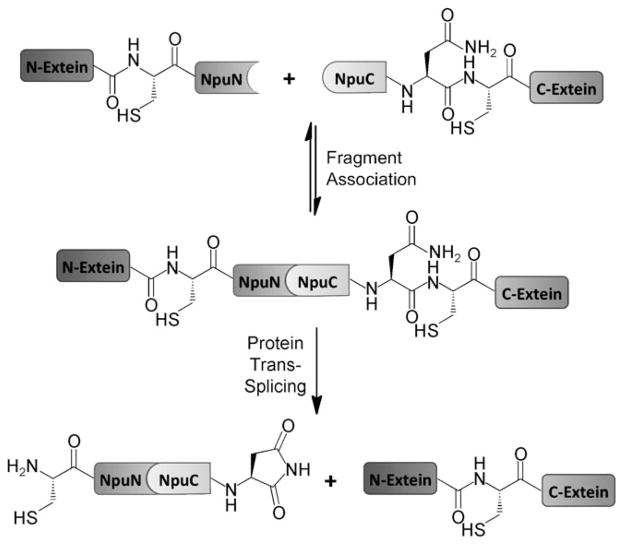
Coulombic forces play an important role in facilitating protein–protein interactions. It has been demonstrated that a strong electrostatic potential between two interacting proteins correlates with a fast rate of association and a strong binding affinity.[1] Indeed, this principle has been exploited to enhance the binding properties of engineered protein interfaces.[2] Nonetheless, little research has focused on utilizing intermolecular ion pairs to modulate specificity in protein–protein interactions.[3–5] Naturally split inteins are a potentially interesting system for engineering electrostatically driven specificity, as the formation of a catalytically competent structure requires the association of two oppositely charged protomers. In their endogenous environment, split inteins catalyze protein trans-splicing (PTS, Scheme 1) of essential gene fragments, and thus are under evolutionary pressure to associate rapidly with high fidelity and dissociate slowly post-catalysis to prevent the re-formation of unproductive complexes. Out of their native context, split inteins have seen widespread use in a number of biotechnological applications, as a result of their capacity to ligate flanking protein sequences (exteins) in trans.[6] Applications of PTS include segmental isotopic labeling of proteins for NMR spectroscopy,[7] protein immobilization,[8] the labeling of proteins with extrinsic probes,[9] protein and peptide cyclization,[10] and control of protein function.[11,12]
Scheme 1.
Protein trans-splicing.
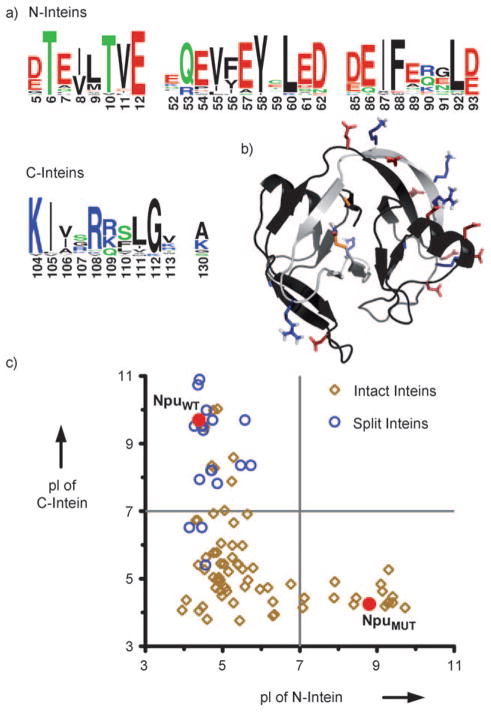
Despite the utility of PTS, little is known about what drives efficient association of split intein fragments. Sequence alignments of naturally split inteins show highly conserved charge segregation, with acidic residues concentrated at specific positions on the N-intein and basic residues conserved on the C-intein (N- and C-terminal protomers, Figure 1a).[13] Furthermore, a bioinformatic sequence analysis of the intein family indicates that this charge segregation is significantly more prevalent in naturally split inteins than intact ones (Figure 1c). Interestingly, when the conserved charged residues are mapped onto the structure determined by NMR spectrocopy of the wild-type DnaE intein from Nostoc punctiforme (NpuWT), many are found to be participating in intermolecular ion pairs and ion triads (Figure 1b).[14]
Figure 1.
