
In situ hybridization
The cellular localization of Jagged1 mRNA was assessed using the in situ hybridization procedure described by Simmons et al. (41) with minor modifications (42). A 445-bp mouse Jagged1 cDNA complementary to nucleotides 3226–3670 in mouse Jagged1 mRNA (NM_013822.4) was generated by PCR amplification of mouse ovary total RNA and cloned into the pGEM-T vector (Promega, Madison, WI). After linearization with NcoI, 500 ng of Jagged1 cDNA template were transcribed with 250 μCi of 35S-uridine triphosphate (PerkinElmer, Boston, MA). Hybridization reactions using this cRNA were performed on 14-μm cryostat sections derived from ovaries collected at postnatal d 4, 6, and 12 and fixed by immersion in 4% paraformaldehyde and 0.1 m sodium borate buffer (pH 9.5) (overnight at 4 C). After an overnight hybridization at 55 C, the sections were washed (36, 42) and exposed to an autoradiography nuclear emulsion (Kodak, Rochester, NY) for 3 wk. After developing the reaction, the sections were counterstained with hematoxylin (Sigma, St. Louis, MO), dehydrated in ascending alcohols, and coverslipped for microscopic examination. Control sections were incubated with a 35S-labeled sense Jagged1 RNA probe transcribed from the same cDNA template used to prepare the antisense probe but in the opposite direction.
Immunohistochemistry
After collection, the ovaries were immersed in Zamboni's fixative overnight at 4 C and processed as described (21, 40), before preparing 14-μm cryostat sections. JAGGED1 was detected with a rabbit polyclonal antibody (SC-6011, diluted 1:400; Santa Cruz Biotechnology, Inc., Santa Cruz, CA). The HA.11 epitope tag used in the LV-Jagged1 construct was detected with a mouse monoclonal antibody (MMS-101R, diluted 1:1000; Covance, Berkeley, CA). After an overnight incubation at 4 C, the immunoreactions were developed by incubating the sections for 1 h at room temperature with Alexa Fluor 488 donkey antirabbit (1:500) and Alexa Fluor 568 donkey antimouse (1:500), respectively. Cell nuclei were stained with the vital dye Hoechst (Molecular Probes, Eugene, OR) as reported (21, 40).
Assessment of cell proliferation
The ovaries from 3-d-old TrkB−/− mice, incubated with LV-Jagged1 or LV-no Jagged1 for 4 d, were fixed in Zamboni's fixative, embedded in paraffin, sectioned at 14 μm, and subjected to immunohistochemistry for proliferating cell nuclear antigen (PCNA), as reported (21, 43), using a monoclonal antibody to PCNA (Mab PC-10, 1:100; Santa Cruz Biotechnology, Inc.) and developing the immunoreaction with a diaminobenzidine, H2O2, and nickel chloride solution, followed by counterstaining with Nuclear Fast Red (undiluted, 10 min at room temperature; Vector Laboratories, Burlingame, CA). Positive cells were counted using the public domain software ImageJ 1.42q (National Institutes of Health, Bethesda, MD; http://rsbweb.nih.gov/ij/index.html).
Morphometric analysis
Ovaries from 3-d-old mice, maintained for 4 d in organ culture, were fixed in Kahle's fixative, embedded in paraffin, serially sectioned at 6 μm, stained with Weigert's iron hematoxylin, and counterstained with picric acid-methyl blue, as reported (21, 38, 43). Every section was imaged as described (21), and the degree of follicle development was subjected to morphometric analysis counting only follicles in which the nucleus of the oocyte was visible (21, 43). Follicles were classified according to well-established criteria (44) that we have previously used (21, 38, 43).
Statistical analysis
Quantitative data were analyzed using SigmaStat 3.1 software (Systat Software, Inc., San Jose, CA). The data were first subjected to a normality test and an equal variance test. Data that passed these two tests were then analyzed as follows: comparison of two groups was performed with the Student's t test, data sets containing more than two groups were analyzed with one-way ANOVA followed by Student-Newman-Keuls multiple test for individual means.
Results
Absence of TRKB receptors results in reduced expression of JAGGED1 and NOTCH2 target genes in postnatal mouse ovaries
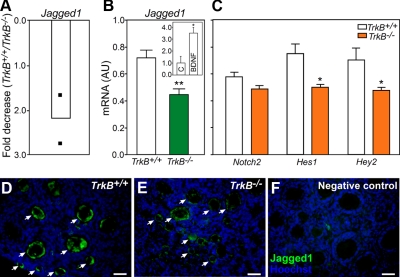
To identify genes that may be controlled by TRKB receptor-dependent signaling in the infantile mouse ovary, we interrogated ovaries from 7-d-old TrkB+/+ and TrkB−/− mice employing DNA microarrays and real-time PCR. Among the genes whose expression decreased in the absence of TRKB signaling, we identified Jagged1 (Fig. 1A), one of the Notch ligands. Using real-time PCR, we confirmed the array result (Fig. 1B) and found that the expression of Hes1 and Hey2, two of the well-characterized NOTCH target genes (45, 46), was also decreased (Fig. 1C). In contrast, there were no differences in Notch2 mRNA expression between WT and KO ovaries (Fig. 1C). Incubation of WT ovaries from 7-d-old mice with the TRKB ligand BDNF (100 ng/ml, 8 h) resulted in a 3-fold increase in Jagged1 mRNA content (Fig. 1B, inset), suggesting that Jagged1 expression decreases in TrkB−/− ovaries due to the absence of TRKB-mediated signaling. Next, we performed immunohistochemical experiments to verify the cellular sites of JAGGED1 expression and to determine whether the content of JAGGED1 protein is also decreased in TrkB−/− ovaries. JAGGED1 was seen exclusively in oocytes, and the intensity of the fluorescence signal was distinctly lower in TrkB-null than in WT ovaries (Fig. 1, D and E). No specific staining was observed in sections incubated without the primary antibody (Fig. 1F).
Fig. 1.
