Abstract
Circadian dyssynchrony of an organism (at the whole body level) with its environment, either through light/dark cycle or genetic manipulation of clock genes, augments various cardiometabolic diseases. The cardiomyocyte circadian clock has recently been shown to influence multiple myocardial processes, ranging from transcriptional regulation and energy metabolism, to contractile function. We therefore reasoned that chronic dyssychrony of the cardiomyocyte circadian clock with its environment would precipitate myocardial maladaptation to a circadian challenge (simulated shift work; SSW). To test this hypothesis, 2 and 20 month old wild-type and CCM (Cardiomyocyte Clock Mutant; a model with genetic temporal suspension of the cardiomyocyte circadian clock at the active-to-sleep phase transition) mice were subjected to chronic (16-wks) bi-weekly 12-hr phase shifts in the light/dark cycle (i.e., SSW). Assessment of adaptation/maladaptation at whole body homeostatic, gravimetric, humoral, histological, transcriptional, and cardiac contractile function levels revealed essentially identical responses between wild-type and CCM littermates. However, CCM hearts exhibit increased bi-ventricular weight, cardiomyocyte size, and molecular markers of hypertrophy (anf, mcip1) independent of aging and/or SSW. Similarly, a second genetic model of selective temporal suspension of the cardiomyocyte circadian clock (Cardiomyocyte-specific BMAL1 Knockout [CBK] mice) exhibits increased bi-ventricular weight and mcip1 expression. Wild-type mice exhibit 5-fold greater cardiac hypertrophic growth (and 6-fold greater anf mRNA induction) when challenged with the hypertrophic agonist isoproterenol at the active-to-sleep phase transition, relative to isoproterenol administration at the sleep-to-active phase transition. This diurnal variation was absent in CCM mice. Collectively, these data suggest that the cardiomyocyte circadian clock likely influences responsiveness of the heart to hypertrophic stimuli.
Keywords: Aging, Circadian Clocks, Hypertrophy, Shift Work
INTRODUCTION
Living organisms exhibit physiological time-of-day-dependent oscillations at multiple levels, both in anticipation of, and in response to, changes in the environment. Cell autonomous circadian clocks allow a cell/organ/organism to anticipate a forthcoming stimulus/stress, thereby facilitating a response of appropriate timing and intensity (Edery 2000). At the core of the mammalian clock mechanism are two transcription factors, CLOCK and BMAL1 (Gekakis, et al. 1998; Hogenesch, et al. 1998; Takahashi, et al. 2008). Upon heterodimerization, CLOCK/BMAL1 activate a number of target genes, many of which will feedback in either a positive or negative manner, ultimately resulting in oscillations of target genes with a periodicity of approximately 24-hrs (Takahashi, et al. 2008). Target genes that are not integral clock components (i.e. do not feedback on CLOCK/BMAL1) are termed clock output genes (Edery 2000; Takahashi, et al. 2008). Many of these output genes influence cellular processes, including metabolism, in a time-of-day-dependent manner (Marcheva, et al. 2009; Wijnen, et al. 2006).
Circadian clocks have been identified in nearly all mammalian cells investigated, including cardiovascular related cell types, such as cardiomyocytes, vascular smooth muscle cells, endothelial cells, and fibroblasts (Balsalobre, et al. 1998; Durgan, et al. 2005; McNamara, et al. 2001; Nonaka, et al. 2001; Takeda, et al. 2007). Although time-of-day-dependent rhythms in heart rate, blood pressure, and cardiac output have classically been attributed to rhythms in the neurohumoral milieu, recent studies have begun to highlight a role for circadian clocks as modulators of these cardiovascular parameters in both animal models and humans (Paschos, et al. 2010; Rudic, et al. 2009; Smolensky 1996; Young 2006). For example, genetic manipulation of circadian clock components, such as CLOCK and BMAL1, abolishes time-of-day-dependent oscillations in both blood pressure and heart rate (Curtis, et al. 2007). Similarly, variations within a tandem repeat of the human clock gene Per3 is associated with alterations in heart rate (Viola, et al. 2008). More recently, several laboratories have initiated studies to elucidate the roles of cell-type specific circadian clocks on cardiovascular physiology. Genetic ablation of the circadian clock within endothelial or vascular smooth muscle cells results in marked attenuation of time-of-day-dependent rhythms in blood pressure and heart rate (Wang, et al. 2008). Through the use of a mouse model in which the cardiomyocyte circadian clock is temporally suspended at the beginning of the inactive/sleep phase (i.e., Cardiomyocyte-specific Clock Mutant [CCM] mice), we have shown that this molecular mechanism directly regulates myocardial gene expression, metabolism (e.g., triglyceride turnover), and contractile function (e.g., heart rate, cardiac output), in a time-of-day-dependent manner (Bray, et al. 2008; Durgan, et al. 2010b; Tsai, et al. 2010; Young 2009).
Given their role in a spectrum of cardiovascular functions, it is not surprising that disruption of circadian clocks has been implicated in the pathogenesis of various cardiovascular diseases (Rudic, et al. 2009; Young, et al. 2007). Circadian clocks are altered in numerous animal models of increased cardiovascular disease risk, including aging, diet-induced obesity, diabetes mellitus, hypertension- and pressure overload-induced hypertrophy, simulated shift work, and ischemia/reperfusion (i.e., myocardial infarction) (Durgan, et al. 2006; Kohsaka, et al. 2007; Kung, et al. 2007; Kunieda, et al. 2006; Mohri, et al. 2003; Young, et al. 2001; Young, et al. 2002). Importantly, circadian dyssynchrony of an organism (at the whole body level) with its environment, either through light/dark cycle or genetic manipulation, has been shown to augment the development of cardiovascular disease (Martino, et al. 2007; Martino, et al. 2008; Sole, et al. 2009). This includes shift work in humans (Knutsson, et al. 1986). What is less apparent is the relative role of cell-type specific circadian clocks in the pathogenesis of cardiovascular dysfunction. Initial studies designed to address this issue revealed that the cardiomyocyte circadian clock directly modulates myocardial ischemia/reperfusion tolerance in a time-of-day-dependent manner (Durgan, et al. 2010a). Whether this molecular mechanism influences responsiveness of the heart to additional adverse stresses is currently unknown.
The purpose of the present study was to test the hypothesis that chronic dyssychrony of the cardiomyocyte circadian clock with its environment would augment myocardial maladaptation to aging and/or a circadian challenge (simulated shift work [SSW]). Somewhat surprisingly, wild-type and CCM mice responded to aging and SSW in essentially identical manners. However, in-depth phenotypic analysis revealed that CCM hearts exhibit greater mass, cardiomyocyte size, and expression of hypertrophic markers (anf, mcip1), independent of aging and/or SSW. Similarly, a second genetic model of selective temporal suspension of the cardiomyocyte circadian clock (Cardiomyocyte-specific BMAL1 Knockout [CBK] mice) exhibits a pro-hypertrophic phenotype. Given that CCM and CBK hearts are temporally suspended at the beginning of the inactive/sleep phase, we hypothesized that the normal heart exhibits greatest responsiveness to pro-hypertrophic stimuli at this time. Consistent with this hypothesis, we observed that wild-type mice treated with isoproterenol at the awake-to-sleep transition exhibit greater cardiac hypertrophy and anf mRNA induction, relative to mice treated with isoproterenol at the sleep-to-awake transition. This diurnal variation was absent in CCM mice. Collectively, these data suggest that the cardiomyocyte circadian clock likely influences responsiveness of the heart to pro-hypertrophic stimuli.
MATERIALS AND METHODS
Animals
Specific disruption of the cardiomyocyte circadian clock was achieved through the use of two distinct genetic models; CCM and CBK mice. CCM mice express the ClockΔ19 mutant protein within cardiomyocytes, through use of the MHCα promoter (Durgan, et al. 2006). CCM (MHCα-ClockΔ19+/−) mice are compared to wild-type (MHCα-ClockΔ19−/−) littermates. BMAL1 was ablated in CBK mice, through crossing BMAL1-floxed mice (Jackson Labs) with MHCα-CRE mice (kind gift from Michael Schneider). CBK (BMAL1flox/flox/MHCα-CRE+/−) mice are compared to wild-type (BMAL1flox/flox/MHCα-CRE−/−) littermates. Male CCM mice (on the FVB/N background), CBK mice (on the C57/Bl6J background), and appropriate littermate controls were housed at the Centers for Comparative Medicine at Baylor College of Medicine (aging, and simulated shift work studies) or at the University of Alabama at Birmingham (isoproterenol administration studies). Male wild-type C57/Bl6J mice were also housed at the Comparative Biology Core Facility at Pennington Biomedical Research Center (PBRC; circadian aging studies). All studies were reviewed and approved by the respective local IACUC committees prior to the conduct of these studies. All mice were housed under controlled conditions (23±1°C; 12-h light/12-h dark cycle), and received standard laboratory chow and water ad libitum, with the exception of the C57/Bl6J circadian aging study and the simulated shift work study. For the latter, mice were subjected to a bi-weekly 12-hr phase shift in the light/dark cycle for a total of 16 weeks. Accordingly, on the 4th day of the week the light phase was extended to 24-hrs, while on the 7th day of the week the dark phase was extended to 24-hrs. For these studies, measurements were made in the middle of the light phase (i.e., zeitgeber time 6; ZT6), 3 days following the last reversal of the light/dark cycle. For the C57/Bl6J aging study performed at PBMC, mice were transferred from 12-h light/12-h dark cycle to constant darkness (dim red lighting) for the five days immediately prior to euthanasia, to facilitate identification of the effects of aging on intrinsic circadian clocks. These studies conform to international ethical standards (Portaluppi, et al. 2010).
