Abstract
The Wnt signaling pathway is capable of self-regulation through positive and negative feedback mechanisms. For example, the oncoprotein c-Myc, which is upregulated by Wnt signaling activity, participates in a positive feedback loop of canonical Wnt signaling through repression of Wnt antagonists DKK1 and SFRP1. In this study, we investigated the mechanism of Wnt inhibitory factor-1 (WIF-1) silencing. Mapping of CpG island methylation of the WIF-1 promoter reveals regional methylation (–295 to –95 bp from the transcription start site) that correlates with transcriptional silencing. We identified Miz-1 as a direct activator of WIF-1 transcriptional activity, which is found at WIF-1 promoter. In addition, we show that c-Myc contributes to WIF-1 transcriptional repression in a Miz-1-dependent manner. Although the transient repression mediated by Miz-1/c-Myc is independent of de novo methylation, the stable repression by this complex is associated with CpG island methylation of the critical –295 to –95-bp region of the WIF-1 promoter. Importantly, Miz-1 and c-Myc are found at WIF-1 promoter in WIF-1 non-expressing cell lines DLD-1 and 209myc. Transient knockdown or somatic knockout of c-Myc in DLD-1 failed to restore WIF-1 expression suggesting that c-Myc is involved in initiating rather than maintaining WIF-1 epigenetic silencing. In a genome-wide screen, DNAJA4, TGFβ-induced and TRIM59 were repressed by c-Myc overexpression and DNA promoter hypermethylation. Our data reveal novel insights into c-Myc-mediated DNA methylation-dependent transcriptional silencing, a mechanism that might contribute to the dysregulation of Wnt signaling in cancer.
Keywords: WIF-1, Wnt signaling, c-Myc, Miz-1, DNA methylation
Introduction
Transcriptional silencing of tumor-suppressor genes associated with promoter hypermethylation is a key epigenetic event that contributes to the hallmarks of cancer (Herman and Baylin, 2003). However, the exact process through which this phenomenon occurs remains elusive for most genes. Transcriptional repression of RARβ by PML–RARα, the defining molecular change in acute promyelocytic leukemia, is thus far the best described example of epigenetic silencing of a target gene (Di Croce et al., 2002). The leukemia-promoting PML–RAR fusion protein directly recruits the DNA methyltransferases (Dnmts) 1 and 3a to the RARβ promoter, leading to DNA promoter hypermethylation and gene silencing of RARβ (Di Croce et al., 2002). The polycomb repressive complex 2 has also been implicated in this process, suggesting that Suz12 not only directly modifies histone tails of PML–RAR target genes but also mediates promoter hypermethylation of target genes (Villa et al., 2007).
MIZ-1 (Myc-interacting Zn finger protein-1) is a member of the POZ domain/zinc-finger transcription factor family (Peukert et al., 1997), which acts as a transcription activator of the cell cycle arrest genes P21CIP1 (Seoane et al., 2002; Wu et al., 2003) and P15INK4b (Seoane et al., 2001; Staller et al., 2001). Through its direct interaction with Myc, Miz-1 can also recruit the Myc/Max complex and prevent the recruitment of transcription activator such as P300 (Staller et al., 2001). In addition, epigenetic mechanisms might also mediate the repressive activity of the Miz-1/c-Myc complex, for example, through the recruitment of the Dnmt3a to gene promoter (Brenner et al., 2005).
Targets of the Miz-1/c-Myc repressive complex include P21CIP1 (Peukert et al., 1997; Seoane et al., 2002; van de Wetering et al., 2002; Wu et al., 2003), P15INK4b (Seoane et al., 2001; Staller et al., 2001), α6 and β1 integrins (Gebhardt et al., 2006). Nevertheless, Miz-1-independent mechanisms of gene silencing mediated by c-Myc also exist. For example, P21CIP1 gene silencing also occurs through the displacement of Sp1 from P21CIP1 promoter by c-Myc, independently of Miz-1 (Gartel et al., 2001). In addition, c-Myc was shown to induce apoptosis through the inactivation of Miz-1 and its target BCL2 (Patel and McMahon, 2006, 2007).
Many of the Wnt target genes are capable of enhancing or antagonizing Wnt pathway activity suggesting that feedback loop mechanisms contribute to its regulation (He et al., 1998; Gujral and MacBeath, 2009). Among these, c-Myc participates in a positive feedback loop in Wnt signaling through the repression of Wnt antagonists DKK1 and SFRP1, resulting in the transformation of mammary epithelial cells (Cowling et al., 2007). WIF-1 (Wnt inhibitor factor-1) encodes for a Wnt pathway antagonist (Hsieh et al., 1999) frequently methylated in lung (Mazieres et al., 2004), colon (Taniguchi et al., 2005), breast (Ai et al., 2006), bladder (Urakami et al., 2006) and nasopharyngeal cancers (Chan et al., 2007). In addition, c-Myc and Wif-1 protein expression inversely correlate in bladder cancer cells, which further argues for the existence of such feedback mechanism in Wnt signaling (Urakami et al., 2006; Tang et al., 2009).
In this study, we examined the mechanism through which WIF-1 transcriptional silencing occurs. We carried out an extensive characterization of the DNA methylation status of the WIF-1 promoter and determined the roles of Miz-1 and c-Myc on WIF-1 promoter activity and transcription. Our study defines a role for c-Myc in the epigenetic silencing of WIF-1 and other genes in cancer.
Results
WIF-1 silencing is associated with DNA methylation of a specific region within WIF-1 promoter
In contrast to most colorectal cancer cell lines that lack basal WIF-1 expression (Taniguchi et al., 2005) (Figure 1a) and have methylation of the entire WIF-1 promoter (data not shown), non-small cell lung cancer (NSCLC) cell lines differed in WIF-1 expression levels (Figure 1a). Specifically, H1703, H920, H1435, H1299 and H358 express WIF-1 at high level. U1752, H125, H157, A549, H460 and normal human bronchial epithelial cells have detectable, but lower levels of WIF-1 (Figure 1a). H23, H1666, H1395, H838, H1993 and H1155 lack WIF-1 expression. Treatment with the demethylating agent 5-aza-2′-deoxycitidine led to an increase in WIF-1 expression for cell lines with low or absent basal WIF-1 expression (Figure 1a).
Figure 1.