Absence of TRKB receptors result in reduced expression of Jagged1 and NOTCH target genes in the mouse ovary. Panel A, Decrease in Jagged1 mRNA content in the ovary of 7-d-old TrkB−/− mice detected using cDNA microarrays. Changes in mRNA content are expressed as fold-decrease with respect to mRNA values in TrkB+/+ mice of the same age. Filled squares represent the values detected in independent microarray determinations. Panel B, Jagged1 mRNA content was reduced in the ovary of 7-d-old TrkB−/− mice, as assessed by real-time PCR. Inset, Jagged1 mRNA content increased in 7-d-old WT ovaries treated in vitro with BDNF (100 ng/ml, 8 h) compared to control (C) ovaries incubated with vehicle. Relative mRNA values are expressed as arbitrary units (AU), normalized using 18s RNA or Ppia mRNA values as the normalizing unit. Panel C, Hes1 and Hey2, but not Notch, mRNA abundance, was also reduced in TrkB-null ovaries. Panel D, JAGGED1 immunoreactive material (green color) mostly localizes to oocytes in the ovary from 7-d-old TrkB+/+ mice. Panel E, JAGGED1 immunoreactivity was noticeably decreased in oocytes of TrkB−/− mice. Panel F, Section incubated without JAGGED1 antibodies. Cell nuclei stained with the DNA-binding dye Hoechst are shown in blue. White arrows point to examples of oocytes showing JAGGED1 staining. Columns in B and C represent means from four to five animals per group, and vertical lines are sem. *, P < 0.05; **, P < 0.01 vs. WT controls. Scale bars, 50 μm.
Ontogeny of Jagged1, Notch2, Hes1, and Hey2 mRNA expression in the postnatal mouse ovary
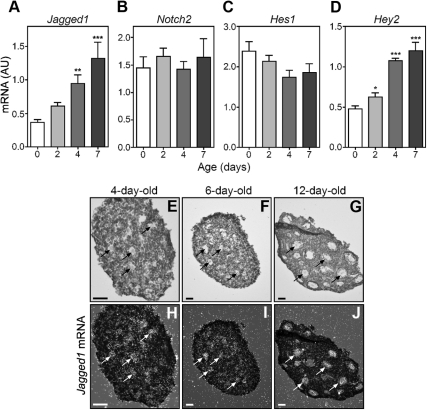
We next performed real-time PCR measurements to determine whether the expression of Jagged1, Notch2, and Notch2 target genes changes during the first 7 d of life, i.e. the period of time when the primordial, primary, and secondary follicles form. In agreement with an earlier report (29), we observed that Jagged1 and Hey2 mRNA abundance increase about 3-fold between the day of birth and the end of the first postnatal week of life (Fig. 2, A and D). In contrast, there were no significant changes in Notch2 and Hes1 mRNA expression (Fig. 2, B and C). Hybridization histochemistry demonstrated negligible Jagged1 expression on the day of birth (data not shown), before follicular assembly, and a gradual increase in Jagged1 mRNA abundance restricted to oocytes between 4 and 12 d of age (Fig. 2, E–G, bright field images, and H–J, dark field images), a time during which primary follicles (Fig. 2, E and H) reach the large preantral stage (Fig. 2, G and J).
Fig. 2.
Changes in ovarian content of Jagged1, Notch2, Hes1, and Hey2 mRNA during the first postnatal week of life of the mouse, as assessed by real-time PCR. A, Jagged1 mRNA. B, Notch2 mRNA. C, Hes1 mRNA. D, Hey2 mRNA. Relative mRNA values are expressed as arbitrary units (AU), normalized using 18s RNA values as the normalizing unit. E–J, In situ hybridization, using a mouse-specific 35S-uridine triphosphate-labeled Jagged1 cRNA probe, shows that Jagged1 mRNA is exclusively expressed in oocytes and that the abundance of Jagged1 mRNA increases during the first 12 d of postnatal life. Bright field images are shown in E–G and dark field images in H–J. Black and white arrows point to examples of Jagged1 mRNA-containing oocytes. Bars represent the mean of four to five mice per group, and vertical lines are sem. *, P < 0.05; **, P < 0.01; ***, P < 0.001 vs. 0-d-old group. Scale bar, 50 μm.
Oocyte-specific increase in JAGGED1 expression rescues deficits in secondary follicle development and GC proliferation in TrkB-null mice
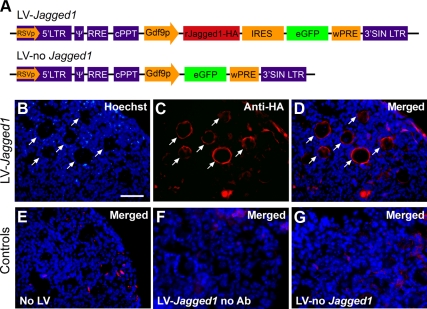
A previous study showed that 7-d-old ovaries lacking all isoforms of the TRKB receptor have a deficiency in secondary follicle development and GC proliferation (21). To determine whether this deficit is related to decreased Jagged1 expression, we exposed ovaries from 3-d-old TrkB−/− mice for 4 d to a lentiviral construct expressing Jagged1, tagged with the human influenza HA epitope tag, under the control of the Gdf9 promoter (Fig. 3A). Immunohistofluorescence analysis of the ovaries 4 d later using antibodies to HA revealed that the JAGGED1-HA protein was exclusively expressed in oocytes, where it was correctly targeted to the cell membrane (Fig. 3, B–D). We verified the specificity of this localization using three different negative controls: 1) ovaries incubated without LV (Fig. 3E), 2) ovaries incubated in presence of LV-Jagged1-HA but without adding the first antibody to the immunohistochemical reaction (Fig. 3F), and 3) ovaries incubated with the LV vector alone (Fig. 3G). In no case was HA oocyte staining observed.
Fig. 3.
Lentiviral-mediated delivery of Jagged1, using the Gdf9 promoter to target expression of a JAGGED1-HA fusion protein to oocytes, correctly targets JAGGED1 to the cell membrane of oocytes. A, Map of the lentiviral delivery construct (LV-Jagged1) used in this study. The lentiviral vector employed has been previously described (79). The 3′LTR of this vector contains a 400-bp deletion that results in the self-inactivation (SIN) of the vector. The other components include the packaging signal (ψ), the Rev response element binding site (RRE), the central polypurine tract (cPPT), and the woodchuck-hepatitis-virus posttranslational regulatory element (wPRE). The LV-Jagged1 construct contains a bicistronic transgene cassette in which expression of a Jagged1-HA cDNA is driven by the rat Gdf9 promoter (Gdf9p). The Jagged1-HA cDNA is linked to an enhanced green fluorescent protein (eGFP) cDNA via an internal ribosome entry site (IRES). A construct lacking Jagged1-HA (LV-no Jagged1) was used as a negative control. B–D, Immunohistofluorescent images of sections from 3-d-old mouse ovaries cultured for 4 d in the presence of LV-Jagged1 and stained with monoclonal antibodies against the HA epitope. E, Section from an ovary not infected with LV. F, Section from an ovary infected with LV-Jagged1 and incubated without HA antibodies. G, Section from an ovary infected with LV-no Jagged1. JAGGED1 immunoreactive cells are seen in red, and cell nuclei stained with the DNA-binding dye Hoechst are shown in blue. Scale bar, 50 μm.