Charge segregation in split inteins. a) Sequence logo based on a sequence alignment of 24 naturally split inteins highlighting the highly conserved charged residues. b) Structure of NpuWT determined by NMR spectroscopy (PDB code: 2KEQ) highlighting intermolecular electrostatic interactions. NpuNWT and NpuCWT are shown in black and gray, respectively. Acidic residues are shown in red, basic residues are shown in blue, and terminal catalytic residues are shown in orange. c) Comparison of the calculated isoelectric points (pI) for the N- and C-terminal fragments of 24 naturally split inteins, the N- and C-terminal sequences of 76 intact inteins, and the mutant split intein (NpuMUT) engineered in this study. NpuWT and NpuMUT are both indicated on the plot as red circles.
While ionic interactions have previously been postulated to play a role in split intein assembly,[13,15] the involvement of electrostatic forces has not been validated experimentally. In this study, we set out to test the hypothesis that ionic interactions facilitate the association of split intein fragments and thus could be manipulated to control the relative reactivities of different N- and C-intein complexes.
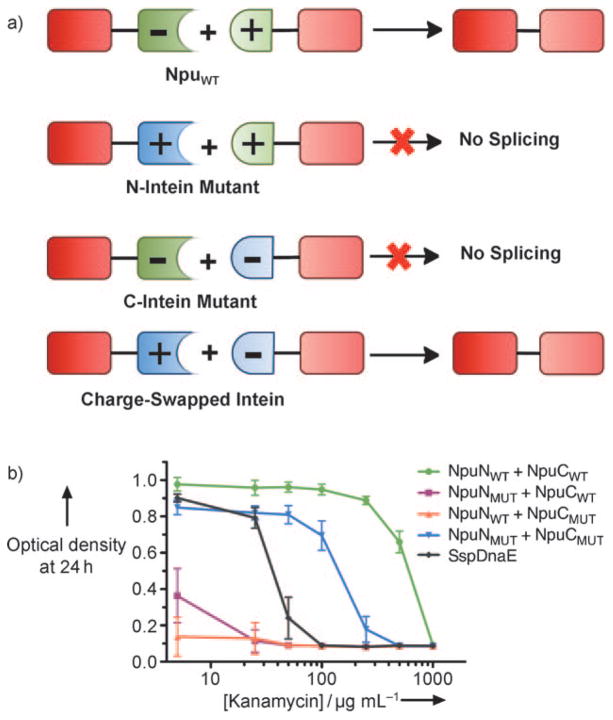
To evaluate the role of specific ionic interactions for split intein splicing and specificity we screened a series of charge-swapped Npu intein mutants and tested their splicing activity in vivo. Starting from the wild-type protomers, NpuNWT and NpuCWT, mutation positions were chosen based on two criteria: 1) residues should be involved in an intermolecular ion pair or triad, and 2) these residues should be moderately isolated from intramolecular ionic interactions. Based on these criteria, we identified four ion clusters comprised of one pair and three triads. For each cluster and several combinations of clusters, three variants were generated: an N-intein mutant in which native acidic residues were mutated to basic residues, a C-intein mutant in which basic residues were mutated to acidic residues, and a charge-swapped intein that combined all N- and C-intein mutations (Figure 2a). The activity of each set was analyzed using an in vivo splicing assay where the kanamycin resistance in E. coli was dependent on the intein splicing activity.[16] Of all the combinations tested (see the Supporting Information), the intein fragments in which all the possible ion clusters were charged swapped, NpuNMUT and NpuCMUT, showed the least cross-reactivity with wild-type fragments in vivo (Figure 2b). Specifically, NpuNWT did not react with NpuCMUT, NpuNMUT reacted minimally with NpuCWT, and both mutant fragments displayed substantial splicing activity when combined in vivo. Importantly, the new charge-swapped intein, NpuMUT, catalyzed protein splicing more efficiently than the widely used SspDnaE intein.
Figure 2.
Rational design strategy for charge-swapped inteins. a) Scheme depicting the desired specificity in charge-swapped inteins. b) Dose–response curves for NpuWT and NpuMUT fragment combinations in an in vivo intein activity-coupled kanamycin resistance assay. Mutation clusters in NpuMUT are (E7K, K130E), (E52K, E54K, K113E), (D85K, E89R, R108E), and (E61K, E91K, K104E).