WIF-1 promoter methylation status. (a) Upper panel: WIF-1 expression was analyzed by RT–PCR in lung and colorectal cancer cell lines either untreated, treated with dimethylsulphoxide (DMSO) (m) or with 2 μm of 5-aza-2′-azadeoxycitidine treatment for 56 h (A56), 82 h (A82) or 96 h (A96). Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as a control. Lower panel: real-time RT–PCR analysis of WIF-1 transcript in NSCLC cell lines. WIF-1 expression was highest in H920 followed by H1435, H1299, H1703, U1752, normal human bronchial epithelial (NHBE), H460, H157, A549 and H125 and absent in H1155, H1666, H1395 and H1993. (b, c) Genomic bisulfite sequencing analysis of WIF-1 promoter region (b) –21/+209 bp, (c, left panel) –401/–350 and (c, right panel) –320/–72 bp. White-filled circles represent unmethylated CGs and black-filled circle methylated CGs. All regions are relative to the TSS.
Genomic bisulfite sequencing was used to identify DNA methylation pattern associated with WIF-1 silencing in NSCLC cell lines. The regions –21/+209 bp from the transcription start site or TSS, and –910/–619 bp both showed discordance between methylation status and WIF-1 expression level (Figure 1b and Supplementary Figure 1, respectively). In contrast, bisulfite sequencing of the –401/–350 bp and the –320/–72-bp regions revealed that cell lines heavily methylated in both regions had no detectable levels of the WIF-1 transcript (Figure 1c), while cell lines with WIF-1 expression had incomplete methylation. More specifically, DNA methylation of the region –295 to –95 bp of WIF-1 promoter seems essential for WIF-1 transcriptional silencing.
Characterization of WIF-1 proximal promoter
Five deletion constructs (WIF-1-C1 to C5) were designed for functional characterization of the WIF-1 proximal promoter (Figure 2a). WIF-1-C3 had the highest relative luciferase activity (Figure 2b) providing a potential explanation for the importance of methylation of the –401/–350 bp and –320/–72-bp regions of WIF-1 promoter on the control of WIF-1 transcription. The minimal difference in luciferase activity between WIF-1-C3 and C4 constructs further refines this to –320/–72 bp as the key region for WIF-1 transcriptional regulation. As a direct evidence for the role of methylation in this region, in vitro methylation of WIF-1-C3 led to a 90% decrease in luciferase activity (Figure 2c).
Figure 2.
Characterization of WIF-1 proximal promoter. (a) Diagram showing the regions of WIF-1 promoter which were analyzed for their ability to induce luciferase activity (upper panel), or by genomic bisulfite sequencing (lower panel). Vertical bars represent individual CGs. A candidate Sp1 binding site and a possible Inr sequence are also shown. (b) Luciferase activity of promoter deletion constructs, as shown in (b). pGL3-construct vector was used as a positive (POS), and empty pGL3-basic vector as a negative control (EV). (c) The promoter region –436 to +120 bp was in vitro methylated, cloned into pGL3 vector and luciferase activity was determined and compared with that of unmethylated WIF-1-C3. (d, e) Real-time ChIP was used to determine the enrichment of H3K4me2 (d) and H3K27me3 (e) at WIF-1 promoter (–836/–608 bp and –250/–50 bp) in H460, H1703 and H838. Enrichment was normalized to the value obtained for the H838 cell line (–836/–608 bp).
The presence of active (H3K4me2) and inactive (H3K27me3) histone modifications at WIF-1 promoter was examined by real-time chromatin immunoprecipitation (ChIP) in H1703, H460 and H838 (Figure 2d). Enrichment of the active mark H3K4me2 (Figure 2d) correlates with WIF-1 expression status and extent of DNA methylation of the WIF-1 promoter. Specifically, the WIF-1-expressing cell lines H1703 and to lesser extent H460 have a higher enrichment of H3K4me2 than the WIF-1 non-expressing cell line H838. In contrast, the repressive mark H3K27me3 (Figure 2e) was most enriched in H838 and least enriched in H1703, with these changes most different at –250 to –50 bp.
Miz-1 directly activates WIF-1 promoter activity
Data describing the repression of Wnt antagonists SFRP1 and DKK1 by Miz-1/c-Myc (Cowling et al., 2007) and the reverse correlation between c-Myc and Wif-1 protein expression in bladder cancer (Urakami et al., 2006) prompted us to determine whether the Miz-1/c-Myc complex may contribute to WIF-1 silencing. We first assessed the effect of full-length Miz-1 on WIF-1 promoter activity in a luciferase-based assay. Cotransfection of 1 and 10 ng of a construct encoding full-length Miz-1 increased the relative luciferase activity of WIF-1-C3 by 1.4- and 4-fold, respectively, when compared with empty vector (Figure 3a). Similarly, Miz-1 overexpression in H1299 increased endogeneous WIF-1 mRNA level by 3.5-fold compared with empty vector (Figure 3b), and led to the enrichment of Miz-1 at WIF-1 promoter (Figure 3c). Transient knockdown of Miz-1 with small interfering RNA (siRNA) in H1299 led to a decrease in endogeneous WIF-1 mRNA level (Figure 3d). This data therefore show that Miz-1 functions as a direct activator of WIF-1 transcription.
Figure 3.
Miz-1 directly increases WIF-1 transcription. (a) Increasing amount of an empty vector (EV) or a plasmid encoding full-length Miz-1 were transiently transfected in 293 cells together with a luciferase construct driven by WIF-1 proximal promoter (WIF-1-C3) and pRL-TK. Luciferase values were normalized to renilla and are shown relative to empty vector. (b) Empty vector or a plasmid encoding full-length Miz-1 were transiently transfected in H1299 and after 24–48 h endogeneous WIF-1 transcript level was measured by Taqman RT–PCR (Miz-1 protein level is shown in the inset). Values for WIF-1 were normalized to a Tata box-binding protein (TBP) standard curve. (c) EV or a plasmid encoding full-length Miz-1 were transiently transfected in H1299 and after 24 samples were cross-linked with DSG and formaldehyde for ChIP analysis. Chromatin was immunoprecipitated with either a normal immunoglobulin G (IgG), Miz-1 or c-Myc antibodies and ChIP DNA was amplified using primers specific for WIF-1 promoter (region –50 to + 120 relative to the TSS). (d) SMARTpool ON-TARGETplus for Miz-1 (ZBTB17) was used to transiently knockdown Miz-1 level in H1299. Forty-eight hours following siRNA transfection, endogeneous WIF-1 transcript levels were measured by Taqman RT–PCR. Miz-1 protein levels were also determined (inset).