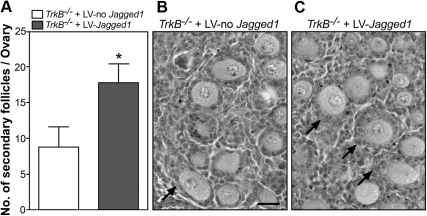
Next, we performed a morphometric analysis of cultured TrkB−/− ovaries to compare the degree of follicular growth achieved after infection with LV-Jagged1 in comparison with ovaries infected with LV-no Jagged1. We observed that the number of secondary follicles per ovary was significantly (P < 0.05) increased in the mutant ovaries infected with LV-Jagged1 as compared with ovaries infected with LV-no Jagged1 (Fig. 4A). Representative images showing this difference are shown in Fig. 4, B and C. No overt changes in other ovarian structures were observed between the two groups.
Fig. 4.
Oocyte-specific restoration of JAGGED1 expression, via lentiviral-mediated gene transfer, rescues the deficit in follicle growth of TrkB−/− ovaries. A, Increased number of secondary follicles in TrkB−/− ovaries incubated for 4 d with LV-Jagged1 in comparison with TrkB−/− ovaries infected with LV-no Jagged1. B, Section from a TrkB−/− ovary infected with LV-no Jagged1. C, Section from a TrkB−/− ovary infected with LV-Jagged1. Arrows point to secondary follicles, which contain an oocyte surrounded by two layers of GC. Scale bar, 50 μm. Columns represent the mean of four mice per group, and vertical lines are sem. One ovary from each animal was infected with LV-Jagged1 and the contralateral ovary from the same animal with LV-no Jagged1. *, P < 0.05.
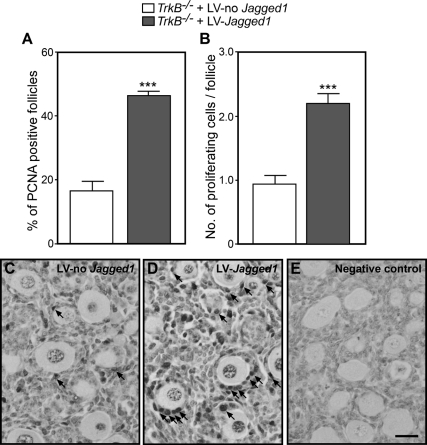
To determine whether this increase in follicle number is accompanied by changes in GC proliferation, we estimated the number of GC immunopositive for the proliferation marker PCNA 4 d after infecting TrkB−/− ovaries with LV-Jagged1 or LV-no Jagged1. We found a 2.5-fold increase in the number of follicles containing PCNA-positive cells in TrkB−/− ovaries infected with LV-Jagged1 compared with controls (Fig. 5A) and a similar increase in the number of PCNA-immunopositive cells per follicle (Fig. 5B). The microphotographs depicted in Fig. 5, C–E, illustrate these changes (examples denoted by arrows).
Fig. 5.
Oocyte-specific restoration of JAGGED1 expression, via lentiviral-mediated gene transfer, rescues the deficit in GC proliferation of TrkB−/− ovaries. A, Percent of follicles showing at least one PCNA-positive GC. B, Number of PCNA-positive GC per follicle (primary and secondary). C, Image of a section from a TrkB−/− ovary incubated for 4 d with LV-no Jagged1. D, A section from a TrkB−/− ovary incubated for 4 d with a LV-Jagged1. E, Ovarian section immunostained in absence of primary antibodies. Columns represent the mean of four mice per group, and vertical lines are sem. In each group, four sections per ovary were used for quantification. One ovary from each animal was infected with LV-Jagged1 and the contralateral ovary from the same animal with LV-no Jagged1. Scale bar, 50 μm. ***, P < 0.001 vs. LV-no Jagged1.
Loss of TRKB signaling results in decreased expression of c-Myc
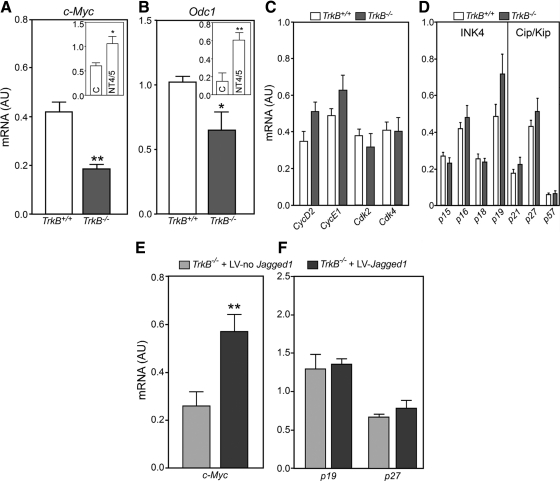
Follicular growth requires proliferation of GC (47–49), a process that involves the coordinated participation of cyclins, cyclin-dependent kinases (CDK), and CDK inhibitors (CKI) (50, 51). In addition, early follicular growth appears to involve the participation of the oncoprotein c-MYC (27, 52, 53). It was, therefore, important to determine whether the reduction in GC proliferation observed in TrkB−/− ovaries (21) is related to changes in expression of these genes. In the absence of TRKB receptors, c-Myc mRNA abundance was significantly reduced, as assessed by qPCR on postnatal d 7 (Fig. 6A). This decrease is, to a significant extent, due to the absence of TRKB-mediated signaling, because WT ovaries responded to NT4/5, a TRKB ligand, with increased c-Myc expression after 8 h in organ culture (Fig. 6A, inset).
Fig. 6.
TrkB signaling sustains c-Myc and Odc1 expression but not the expression of core regulatory components of the cell cycle. Panel A, c-Myc mRNA content was reduced in 7-d-old TrkB−/− ovaries as compared with WT animals. Inset, In vitro exposure of WT ovaries to NT4/5 (100 ng/ml, 8 h) increased c-Myc mRNA abundance as compared to control (C) ovaries incubated with vehicle. Panel B, Odc1 mRNA abundance was also decreased in TrkB−/− ovaries. Inset, NT4/5 increased Odc1 mRNA abundance in WT ovaries. Panels C and D, The content of mRNA encoding cyclins (CycD2 and CycE1), CDK (Cdk2 and Cdk4), and the CKI of the INK4 family (p15INK4b, p16INK4a, p18INK4c, and p19INK4d) and CIP/KIP family (p21Cip1, p27Kip1, and p57Kip2) remain unaltered in TrkB−/− ovaries as compared with TrkB+/+ mice. Panel E, c-Myc mRNA levels were increased in TrkB−/− ovaries after oocyte-specific restoration of JAGGED1 synthesis. The ovaries from 3-d-old mice were incubated for 4 d with a lentiviral construct carrying the Jagged1-coding region under the control of the Gdf9 promoter. Panel F, Neither p19INK4D nor p27Kip1 mRNA levels changed after lentiviral-mediated restoration of JAGGED1 synthesis. Control ovaries were infected with a LV lacking Jagged1 cDNA (LV-no Jagged1). Each column represents the mean of four to five mice per group, and vertical lines are sem. One ovary from each animal was infected with LV-Jagged1 and the contralateral ovary with LV-no Jagged1. *, P < 0.05; **, P < 0.01 vs. their respective controls. AU, Arbitrary units.