To determine if the observed trend for in vivo splicing activities was a direct result of relative fragment binding affinities we developed an in vitro binding assay. Using ubiquitin (Ub) and the small ubiquitin-like modifier (SUMO) as model N- and C-extein domains, we expressed and purified Ub-NpuN and NpuC-SUMO fusion proteins bearing wild-type or mutant intein sequences. The Ub-NpuN fusions contained a cysteine to alanine mutation (C1A) to prevent trans-splicing. To measure the binding affinities, we took advantage of the single tryptophan residue (W47) in the Npu intein and measured a decrease in the intrinsic fluorescence of NpuN in the presence of increasing concentrations of NpuC-SUMO (Table 1 and see the Supporting Information). As expected, the wild-type fragments had a low-nanomolar binding affinity, consistent with previous measurements for the highly homologous SspDnaE intein.[15] The fully charged-swapped NpuMUT N- and C-inteins associated with a 40-fold weaker affinity than the NpuWT protomers (Kd = 118.4 nM), which may explain their slightly diminished splicing activity in vivo. Surprisingly, both wild-type/mutant hybrids had measurable binding affinities only two- to threefold weaker than NpuNMUT + NpuCMUT, despite their extremely low or undetectable levels of splicing in vivo. These data indicated that while charge-swapping modulated the intein fragment affinities with the expected trend, the magnitude of these energetic effects do not fully explain the splicing selectivity observed in vivo. Notably, we observed that the NpuNWT + NpuCMUT combination showed only a 29% decrease in N-intein fluorescence upon binding, while all other fragment combinations showed a 60–70% decrease in fluorescence (see the Supporting Information). This anomalous fluorescence strongly suggests that the NpuNWT + NpuCMUT complex adopts a unique conformation which could explain its lack of splicing activity in vivo.
Table 1.
In vitro characterization of NpuWT and NpuMUT.
| Fragment pair | Kd [nM]a | Initial t1/2 of trans-splicing [min]a,b | |||
|---|---|---|---|---|---|
| NpuN | NpuC | 1.0 μM | 0.1 μM | 0.05 μM | |
| WT | WT | 2.9 ± 1.8 | 0.9 ± 0.1 | 1.1 ± 0.2 | 2.4 ± 0.4 |
| MUT | WT | 211.6 ± 21.0 | 8.2 ± 0.4 | 71.0 ± 4.9 | 350.3± 41.2 |
| WT | MUT | 281.8 ± 51.5 | n/ac | n/ac | n/ac |
| MUT | MUT | 118.4 ± 11.4 | 6.5 ± 0.4 | 43.7 ± 2.1 | 103.0± 8.7 |
Error values indicate the standard error from a global fit of three replicate data sets.
Initial half-lives are calculated from a second-order rate constant.
No detectable trans-splicing was observed for the NpuNWT + NpuCMUT combination.
n/a = not active.
We next examined the relative rates of trans-splicing for NpuWT, NpuMUT, and their N- and C-intein combinations in vitro. Splicing kinetics at 30°C were measured by pairwise mixing of Ub-NpuN and NpuC-SUMO fusions at equimolar ratios and monitoring the formation of the Ub-SUMO spliced product by gel electrophoresis (Table 1). NpuWT showed extremely rapid splicing (t1/2 ≈ 1 min) as previously observed.[17] As expected, given the low-nanomolar affinity for the wild-type fragments, this rapid rate of splicing was relatively independent of intein concentrations from 1.0 μM to 0.05 μM. Consistent with the in vivo splicing and in vitro binding experiments, NpuMUT spliced slightly slower than NpuWT at 1.0 μM (t1/2 = 6.5 min), but its rate decreased roughly tenfold at intein concentrations near its Kd value. Surprisingly, the NpuNMUT + NpuCWT combination catalyzed splicing almost as quickly as NpuNMUT + NpuCMUT at high concentrations, but this difference in trans-splicing rates increased dramatically at lower concentrations, because of the weaker binding affinity for NpuNMUT + NpuCWT. The NpuNWT + NpuCMUT combination showed no detectable trans-splicing at all three concentrations tested. Not only is this observation consistent with the in vivo results, it also supports the fluorescence data that suggest that this N- and C-intein bind to form a catalytically incompetent complex. The in vitro binding and splicing assays collectively demonstrate that charge complementation or repulsion can bias relative split intein binding affinities, which in turn bias relative splicing kinetics. These experiments also shed light on our in vivo splicing assays. Specifically, the degree of concentration dependence on splicing rates observed in vitro indicates that the intein concentration in vivo is probably at the low nanomolar range, at which level the in vivo and in vitro results would be consistent.