Isoproterenol-Induced Cardiac Hypertrophy
Cardiac hypertrophy was induced pharmacologically, through administration of the β-adrenergic agonist isoproterenol. Mice (6 months of age) were injected intraperitoneally with either isoproterenol (5mg/Kg/day) or saline (vehicle control) on a daily basis, for a 1 week period. Injections were made either at the dark-to-light phase transition (i.e., ZT0) or at the light-to-dark phase transition (i.e., ZT12). Hearts were isolated exactly 24-hrs following the last injection, and stored at −80°C prior to subsequent biochemical analysis.
Non-invasive Mouse Monitoring
Body weight was monitored in mice at one-week intervals. Twenty-four hour patterns of body temperature (telemetry), energy expenditure (indirect calorimetry), and physical activity (beam breaks) were measured using a Comprehensive Laboratory Animal Monitoring System (CLAMS; Columbus Instruments Inc., Columbus, OH). For body temperature measurements, mice were allowed to recover for 2 weeks following intra-abdominal implantation of radiotelemetric probes, prior to data acquisition (using the Mini-Mitter system).
Humoral Factor Analysis
Non-fasted (i.e., fed state) plasma glucose, triglyceride, insulin, leptin, adiponectin, and corticosterone concentrations were measured using commercially available kits (Stanbio, Boerne, TX; Millipore, St. Charles, MO; MP Biomedicals, LLC, Orangeburg, NY).
Histological Analysis
Following perfusion with ice cold cardioplegic solution, hearts underwent Z-fix aqueous buffered zinc formalin perfusion fixation at constant pressure (approximately 16mmHg for 10 minutes). 5μm longitudinal four-chamber view sections were subsequently obtained, mounted on slides, deparaffinized in xylene and rehydrated in 100%, 95%, 85%, 70% solutions of ethanol. For enzyme antigen retrieval, sections were treated with Proteinase K, 20μg/ml in 10mM Tris/HCl for 30 minutes at 37°C. After blocking with 5% goat serum in 1% bovine serum/PBS, sections were stained with laminin rabbit polyclonal antibody (Abcam #11575) at 4°C overnight, followed with goat anti-rabbit AlexaFluor 488 secondary antibody (Molecular Probes #11034), to demonstrate the basement membrane. Images of tissue in cross-sectional orientation were acquired (40x objective), total field size was measured, and myocytes were counted within the field to determine the average myocyte cross-sectional area. At least 100 myocytes were counted per heart. Picosirius Red staining of collagen fibers was used for semi-quantitative assessment of outer left ventricular wall fibrosis, using ImagePro software.
RNA Isolation and Real-Time RT-PCR
RNA was extracted from hearts using standard procedures (Chomczynski, et al. 1987). Candidate gene expression analysis was performed by quantitative RT-PCR using previously described methods (Gibson, et al. 1996; Heid, et al. 1996). Specific assays were designed for each gene from mouse sequences available in GenBank. Primer and probe sequences have been reported previously (Bray, et al. 2008; Young, et al. 2001). RT-PCR data are represented as fold change from an appropriate reference group.
In Vivo Assessment of Cardiovascular Function
Myocardial contractile function was assessed in vivo in mice under anesthesia (isoflurane) through MRI (using a Bruker BioSpin MRI PharmaScan 70/16; 7.0 Tesla). Blood pressure was assessed through tail cuff measurements (using a Facility - IITC Inc./Life Science Non-Invasive Blood Pressure system).
Western Blotting
Qualitative analysis of protein expression was performed using standard Western blotting techniques. Briefly, powdered tissue (~20mg) was homogenized in tissue protein extraction reagent containing 2mM PMSF, 2mM EDTA pH 8.0, 0.25% NP-40 alternative with protease and phosphatase inhibitor cocktails. Lysates (10ug) were separated on a 7.5% bis-acrylamide gel by sodium dodecyl sulfate–polyacrylamide gel electrophoresis, transferred to a PVDF membrane and probed with Bmal-1 (1:2000 – Millipore), Cry-2 (1:2000 – Millipore) or Calsequestrin (1:4000 – Abcam) diluted in 1% Casein/PBS for 1.5 hours at room temperature. Membranes were incubated with goat anti-rabbit horse radish peroxidase-conjugated secondary antibody (1:10,000–1:40,000 – Santa Cruz). Bands were visualized with Immunstar Western C detection kit (Biorad) on x-ray film, scanned and quantified using Scion Image version 4.0.3.2 and normalized to Calsequestrin.
Statistical Analysis
For circadian aging studies, Cosinor analysis (Nelson, et al. 1979; Xu, et al. 2009) was performed using the Nonlinear Regression function of PASW 18 software with the following equation where the initial values and default parameter ranges were set manually:
Mesor (acronym for midline estimating statistic of rhythm) = the mean of the oscillation
A = the amplitude
T = the period (24 h)
Acrophase = the timing of the cosine maximum
t = a timepoint
R2 is the resulting statistic that measures the percent variance accounted for by a 24 h rhythm (or rhythm of any period τ). When the cosinor analyses were performed based on the assumption that the free-running period is fixed at 24 h, the R2 values for the expression levels of the three genes were as follows:
Individual cosinor parameters (mesor, amplitude, and acrophase) for each age group were compared for each gene with t-tests with significance ascribed at p < 0.05.
Two- and three- way analysis of variance (ANOVA) was performed to investigate the main effects of group (e.g., genotype, age, intervention) and time, using Stata version 10.0 (Stata Corp., San Antonio, TX). The null hypothesis of no model effects was rejected at p<0.05. For analyses in which the full model was statistically significant (i.e., p<0.05), Bonferroni post-hoc analyses were performed to examine all possible pair-wise comparisons. A full model including second-order interactions was conducted for each experiment. Statistically significant differences were determined using Type III sums of squares. Main effects of groups are reported in the top left hand corner of the figures, as well as in supplemental tables. Significant pair-wise comparisons are indicated directly within tables and figures, using appropriate symbols (as defined in legends).
RESULTS
Aging Influences Myocardial Circadian Clock Gene Expression in a Time-of-Day-Dependent Manner
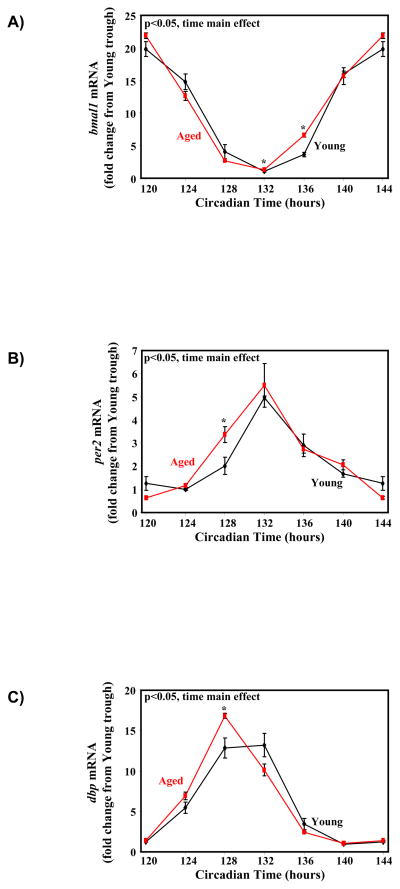
We have previously investigated the rate of resynchronization of the circadian clock within the rat heart following reversal of the light/dark cycle; full resynchronization takes between 5 and 8 days (Durgan, et al. 2006). Although previously published studies have shown that the circadian clock in extra-cardiac tissues is altered with aging (Kunieda, et al. 2006), whether aging influences the circadian clock within the heart is unknown. To address this issue, circadian oscillations in myocardial circadian clock components were investigated in young (6 months) versus aged (24 months) wild-type mice. Cosinor analysis did not reveal any significant differences in amplitude, acrophase, or mesor between young versus aged hearts, for the circadian clock (bmal1, per2) and clock output (dbp) genes investigated, although a trend for a slight (≤1 hour) phase advance is noteworthy in aged hearts (e.g., p=0.066 for a 0.68 hour phase advance in bmal1; Supplemental Table 1). Analysis of variance revealed significant time main effects for all genes investigated, independent of age (Supplemental Table 2). Subsequent post hoc pair-wise comparisons identified significant time-of-day-dependent age effects in expression of the genes investigated (Figure 1).
Figure 1.
Circadian rhythms in expression of clock genes (bmal1 (A) and per2 (B)), as well as a clock output gene (dbp; C) in young (6 month old) versus aged (24 month old) mouse hearts. Values are expressed as mean ± SEM (n=3). * denotes p<0.05 for young versus aged mice at the same zeitgeber time.
Adaptation of Wild-type and CCM Littermates to Aging and Simulated Shift Work
In order to investigate whether genetic disruption of the cardiomyocyte circadian clock predisposes towards development of myocardial contractile dysfunction in response to aging and/or manipulation of the light/dark cycle, 2 month old (young) and 18 month old (aged) wild-type and CCM mice were randomized into two major groups; control and SSW (simulated shift work). Control mice were housed in a normal 12-hr/12-hr light/dark cycle. SSW mice were subjected to a bi-weekly 12-hr phase shift in the light/dark cycle for a total of 16-wks (as described in the Materials and Methods section). Mice were therefore 6 months (young) and 22 months (aged) old at the end of the study. For 6 out of the 8 groups investigated, no mice died before the end of the study protocol. However, 1 mouse (out of 11) died in the aged wild-type SSW group, while 3 mice (out of 10) died in the aged CCM SSW group; in all four cases, the cause of death was euthanasia following development of overt infections.