Miz-1/c-Myc represses WIF-1 transcription
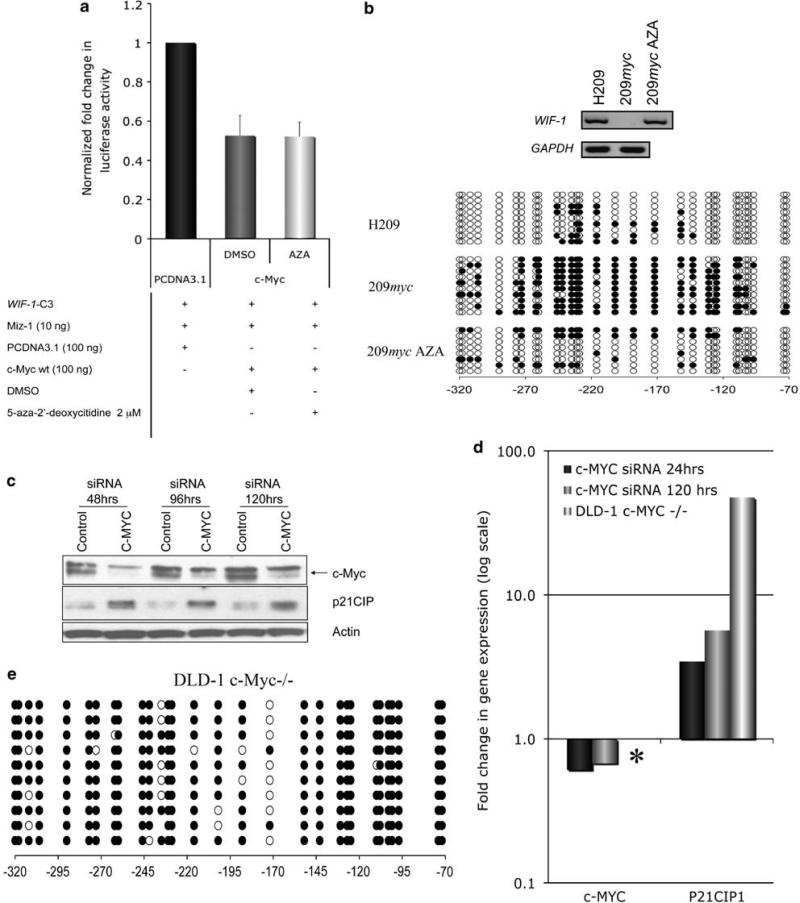
Although c-Myc alone led to a 52% reduction in P21CIP1 promoter activity (Supplementary Figure 2A), as previously reported (Brenner et al., 2005), it had no effect on WIF-1-C3 luciferase activity (Figure 4a). We therefore hypothesized that c-Myc-mediated repression of WIF-1 may require co-expression of Miz-1 in this assay. Indeed, overexpression of c-Myc led to a 50% reduction in WIF-1-C3 promoter activity driven by the overexpression of Miz-1 (Figure 4a). Repression by c-Myc was lost when using a mutant version of c-Myc (c-MycV394D), which cannot bind Miz-1. Transient expression of c-Myc in H1299 also modestly repressed endogenous WIF-1 mRNA, suggesting that this repression was present at the native promoter (Supplementary Figure 2B). WIF-1 repression was also observed in cells, which stably express or constitutively express c-Myc, 209myc and DLD-1, respectively (Figure 1a and Supplementary Figure 2C). Importantly, both Miz-1 and c-Myc were present at WIF-1 promoter in these WIF-1 non-expressing cell lines (Figure 4b).
Figure 4.
Miz-1/c-Myc co-operate to silence WIF-1 promoter activity. (a) In all, 100 ng of an empty vector (EV), c-Myc or c-Myc 394D were co-transfected with WIF-1-C3, pRL-TK together with or without 10 ng of plasmid encoding full-length Miz-1. At 48 h after transfection, luciferase and renilla was measured. Luciferase values were normalized to renilla and are relative to their corresponding empty vector. (b) ChIP was carried out with Miz-1, c-Myc or an IgG control in 209myc and DLD-1 cells were. ChIP DNA was amplified with primers specific for WIF-1 promoter (–50/+ 120 bp).
Given that the Miz-1/c-Myc repressive complex was shown to physically interact with Dnmt3a at P21CIP1 promoter (Brenner et al., 2005), we next analyzed the contribution of de novo methylation to the c-Myc-mediated transient repression of WIF-1. Treatment with 5-aza-2′-deoxycitidine had no effect on c-Myc-mediated transient repression of Miz-1-driven WIF-1-C3 luciferase activity (Figure 5a). We also analyzed the effect of 5-aza-2′-deoxycitidine on WIF-1 expression and promoter hypermethylation in H209 and the stably derived c-Myc overexpressing cell line 209myc. WIF-1 was expressed in H209, lacking c-Myc expression, but fully repressed in 209myc cells (Figure 5b, upper panel). WIF-1 expression was restored in 209myc cells after treatment with 5-aza-2′-deoxycitidine (209myc AZA), showing that WIF-1 repression in 209myc is conferred by promoter hypermethylation. Bisulfite genomic sequencing showed that the –401/–350-bp region of WIF-1 promoter was methylated in both H209 and 209myc cells (Supplementary Figure 3A) while the –320/–72-bp region was heavily methylated only in 209myc (Figure 5b, lower panel). In particular, the increase in density of methylation of the –295 to –95 bp of WIF-1 promoter is highly similar to the methylation patterns observed in NSCLC cell lines not expressing WIF-1 (Figure 1c). The increase in methylation of –21/+209 bp region in 209myc (Supplementary Figure 3B) was less prominent. Finally, the re-expression of WIF-1 in 209myc AZA was associated with demethylation of the WIF-1 promoter (Figure 5b, lower panel and Supplementary Figure 3B).
Figure 5.