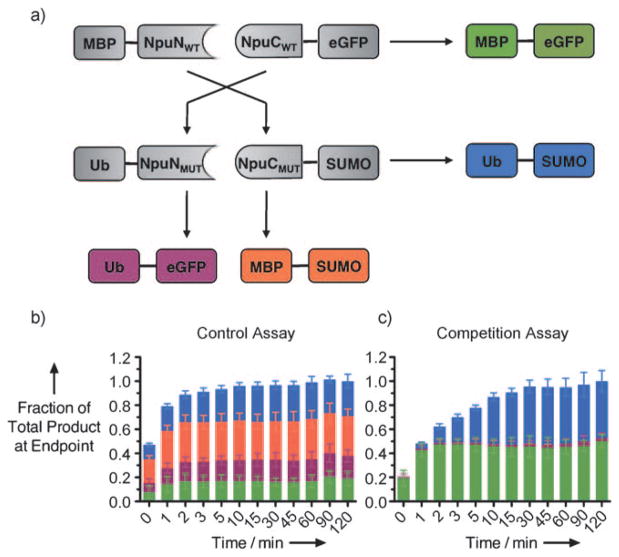
Given the differences in splicing kinetics observed in vitro, we envisioned that NpuWT and NpuMUT could be used simultaneously to catalyze multiple trans-splicing reactions with kinetically controlled selectivity.[18,19] To test this, we developed an in vitro competition assay (Figure 3a) using our splicing assay fusion proteins and two additional fusions to the NpuWT fragments bearing unique exteins—maltose binding protein (MBP) and enhanced green fluorescent protein (eGFP). As a control reaction, we mixed two NpuNWT fusions and two NpuCWT fusions bearing unique exteins at equimolar concentrations (0.5 μM) at 30°C. The formation of all four possible products was monitored by Western blotting (see the Supporting Information). As expected, all the products formed to a similar extent in the control reaction (Figure 3b).
Figure 3.
Competition assays with NpuWT and NpuMUT. a) Scheme showing four possible splicing reactions in the competition assay. b) Control reaction with wild-type intein fragments. c) Competition reaction with wild-type and mutant fragments.
When Ub-NpuNMUT and NpuCMUT-SUMO were used in place of their wild-type counterparts, a clear bias in product formation was observed. The NpuWT product (MBP-eGFP) formed rapidly, and the NpuMUT product (Ub-SUMO) emerged at a slightly slower rate, consistent with our in vitro splicing assays. These products formed almost exclusively, close to 50% each, while the NpuNMUT + NpuCWT product (Ub-eGFP) accounted for less than 5% of the total product formed, and the NpuNWT + NpuCMUT product (MBP-SUMO) was not observed (Figure 3c). These results were reproducible over a range of temperatures—from 25°C to 37°C—and at higher ionic strength, thus indicating that the electrostatically driven selectivity between NpuWT and NpuMUT is extremely robust (see the Supporting Information).
Next, we explored the utility of these inteins for one-pot three-piece ligations of proteins (Figure 4a). Previously, orthogonal intein systems have been developed by combining naturally and artificially split inteins or by using a wild-type and linearly permuted split intein.[15,20] While these systems could catalyze three-piece ligations, the prior system showed low efficiency, and the latter could not be carried out in one pot. We envisioned that our NpuWT and NpuMUT pair could efficiently catalyze one-pot three-piece ligations, given the high degree of kinetic control seen in our competition assay. To test this, we designed a model system to ligate a Src-homology 3 (SH3) domain, domain B1 of protein G (GB1), and eGFP. The domains were fused to split intein sequences to generate an N-terminal fragment (N: SH3-NpuNMUT), a middle fragment (M: NpuCMUT-GB1-NpuNWT), and a C-terminal fragment (C: NpuCWT-eGFP). Importantly, we designed the middle domain flanked by the N- and C-inteins that do not react in trans since this would preclude GB1 cyclization or oligomerization (Figure 4a). To test our three-piece ligation system, the N, M, and C fragments were mixed with a slight excess of the middle fragment. Analysis of the reaction mixture by gel electrophoresis indicated that the reaction between NpuNWT and NpuCWT (M + C) occurred extremely fast to yield one intermediate product. This intermediate was more slowly converted into the full-length three-piece-ligated product, SH3-GB1-eGFP, upon reaction with the N fragment, consistent with the kinetically controlled trans-splicing paradigm (see the Supporting Information).