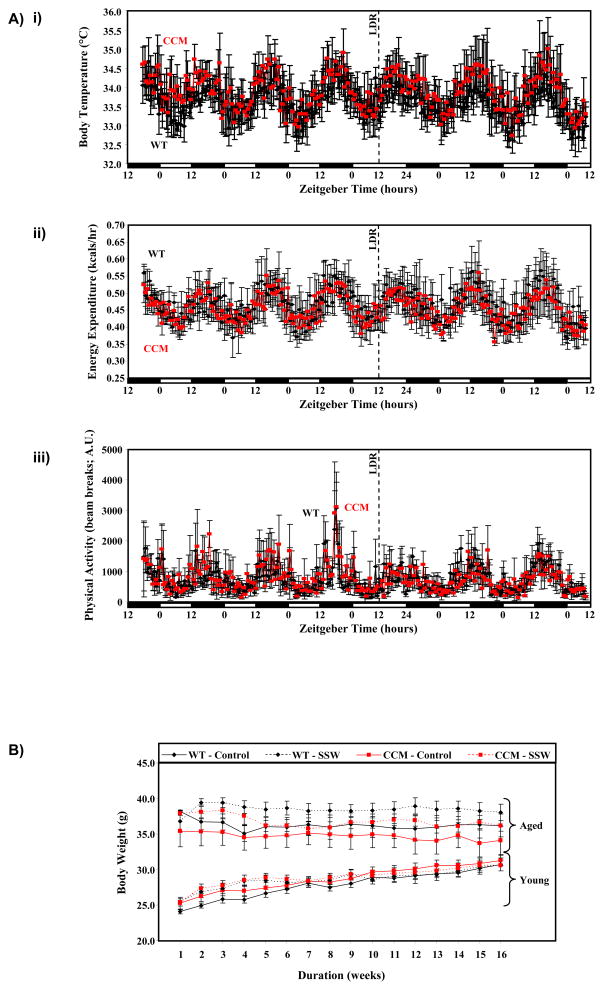
The rate of re-entrainment of body temperature, energy expenditure, and physical activity rhythms following a 12-hr phase shift in the light/dark cycle were identical in wild-type versus CCM littermates (Figure 2A), consistent with the cardiomyocyte-restricted nature of the CCM model. Body weight during the 16-wk study was significantly higher in aged versus young animals, independent of genotype or light/dark cycle manipulation (Figure 2B). Significant main effects for age were also observed for body weight, lung weight, kidney weight, epididymal fat pad weight and bi-ventricular weight (all were higher in the aged mice compared to young mice, independent of SSW and genotype; Supplemental Table 3). A main effect of SSW was observed only for epididymal fat pad weight (higher in the SSW compared to control group, independent of age and genotype; Supplemental Table 3). Main effects of genotype were observed for epididymal fat pad weight, bi-ventricular weight, and bi-ventricular weight-to-body weight ratio in CCM mice independent of age and SSW (Supplemental Table 3). Interestingly, post hoc analyses (Table 1) demonstrated that bi-ventricular weight-to-body weight ratio is significantly higher in CCM hearts relative to wild-type hearts for 3 out of 4 possible pair-wise comparisons.
Figure 2.
Re-synchronization of body temperature (i), energy expenditure (ii), and physical activity (iii) diurnal variations in wild-type versus CCM mice following a reversal of the light/dark cycle (LDR; panel A). Changes in body weight between young/aged, wild-type/CCM, and control/SSM mice during the 16 week protocol (panel B). Values are expressed as mean ± SEM (n=3–16).
Table 1.
Mean values for gravimetric measures in post hoc comparison groups.
| Parameter | Group [(Young/Aged)-(Control/SSW)-(Wild-type/CCM)] | |||||||
|---|---|---|---|---|---|---|---|---|
| Y-C-W | Y-S-W | A-C-W | A-S-W | Y-C-C | Y-S-C | A-C-C | A-S-C | |
| BW (g) | 32.4 ± 1.6 | 31.0 ± 2.2 | 38.1 ± 2.5 | 36.6 ± 1.6# | 27.8 ± 1.7 | 32.4 ± 1.6$ | 34.0 ± 2.6# | 35.8 ± 1.6 |
| LW (mg) | 156 ± 8 | 147 ± 11 | 197 ± 23# | 241 ± 57 | 170 ± 6 | 150 ± 10 | 197 ± 11# | 166 ± 19 |
| KW (mg) | 226 ± 15 | 241 ± 22 | 269 ± 11 | 278 ± 11 | 231 ± 8 | 220 ± 16 | 269 ± 23 | 243 ± 11 |
| EFW (g) | 0.94 ± 0.12 | 1.01 ± 0.11 | 1.35 ± 0.15 | 1.53 ± 0.18# | 0.49 ± 0.15* | 1.00 ± 0.15$ | 1.06 ± 0.19# | 1.41 ± 0.17 |
| BVW (mg) | 173 ± 11 | 145 ± 6 | 183 ± 16 | 193 ± 14# | 198 ± 9 | 198 ± 19* | 215 ± 19 | 212 ± 11 |
| BVW/BW (mg/g) | 5.36 ± 0.31 | 4.69 ± 0.17 | 4.78 ± 0.11 | 5.36 ± 0.55 | 7.27 ± 0.55* | 6.08 ± 0.40* | 6.37 ± 0.59* | 5.93 ± 0.32 |
Definitions include: Y, young (6 month old) mice; A, aged (22 month old) mice; C, control light/dark cycle; S, simulated shift work (i.e., repetitive reversal of the light/dark cycle for 4 months); W, wild-type mice; C, cardiomyocyte-specific circadian clock mutant (CCM) mice; SSW, simulated shift work, light/dark cycle manipulation; BW, body weight; LW, lung weight; KW, kidney weight; EFW, epididymal fat weight, BVW, bi-ventricular weight; NS, not significant. Values are expressed as mean ± SEM (n=3–7).
denotes p<0.05 for young versus aged hearts within the same light/dark cycle and genotype groups.
denotes p<0.05 for control versus SSW hearts within the same age and genotype groups.
denotes p<0.05 for wild-type versus CCM hearts within the same age and light/dark cycle groups.
We next investigated whether age, SSW, and/or genotype altered distinct humoral factors known to influence whole body metabolic homeostasis and/or heart function (Supplemental Table 3 [main effects] and Table 2 [individual group data and post hoc pair-wise comparisons]). Of the humoral factors investigated, main effects of age were observed only for plasma glucose (higher in the aged versus young animals, independent of SSW and genotype; Supplemental Table 3). SSW had main effects on plasma triglyceride and corticosterone levels (both higher in the SSW group compared to the control group, independent of age and genotype; Supplemental Table 3). In contrast, none of the humoral factors investigated were influenced by genotype (Supplemental Table 3 and Table 2).
Table 2.
Mean values for humoral factors in post hoc comparison groups.
| Parameter | Group [(Young/Aged)-(Control/SSW)-(Wild-type/CCM)] | |||||||
|---|---|---|---|---|---|---|---|---|
| Y-C-W | Y-S-W | A-C-W | A-S-W | Y-C-C | Y-S-C | A-C-C | A-S-C | |
| Glucose (mg/dl) | 401 ± 29 | 238 ± 38$ | 370 ± 36 | 366 ± 31# | 327 ± 32 | 243 ± 42 | 335 ± 47 | 313 ± 35 |
| Triglyceride (mg/dl) | 99 ± 18 | 100 ± 24 | 93 ± 23 | 237 ± 47$ | 90 ± 31 | 217 ± 71 | 155 ± 24 | 175 ± 22 |
| Insulin (ng/ml) | 0.65 ± 0.10 | 0.38 ± 0.16 | 0.75 ± 0.26 | 1.72 ± 0.73 | 0.52 ± 0.11 | 0.74 ± 0.25 | 0.54 ± 0.11 | 0.49 ± 0.13 |
| Leptin (ng/ml) | 7.9 ± 1.3 | 9.8 ± 3.1 | 18.5 ± 9.2 | 13.9 ± 3.4 | 5.3 ± 1.0 | 20.4 ± 6.7 | 8.8 ± 1.2 | 11.3 ± 3.4 |
| Adiponectin (μg/ml) | 5.0 ± 0.6 | 4.5 ± 0.5 | 5.5 ± 0.8 | 4.4 ± 0.3 | 4.6 ± 0.6 | 5.0 ± 0.6 | 4.7 ± 0.6 | 4.3 ± 0.7 |
| Corticosterone (ng/ml) | 282 ± 16 | 312 ± 22 | 249 ± 23 | 327 ± 22$ | 246 ± 24 | 344 ± 35 | 257 ± 18 | 280 ± 37 |
Definitions include: Y, young (6 month old) mice; A, aged (22 month old) mice; C, control light/dark cycle; S, simulated shift work (i.e., repetitive reversal of the light/dark cycle for 4 months); W, wild-type mice; C, cardiomyocyte-specific circadian clock mutant (CCM) mice; SSW, simulated shift work, light/dark cycle manipulation; NS, not significant. Values are expressed as mean ± SEM (n=3–7).
denotes p<0.05 for young versus aged hearts within the same light/dark cycle and genotype groups.
denotes p<0.05 for control versus SSW hearts within the same age and genotype groups.