DNA methylation is involved in the stable but not transient repression of WIF-1. (a) The effect 5-aza-2′-deoxycitidine (48 h) or vehicle (dimethylsulphoxide (DMSO)) on c-Myc-mediated repression of Miz-1-driven WIF-1-C3 promoter activity was determined by luciferase assay. (b, upper panel) Expression of Wnt antagonist WIF-1 was determined by RT–PCR in H209, 209myc cells or 209myc cells treated with 5-aza-2′-deoxycitidine (209myc AZA) for 48 h. The expression of glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was also monitored in these cell lines as a control. (b, lower panel) Genomic bisulfite sequencing was determined for WIF-1 promoter region –320/–72 bp. Other regions (–401/–350 bp; –21/+ 209 bp) of WIF-1 promoter were also analyzed (Supplementary Figure 3A and 3B). c-Myc depletion has no effect on WIF-1 promoter methylation status or expression level. (c) Western blot analysis of c-Myc after the transient treatment (48, 96 and 120 h) of colorectal cancer cell line DLD-1 with c-Myc SMARTpool siRNA. The expression of p21CIP1, a known target of c-Myc repression and the loading control β-actin was also analyzed. (d) Real-time RT–PCR was used to quantify c-MYC and P21CIP levels in wt DLD-1, DLD-1 –/– as well as DLD-1 cells transiently treated for 24 or 120 h with c-Myc siRNA. c-MYC was not detected in the knockout DLD-1 –/– cells (*). WIF-1 expression was not detected in either DLD-1 wt, DLD-1 c-Myc siRNA or c-Myc –/– (data not shown). Fold change in expression is relative to GAPDH. The graph is on a log scale. (e) Bisulfite genomic sequencing showing that WIF-1 promoter (–320/–72 bp) remains fully methylated in the knockout DLD-1 c-Myc –/– cells.
Loss of c-Myc does not affect WIF-1 expression in DLD-1 cells
c-Myc was depleted through transient siRNA knockdown (Figure 5c) or somatic gene knockout in the colorectal cancer cell DLD-1 (DLD-1 c-Myc–/–) and the expression of WIF-1 and two other c-Myc targets, P21CIP1 and GADD45, analyzed. Although c-Myc depletion by siRNA was incomplete, we observed a 3.4- and 5.6-fold induction in p21CIP1 mRNA after c-Myc siRNA for 24 and 120 h, respectively (Figure 5d). Complete loss of c-Myc expression in c-Myc–/– resulted in a 47-fold increase in P21CIP1 transcript compared with parental DLD-1 (Figure 5d). GADD45, another gene repressed by c-Myc (Bush et al., 1998), showed a 30-fold increase in expression relative to wildtype DLD-1 cells (data not shown). However, WIF-1 expression remained undetectable by real-time reverse transcriptase (RT)–PCR and western blot analysis in DLD-1 cell treated with c-Myc siRNA for 24 or 120 h and in the complete genetic c-Myc knockout (DLD-1 c-Myc–/–) (data not shown). In addition, WIF-1 promoter remained heavily methylated in DLD-1 c-Myc–/– (Figure 5e) cells. This data therefore suggest that c-Myc might initiate but not maintain WIF-1 transcriptional silencing.
New targets of c-Myc-mediated DNA methylation-dependent gene silencing
Although the mode of repression of already known Miz-1/c-Myc targets seems to be transient and DNA methylation-independent, our data argue that the repression of some genes such as WIF-1 might be locked in place by DNA promoter hypermethylation. We therefore used a gene expression microarray to discover additional genes repressed by c-Myc and DNA promoter hypermethylation (Figure 6). 209myc was compared with parental H209 and to 209myc AZA, resulting in 185 Myc-repressed genes that could be reversed by DNA demethylation treatment (Figure 6a and Supplementary Table 3). Thirty-six were then validated for expression differences by gel-based and/or real-time RT–PCR (Figure 6b and data not shown), with many genes having only minimal changes in expression levels (Figure 6b, left panel). However, transforming growth factor (TGF)β-induced, DNAJA4 and TRIM59 were expressed in H209, silenced (that is, undetectable by gel-based RT–PCR) in 209myc and re-expressed in 209myc AZA (Figure 6b, right panel).
Figure 6.
Identification of novel genes repressed by c-Myc and DNA promoter hypermetylation. (a) Scatter plot showing the fold change in expression in 209myc AZA relative to 209myc (Y axis) and H209 relative to 209myc cells (X axis). Each spot on the scatter plot represents one probe on the microarray. Probes for which the signal decreased because of c-Myc and increased as a result of 5-aza-2′-azadeoxycitidine treatment (fold change of at least 1.41) are located in the box on the top right hand side of the scatter plot. DNAJA4, TRIM59 and TGFβi are highlighted. (b, left panel) Real-time RT–PCR analysis of 36/185 CpG island-containing genes. RHO and BCAM were also included as controls (both genes were in the original list of 264-candidate genes) but have no CpG islands. Data are expressed as fold change in expression (log scale) relative to glyceraldehyde 3-phosphate dehydrogenase (GAPDH). (b, right panel) Gel-based RT–PCR shows that DNAJA4, TRIM59 and TGFβi were expressed in H209, silenced in 209myc and re-expressed in 209myc AZA. SP5, DKK3, ITM2C are control genes that showed only mild change in the real-time RT–PCR. GAPDH was used as a control. A negative control in which no reverse transcriptase was used in the cDNA synthesis is included (no RT).
Although we were able to see individual genes repressed by c-Myc and reactivated by 5-aza-2′-deoxyciditine, WIF-1 was not detectable above background signal using the Agilent microarray at baseline or in 209myc AZA. This has previously been observed for other known silenced genes in cancer (Schuebel et al., 2007). Methylation-specific PCR (Figure 7a) and bisulfite sequencing (Figures 7b–d) demonstrated that all three genes were unmethylated in H209, methylated in 209myc and had reduced methylation in 209myc AZA. There was no evidence of promoter hypermethylation of control genes (TP53INP1, SP5, DKK3 and ITM2C) in 209myc cells (data not shown).
Figure 7.