A well-established target of c-MYC is Odc1 (54, 55), which encodes ODC, the rate-limiting step in polyamine biosynthesis. ODC plays an essential role in cell proliferation (56). DNA microarrays revealed that Odc1 expression was reduced (2-fold decrease) in the ovaries from 7-d-old TrkB−/− mice compared with WT ovaries (data not shown). Using real-time PCR, we confirmed the array result (Fig. 6B). WT ovaries treated in vitro with NT4/5 (100 ng/ml, 8 h) showed an increase in Odc1 mRNA abundance (Fig. 6B, inset), suggesting that, as is the case of c-Myc, Odc1 expression is also enhanced by TRKB-mediated signaling. In contrast, no changes were observed in the gene expression of several core regulators of the cell cycle, including two mRNA encoding cyclins (CycD2 and CycE1), two mRNA encoding CDK (Cdk2 and Cdk4), and mRNA encoding CKI of either the inhibitors of CDK4 (INK4) gene family (p15INK4B, p16INK4A, p18INK4C, and p19INK4D) or the cyclin and CDK inhibitors (CIP/KIP) family (p21Cip1, p27Kip1, and p57Kip2) (Fig. 6, C and D).
Restoring JAGGED1 expression in oocytes of TrkB-null ovaries rescues c-Myc expression
TrkB−/− ovaries infected with LV-Jagged1-HA showed increased levels of c-Myc mRNA after 4 d of treatment in organ culture (Fig. 6E). This change is not due to a general effect of JAGGED1 on cell cycle regulators, because neither p19INK4D nor p27Kip1 mRNA levels, selected as examples of each class of genes, changed in LV-Jagged1-HA-infected ovaries (Fig. 6F). These results suggest that NOTCH signaling supports GC proliferation by activating a c-MYC-dependent pathway and not by affecting the expression of core regulators of cell-cycle progression.
Discussion
Studies in several species, including rodents, humans, cattle, and pigs, have established the concept that NT are physiological components of the intraovarian machinery controlling both the assembly of primordial follicles and the growth of newly formed follicles (reviewed in Ref. 57). The mechanisms underlying the supportive actions of NT on these two developmental events have not been yet elucidated. The present results identify the NOTCH signaling pathway as a mediator of the process by which NT acting via TRKB receptors facilitate early follicle development and stimulate GC proliferation in primary follicles. Our results indicate that activation of this pathway involves a NOTCH ligand (JAGGED1) produced in oocytes and NOTCH receptors, presumably located in GC. Although Jagged1 expression is reduced in the absence of TRKB receptors, oocyte-specific restoration of JAGGED1 synthesis in TrkB-null ovaries reinvigorates follicle growth and GC proliferation, suggesting that JAGGED1 produced in oocytes is crucial for NT4/5-BDNF (the TRKB ligands) to promote early follicle development. We also observed loss of c-Myc expression in the ovaries of TrkB-null mice and rescue of this deficit by the oocyte-specific restoration of JAGGED1 synthesis. Considering that c-MYC, which drives cell proliferation by promoting entry into the cell cycle (32), is a direct target of the NOTCH signaling system (30, 31), our results implicate c-MYC as a downstream mediator of TRKB-dependent stimulation of GC proliferation. None of the aforementioned deficits can be attributed to a general effect related to the absence of TRKB receptors, because all of these deficits were rescued by specifically recovering JAGGED1 synthesis in oocytes of the mutant mice.
NOTCH signaling is an evolutionarily conserved mechanism that regulates cell fate, differentiation, and growth in a vast array of tissues (45, 58, 59). There are at least four NOTCH receptors (NOTCH1–NOTCH4) and at least five ligands, including JAGGED1, JAGGED2, Δ-like 1, Δ-like 2, and Δ-like 3 (59, 60). A major feature of NOTCH signaling is that it mediates communication between adjacent cells. Upon ligand binding, the intracellular domain of NOTCH receptors (NICD) is released by proteolytic cleavage (61) and translocates to the nucleus, where it binds to a repressor of the C-promoter binding factor 1/suppressor of hairless/Lag-1 family to convert it into a trans-activating complex, which then promotes the transcription of target genes, including Hes1, Hey2, c-Myc, and others (45, 46, 59).
Several components of the NOTCH signaling pathway have been previously described in the ovary. Although JAGGED1 is expressed exclusively in germ cells, and oocytes of primordial, primary, and secondary follicles, NOTCH2, HES1, and HEY2 are expressed mainly in GC (27–29). Although it has been known for some time that the NOTCH system plays a crucial role in the control of follicle cell and oocyte development in Drosophila (23–25), it is only recently that the importance of NOTCH signaling in primordial follicle formation and GC proliferation during early follicle development in mammals has been documented (27, 29). An essential requirement for NOTCH activity is the addition of N-acetylglucosamine to its extracellular domain by Fringe proteins (62). Lunatic Fringe (Lfng), one of these proteins, specifically facilitates JAGGED1/NOTCH2 signaling (63). The finding that mice lacking lunatic fringe are infertile and that the infertility is due to an ovarian defect (26) adds further credence to the notion that NOTCH signaling is required for normal ovarian development.
It is surprising that the ability of NT to activate Hes1 transcription has been known for more than 15 yr (64), but only recently evidence has been provided demonstrating the existence of a functional relationship between NT and the NOTCH signaling pathway (65). Our results suggest that a mechanism by which activation of TRKB receptors enhances NOTCH signaling in the ovary is by stimulating the synthesis of JAGGED1 in oocytes. Whether this is a direct effect or involves the production of an intermediate molecule produced in GC is unclear. Because expression of full-length TRKB receptors is minimal in oocytes of neonatal-infantile animals (12, 21), a direct effect would imply a role for truncated TRKB receptors in inducing Jagged1 expression. These receptors are the most abundant TRKB isoform expressed in infantile mouse ovaries (12, 21), but they lack canonical signaling motifs. They may, however, be able to initiate intracellular signaling via pathways other than those activated by full-length TrkB receptors (66, 67). A more detailed examination of this issue is warranted.