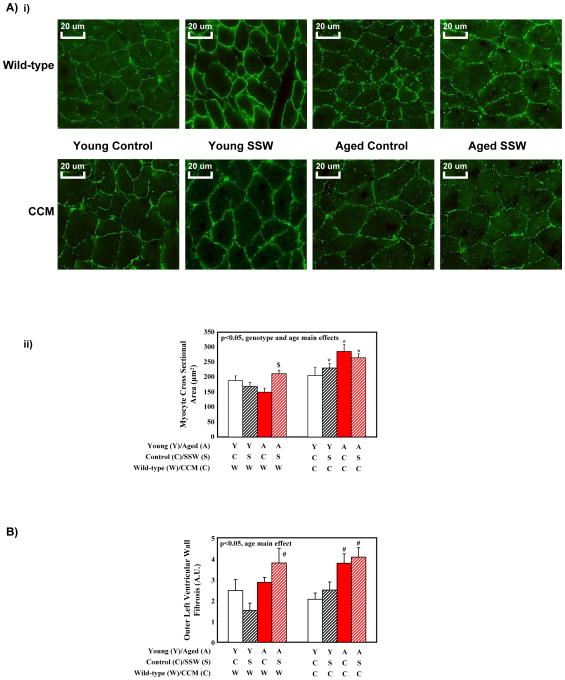
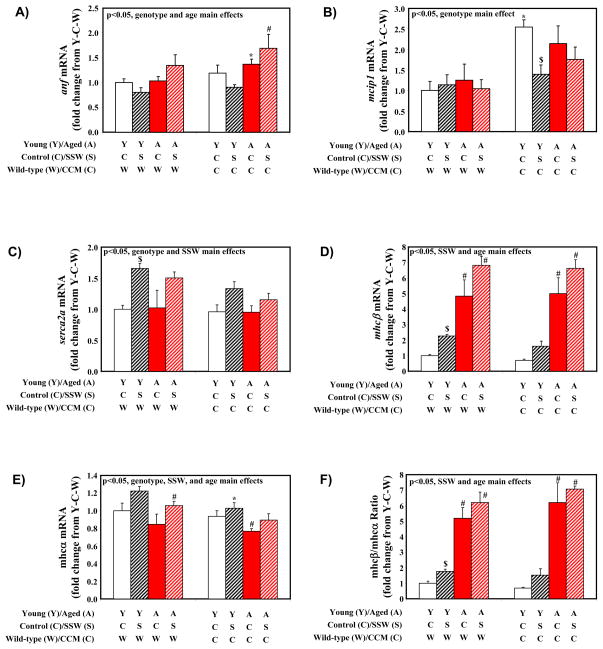
To characterize further wild-type versus CCM hearts following aging and/or SSW, histological and gene expression analyses were next performed. Consistent with bi-ventricular weight, three-way analysis of variance revealed main effects of age and genotype for myocyte cross sectional area (i.e., higher in aged and in CCM mice; Supplemental Table 3 and Figure 3A). However, only a main effect of age was observed for fibrosis; no significant main effects were detected in fibrosis for genotype or SSW (Supplemental Table 3 and Figure 3B). Gene expression analysis included measurements of established molecular markers of cardiac hypertrophy (wherein anf, mcip1, mhcβ are generally induced, while serca2a and mhcα are generally repressed, in hypertrophied rodent hearts (Depre, et al. 1998)). Significant main effects for age were observed for anf, mhcβ, and mhcα independent of SSW and genotype (Supplemental Table 3 and Figure 4). Main effects of SSW were also observed for serca2a, mhcβ, and mhcα (increased with SSW, independent of age and genotype; Supplemental Table 3). Consistent with increased myocyte cross sectional area in CCM hearts (Figure 3A), three-way analysis of variance revealed main effects of genotype on anf, mcip1, serca2a, and mhcα independent of age and SSW (Supplemental Table 3 and Figure 4). Importantly, Figure 4 shows similar transcriptional response patterns of wild-type and CCM hearts to aging and SSW.
Figure 3.
Cardiomyocyte size (A) and fibrosis (B) assessment. Images (i) are representative of between 3 and 6 independent observations per group. Semi-quantitative values are expressed as mean ± SEM (n=3–6). # denotes p<0.05 for young versus aged hearts within the same light/dark cycle and genotype groups. $ denotes p<0.05 for control versus SSW hearts within the same age and genotype groups. * denotes p<0.05 for wild-type versus CCM hearts within the same age and light/dark cycle groups.
Figure 4.
Gene expression markers of cardiac hypertrophy in young versus old wild-type and CCM mice subjected to simulated shift work (SSW). Values are expressed as mean ± SEM (n=3–7). # denotes p<0.05 for young versus aged hearts within the same light/dark cycle and genotype groups. $ denotes p<0.05 for control versus SSW hearts within the same age and genotype groups. * denotes p<0.05 for wild-type versus CCM hearts within the same age and light/dark cycle groups.
MRI analysis was performed next, to investigate whether distinct adaptation of CCM hearts at gravimetric, histological, and transcriptional levels resulted in differences in myocardial contractile function. Somewhat surprisingly, only septal wall thickness was significantly different between CCM and wild-type groups, independent of age and SSW (Supplemental Table 3 and Table 3). Neither aging nor SSW significantly influenced any measures of myocardial contractile function, as assessed by MRI (Supplemental Table 3 and Table 3). Blood pressure measurements revealed no significant differences in this parameter between the investigated groups (Supplemental Table 3 and Table 3).
Table 3.
Mean values for in vivo cardiovascular function measures in post hoc comparison groups.
| Parameter | Group [(Young/Aged)-(Control/SSW)-(Wild-type/CCM)] | |||||||
|---|---|---|---|---|---|---|---|---|
| Y-C-W | Y-S-W | A-C-W | A-S-W | Y-C-C | Y-S-C | A-C-C | A-S-C | |
| LVEF | 0.62 ± 0.03 | 0.60 ± 0.05 | 0.68 ± 0.04 | 0.69 ± 0.05 | 0.65 ± 0.07 | 0.69 ± 0.05 | 0.56 ± 0.10 | 0.66 ± 0.03 |
| RVEF | 0.53 ± 0.02 | 0.51 ± 0.04 | 0.55 ± 0.05 | 0.55 ± 0.05 | 0.49 ± 0.07 | 0.61 ± 0.04 | 0.51 ± 0.08 | 0.65 ± 0.03 |
| LVFS | 0.39 ± 0.04 | 0.33 ± 0.04 | 0.40 ± 0.03 | 0.43 ± 0.05 | 0.45 ± 0.05 | 0.48 ± 0.08 | 0.40 ± 0.05 | 0.41 ± 0.03 |
| RVFS | 0.40 ± 0.07 | 0.42 ± 0.07 | 0.48 ± 0.12 | 0.35 ± 0.10 | 0.32 ± 0.14 | 0.26 ± 0.06 | 0.32 ± 0.07 | 0.44 ± 0.06 |
| LVEDD (mm) | 4.54 ± 0.05 | 4.31 ± 0.33 | 4.65 ± 0.18 | 4.62 ± 0.15 | 4.73 ± 0.32 | 4.66 ± 0.33 | 5.02 ± 0.20 | 4.74 ± 0.09 |
| RVEDD (mm) | 1.83 ± 0.11 | 2.04 ± 0.18 | 1.74 ± 0.18 | 1.82 ± 0.14 | 2.02 ± 0.11 | 1.94 ± 0.26 | 1.96 ± 0.16 | 1.82 ± 0.18 |
| SWT (mm) | 1.25 ± 0.07 | 1.13 ± 0.09 | 1.30 ± 0.10 | 1.20 ± 0.09 | 1.73 ± 0.16* | 1.54 ± 0.26 | 1.56 ± 0.16 | 1.71 ± 0.19* |
| OWT (mm) | 1.02 ± 0.17 | 1.16 ± 0.11 | 1.12 ± 0.23 | 1.38 ± 0.22 | 1.07 ± 0.12 | 1.11 ± 0.08 | 1.32 ± 0.06 | 1.15 ± 0.25 |
| BP (mmHg) | 129 ± 4 | 132 ± 4 | 133 ± 5 | 121 ± 6 | 129 ± 3 | 131 ± 6 | 139 ± 5 | 135 ± 10 |
Definitions include: Y, young (6 month old) mice; A, aged (22 month old) mice; C, control light/dark cycle; S, simulated shift work (i.e., repetitive reversal of the light/dark cycle for 4 months); W, wild-type mice; C, cardiomyocyte-specific circadian clock mutant (CCM) mice; SSW, simulated shift work, light/dark cycle manipulation; LVEF, left ventricular ejection fraction; RVEF, right ventricular ejection fraction; LVFS, left ventricular fractional shortening; RVFS, right ventricular fractional shortening; LVEDD, left ventricular end diastolic diameter; RVEDD, right ventricular end diastolic diameter; SWT, septal wall thickness; OWT, outer wall thickness; BP, peak systolic blood pressure; NS, not significant. Values are expressed as mean ± SEM (n=3–6).
denotes p<0.05 for wild-type versus CCM hearts within the same age and light/dark cycle groups.
Selective Genetic Ablation of BMAL1 in Cardiomyocytes Results in a Pro-Hypertrophic Phenotype
Data presented thus far are consistent with the hypothesis that temporal suspension of the cardiomyocyte circadian clock at the beginning of the light phase (i.e., ZT0) results in a hypertrophic phenotype. These data are based on expression of the CLOCKΔ19 mutant protein in the cardiomyocytes of mice on the FVB/N background (i.e., CCM mice; see above). To gain further insight, we investigated a second novel mouse model of temporal suspension of the cardiomyocyte circadian clock through selective genetic ablation of BMAL1 from cardiomyocytes (i.e., CBK mice) on the C57/Bl6J background. As anticipated, hearts from CBK mice show decreased expression of bmal1 mRNA (Figure 5Ai) and BMAL1 protein (Figure 5Aii), versus wild-type hearts; residual expression is likely from non-cardiomyocyte cells present in intact hearts. Similar to CCM hearts (Figure 5Bi), CBK hearts (Figure 5Bii) exhibit attenuated dbp mRNA diurnal variations (Supplemental Table 4). Importantly, CBK hearts exhibit a significant increase in bi-ventricular weight-to-body weight ratio relative to wild-type littermates (Figure 5Ci), as well as increased expression of the hypertrophic marker mcip1 (Figure 5Cii).
Figure 5.
Phenotypic characterization of CBK, versus wild-type littermate, hearts. Gene (A) and protein (B) expression of BMAL1. Diurnal variations in dbp mRNA in CCM (C) and CBK (D) hearts. Biventricular weight-to-body weight ratio (E) and mcip1 mRNA (F). Values are expressed as mean ± SEM (n=3–19). # denotes p<0.05 for ZT0 versus ZT12 hearts within the same genotype group. * denotes p<0.05 for wild-type versus CCM/CBK hearts within the same ZT group.