Newly identified genes are silenced by DNA promoter hypermethylation. (a) Methylation-specific PCR (MSP) was used to assess the methylation status of DNAJA4, TRIM59 and TGFβi in cells lacking de novo methylation (DKO), an in vitro methylated DNA control (IVD), H209, 209myc and 209myc AZA as well as a water control. A signal in the U lane indicates an unmethylated promoter, a signal in the M lane identifies methylated promoter and a signal in both lanes suggests that the promoter is either hemimethylated or that a portion of the alleles are fully unmethylated while the rest is fully unmethylated. (b–d) Genomic bisulfite sequencing was employed to map out promoter methylation of CGs at DNAJA4, TRIM59 and TGFβi promoters in H209, 209myc and 209myc AZA cells. Arrows indicate the CGs, which were assessed by MSP analysis.
Discussion
Through a comprehensive bisulfite sequencing mapping of WIF-1 CpG island, we found a complex pattern of methylation in NSCLC not previously reported (Mazieres et al., 2004). In particular, DNA methylation at the WIF-1 promoter in some NSCLC cell lines remains incomplete compared with colorectal carcinoma (Taniguchi et al., 2005; Ai et al., 2006). This heterogeneous promoter hypermethylation defines a region within WIF-1 promoter spanning –295 to –95 bp relative to WIF-1 TSS as critical for complete transcriptional repression. Hence, while a previous study has suggested that a region 1.5 Kbp from the TSS was required for expression (Reguart et al., 2004), we found the WIF-1 proximal promoter spanning –436 to + 120 bp to have maximal activity. The critical role of this region is consistent with our mapping of promoter hypermethylation at WIF-1 promoter and histone marks revealing active or repressive chromatin associated with variable WIF-1 expression. The mosaic methylation pattern observed in NSCLC cell lines suggests that distinct regions of methylation within gene promoters exist and that such regions of methylation have functional significance. This might also be the case in breast cancer, where a similar methylation pattern was reported (Ai et al., 2006).
We found that Miz-1 is recruited to WIF-1 promoter where it activates WIF-1 transcription. This is particularly interesting given that Miz-1 expression suppresses growth of neuroblastoma cell lines in vitro (Ikegaki et al., 2007). It would be interesting to see whether this effect might be mediated through the increase in WIF-1 therefore resulting in a decrease in Wnt signaling activity, which contributes to neuroblastoma progression (Liu et al., 2008).
We next showed that Miz-1/c-Myc represses WIF-1 transcription. First, in a transient assay we found that c-Myc repression of WIF-1 is Miz-1-dependent but is independent of DNA promoter hypermethylation. The latter is further suggested by our preliminary data showing that Dnmt3a reduces Miz-1-driven WIF-1-C3 luciferase activity independently of its Dnmt activity (Supplementary Figure 3C). This is particularly interesting given the previously reported ability of another Dnmt, Dnmt1, to participate in transcriptional repression through interactions with HDAC2 (Rountree et al., 2000).
Second, we found that Miz-1/c-Myc are recruited to WIF-1 in cells stably overexpressing c-Myc (that is, 209myc) but also in cells constitutively expressing c-Myc through activated β-catenin/TCF (that is, DLD-1). The latter therefore suggests that WIF-1 repression by Miz-1/c-Myc might indeed reflect a more physiological mechanism. However, and in contrast to what we observed in our transient assay, WIF-1 repression in 209myc cells is accompanied by extensive promoter hypermethylation, particularly in the –295 to –95-bp region. Interestingly, c-Myc-mediated immortalization of human fibroblasts is associated with DNA hypermethylation and silencing of ARF, and is found in immortalized but not early-passage cells (Benanti et al., 2007).
The absence of WIF-1 re-expression or promoter demethylation after c-Myc depletion in DLD-1 suggests that c-Myc is involved in the initiation, but not the maintenance of WIF-1 silencing. This supports a model where recruitment of c-Myc to the WIF-1 promoter by Miz-1 might, if sustained, trigger epigenetic events leading to the irreversible silencing of WIF-1. These include the targeted hypermethylation of the critical region of the WIF-1 promoter, mediated by Dnmts (Ai et al., 2006) and possibly changes in chromatin conformation as well (data not shown) (Tiwari et al., 2008). Hence, similar to the way that c-Myc dynamically relocates to preassembled transcription factories (Osborne et al., 2007), one could hypothesize that c-Myc might also be involved at repressive factories, which might preferentially locate to the nuclear periphery (Finlan et al., 2008), where the methyl-CpG-binding protein MeCP2 and the inner nuclear membrane protein LBR interact (Guarda et al., 2009).
Through genomic studies, we find that this process is likely not limited to WIF-1. Indeed we identified additional genes repressed by c-Myc and promoter hypermethylation include TGFβ-induced (TGFβi), the heat shock protein DNAJA4 and the RING finger 1-like TRIM59. The fact that WIF-1 was not identified in this screen suggests that there are likely additional Myc-repressed genes subsequently silenced by DNA methylation, which may have also been missed using this screening strategy.
Preliminary analysis suggests that the consensus Inr sequence (YYAN(T/A)YY), a candidate for Miz-1 binding, might be found at the promoter of TGFβi, TRIM59, DNAJA4 and WIF-1 which, if confirmed, would further suggest that Miz-1 is indeed a bona fide transcriptional regulator of these genes (Smale and Baltimore, 1989; Peukert et al., 1997; Bowen et al., 2002). These genes may have important roles in carcinogenesis. For example, given the prominent role of ubiquitylation in Wnt signaling, it would be interesting to further characterize the biological significance of c-Myc transcriptional repression on TRIM59 in this context. Loss of TGFβi was recently shown to predis-pose mice to tumor development (Zhang et al., 2009), and TGFβi downregulation is associated with promoter hypermethylation in lung and prostate cancer (Shah et al., 2008) as well as in MLL rearrangement leukemia (Li et al., 2009). Finally, TGFβi also appears to be a marker for drug sensitivity for paclitaxel in ovarian cancer cells (Ahmed et al., 2007) and decitabine in melanoma cells (Halaban et al., 2009).
Materials and methods
Cell culture
NSCLC and colorectal cancer cell lines used were obtained from ATCC (ATCC, Manassas, VA, USA). DKO, a double knockout for DNMT3b and DNMT1 has been previously described (Rhee et al., 2002). Normal human bronchoepithelial cell line was purchased from Cambrex Bio Science and grown in the Bullet kit media (Cambrex Bio Science, Walkersville, MD, USA). The DLD-1 homozygote (c-Myc–/–) DNA and cDNA were a gift from Dr Kurt E Bachman. The H209 and 209myc cell lines have been previously described (Barr et al., 2000). Treatment of cells with 2 μm of 5-aza-2′-deoxycitidine (Sigma-Aldrich, St Louis, MO, USA), DNA, RNA lands whole cell protein lysate extractions were carried out as previously described (Licchesi et al., 2008).