An involvement of the NOTCH system in stimulating GC proliferation of primary follicles was recently demonstrated by Zhang et al. (27), who exposed mouse primary ovarian follicles cultured in vitro to γ-secretase inhibitors to block the proteolytic release of NICD and observed arrest of follicle growth and inhibition of GC proliferation. These authors also showed that overexpression of NICD promotes GC proliferation (27). By showing that GC proliferation is diminished in the absence of TRKB-mediated signaling (21) and that selective restoration of JAGGED1 production in oocytes rescues the defect in follicle growth and GC proliferation seen in TrkB-null mutants, our results provide a functional link between the NT and NOTCH signaling systems.
NOTCH signaling promotes progression of the cell cycle in all species thus far examined (25, 68–71), suggesting that the loss of GC proliferation observed in TrkB-null ovaries may be related to loss-of-function of one or more regulatory components of the cell cycle. Cell-cycle progression is promoted by phase-specific kinase complexes composed of cyclin and CDK. Although cyclin D-CDK4/6 complexes promote G1 progression, cyclin E-CDK2 complexes facilitate completion of the G1 phase (72–74). CDK activity is, in turn, regulated by CKI, which induce cell-cycle arrest by blocking the activity of cyclin-CDK complexes (75–78). Two CKI families have been described: members of the INK4 family (p16INK4a, p15INK4b, p18INK4c, and p19INK4d) bind to and inhibit cyclin D-CDK4/6 complexes; members of the CIP/KIP family (p21Cip1, p27Kip1, and p57Kip2) bind to and inhibit the cyclin E-CDK2 complex, as well as other cyclin-CDK complexes operating throughout the cell cycle (77, 78).
We measured two mRNA encoding cyclins (CycD2 and CycE1), two encoding CDK (Cdk2 and Cdk4), four encoding CKI of the INK4 family (p16INK4a, p15INK4b, p18INK4c, and p19INK4d), and three encoding members of the CIP/KIP family (p21Cip1, p27Kip1, and p57Kip2) in ovaries from TrkB-null mice and found that their abundance was similar to that of WT ovaries. In addition, we measured two mRNA encoding CKI (p19INK4d and p27Kip1) after restoring Jagged1 expression in oocytes and again found the abundance of these mRNA to be unaltered. These results suggest that the deficit in GC proliferation seen in TrkB-null ovaries is not due to decreased expression of core regulatory components of the cell cycle.
In contrast to these results, expression of the proto-oncogene c-Myc, a bona fide NOTCH target (30, 31), decreased in the absence of TRKB signaling and increased after oocyte-specific restoration of JAGGED1 synthesis. These findings suggest that the proliferative and growth-inducing actions of TRKB-activated NOTCH signaling are mediated by c-MYC. Earlier studies support this conclusion. For instance, several years ago, it was reported that expression of both c-Myc mRNA and c-MYC is restricted to preantral follicles (52, 53). A very recent report showed that inhibition of NOTCH2 signaling in primary follicles decreased GC proliferation and reduced c-Myc expression and that overexpression of the NOTCH2 intracellular domain increases GC proliferation and induces c-Myc expression (27). Further supporting an involvement of c-Myc in mediating TRKB-activated Notch-dependent GC proliferation is provided by the reduction in Odc1 mRNA abundance observed in TrkB KO ovaries and the increase in Odc1 expression elicited in WT ovaries by the ligand-dependent activation of TRKB receptors. Odc1 encodes ODC, an enzyme that plays an essential role in cell proliferation (56) and whose transcription is activated by c-MYC (54, 55).
It has been shown that c-MYC activates the Cdc25A gene, which encodes a phosphatase controlling CDK2 activity, in addition to the genes encoding cyclin D2 and CDK4. It has also been reported that c-MYC represses p21Cip1, suggesting that c-MYC drives the cell cycle by prolonging activation of cyclin E/CDK2 complexes (32). The lack of changes in CycD2, Cdk4, and p21Cip1 expression that we observed in our studies may be interpreted as indicating a lack of c-MYC effect on the expression of these genes in the infantile ovary. However, in several instances, c-MYC appears to act via a hit-and-run mechanism (33), raising the possibility that changes in expression of these cell cycle components in our model are evanescent and no longer detected after several days in culture.
In sum, our results suggest that NOTCH signaling is one of the cell-cell communication pathways used by NT to control early follicular growth and that the coordinated activation of TRKB and NOTCH signaling represents one of the mechanisms of reciprocal oocyte-GC communication underlying the initiation of follicular growth. Our results are consistent with a model in which BDNF/NT4/5 produced by GC would activate (directly or indirectly) Jagged1 expression. In turn, JAGGED1 would activate NOTCH2 receptors in GC, which would promote GC proliferation and follicle growth by inducing expression of the cell cycle regulator factor c-MYC. Although the early stages of follicle development may be the most sensitive to NOTCH signaling, it is possible that NT-initiated cell-cell communication mediated through JAGGED1-NOTCH also plays a role in later stages of follicle development. Studies using mice in which TRKB receptors are conditionally deleted from the ovary in a cell-specific manner will be useful to address this question.
Acknowledgments
We thank Ms. Maria Costa for expert technical help with the in situ hybridization procedure.
Present address for B.K.: Centro de Estudios Científicos, Valdivia, Chile 5110466.
Present address for C.G.-R.: Department of Physiology, Monash University and Pediatric Endocrinology Unit, Monash Medical Centre, Clayton VIC 3168, Australia.
Present address for A.H.P.: Laboratory of Neurobiochemistry, Faculty of Chemistry and Pharmaceutical Sciences, Universidad de Chile, Santiago, Chile 8380492.
This work was supported by National Institutes of Health (NIH) Grants HD24870 (to S.R.O.), the Eunice Kennedy Shriver National Institute of Child Health and Human Development/NIH through cooperative agreement HD18185 as part of the Specialized Cooperative Centers Program in Reproduction and Infertility Research (to S.R.O.), and RR000163 for the operation of the Oregon National Primate Research Center (to G.A.D. and S.R.O.).
Disclosure Summary: The authors have nothing to disclose.
Footnotes
- BDNF
- Brain-derived neurotrophic factor
- CDK
- cyclin-dependent kinase
- CIP/KIP
- cyclin and CDK inhibitors
- CKI
- CDK inhibitor
- GC
- granulosa cell
- HA
- hemagglutinin
- INK4
- inhibitors of CDK4
- KO
- knockout
- LV
- lentivirus
- NICD
- intracellular domain of NOTCH receptors
- NT
- neurotrophin
- Odc1
- ornithine decarboxylase1
- PCNA
- proliferating cell nuclear antigen
- qPCR
- quantitative PCR
- TRK
- tropomyosin-related kinase
- WT
- wild type.