Diurnal Variation in Responsiveness of the Heart to the Hypertrophic Stimulus Isoproterenol
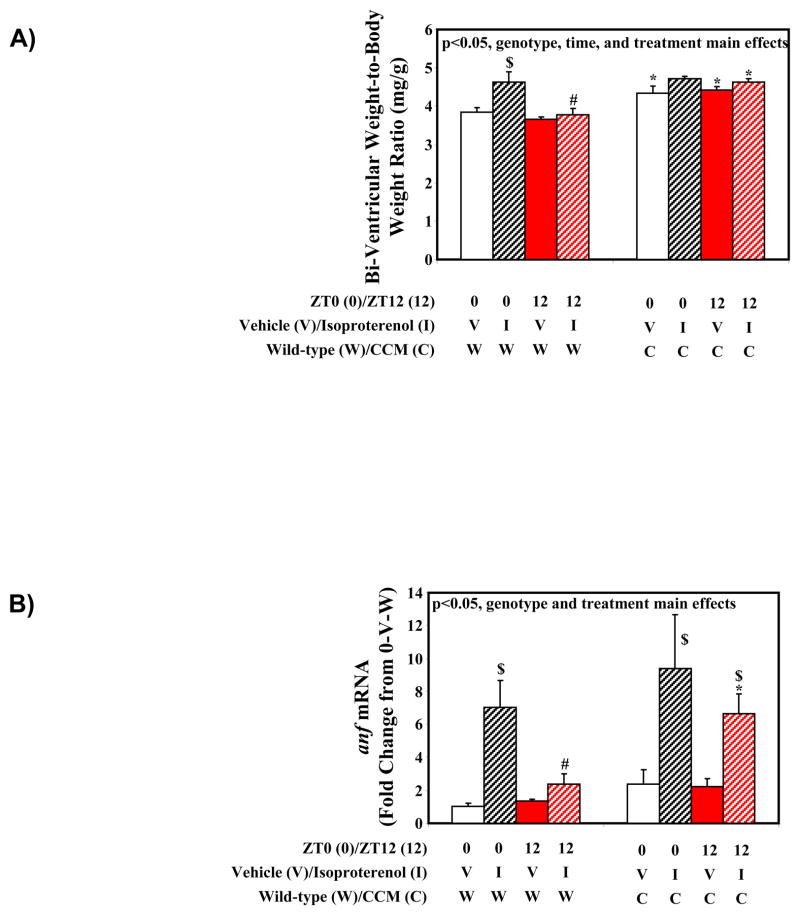
Results presented thus far suggest that temporal suspension of the cardiomyocyte circadian clock at ZT0 results in a pro-hypertrophic phenotype. These observations led to the hypothesis that the cardiomyocyte circadian clock increases responsiveness of the heart to pro-hypertrophic stimuli at the awake-to-sleep phase transition (i.e., ZT0). To address this hypothesis, wild-type and CCM mice were treated with the pro-hypertrophic agonist isoproterenol at one of two distinct times of the day; the awake-to-sleep phase transition (ZT0) or the sleep-to-awake phase transition (ZT12). Following seven consecutive days of isoproterenol treatment, three-way analysis of variance revealed main effects of time, treatment and genotype for bi-ventricular weight-to-body weight ratio (i.e., highest at ZT0 following treatment with isoproterenol, in CCM mice; Supplemental Table 5 and Figure 6A). The effect of isoproterenol treatment on bi-ventricular weight-to-body weight ratio was approximately 5-fold greater for wild-type mice treated with isoproterenol at ZT0 (21% increase; p<0.05) versus ZT12 (4% increase; NS) (Figure 6A). In contrast, this time-of-day-dependence in the isoproterenol response was not observed in CCM hearts (9% increase at ZT0 [NS] versus 5% increase at ZT12 [NS]) (Figure 6A). Similarly, gene expression analysis revealed main effects of treatment and genotype for anf mRNA (increased with isoproterenol and in CCM hearts; Supplemental Table 5 and Figure 6B). An approximate 6-fold greater induction of anf mRNA was observed in wild-type hearts treated with isoproterenol at ZT0 (604% increase; p<0.05) versus ZT12 (101% increase; NS), a diurnal variation that was again absent in CCM hearts (Figure 6B).
Figure 6.
Gravimetric (A) and gene expression (B) markers of cardiac hypertrophy in wild-type and CCM mice administered with isoproterenol in a time-of-day-dependent manner. Values are expressed as mean ± SEM (n=3–9). # denotes p<0.05 for ZT0 versus ZT12 hearts within the same treatment and genotype groups. $ denotes p<0.05 for vehicle versus isoproterenol treated hearts within the same ZT and genotype groups. * denotes p<0.05 for wild-type versus CCM hearts within the same ZT and treatment groups.
Diurnal Variation in Cryptochrome 2 in Wild-type, but not CCM and CBK, Hearts
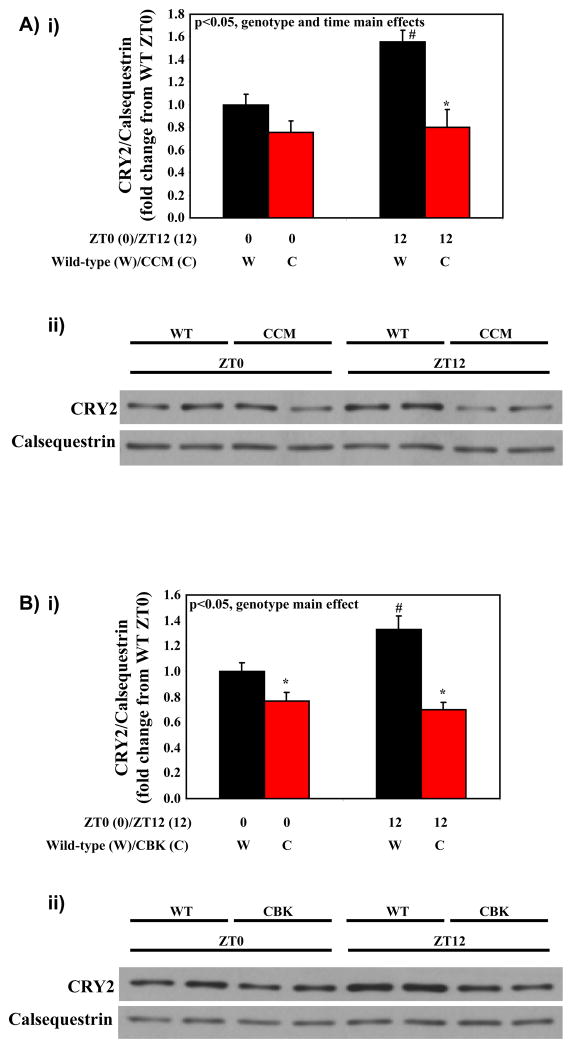
We next decided to investigate the possible mechanism by which the circadian clock influences time-of-day-dependent oscillations in the hypertrophic response of the heart to the β-adrenergic agonist, isoproterenol. Possible mediators include the cryptochromes (CRY), integral clock proteins, of which CRY2 is the dominant isoform in the heart (Young 2006). CRY proteins have recently been shown to directly bind to GSα in the liver, thereby reducing adrenergic responsiveness (Zhang, et al. 2010). We report that CRY2 protein oscillates in a diurnal manner in wild-type hearts, with higher expression at ZT12 (Figures 7A and 7B). These time-of-day-dependent oscillations are absent in CCM and CBK hearts, wherein CRY2 protein expression is essentially suspended at ZT0 (Figures 7A and 7B).
Figure 7.
Diurnal variations in CRY2 protein in CCM (A) and CBK (B) hearts. Values are expressed as mean ± SEM (n=5–6). # denotes p<0.05 for ZT0 versus ZT12 hearts within the same genotype group. * denotes p<0.05 for wild-type versus CCM/CBK hearts within the same ZT group.
DISCUSSION
The purpose of the present study was to investigate whether chronic circadian dyssynchrony of the heart with its environment (through genetic temporal suspension of the cardiomyocyte circadian clock) would augment maladaptation to a circadian challenge (i.e., aging and/or simulated shift work). Counter to this hypothesis, CCM mice exhibit essentially identical responsiveness to aging and SSW relative to wild-type littermates, at whole body homeostatic, gravimetric, humoral, histological, transcriptional, and cardiac contractile function levels. However, CCM hearts appear to have a pro-hypertrophic phenotype, as observed through gravimetric, histological, transcriptional, and imaging analyses. Similarly, a second genetic model of selective disruption of the cardiomyocyte circadian clock (namely CBK mice) exhibits a pro-hypertrophic phenotype. To investigate further the potential role of the cardiomyocyte circadian clock in modulating responsiveness of the heart to hypertrophic stimuli, wild-type and CCM mice were treated with isoproterenol in a time-of-day-dependent manner. We report that isoproterenol induced cardiac hypertrophy and anf expression to the greatest extent when administered to wild-type mice at the awake-to-sleep transition (relative to administration at the sleep-to-wake transition). This diurnal variation in isoproterenol responsiveness is absent in CCM mice. Collectively, these data suggest that the cardiomyocyte circadian clock likely influences responsiveness of the heart to hypertrophic stimuli.