Figure 4.
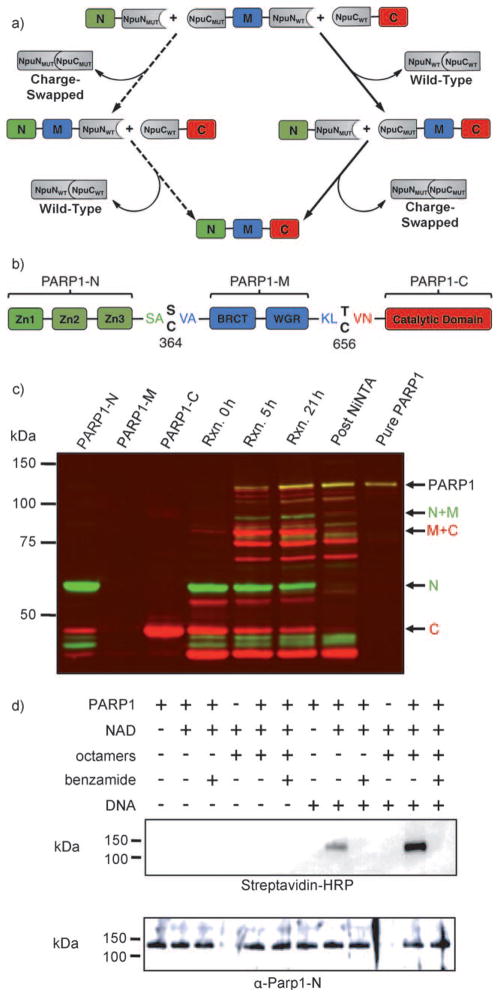
Three-piece ligation of human PARP1. a) Scheme depicting a three-piece ligation using NpuWT and NpuMUT. The major reaction pathway is indicated by solid arrows, and the minor reaction pathway is indicated by dashed arrows. b) Domain organization and splice junctions in PARP1. c) Western blot of PARP1 three-piece ligation and purification. Color scheme: green bands (α-PARP1-N), red bands (α-PARP1-C), and yellow bands (α-PARP1-N + α-PARP1-C). Un-annotated bands are identified in Figure S20 in the Supporting Information. d) Biotinylated-NAD blot showing automodification of PARP1 (streptavidin-HRP) and PARP1 blot loading control (α-PARP1-N).
Next, we sought to apply this three-piece ligation technology towards the semisynthesis of human poly(ADP-ribose) polymerase 1 (PARP1). This approximately 115 kDa enzyme, which catalyzes the transfer of ADP-ribose from nicotinamide adenine dinucleotide (NAD) onto protein side chains in the form of monomers or polymers, is involved in DNA-damage pathways and is a promising target for chemo-therapeutics.[21,22] Given its large size and its capacity to automodify itself,[23] full-length PARP1 is not readily isolable by over-expression in E. coli.[24,25] We envisioned that our orthogonal inteins could be used to generate full-length PARP1 by expression of its fragments separately in E. coli followed by in vitro three-piece ligation. Based on known domain boundaries,[26,27] we chose two ligation sites in putatively flexible regions that separated PARP1 into three fragments: the N-terminal zinc finger DNA-binding domains (PARP1-N), the central dimerization and automodification domains (PARP1M), and the C-terminal catalytic domain (PARP1-C) (Figure 4b). Importantly, each of these functional domains is required for activation and regulation of PARP1 catalysis,[28] and thus their efficient and accurate assembly is a rigorous test of our three-piece ligation system.
As a consequence of the lack of cysteine residues near the desired splice junctions, we introduced two modest mutations, S364C and T656C, to allow for split intein catalysis and fused the PARP1 fragments to our split inteins. In addition, we designed a traceless tagging strategy in which His6 tags were placed at the free termini of every intein sequence (see the Supporting Information). These tags could be used not only to facilitate enrichment of the proteins after over-expression but also to trap any remaining starting materials, intermediates, and spliced intein fragments after a three-piece ligation. To generate full-length PARP1 we expressed the three intein-fusion fragments separately in E. coli and enriched the proteins over nickel columns. The semipure proteins were mixed and allowed to react at room temperature for 21 h. By following the reaction by Western blotting against PARP1-N and PARP1-C, we observed the formation of both reaction intermediates as well as full-length PARP1. The reaction mixture was passed through a nickel affinity column to trap residual starting materials, intermediates, and free intein fragments. The flow-through, containing the full-length product, was then purified by size-exclusion chromatography to yield pure, three-piece-ligated PARP1 (Figure 4c). Mass spectrometric analysis of tryptic fragments confirmed the identity of the purified product, and peptides spanning the ligation junctions were further analyzed by MS/MS to confirm their sequences (see the Supporting Information).