References
- 1. Epifano O, Dean J. 2002. Genetic control of early folliculogenesis in mice. Trends Endocrinol Metab 13:169–173 [DOI] [PubMed] [Google Scholar]
- 2. Matzuk MM, Burns KH, Viveiros MM, Eppig JJ. 2002. Intercellular communication in the mammalian ovary: oocytes carry the conversation. Science 296:2178–2180 [DOI] [PubMed] [Google Scholar]
- 3. Kezele P, Nilsson E, Skinner MK. 2002. Cell-cell interactions in primordial follicle assembly and development. Front Biosci 7:d1990–d1996 [DOI] [PubMed] [Google Scholar]
- 4. Skinner MK. 2005. Regulation of primordial follicle assembly and development. Hum Reprod Update 11:461–471 [DOI] [PubMed] [Google Scholar]
- 5. Dechant G, Barde YA. 2002. The neurotrophin receptor p75(NTR): novel functions and implications for diseases of the nervous system. Nat Neurosci 5:1131–1136 [DOI] [PubMed] [Google Scholar]
- 6. Patapoutian A, Reichardt LF. 2001. Trk receptors: mediators of neurotrophin action. Curr Opin Neurobiol 11:272–280 [DOI] [PubMed] [Google Scholar]
- 7. Barker PA. 2004. p75NTR is positively promiscuous: novel partners and new insights. Neuron 42:529–533 [DOI] [PubMed] [Google Scholar]
- 8. Snider WD. 1994. Functions of the neurotrophins during nervous system development: what the knockouts are teaching us. Cell 77:627–638 [DOI] [PubMed] [Google Scholar]
- 9. Davies AM. 2000. Neurotrophins: neurotrophic modulation of neurite growth. Curr Biology 10:R198–R200 [DOI] [PubMed] [Google Scholar]
- 10. Tessarollo L. 1998. Pleiotrophic functions of neurotrophins in development. Cytokine Growth Factor Rev 9:125–137 [DOI] [PubMed] [Google Scholar]
- 11. Dissen GA, Hirshfield AN, Malamed S, Ojeda SR. 1995. Expression of neurotrophins and their receptors in the mammalian ovary is developmentally regulated: changes at the time of folliculogenesis. Endocrinology 136:4681–4692 [DOI] [PubMed] [Google Scholar]
- 12. Spears N, Molinek MD, Robinson LL, Fulton N, Cameron H, Shimoda K, Telfer EE, Anderson RA, Price DJ. 2003. The role of neurotrophin receptors in female germ-cell survival in mouse and human. Development 130:5481–5491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Anderson RA, Robinson LL, Brooks J, Spears N. 2002. Neurotropins and their receptors are expressed in the human fetal ovary. J Clin Endocrinol Metab 87:890–897 [DOI] [PubMed] [Google Scholar]
- 14. Anesetti G, Lombide P, D'Albora H, Ojeda SR. 2001. Intrinsic neurons in the human ovary. Cell Tissue Res 306:231–237 [DOI] [PubMed] [Google Scholar]
- 15. Abir R, Fisch B, Jin S, Barnnet M, Ben-Haroush A, Felz C, Kessler-Icekson G, Feldberg D, Nitke S, Ao A. 2005. Presence of NGF and its receptors in ovaries from human fetuses and adults. Mol Hum Reprod 11:229–236 [DOI] [PubMed] [Google Scholar]
- 16. Harel S, Jin S, Fisch B, Feldberg D, Krissi H, Felz C, Freimann S, Tan SL, Ao A, Abir R. 2006. Tyrosine kinase B receptor and its activated neurotrophins in ovaries from human fetuses and adults. Mol Hum Reprod 12:357–365 [DOI] [PubMed] [Google Scholar]
- 17. Childs AJ, Bayne RA, Murray AA, Martins Da Silva SJ, Collins CS, Spears N, Anderson RA. 2010. Differential expression and regulation by activin of the neurotrophins BDNF and NT4 during human and mouse ovarian development. Dev Dyn 239:1211–1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Oron G, Ao A, Friedman O, Fisch B, Zhang XY, Ben-Haroush A, Peled Y, Abir R. 2011. Expression of neurotrophin 3 and its tropomyosin-related kinase receptor C in human preantral follicles. Fertil Steril 95:2056–2062 [DOI] [PubMed] [Google Scholar]
- 19. Kawamura K, Kawamura N, Mulders SM, Sollewijn Gelpke MD, Hsueh AJ. 2005. Ovarian brain-derived neurotrophic factor (BDNF) promotes the development of oocytes into preimplantation embryos. Proc Natl Acad Sci USA 102:9206–9211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Farhi J, Fisch B, Garor R, Peled Y, Pinkas H, Abir R. 2011. Neurotrophin 4 enhances in vitro follicular assembly in human fetal ovaries. Fertil Steril 95:1267–1271 [DOI] [PubMed] [Google Scholar]
- 21. Paredes A, Romero C, Dissen GA, DeChiara TM, Reichardt L, Cornea A, Ojeda SR, Xu B. 2004. TrkB receptors are required for follicular growth and oocyte survival in the mammalian ovary. Dev Biol 267:430–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kerr B, Garcia-Rudaz C, Dorfman M, Paredes A, Ojeda SR. 2009. NTRK1 and NTRK2 receptors facilitate follicle assembly and early follicular development in the mouse ovary. Reproduction 138:131–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ruohola H, Bremer KA, Baker D, Swedlow JR, Jan LY, Jan YN. 1991. Role of neurogenic genes in establishment of follicle cell fate and oocyte polarity during oogenesis in Drosophila. Cell 66:433–449 [DOI] [PubMed] [Google Scholar]
- 24. Grammont M, Irvine KD. 2001. fringe and Notch specify polar cell fate during Drosophila oogenesis. Development 128:2243–2253 [DOI] [PubMed] [Google Scholar]
- 25. López-Schier H, St Johnston D. 2001. Δ signaling from the germ line controls the proliferation and differentiation of the somatic follicle cells during Drosophila oogenesis. Genes Dev 15:1393–1405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hahn KL, Johnson J, Beres BJ, Howard S, Wilson-Rawls J. 2005. Lunatic fringe null female mice are infertile due to defects in meiotic maturation. Development 132:817–828 [DOI] [PubMed] [Google Scholar]
- 27. Zhang CP, Yang JL, Zhang J, Li L, Huang L, Ji SY, Hu ZY, Gao F, Liu YX. 2011. Notch signaling is involved in ovarian follicle development by regulating granulosa cell proliferation. Endocrinology 152:2437–2447 [DOI] [PubMed] [Google Scholar]