Expression constructs
PCDNA3-c-Myc (wt), a Myc mutant that abolishes binding of c-Myc to Miz-1 (c-Myc V394D) and PCDNA3-Miz-1 were kindly provided by Dr Martin Eilers (University of Marburg, Germany). PCDNA3.1-Myc-Dnmt3a was obtained from Dr Riggs AD (Beckman Research Institute, Duarte, CA, USA). Dnmt3a was subcloned in pCMV-3xFLAG (Stratagene, La Jolla, CA, USA) (Li et al., 2006). A mutant version of this construct, targeting a catalytic cysteine important for its DNMTase activity (Chen et al., 2005), was also made (pCMV-3xFLAG-Dnmt3a-C487W).
WIF-1 luciferase constructs
Each of the five constructs engineered had the same 3′ end, ending at +120 bp from the TSS. Construct WIF-1-C1 spanned the –1391 to +120 bp; construct WIF-1-C2, –929 to +120; construct WIF-1-C3, –436 to +120; construct WIF-1-C4, –348 to +120 and construct WIF-1-C5, –50 to +120. All distance are relative to the TSS. It is to be noted that–construct WIF-1-C1 covers the same region of WIF-1 promoter as WIF-1 construct 5 described by others (Reguart et al., 2004). Restriction sites NheI (5′) and HindIII (3′) were used to clone WIF-1 promoters constructs into the pGL3 basic vector (Promega, Madison, WI, USA). The full-length P21CIP1 luciferase construct was kindly provided by Dr Bert Vogelstein (Johns Hopkins University, Baltimore, MD, USA).
Western blot
Primary antibodies used include c-Myc (N262), Miz-1 (H190), Wif-1 (K16), (Santa Cruz Biotechnology, Santa Cruz, CA, USA), p21CiP1 (SXM30) (BD Biosciences, San Jose, CA, USA) and β-actin (A5441) (Sigma-Aldrich). The enhanced chemiluminescence or enhanced chemiluminescence femto were used as detection reagents (Pierce Biotechnology, Rockford, IL, USA).
Gel-based RT–PCR
RT–PCR primers for WIF-1 and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) have been previously described (Mazieres et al., 2004; Licchesi et al., 2008). Primers for genes discovered through microarray experiments and for c-MYC and P21CIP1 were designed with DNASTAR software (DNASTAR, Inc., Madison, WI, USA) (Supplementary Table 1). Real time RT-PCR: real-time RT–PCR was carried out in triplicate in 20 μl reactions containing 10 × PCR buffer, 10 μm dNTPs, 10 pmol of each of the primer, 0.1 unit of Hot Start DNA polymerase, 1 μl of Syber Green (Molecular Probes, Carlsbad, CA, USA) and 0.6 μl of fluorescein (Bio-Rad, Hercules, CA, USA) in an iCycler Optical Module (Bio-Rad). Data are reported as relative expression (1/(CtWIF-1-CtGAPDH) (Figure 1a) or as the relative fold change in expression and is equal to 2–(Sample1 ΔCt – Sample2 ΔCt), where ΔCt = average Ct (gene) – average Ct (GAPDH) (Figure 6b). Taqman RT–PCR: Taqman RT–PCR was carried out with the ABI7900 Taqman thermocycler (Applied Biosystems, Foster City, CA, USA). Taqman gene expression assays for WIF-1 (Hs00183662) (Applied Biosystems), and Tata box-binding protein were used (de la Roche et al., 2008). Values for gene expression in each case were calculated relative to a standard curve of Tata box-binding protein expression.
Chromatin immunoprecipitation
ChIP for Miz-1 and c-Myc was performed using a dual cross-linking ChIP protocol as previously described (Perini et al., 2005). H1299 transfected with 2 μg/T75 flask of empty vector or Miz-1 for 24 h, 209myc and DLD-1 cells were cross-linked with disuccinimidyl glutarate (2 μm final concentration) for 45 min at room temperature followed by 15 min with formaldehyde (1% final concentration). Cross-linking was stop with glycine (125 μm final concentration). After washing, samples were sonicated, centrifuged and the supernatant was pre-cleared for 2 h with Dynabeads protein G (Invitrogen Corporation, Carlsbad, CA, USA). In all, 50 μg of chromatin were used for each ChIP condition and incubated overnight with either 5 μg/ml of Miz-1 (H190) or Myc (N262) antibody (Santa Cruz Biotechnology). An immunoglobulin G control was also carried out as a negative control. The following day complexes were captured with magnetic beads, washed several times and the cross-linking was reversed overnight at 65 °C. DNA was then precipitated and resuspended in 50 μl of water. Input was diluted 1/100 and PCR was performed using the following condition. WIF-1 (–50/+ 120; annealing 69 °C, 33 cycles). Finally, PCR products were resolved on 2% agarose gel and stained with ethidium bromide.
Real-time ChIP
Real-time ChIP was carried out as previously described (McGarvey et al., 2006). Antibodies against H3K4-me2 and H3K27me3 were purchased from Upstate (Millipore, MA, USA). ChIP primers for WIF-1 region –836/–608 region and –250/–50 were manually designed (Supplementary Table 1). Amplifications were performed in triplicate, and the enrichment was determined compared with input. Data were presented normalized to the value obtained for the H838 cell line (–836/–608 region).
Luciferase assay
293 cells were seeded in 24-well dish 24 h before transfection. In all, 200 ng of WIF-1 deletion construct (WIF-1-C1 to C5), pGL3-control vector or pGL3-basic vector constructs was cotransfected with 10 ng of pRL-TK vector using lipofectamine 2000 (Invitrogen). For co-transfection, various quantity of Miz-1 (1, 10, 100 ng), c-Myc (1, 10, 100 ng), Dnmts (1, 10, 50 ng) were used. In each experiment, the amount of plasmid transfected was kept constant by adding empty vector, and each data point was done is triplicate. Transfections were carried out for 24–48 h. Luciferase and renilla activity were determined using dual-luciferase (Promega) and is shown relative to control, which was set to 1.