To determine whether our semisynthetic PARP1 was active, we conducted ADP-ribosylation assays using biotinylated-NAD and streptavidin blotting. We observed that our three-piece ligated PARP1 could catalyze automodification (Figure 4d). Importantly, this activity required activated DNA, could be inhibited by benzamide, and was stimulated by histone octamers, all of which are hallmarks of full-length PARP1 activity.[29,30] Furthermore, our enzyme could catalyze the formation of poly(ADP-ribose) chains on histones, as previously reported for endogenous PARP1 (see the Supporting Information).[31] These enzymatic data unequivocally demonstrate that our three-piece ligated PARP1 is full-length and properly folded, as the DNA-dependent activity of this enzyme requires allosteric communication between all three segments. Consistent with this requirement, the PARP1-C fragment, bearing only the catalytic domain of PARP1, did not catalyze poly(ADP-ribosylation) of histones (see the Supporting Information).
In this study, we probed the role of intermolecular ion clusters for fragment assembly and splicing in the NpuWT split intein both in vivo and in vitro. Through these experiments, we rationally designed a new split intein, NpuMUT, which displays low cross-reactivity with NpuWT. These orthogonal inteins were used to generate the large, full-length, active mammalian protein PARP1 through a one-pot three-piece ligation. Collectively, our results demonstrate that electrostatic interactions can engender kinetic control in a complex enzymatic system. Furthermore, these results provide insight into the molecular requirements for efficient and specific protein trans-splicing.
Supplementary Material
Footnotes
We thank Dr. P. Moyle for valuable discussions and Dr. J. Fernandez from The Rockefeller University Proteomics Resource Center for the MS/MS analysis of PARP1. This work was supported by the US National Institutes of Health (NIH grant GM086868).
Contributor Information
Dr. Miquel Vila-Perelló, Email: mvila@rockefeller.edu.
Prof. Tom W. Muir, Email: muirt@rockefeller.edu.
References
- 1.Schreiber G, Fersht AR. Nat Struct Biol. 1996;3:427. doi: 10.1038/nsb0596-427. [DOI] [PubMed] [Google Scholar]
- 2.Selzer T, Albeck S, Schreiber G. Nat Struct Biol. 2000;7:537. doi: 10.1038/76744. [DOI] [PubMed] [Google Scholar]
- 3.Graddis TJ, Myszka DG, Chaiken IM. Biochemistry. 1993;32:12664. doi: 10.1021/bi00210a015. [DOI] [PubMed] [Google Scholar]
- 4.O’Shea EK, Lumb KJ, Kim PS. Curr Biol. 1993;3:658. doi: 10.1016/0960-9822(93)90063-t. [DOI] [PubMed] [Google Scholar]
- 5.Gunasekaran K, Pentony M, Shen M, Garrett L, Forte C, Woodward A, Ng SB, Born T, Retter M, Manchulenko K, Sweet H, Foltz IN, Wittekind M, Yan W. J Biol Chem. 2010;285:19637. doi: 10.1074/jbc.M110.117382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vila-Perelló M, Muir TW. Cell. 2010;143:191. doi: 10.1016/j.cell.2010.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yamazaki T, Otomo T, Oda N, Kyogoku Y, Uegaki K, Ito N, Ishino Y, Nakamura H. J Am Chem Soc. 1998;120:5591. [Google Scholar]
- 8.Kwon Y, Coleman MA, Camarero JA. Angew Chem. 2006;118:1758. doi: 10.1002/anie.200503475. [DOI] [PubMed] [Google Scholar]; Angew Chem Int Ed. 2006;45:1726. [Google Scholar]