- 28. Johnson J, Espinoza T, McGaughey RW, Rawls A, Wilson-Rawls J. 2001. Notch pathway genes are expressed in mammalian ovarian follicles. Mech Dev 109:355–361 [DOI] [PubMed] [Google Scholar]
- 29. Trombly DJ, Woodruff TK, Mayo KE. 2009. Suppression of Notch signaling in the neonatal mouse ovary decreases primordial follicle formation. Endocrinology 150:1014–1024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Weng AP, Millholland JM, Yashiro-Ohtani Y, Arcangeli ML, Lau A, Wai C, Del Bianco C, Rodriguez CG, Sai H, Tobias J, Li Y, Wolfe MS, Shachaf C, Felsher D, Blacklow SC, Pear WS, Aster JC. 2006. c-Myc is an important direct target of Notch1 in T-cell acute lymphoblastic leukemia/lymphoma. Genes Dev 20:2096–2109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sharma VM, Calvo JA, Draheim KM, Cunningham LA, Hermance N, Beverly L, Krishnamoorthy V, Bhasin M, Capobianco AJ, Kelliher MA. 2006. Notch1 contributes to mouse T-cell leukemia by directly inducing the expression of c-myc. Mol Cell Biol 26:8022–8031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Grandori C, Cowley SM, James LP, Eisenman RN. 2000. The Myc/Max/Mad network and the transcriptional control of cell behavior. Annu Rev Cell Dev Biol 16:653–699 [DOI] [PubMed] [Google Scholar]
- 33. Levens DL. 2003. Reconstructing MYC. Genes Dev 17:1071–1077 [DOI] [PubMed] [Google Scholar]
- 34. Kerr B, Paredes A, Garcia-Rudaz C, de la Chesnaye E, Dissen GA, Ojeda SR. Neurotrophins acting via TrkB receptors use the Notch-Jagged1 cell-cell communication pathway to facilitate early ovarian development. 88th Annual Meeting of The Endocrine Society, Boston, MA, June 24–28, 2006 (Abstract OR38-1) [Google Scholar]
- 35. Rohrer B, Korenbrot JI, LaVail MM, Reichardt LF, Xu B. 1999. Role of neurotrophin receptor TrkB in the maturation of rod photoreceptors and establishment of synaptic transmission to the inner retina. J Neurosci 19:8919–8930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. De La Chesnaye E, Kerr B, Paredes A, Merchant-Larios H, Méndez JP, Ojeda SR. 2008. Fbxw15/Fbxo12J is an F-box protein-encoding gene selectively expressed in oocytes of the mouse ovary. Biol Reprod 78:714–725 [DOI] [PubMed] [Google Scholar]
- 37. Yang YH, Dudoit S, Luu P, Lin DM, Peng V, Ngai J, Speed TP. 2002. Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res 30:1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Romero C, Paredes A, Dissen GA, Ojeda SR. 2002. Nerve growth factor induces the expression of functional FSH receptors in newly formed follicles of the rat ovary. Endocrinology 143:1485–1494 [DOI] [PubMed] [Google Scholar]
- 39. Follenzi A, Ailles LE, Bakovic S, Geuna M, Naldini L. 2000. Gene transfer by lentiviral vectors is limited by nuclear translocation and rescued by HIV-1 pol sequences. Nat Genet 25:217–222 [DOI] [PubMed] [Google Scholar]
- 40. Garcia-Rudaz C, Luna F, Tapia V, Kerr B, Colgin L, Galimi F, Dissen GA, Rawlings ND, Ojeda SR. 2007. Fxna, a novel gene differentially expressed in the rat ovary at the time of folliculogenesis, is required for normal ovarian histogenesis. Development 134:945–957 [DOI] [PubMed] [Google Scholar]
- 41. Simmons DM, Arriza JL, Swanson LW. 1989. A complete protocol for in situ hybridization of messenger RNAs in brain and other tissues with radiolabeled single-stranded RNA probes. J Histotechnol 12:169–181 [Google Scholar]
- 42. Dissen GA, Hill DF, Costa ME, Ma YJ, Ojeda SR. 1991. Nerve growth factor receptors in the peripubertal rat ovary. Mol Endocrinol 5:1642–1650 [DOI] [PubMed] [Google Scholar]
- 43. Dissen GA, Romero C, Hirshfield AN, Ojeda SR. 2001. Nerve growth factor is required for early follicular development in the mammalian ovary. Endocrinology 142:2078–2086 [DOI] [PubMed] [Google Scholar]
- 44. Peters H. 1969. The development of the mouse ovary from birth to maturity. Acta Endocrinol 62:98–116 [DOI] [PubMed] [Google Scholar]
- 45. Yoon K, Gaiano N. 2005. Notch signaling in the mammalian central nervous system: insights from mouse mutants. Nat Neurosci 8:709–715 [DOI] [PubMed] [Google Scholar]
- 46. Fortini ME. 2002. γ-Secretase-mediated proteolysis in cell-surface-receptor signalling. Nat Rev Mol Cell Biol 3:673–684 [DOI] [PubMed] [Google Scholar]
- 47. Pedersen T. 1970. Determination of follicle growth rate in the ovary of the immature mouse. J Reprod Fertil 21:81–93 [DOI] [PubMed] [Google Scholar]
- 48. Hirshfield AN. 1985. Comparison of granulosa cell proliferation in small follicles of hypophysectomized, prepubertal, and mature rats. Biol Reprod 32:979–987 [DOI] [PubMed] [Google Scholar]
- 49. McGee EA, Hsueh AJ. 2000. Initial and cyclic recruitment of ovarian follicles. Endocr Rev 21:200–214 [DOI] [PubMed] [Google Scholar]
- 50. Bayrak A, Oktay K. 2003. The expression of cyclin-dependent kinase inhibitors p15, p16, p21, and p27 during ovarian follicle growth initiation in the mouse. Reprod Biol Endocrinol 1:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Cannon JD, Cherian-Shaw M, Lovekamp-Swan T, Chaffin CL. 2007. Granulosa cell expression of G1/S phase cyclins and cyclin-dependent kinases in PMSG-induced follicle growth. Mol Cell Endocrinol 264:6–15 [DOI] [PubMed] [Google Scholar]
- 52. Sato A, Bo M, Maruo T, Yoshida S, Mochizuki M. 1994. Stage-specific expression of c-myc messenger ribonucleic acid in porcine granulosa cells early in follicular growth. Eur J Endocrinol 131:319–322 [DOI] [PubMed] [Google Scholar]
- 53. Li S, Maruo T, Ladines-Llave CA, Kondo H, Mochizuki M. 1994. Stage-limited expression of myc oncoprotein in the human ovary during follicular growth, regression and atresia. Endocr J 41:83–92 [DOI] [PubMed] [Google Scholar]