The primary function of cell autonomous circadian clocks is to confer anticipation of extracellular/environmental stimuli, thereby promoting synchrony of the cell/organ/organism with its environment (Edery 2000). As such, impairment or loss of this mechanism would likely promote maladaptive responses. With regards to the cardiovascular system, the recent exposure of circadian clocks as critical regulators of numerous aspects of cardiovascular function over the course of the day has led to speculation that impairment of these molecular timekeepers may precipitate cardiovascular disease (Young, et al. 2007). Evidence in support of this hypothesis is several fold: 1) circadian clocks are altered/impaired in many animal models of increased cardiovascular disease risk (e.g., DIO, diabetes, hypertension, shift-work, etc.) (Durgan, et al. 2006; Kohsaka, et al. 2007; Kung, et al. 2007; Kunieda, et al. 2006; Mohri, et al. 2003; Young, et al. 2001; Young, et al. 2002); 2) common environmental factors that are implicated as significant contributors towards cardiovascular disease in Western society (i.e., quality and quantity of calories ingested, physical activity, and sleep) are also known to be strong entrainment factors regulating circadian clocks (Burioka, et al. 2008; Damiola, et al. 2000; Kohsaka, et al. 2007; Zambon, et al. 2003); 3) genetic polymorphisms in human circadian clock genes have been shown to be associated with cardiovascular disease development, such as hypertension (Woon, et al. 2007); and 4) multiple animal models of environment- and genetic- induced perturbations in molecular clocks generally exhibit increased susceptibility towards cardiovascular disease development (Sole, et al. 2009). Several examples exist for the latter. This includes increased mortality in cardiomyopathic hamsters following light/dark cycle manipulation (Penev, et al. 1998). Similarly, Martino et al have reported that light/dark cycle manipulation augments myocardial dysfunction in mice subjected to pressure overload, an effect that could be reversed following re-entrainment to a normal light/dark cycle (Martino, et al. 2007). More recently, the same group has shown that housing heterozygous tau hamsters (that possess an altered circadian clock, with a periodicity of 22 hours) in a normal 24-hr light/dark cycle results in profound cardiorenal disease, which is absent when these rodents remain synchronized with their environment (by housing in a 22-hr light/dark cycle) (Martino, et al. 2008). Targeted genetic ablation of circadian clock components has also been shown to influence susceptibility of mice to adverse cardiovascular stresses, such as vascular injury and high salt intake (Anea, et al. 2009; Doi, et al. 2010).
Although a number of studies have been performed investigating whether disruption/impairment of circadian clocks in a ubiquitous fashion influences the etiology of cardiovascular disease, few studies have similarly reported a role for distinct cell-type specific clocks. Through the use of CCM mice, we recently revealed that the cardiomyocyte circadian clock directly influences myocardial ischemia/reperfusion tolerance, in a time-of-day-dependent manner (Durgan, et al. 2010a). However, the role of this clock in the etiology of cardiovascular disease in response to other clinically-relevant stressors remains unknown. To address whether the cardiomyocyte circadian clock plays a significant role in the development of cardiovascular disease in response to aging and/or light/dark cycle manipulations (i.e., simulated shift work), we utilized a genetic mouse model wherein the heart is dyssynchronized with the rest of the organism, through temporal suspension of the cardiomyocyte circadian clock (i.e., CCM mice) (Young 2009). Contrary to expectation, wild-type and CCM littermates respond to aging and SSW in essentially identical manners (at whole body homeostatic, gravimetric, humoral, histological, transcriptional, and cardiac contractile function levels; Tables 1–3 and Figures 2–4). These observations suggest that extra-cardiac factors likely play a dominant role in the adaptation/maladaptation of the myocardium to these distinct stresses (i.e., aging and light/dark cycle manipulation). Such factors will likely include paracrine and endocrine factors, many of which are themselves under direct control of non-cardiomyocyte circadian clocks. For this reason, ubiquitous disruption of circadian clocks likely augments cardiovascular disease progression to a greater extent then selective temporal suspension of the cardiomyocyte circadian clock.
One potential limitation of this aspect of the present study is the fact that neither aging nor simulated shift work precipitated cardiac dysfunction (in wild-type or CCM mice). It is conceivable that further aging and/or extending the light/dark cycle manipulation regime for greater than 16 weeks, might induce contractile dysfunction. However, approximately 20% of the aged mice died within 16 weeks of the SSW regime, thereby limiting justification for prolongation. Importantly, previous studies have shown that manipulation of the light/dark cycle for this duration is a significant stress, particularly for aged mice (Davidson, et al. 2006).
During extensive characterization of wild-type and CCM littermates in the present study, we revealed a pro-hypertrophic phenotype in CCM mice, independent of age and SSW. More specifically, CCM mice exhibit a greater bi-ventricular weight-to-body weight ratio (Table 1), increased cardiomyocyte cross sectional area (Figure 3), increased septal wall thickness (Table 3), and a transcriptional profile indicative of cardiac hypertrophy (i.e., induction of anf and mcip1, as well as a repression of mhcα and serca2a; Figure 4). Previous studies have shown that the genetic manipulation performed to disrupt the circadian clock can dramatically influence the resulting phenotype. For example, ubiquitous expression of the CLOCKΔ19 mutant protein has been reported to result in a pro-obesity phenotype, while knockout of BMAL1 in a ubiquitous manner results in a lean phenotype (Bunger, et al. 2005; Turek, et al. 2005). Genetic background of the mouse model also significantly influences the phenotype observed; CLOCKΔ19 mutant mice on the C57/Bl6J background are obesity-prone, while the same mice on either the ICR or CBA background are obesity-resistant (Kennaway, et al. 2007; Oishi, et al. 2006; Turek, et al. 2005). To ensure the pro-hypertrophic phenotype observed in CCM mice was neither model nor background specific, we investigated a novel, second mouse model of temporal suspension of the cardiomyocyte circadian clock on a distinct genetic background (i.e., CBK mice on C57/Bl6J background). Similar to CCM hearts (Young 2009), clock output from CBK hearts is essentially suspended at ZT0 (Figures 5Bi and 5Bii). Importantly, genetic disruption of the cardiomyocyte circadian clock in CBK mice was associated with a pro-hypertrophic phenotype (Figures 5Ci and 5Cii).
Given that CCM and CBK hearts both appear to be temporally suspended at the awake-to-sleep phase transition, we hypothesized that wild-type hearts may be more responsive to pro-hypertrophic stimuli at this time of the day. To test this hypothesis, mice were treated with the hypertrophic agonist isoproterenol at either the awake-to-sleep phase transition or the sleep-to-awake phase transition. Consistent with the hypothesis, wild-type mice exhibited greatest hypertrophic growth and anf induction when treated with isoproterenol at the awake-to-sleep phase transition (Figure 6). In contrast, this diurnal variation in responsiveness to isoproterenol was absent in CCM mice (Figure 6). Collectively, these data support the hypothesis that the cardiomyocyte circadian clock modulates responsiveness of the heart to pro-hypertrophic stimuli in a time-of-day-dependent manner.
An important question relates to the identity of potential mechanisms by which the cardiomyocyte circadian clock modulates responsiveness of the myocardium to hypertrophic stimuli. Similar to our data regarding hypertrophic responsiveness, Collins and Rodrigo reported that the inotropic response of adult cardiomyocytes to isoproterenol exhibits a time-of-day-dependent variation (Collins, et al.). Isoproterenol is a β-adrenergic agonist (Lefkowitz, et al. 2000). Zhang et al recently revealed a mechanistic link between β-adrenergic signaling and the circadian clock in hepatocytes (Zhang, et al. 2010). Through a series of elegant studies, the investigators reported that cryptochromes (an integral clock component) can bind directly to Gsα, resulting in subsequent decreases in G-protein coupled receptor signaling (Zhang, et al. 2010). Consistent with such a model, we report that CRY2 protein levels exhibit a diurnal variation in wild type, but not CCM or CBK hearts, with approximate 1.5-fold greater levels at ZT12 (the time of day at which isoproterenol-induced hypertrophy was lowest; Figure 7A–B). These data are consistent with the hypothesis that the cardiomyocyte circadian clock influences myocardial β-adrenergic responsiveness, through time-of-day-dependent attenuation of Gsα by cryptochromes. However, it is important to note that additional potential molecular links may exist. For example, we have recently shown that the phosphorylation status of the growth signaling kinases Akt and GSK3β oscillates in wild-type hearts, in a cardiomyocyte circadian clock dependent manner (Durgan, et al. 2010a); both Akt and GSK3β are known modulators of cardiac hypertrophic growth (Heineke, et al. 2006). Whether CRY2, Akt and/or GSK3β serve as molecular links between the cardiomyocyte circadian clock and hypertrophic growth requires full elucidation.
The findings of the present study have significant clinical implications. Non-dipping hypertensive patients (whose blood pressure drops <10% during sleep, and therefore exhibit inappropriate stress on the heart during the sleep phase) exhibit increased left ventricular hypertrophy and are at increased risk of cardiovascular and renal disease (compared to dipping hypertensive subjects) (Bianchi, et al. 1994; Hermida, et al. 2005; Palatini, et al. 1992). Similarly, obstructive sleep apnea (OSA) is associated with increased sympathetic stimulation on the heart during the sleep phase, adverse cardiac remodeling and increased risk of heart failure (Bradley, et al. 2009; Kario 2009). Recently, Sole and Martino hypothesized that the sleep phase corresponds to a time of increased myocardial renewal and growth, and as such pressure overload during this phase may accelerate the development of pathological hypertrophy (Sole, et al. 2009). Our findings are consistent with this hypothesis, and also suggest an important mediatory role for the cardiomyocyte circadian clock.
In conclusion, we report that genetic, temporal suspension of the cardiomyocyte circadian clock has little impact on adaptation of the mouse to aging and/or simulated shift work. However, these studies highlight a novel potential role for the cardiomyocyte circadian clock in modulating responsiveness of the heart to pro-hypertrophic stimuli in a time-of-day-dependent manner.