- 9.Giriat I, Muir TW. J Am Chem Soc. 2003;125:7180. doi: 10.1021/ja034736i. [DOI] [PubMed] [Google Scholar]
- 10.Scott CP, Abel-Santos E, Wall M, Wahnon DC, Benkovic SJ. Proc Natl Acad Sci USA. 1999;96:13638. doi: 10.1073/pnas.96.24.13638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mootz HD, Blum ES, Tyszkiewicz AB, Muir TW. J Am Chem Soc. 2003;125:10561. doi: 10.1021/ja0362813. [DOI] [PubMed] [Google Scholar]
- 12.Vila-Perelló M, Hori Y, Ribó M, Muir TW. Angew Chem. 2008;120:7878. doi: 10.1002/anie.200802502. [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew Chem Int Ed. 2008;47:7764. [Google Scholar]
- 13.Dassa B, Amitai G, Caspi J, Schueler-Furman O, Pietrokovski S. Biochemistry. 2007;46:322. doi: 10.1021/bi0611762. [DOI] [PubMed] [Google Scholar]
- 14.Oeemig JS, Aranko AS, Djupsjöbacka J, Heinämäki K, Iwaï H. FEBS Lett. 2009;583:1451. doi: 10.1016/j.febslet.2009.03.058. [DOI] [PubMed] [Google Scholar]
- 15.Shi J, Muir TW. J Am Chem Soc. 2005;127:6198. doi: 10.1021/ja042287w. [DOI] [PubMed] [Google Scholar]
- 16.Lockless SW, Muir TW. Proc Natl Acad Sci USA. 2009;106:10999. doi: 10.1073/pnas.0902964106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zettler J, Schütz V, Mootz HD. FEBS Lett. 2009;583:909. doi: 10.1016/j.febslet.2009.02.003. [DOI] [PubMed] [Google Scholar]
- 18.Zhang Z, Ollmann IR, Ye X-S, Wischnat R, Baasov T, Wong C-H. J Am Chem Soc. 1999;121:734. [Google Scholar]
- 19.Bang D, Pentelute BL, Kent SBH. Angew Chem. 2006;118:4089. doi: 10.1002/anie.200600702. [DOI] [PubMed] [Google Scholar]; Angew Chem Int Ed. 2006;45:3985. [Google Scholar]
- 20.Busche AEL, Aranko AS, Talebzadeh-Farooji M, Bernhard F, Dötsch V, Iwaï H. Angew Chem. 2009;121:6244. doi: 10.1002/anie.200901488. [DOI] [PubMed] [Google Scholar]; Angew Chem Int Ed. 2009;48:6128. [Google Scholar]
- 21.Hassa PO, Haenni SS, Elser M, Hottiger MO. Microbiol Mol Biol Rev. 2006;70:789. doi: 10.1128/MMBR.00040-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martin SA, Lord CJ, Ashworth A. Curr Opin Genet Dev. 2008;18:80. doi: 10.1016/j.gde.2008.01.016. [DOI] [PubMed] [Google Scholar]
- 23.Tao Z, Gao P, Liu H-w. J Am Chem Soc. 2009;131:14258. doi: 10.1021/ja906135d. [DOI] [PubMed] [Google Scholar]
- 24.Knight MI, Chambers PJ. Protein Expression Purif. 2001;23:453. doi: 10.1006/prep.2001.1513. [DOI] [PubMed] [Google Scholar]
- 25.Gagnon SN, Desnoyers S. Mol Cell Biochem. 2003;243:15. doi: 10.1023/a:1021645327079. [DOI] [PubMed] [Google Scholar]
- 26.Tao Z, Gao P, Hoffman DW, Liu H-W. Biochemistry. 2008;47:5804. doi: 10.1021/bi800018a. [DOI] [PubMed] [Google Scholar]
- 27.Lilyestrom W, van der Woerd MJ, Clark N, Luger K. J Mol Biol. 2010;395:983. doi: 10.1016/j.jmb.2009.11.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Simonin F, Höfferer L, Panzeter PL, Muller S, de Murcia G, Althaus FR. J Biol Chem. 1993;268:13454. [PubMed] [Google Scholar]
- 29.Pinnola A, Naumova N, Shah M, Tulin AV. J Biol Chem. 2007;282:32511. doi: 10.1074/jbc.M705989200. [DOI] [PubMed] [Google Scholar]
- 30.Southan G, Szabo C. Curr Med Chem. 2003;10:321. doi: 10.2174/0929867033368376. [DOI] [PubMed] [Google Scholar]
- 31.Messner S, Altmeyer M, Zhao H, Pozivil A, Roschitzki B, Gehrig P, Rutishauser D, Huang D, Caflisch A, Hottiger MO. Nucleic Acids Res. 2010;38:6350. doi: 10.1093/nar/gkq463. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.