In vitro DNA methylation
Construct pGL3-C3 was in vitro methylated as described (Yu et al., 2005). 200 ng of mock or SSI-treated vector were transfected in 293T cells. After 48 h, luciferase and renilla activity were assayed as described above.
Methylation analysis
Bisulfite modification, methylation-specific PCR (Licchesi et al., 2008) and bisulfite sequencing (Zinn et al., 2007) were carried out as previously described. For cloning experiments, 5–10 clones were sequenced at the Johns Hopkins Core sequencing facility. Primers used for methylation-specific PCR and bisulfite sequencing were manually designed (Supplementary Table 1).
Transient knockdown
c-Myc (MYC), Miz-1 (ZBTB17) SMARTpool ON-TARGET-plus siRNA or control siRNA (Dharmacon Inc., Chicago, IL, USA) were transfected with lipofectamine 2000. Samples were harvested at 24, 48 or 72 h. For the time course c-Myc siRNA in DLD-1 cells, transfection with siRNA was repeated at 48 and 96 h. Protein and RNA were extracted at 24, 48, 96 and 120 h after the first transfection and analyzed for c-MYC and P21CIP1 expression by western blot and quantitative real-time RT–PCR or Taqman PCR.
Microarray
The experimental procedure was as previously described (Schuebel et al., 2007). 209myc 5-aza-2′-deoxycitidine (209myc AZA) and 209myc mock samples were co-hybridized on a single array (Human 44K Agilent Technologies array platform, Agilent Technologies Inc, Santa Clara, CA, USA), as were H209 and 209myc mock. Data analysis identified 324 unique probes which had both at least a 1.41-fold increase in the H209 vs 209myc and at least a 1.41-fold increase in 209myc AZA vs 209myc (Supplementary Table 2). In total, 264 out of 324 (81%) of the probes either represented individual genes or had an annotation that could be confirmed by using a modified BLAST algorithm to identify short (nearly) identical matches and map all the probes on a transcriptome database as downloaded from EnsEMBL release 43 http://www.ensembl.org, European Bioinformatics Institute, Hinxton, UK). In all, 185 out of 264 (70%) of these genes were likely to have a CpG island in the upstream region of the promoter (–1 Kbp up to +200 bp from the putative TSS) (Supplementary Table 3).
Supplementary Material
Acknowledgements
We acknowledge Dr Martin Eilers (Marbourg University, Germany), Dr Arthur Riggs (Beckman Research Institute, Duarte, CA, USA) and Dr Bert Vogelstein (Johns Hopkins School of Medicine, USA) for plasmids, Dr Kurt Bachman (University of Maryland, USA) for the DLD-1 c-Myc –/– DNA and cDNA. We are grateful to Dr Wayne Yu at the Johns Hopkins Microarray Core facility, Dr Kornel Schuebel for his help with the microarray experiment and Dr Mariann Bienz for her support. Finally, we thank all the members of the Baylin/Herman laboratory for useful discussions and Kathy Bender for administrative assistance. This study was supported by NCI/SPORE grant CA058184 to James G Herman, AICR grant 07-0040 to Mariann Bienz (MRC-LMB) and NCI grant P30 CA06973-44 to Leslie Cope.
Abbreviations
- ChIP
chromatin immunoprecipitation
- MSP
methylationspecific PCR
- NSCLC
non-small cell lung cancer
- TSS
transcription start site
- WIF-1
Wnt inhibitory factor-1
Footnotes
Conflict of interest
The commercial rights to the MSP technique belong to OncoMethylome Sciences. SBB and JGH serve as consultants to OncoMethylome Sciences and are entitled to royalties from any commercial use of this procedure. SBB and JGH also receive research support from OncoMethylome Sciences. The terms of this arrangement are being managed by the Johns Hopkins University in accordance with its conflict of interest policies.
Supplementary Information accompanies the paper on the Oncogene website (http://www.nature.com/onc)
References
- Ahmed AA, Mills AD, Ibrahim AE, Temple J, Blenkiron C, Vias M, et al. The extracellular matrix protein TGFBI induces microtubule stabilization and sensitizes ovarian cancers to paclitaxel. Cancer Cell. 2007;12:514–527. doi: 10.1016/j.ccr.2007.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ai L, Tao Q, Zhong S, Fields CR, Kim WJ, Lee MW, et al. Inactivation of Wnt inhibitory factor-1 (WIF1) expression by epigenetic silencing is a common event in breast cancer. Carcino-genesis. 2006;27:1341–1348. doi: 10.1093/carcin/bgi379. [DOI] [PubMed] [Google Scholar]
- Barr LF, Campbell SE, Diette GB, Gabrielson EW, Kim S, Shim H, et al. c-Myc suppresses the tumorigenicity of lung cancer cells and down-regulates vascular endothelial growth factor expression. Cancer Res. 2000;60:143–149. [PubMed] [Google Scholar]
- Benanti JA, Wang ML, Myers HE, Robinson KL, Grandori C, Galloway DA. Epigenetic down-regulation of ARF expression is a selection step in immortalization of human fibroblasts by c-Myc. Mol Cancer Res. 2007;5:1181–1189. doi: 10.1158/1541-7786.MCR-06-0372. [DOI] [PubMed] [Google Scholar]
- Bowen H, Biggs TE, Phillips E, Baker ST, Perry VH, Mann DA, et al. c-Myc represses and Miz-1 activates the murine natural resistance-associated protein 1 promoter. J Biol Chem. 2002;277:34997–35006. doi: 10.1074/jbc.M204232200. [DOI] [PubMed] [Google Scholar]
- Brenner C, Deplus R, Didelot C, Loriot A, Vire E, De Smet C, et al. Myc represses transcription through recruitment of DNA methyltransferase corepressor. EMBO J. 2005;24:336–346. doi: 10.1038/sj.emboj.7600509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush A, Mateyak M, Dugan K, Obaya A, Adachi S, Sedivy J, et al. c-myc null cells misregulate cad and gadd45 but not other proposed c-Myc targets. Genes Dev. 1998;12:3797–3802. doi: 10.1101/gad.12.24.3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan SL, Cui Y, van Hasselt A, Li H, Srivastava G, Jin H, et al. The tumor suppressor Wnt inhibitory factor 1 is frequently methylated in nasopharyngeal and esophageal carcinomas. Lab Invest. 2007;87:644–650. doi: 10.1038/labinvest.3700547. [DOI] [PubMed] [Google Scholar]