Supplementary Material
Acknowledgments
This work was supported by the National Heart, Lung, and Blood Institute (HL-074259 [MEY]) and support from the Pennington Biomedical Research Foundation (JMG). Ju-Yun Tsai was supported by the DeBakey Heart Fund at Baylor College of Medicine. David J. Durgan was supported by the NSF GK-12 Fellowship. We wish to acknowledge the DRTC-funded (P30DK56336; P30NS057098; P60DK079626) Small Animal Physiology Core at UAB (Dr. Timothy Nagy, Director) for help with the MRI analysis. We also wish to thank Dr. Michael Schneider for providing MHCα-CRE mice.
References
- Anea CB, Zhang M, Stepp DW, Simkins GB, Reed G, Fulton DJ, Rudic RD. Vascular disease in mice with a dysfunctional circadian clock. Circulation. 2009;119:1510–1517. doi: 10.1161/CIRCULATIONAHA.108.827477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balsalobre A, Damiola F, Schibler U. A serum shock induces circadian gene expression in mammalian tissue culture cells. Cell. 1998;93:929–937. doi: 10.1016/s0092-8674(00)81199-x. [DOI] [PubMed] [Google Scholar]
- Bianchi S, Bigazzi R, Baldari G, Sgherri G, Campese VM. Diurnal variations of blood pressure and microalbuminuria in essential hypertension. Am J Hypertens. 1994;7:23–29. doi: 10.1093/ajh/7.1.23. [DOI] [PubMed] [Google Scholar]
- Bradley TD, Floras JS. Obstructive sleep apnoea and its cardiovascular consequences. Lancet. 2009;373:82–93. doi: 10.1016/S0140-6736(08)61622-0. [DOI] [PubMed] [Google Scholar]
- Bray M, Shaw C, Moore M, Garcia R, Zanquetta M, Durgan D, Jeong W, Tsai J, Bugger H, Zhang D, Rohrwasser A, Rennison J, Dyck J, Litwin S, Hardin P, Chow C, Chandler M, Abel E, Young M. Disruption of the circadian clock within the cardiomyocyte influences myocardial contractile function; metabolism; and gene expression. Am J Physiol Heart Circ Physiol. 2008;294:H1036–H1047. doi: 10.1152/ajpheart.01291.2007. [DOI] [PubMed] [Google Scholar]
- Bunger MK, Walisser JA, Sullivan R, Manley PA, Moran SM, Kalscheur VL, Colman RJ, Bradfield CA. Progressive arthropathy in mice with a targeted disruption of the Mop3/Bmal-1 locus. Genesis. 2005;41:122–132. doi: 10.1002/gene.20102. [DOI] [PubMed] [Google Scholar]
- Burioka N, Koyanagi S, Endo M, Takata M, Fukuoka Y, Miyata M, Takeda K, Chikumi H, Ohdo S, Shimizu E. Clock gene dysfunction in patients with obstructive sleep apnoea syndrome. Eur Respir J. 2008;32:105–112. doi: 10.1183/09031936.00138207. [DOI] [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Collins HE, Rodrigo GC. Inotropic response of cardiac ventricular myocytes to beta-adrenergic stimulation with isoproterenol exhibits diurnal variation: involvement of nitric oxide. Circ Res. 106:1244–1252. doi: 10.1161/CIRCRESAHA.109.213942. [DOI] [PubMed] [Google Scholar]
- Curtis AM, Cheng Y, Kapoor S, Reilly D, Price TS, Fitzgerald GA. Circadian variation of blood pressure and the vascular response to asynchronous stress. Proc Natl Acad Sci U S A. 2007;104:3450–3455. doi: 10.1073/pnas.0611680104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damiola F, Le MN, Preitner N, Kornmann B, Fleury-Olela F, Schibler U. Restricted feeding uncouples circadian oscillators in peripheral tissues from the central pacemaker in the suprachiasmatic nucleus. Genes Dev. 2000;14:2950–2961. doi: 10.1101/gad.183500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson AJ, Sellix MT, Daniel J, Yamazaki S, Menaker M, Block GD. Chronic jet-lag increases mortality in aged mice. Curr Biol. 2006;16:R914–916. doi: 10.1016/j.cub.2006.09.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depre C, Shipley GL, Chen W, Han Q, Doenst T, Moore ML, Stepkowski S, Davies PJ, Taegtmeyer H. Unloaded heart in vivo replicates fetal gene expression of cardiac hypertrophy. Nat Med. 1998;4:1269–1275. doi: 10.1038/3253. [DOI] [PubMed] [Google Scholar]
- Doi M, Takahashi Y, Komatsu R, Yamazaki F, Yamada H, Haraguchi S, Emoto N, Okuno Y, Tsujimoto G, Kanematsu A, Ogawa O, Todo T, Tsutsui K, van der Horst GT, Okamura H. Salt-sensitive hypertension in circadian clock-deficient Cry-null mice involves dysregulated adrenal Hsd3b6. Nat Med. 2010;16:67–74. doi: 10.1038/nm.2061. [DOI] [PubMed] [Google Scholar]
- Durgan D, Hotze M, Tomlin T, Egbejimi O, Graveleau C, Abel E, Shaw C, Bray M, Hardin P, Young M. The intrinsic circadian clock within the cardiomyocyte. Am J Physiol Heart Circ Physiol. 2005;289:H1530–H1541. doi: 10.1152/ajpheart.00406.2005. [DOI] [PubMed] [Google Scholar]
- Durgan D, Trexler N, Egbejimi O, McElfresh T, Suk H, Petterson L, Shaw C, Hardin P, Bray M, Chandler M, Chow C, Young M. The circadian clock within the cardiomyocyte is essential for responsiveness of the heart to fatty acids. J Biol Chem. 2006;281:24254–24269. doi: 10.1074/jbc.M601704200. [DOI] [PubMed] [Google Scholar]
- Durgan DJ, Pulinilkunnil T, Villegas-Montoya C, Garvey ME, Frangogiannis NG, Michael LH, Chow CW, Dyck JR, Young ME. Short communication: ischemia/reperfusion tolerance is time-of-day-dependent: mediation by the cardiomyocyte circadian clock. Circ Res. 2010a;106:546–550. doi: 10.1161/CIRCRESAHA.109.209346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durgan DJ, Young ME. The cardiomyocyte circadian clock: emerging roles in health and disease. Circ Res. 2010b;106:647–658. doi: 10.1161/CIRCRESAHA.109.209957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edery I. Circadian rhythms in a nutshell. Physiol Genomics. 2000;3:59–74. doi: 10.1152/physiolgenomics.2000.3.2.59. [DOI] [PubMed] [Google Scholar]
- Gekakis N, Staknis D, Nguyen H, Davis F, Wilsbacher L, King D, Takahashi J, Weitz C. Role of the CLOCK protein in the mammalian circadian mechanism. Science. 1998;280:1564–1569. doi: 10.1126/science.280.5369.1564. [DOI] [PubMed] [Google Scholar]
- Gibson UE, Heid CA, Williams PM. A novel method for real time quantitative RT-PCR. Genome Res. 1996;6:995–1001. doi: 10.1101/gr.6.10.995. [DOI] [PubMed] [Google Scholar]
- Heid CA, Stevens J, Livak KJ, Williams PM. Real time quantitative PCR. Genome Res. 1996;6:986–994. doi: 10.1101/gr.6.10.986. [DOI] [PubMed] [Google Scholar]
- Heineke J, Molkentin JD. Regulation of cardiac hypertrophy by intracellular signalling pathways. Nat Rev Mol Cell Biol. 2006;7:589–600. doi: 10.1038/nrm1983. [DOI] [PubMed] [Google Scholar]
- Hermida RC, Calvo C, Ayala DE, Lopez JE. Decrease in urinary albumin excretion associated with the normalization of nocturnal blood pressure in hypertensive subjects. Hypertension. 2005;46:960–968. doi: 10.1161/01.HYP.0000174616.36290.fa. [DOI] [PubMed] [Google Scholar]
- Hogenesch J, Gu Y, Jain S, Bradfield C. The basic-helix-loop-helix-PAS orphan MOP3 forms transcriptionally active complexes with circadian and hypoxia factors. Proc Natl Aad Sci USA. 1998;95:5474–5479. doi: 10.1073/pnas.95.10.5474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kario K. Obstructive sleep apnea syndrome and hypertension: mechanism of the linkage and 24-h blood pressure control. Hypertens Res. 2009;32:537–541. doi: 10.1038/hr.2009.73. [DOI] [PubMed] [Google Scholar]
- Kennaway DJ, Owens JA, Voultsios A, Boden MJ, Varcoe TJ. Metabolic homeostasis in mice with disrupted Clock gene expression in peripheral tissues. Am J Physiol Regul Integr Comp Physiol. 2007;293:R1528–1537. doi: 10.1152/ajpregu.00018.2007. [DOI] [PubMed] [Google Scholar]
- Knutsson A, Akerstedt T, Jonsson B, Orth-Gomer K. Increased risk of ischaemic heart disease in shift workers. Lancet. 1986;12:89–92. doi: 10.1016/s0140-6736(86)91619-3. [DOI] [PubMed] [Google Scholar]
- Kohsaka A, Laposky AD, Ramsey KM, Estrada C, Joshu C, Kobayashi Y, Turek FW, Bass J. High-fat diet disrupts behavioral and molecular circadian rhythms in mice. Cell Metab. 2007;6:414–421. doi: 10.1016/j.cmet.2007.09.006. [DOI] [PubMed] [Google Scholar]
- Kung T, Egbejimi O, Cui J, Ha N, Durgan D, Essop M, Bray M, Shaw C, Hardin P, Stanley W, Young M. Rapid attenuation of circadian clock gene oscillations in the rat heart following ischemia-reperfusion. J Mol Cell Cardiol. 2007;43:744–753. doi: 10.1016/j.yjmcc.2007.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunieda T, Minamino T, Katsuno T, Tateno K, Nishi J, Miyauchi H, Orimo M, Okada S, Komuro I. Cellular senescence impairs circadian expression of clock genes in vitro and in vivo. Circ Res. 2006;98:532–539. doi: 10.1161/01.RES.0000204504.25798.a8. [DOI] [PubMed] [Google Scholar]