- 54. Shichiri M, Hanson KD, Sedivy JM. 1993. Effects of c-myc expression on proliferation, quiescence, and the G0 to G1 transition in nontransformed cells. Cell Growth Differ 4:93–104 [PubMed] [Google Scholar]
- 55. Packham G, Cleveland JL. 1997. Induction of ornithine decarboxylase by IL-3 is mediated by sequential c-Myc-independent and c-Myc-dependent pathways. Oncogene 15:1219–1232 [DOI] [PubMed] [Google Scholar]
- 56. Nasizadeh S, Myhre L, Thiman L, Alm K, Oredsson S, Persson L. 2005. Importance of polyamines in cell cycle kinetics as studied in a transgenic system. Exp Cell Res 308:254–264 [DOI] [PubMed] [Google Scholar]
- 57. Dissen GA, Garcia-Rudaz C, Ojeda SR. 2009. Role of neurotrophic factors in early ovarian development. Semin Reprod Med 27:24–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Artavanis-Tsakonas S, Rand MD, Lake RJ. 1999. Notch signaling: cell fate control and signal integration in development. Science 284:770–776 [DOI] [PubMed] [Google Scholar]
- 59. Lai EC. 2004. Notch signaling: control of cell communication and cell fate. Development 131:965–973 [DOI] [PubMed] [Google Scholar]
- 60. Lendahl U. 1998. A growing family of Notch ligands. BioEssays 20:103–107 [DOI] [PubMed] [Google Scholar]
- 61. De Strooper B, Annaert W, Cupers P, Saftig P, Craessaerts K, Mumm JS, Schroeter EH, Schrijvers V, Wolfe MS, Ray WJ, Goate A, Kopan R. 1999. A presenilin-1-dependent γ-secretase-like protease mediates release of Notch intracellular domain. Nature 398:518–522 [DOI] [PubMed] [Google Scholar]
- 62. Johnston SH, Rauskolb C, Wilson R, Prabhakaran B, Irvine KD, Vogt TF. 1997. A family of mammalian Fringe genes implicated in boundary determination and the Notch pathway. Development 124:2245–2254 [DOI] [PubMed] [Google Scholar]
- 63. Hicks C, Johnston SH, diSibio G, Collazo A, Vogt TF, Weinmaster G. 2000. Fringe differentially modulates Jagged1 and Δ1 signalling through Notch1 and Notch2. Nat Cell Biol 2:515–520 [DOI] [PubMed] [Google Scholar]
- 64. Feder JN, Jan LY, Jan YN. 1993. A rat gene with sequence homology to the Drosophila gene hairy is rapidly induced by growth factors known to influence neuronal differentiation. Mol Cell Biol 13:105–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Salama-Cohen P, Arévalo MA, Meier J, Grantyn R, Rodríguez-Tébar A. 2005. NGF controls dendrite development in hippocampal neurons by binding to p75NTR and modulating the cellular targets of Notch. Mol Biol Cell 16:339–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Baxter GT, Radeke MJ, Kuo RC, Makrides V, Hinkle B, Hoang R, Medina-Selby A, Coit D, Valenzuela P, Feinstein SC. 1997. Signal transduction mediated by the truncated trkB receptor isoforms, trkB.T1 and trkB.T2. J Neurosci 17:2683–2690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Rose CR, Blum R, Pichler B, Lepier A, Kafitz KW, Konnerth A. 2003. Truncated TrkB-T1 mediates neurotrophin-evoked calcium signalling in glia cells. Nature 426:74–78 [DOI] [PubMed] [Google Scholar]
- 68. van Es JH, van Gijn ME, Riccio O, van den Born M, Vooijs M, Begthel H, Cozijnsen M, Robine S, Winton DJ, Radtke F, Clevers H. 2005. Notch/γ-secretase inhibition turns proliferative cells in intestinal crypts and adenomas into goblet cells. Nature 435:959–963 [DOI] [PubMed] [Google Scholar]
- 69. Campa VM, Gutiérrez-Lanza R, Cerignoli F, Díaz-Trelles R, Nelson B, Tsuji T, Barcova M, Jiang W, Mercola M. 2008. Notch activates cell cycle reentry and progression in quiescent cardiomyocytes. J Cell Biol 183:129–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Monahan P, Rybak S, Raetzman LT. 2009. The notch target gene HES1 regulates cell cycle inhibitor expression in the developing pituitary. Endocrinology 150:4386–4394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Rao SS, O'Neil J, Liberator CD, Hardwick JS, Dai X, Zhang T, Tyminski E, Yuan J, Kohl NE, Richon VM, Van der Ploeg LH, Carroll PM, Draetta GF, Look AT, Strack PR, Winter CG. 2009. Inhibition of NOTCH signaling by γ secretase inhibitor engages the RB pathway and elicits cell cycle exit in T-cell acute lymphoblastic leukemia cells. Cancer Res 69:3060–3068 [DOI] [PubMed] [Google Scholar]
- 72. Ekholm SV, Reed SI. 2000. Regulation of G(1) cyclin-dependent kinases in the mammalian cell cycle. Curr Opin Cell Biol 12:676–684 [DOI] [PubMed] [Google Scholar]
- 73. Endicott JA, Noble ME, Tucker JA. 1999. Cyclin-dependent kinases: inhibition and substrate recognition. Curr Opin Struct Biol 9:738–744 [DOI] [PubMed] [Google Scholar]
- 74. Reed SI. 1997. Control of the G1/S transition. Cancer Surv 29:7–23 [PubMed] [Google Scholar]
- 75. Sherr CJ, Roberts JM. 1995. Inhibitors of mammalian G1 cyclin-dependent kinases. Genes Dev 9:1149–1163 [DOI] [PubMed] [Google Scholar]
- 76. LaBaer J, Garrett MD, Stevenson LF, Slingerland JM, Sandhu C, Chou HS, Fattaey A, Harlow E. 1997. New functional activities for the p21 family of CDK inhibitors. Genes Dev 11:847–862 [DOI] [PubMed] [Google Scholar]
- 77. Besson A, Dowdy SF, Roberts JM. 2008. CDK inhibitors: cell cycle regulators and beyond. Dev Cell 14:159–169 [DOI] [PubMed] [Google Scholar]
- 78. Nakayama K, Nakayama K. 1998. Cip/Kip cyclin-dependent kinase inhibitors: brakes of the cell cycle engine during development. BioEssays 20:1020–1029 [DOI] [PubMed] [Google Scholar]
- 79. Dissen GA, Lomniczi A, Neff TL, Hobbs TR, Kohama SG, Kroenke CD, Galimi F, Ojeda SR. 2009. In vivo manipulation of gene expression in non-human primates using lentiviral vectors as delivery vehicles. Methods 49:70–77 [DOI] [PMC free article] [PubMed] [Google Scholar]