- Lefkowitz RJ, Rockman HA, Koch WJ. Catecholamines, cardiac beta-adrenergic receptors, and heart failure. Circulation. 2000;101:1634–1637. doi: 10.1161/01.cir.101.14.1634. [DOI] [PubMed] [Google Scholar]
- Marcheva B, Ramsey KM, Affinati A, Bass J. Clock genes and metabolic disease. J Appl Physiol. 2009;107:1638–1646. doi: 10.1152/japplphysiol.00698.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martino TA, Tata N, Belsham DD, Chalmers J, Straume M, Lee P, Pribiag H, Khaper N, Liu PP, Dawood F, Backx PH, Ralph MR, Sole MJ. Disturbed diurnal rhythm alters gene expression and exacerbates cardiovascular disease with rescue by resynchronization. Hypertension. 2007;49:1104–1113. doi: 10.1161/HYPERTENSIONAHA.106.083568. [DOI] [PubMed] [Google Scholar]
- Martino TA, Oudit GY, Herzenberg AM, Tata N, Koletar MM, Kabir GM, Belsham DD, Backx PH, Ralph MR, Sole MJ. Circadian rhythm disorganization produces profound cardiovascular and renal disease in hamsters. Am J Physiol Regul Integr Comp Physiol. 2008;294:R1675–1683. doi: 10.1152/ajpregu.00829.2007. [DOI] [PubMed] [Google Scholar]
- McNamara P, Seo SB, Rudic RD, Sehgal A, Chakravarti D, FitzGerald GA. Regulation of CLOCK and MOP4 by nuclear hormone receptors in the vasculature: a humoral mechanism to reset a peripheral clock. Cell. 2001;105:877–889. doi: 10.1016/s0092-8674(01)00401-9. [DOI] [PubMed] [Google Scholar]
- Mohri T, Emoto N, Nonaka H, Fukuya H, Yagita K, Okamura H, Yokoyama M. Alterations of circadian expressions of clock genes in dahl salt-sensitive rats fed a high-salt diet. Hypertension. 2003;42:189–194. doi: 10.1161/01.HYP.0000082766.63952.49. [DOI] [PubMed] [Google Scholar]
- Nelson W, Tong YL, Lee JK, Halberg F. Methods for cosinor-rhythmometry. Chronobiologia. 1979;6:305–323. [PubMed] [Google Scholar]
- Nonaka H, Emoto N, Ikeda K, Fukuya H, Rohman MS, Raharjo SB, Yagita K, Okamura H, Yokoyama M. Angiotensin II induces circadian gene expression of clock genes in cultured vascular smooth muscle cells. Circulation. 2001;104:1746–1748. doi: 10.1161/hc4001.098048. [DOI] [PubMed] [Google Scholar]
- Oishi K, Atsumi G, Sugiyama S, Kodomari I, Kasamatsu M, Machida K, Ishida N. Disrupted fat absorption attenuates obesity induced by a high-fat diet in Clock mutant mice. FEBS Lett. 2006;580:127–130. doi: 10.1016/j.febslet.2005.11.063. [DOI] [PubMed] [Google Scholar]
- Palatini P, Penzo M, Racioppa A, Zugno E, Guzzardi G, Anaclerio M, Pessina AC. Clinical relevance of nighttime blood pressure and of daytime blood pressure variability. Arch Intern Med. 1992;152:1855–1860. [PubMed] [Google Scholar]
- Paschos GK, FitzGerald GA. Circadian clocks and vascular function. Circ Res. 2010;106:833–841. doi: 10.1161/CIRCRESAHA.109.211706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penev P, Kolker D, Zee P, Turek F. Chronic circadian desynchronization decreases the survival of animals with cardiomyopathic heart disease. Am J Physiol. 1998;275:H2334–H2337. doi: 10.1152/ajpheart.1998.275.6.H2334. [DOI] [PubMed] [Google Scholar]
- Portaluppi F, Smolensky MH, Touitou Y. Ethics and methods for biological rhythm research on animals and human beings. Chronobiol Int. 2010;27:1911–1929. doi: 10.3109/07420528.2010.516381. [DOI] [PubMed] [Google Scholar]
- Rudic RD, Fulton DJ. Pressed for time: the circadian clock and hypertension. J Appl Physiol. 2009;107:1328–1338. doi: 10.1152/japplphysiol.00661.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smolensky MH. Chronobiology and chronotherapeutics. Applications to cardiovascular medicine. Am J Hypertens. 1996;9:11S–21S. doi: 10.1016/0895-7061(95)00405-x. [DOI] [PubMed] [Google Scholar]
- Sole MJ, Martino TA. Diurnal physiology: core principles with application to the pathogenesis, diagnosis, prevention, and treatment of myocardial hypertrophy and failure. J Appl Physiol. 2009;107:1318–1327. doi: 10.1152/japplphysiol.00426.2009. [DOI] [PubMed] [Google Scholar]
- Takahashi JS, Hong HK, Ko CH, McDearmon EL. The genetics of mammalian circadian order and disorder: implications for physiology and disease. Nat Rev Genet. 2008;9:764–775. doi: 10.1038/nrg2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda N, Maemura K, Horie S, Oishi K, Imai Y, Harada T, Saito T, Shiga T, Amiya E, Manabe I, Ishida N, Nagai R. Thrombomodulin is a clock-controlled gene in vascular endothelial cells. J Biol Chem. 2007;282:32561–32567. doi: 10.1074/jbc.M705692200. [DOI] [PubMed] [Google Scholar]
- Tsai JY, Kienesberger PC, Pulinilkunnil T, Sailors MH, Durgan DJ, Villegas-Montoya C, Jahoor A, Gonzalez R, Garvey ME, Boland B, Blasier Z, McElfresh TA, Nannegari V, Chow CW, Heird WC, Chandler MP, Dyck JR, Bray MS, Young ME. Direct regulation of myocardial triglyceride metabolism by the cardiomyocyte circadian clock. J Biol Chem. 2010;285:2918–2929. doi: 10.1074/jbc.M109.077800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turek F, Joshu C, Kohsaka A, Lin E, Ivanova G, McDearmon E, Laposky A, Losee-Olson S, Easton A, Jensen D, Eckel R, Takahashi J, Bass J. Obesity and metabolic syndrome in Clock mutant mice. Science. 2005;308:1043–1045. doi: 10.1126/science.1108750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viola AU, James LM, Archer SN, Dijk DJ. PER3 polymorphism and cardiac autonomic control: effects of sleep debt and circadian phase. Am J Physiol Heart Circ Physiol. 2008;295:H2156–2163. doi: 10.1152/ajpheart.00662.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N, Yang G, Jia Z, Zhang H, Aoyagi T, Soodvilai S, Symons JD, Schnermann JB, Gonzalez FJ, Litwin SE, Yang T. Vascular PPARgamma controls circadian variation in blood pressure and heart rate through Bmal1. Cell Metab. 2008;8:482–491. doi: 10.1016/j.cmet.2008.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijnen H, Young MW. Interplay of circadian clocks and metabolic rhythms. Annu Rev Genet. 2006;40:409–448. doi: 10.1146/annurev.genet.40.110405.090603. [DOI] [PubMed] [Google Scholar]
- Woon PY, Kaisaki PJ, Braganca J, Bihoreau MT, Levy JC, Farrall M, Gauguier D. Aryl hydrocarbon receptor nuclear translocator-like (BMAL1) is associated with susceptibility to hypertension and type 2 diabetes. Proc Natl Acad Sci U S A. 2007;104:14412–14417. doi: 10.1073/pnas.0703247104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Graeff R, Xie Q, Gamble KL, Mori T, Johnson CH. Comment on “The Arabidopsis circadian clock incorporates a cADPR-based feedback loop”. Science. 2009;326:230. doi: 10.1126/science.1169503. author reply 230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young M, Razeghi P, Taegtmeyer H. Clock genes in the heart: characterization and attenuation with hypertrophy. Circ Res. 2001;88:1142–1150. doi: 10.1161/hh1101.091190. [DOI] [PubMed] [Google Scholar]
- Young M, Wilson C, Razeghi P, Guthrie P, Taegtmeyer H. Alterations of the Circadian Clock in the Heart by Streptozotocin-induced Diabetes. J Mol Cell Cardiol. 2002;34:223–231. doi: 10.1006/jmcc.2001.1504. [DOI] [PubMed] [Google Scholar]
- Young M. The circadian clock within the heart: potential influence on myocardial gene expression; metabolism; and function. Am J Physiol Heart Circ Physiol. 2006;290:H1–H16. doi: 10.1152/ajpheart.00582.2005. [DOI] [PubMed] [Google Scholar]
- Young M, Bray M. Potential role for peripheral circadian clock dyssynchrony in the pathogenesis of cardiovascular disease. Sleep Med. 2007 doi: 10.1016/j.sleep.2006.12.010. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young ME. Anticipating anticipation: pursuing identification of cardiomyocyte circadian clock function. J Appl Physiol. 2009;107:1339–1347. doi: 10.1152/japplphysiol.00473.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zambon AC, McDearmon EL, Salomonis N, Vranizan KM, Johansen KL, Adey D, Takahashi JS, Schambelan M, Conklin BR. Time- and exercise-dependent gene regulation in human skeletal muscle. Genome Biol. 2003;4:R61. doi: 10.1186/gb-2003-4-10-r61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang EE, Liu Y, Dentin R, Pongsawakul PY, Liu AC, Hirota T, Nusinow DA, Sun X, Landais S, Kodama Y, Brenner DA, Montminy M, Kay SA. Cryptochrome mediates circadian regulation of cAMP signaling and hepatic gluconeogenesis. Nat Med. 2010;16:1152–1156. doi: 10.1038/nm.2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.