- Chen ZX, Mann JR, Hsieh CL, Riggs AD, Chedin F. Physical and functional interactions between the human DNMT3L protein and members of the de novo methyltransferase family. J Cell Biochem. 2005;95:902–917. doi: 10.1002/jcb.20447. [DOI] [PubMed] [Google Scholar]
- Cowling VH, D'Cruz CM, Chodosh LA, Cole MD. c-Myc transforms human mammary epithelial cells through repression of the Wnt Inhibitors DKK1 and SFRP1. Mol Cell Biol. 2007;27:5135–5146. doi: 10.1128/MCB.02282-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Roche M, Worm J, Bienz M. The function of BCL9 in Wnt/beta-catenin signaling and colorectal cancer cells. BMC Cancer. 2008;8:199. doi: 10.1186/1471-2407-8-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Croce L, Raker VA, Corsaro M, Fazi F, Fanelli M, Faretta M, et al. Methyltransferase recruitment and DNA hypermethylation of target promoters by an oncogenic transcription factor. Science. 2002;295:1079–1082. doi: 10.1126/science.1065173. [DOI] [PubMed] [Google Scholar]
- Finlan LE, Sproul D, Thomson I, Boyle S, Kerr E, Perry P, et al. Recruitment to the nuclear periphery can alter expression of genes in human cells. PLoS Genet. 2008;4:e1000039. doi: 10.1371/journal.pgen.1000039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gartel AL, Ye X, Goufman E, Shianov P, Hay N, Najmabadi F, et al. Myc represses the p21(WAF1/CIP1) promoter and interacts with Sp1/Sp3. Proc Natl Acad Sci USA. 2001;98:4510–4515. doi: 10.1073/pnas.081074898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebhardt A, Frye M, Herold S, Benitah SA, Braun K, Samans B, et al. Myc regulates keratinocyte adhesion and differentiation via complex formation with Miz1. J Cell Biol. 2006;172:139–149. doi: 10.1083/jcb.200506057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarda A, Bolognese F, Bonapace IM, Badaracco G. Interaction between the inner nuclear membrane lamin b receptor and the heterochromatic methyl binding protein, MeCP2. Exp Cell Res. 2009;315:18903. doi: 10.1016/j.yexcr.2009.01.019. [DOI] [PubMed] [Google Scholar]
- Gujral TS, MacBeath G. Emerging miniaturized proteomic technologies to study cell signaling in clinical samples. Sci Signal. 2009;2:pe65. doi: 10.1126/scisignal.293pe64. [DOI] [PubMed] [Google Scholar]
- Halaban R, Krauthammer M, Pelizzola M, Cheng E, Kovacs D, Sznol M, et al. Integrative analysis of epigenetic modulation in melanoma cell response to decitabine: clinical implications. PLoS One. 2009;4:e4563. doi: 10.1371/journal.pone.0004563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He TC, Sparks AB, Rago C, Hermeking H, Zawel L, da Costa LT, et al. Identification of c-MYC as a target of the APC pathway. Science. 1998;281:1509–1512. doi: 10.1126/science.281.5382.1509. [DOI] [PubMed] [Google Scholar]
- Herman JG, Baylin SB. Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med. 2003;349:2042–2054. doi: 10.1056/NEJMra023075. [DOI] [PubMed] [Google Scholar]
- Hsieh JC, Kodjabachian L, Rebbert ML, Rattner A, Smallwood PM, Samos CH, et al. A new secreted protein that binds to Wnt proteins and inhibits their activities. Nature. 1999;398:431–436. doi: 10.1038/18899. [DOI] [PubMed] [Google Scholar]
- Ikegaki N, Gotoh T, Kung B, Riceberg JS, Kim DY, Zhao H, et al. De novo identification of MIZ-1 (ZBTB17) encoding a MYC-interacting zinc-finger protein as a new favorable neuroblastoma gene. Clin Cancer Res. 2007;13:6001–6009. doi: 10.1158/1078-0432.CCR-07-0071. [DOI] [PubMed] [Google Scholar]
- Li H, Rauch T, Chen ZX, Szabo PE, Riggs AD, Pfeifer GP. The histone methyltransferase SETDB1 and the DNA methyltransferase DNMT3A interact directly and localize to promoters silenced in cancer cells. J Biol Chem. 2006;281:19489–19500. doi: 10.1074/jbc.M513249200. [DOI] [PubMed] [Google Scholar]
- Li Z, Luo RT, Mi S, Sun M, Chen P, Bao J, et al. Consistent deregulation of gene expression between human and murine MLL rearrangement leukemias. Cancer Res. 2009;69:1109–1116. doi: 10.1158/0008-5472.CAN-08-3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licchesi JD, Westra WH, Hooker CM, Machida EO, Baylin SB, Herman JG. Epigenetic alteration of Wnt pathway antagonists in progressive glandular neoplasia of the lung. Carcinogenesis. 2008;29:895–904. doi: 10.1093/carcin/bgn017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Mazanek P, Dam V, Wang Q, Zhao H, Guo R, et al. Deregulated Wnt/beta-catenin program in high-risk neuroblastomas without MYCN amplification. Oncogene. 2008;27:1478–1488. doi: 10.1038/sj.onc.1210769. [DOI] [PubMed] [Google Scholar]
- Mazieres J, He B, You L, Xu Z, Lee AY, Mikami I, et al. Wnt inhibitory factor-1 is silenced by promoter hypermethylation in human lung cancer. Cancer Res. 2004;64:4717–4720. doi: 10.1158/0008-5472.CAN-04-1389. [DOI] [PubMed] [Google Scholar]
- McGarvey KM, Fahrner JA, Greene E, Martens J, Jenuwein T, Baylin SB. Silenced tumor suppressor genes reactivated by DNA demethylation do not return to a fully euchromatic chromatin state. Cancer Res. 2006;66:3541–3549. doi: 10.1158/0008-5472.CAN-05-2481. [DOI] [PubMed] [Google Scholar]
- Osborne CS, Chakalova L, Mitchell JA, Horton A, Wood AL, Bolland DJ, et al. Myc dynamically and preferentially relocates to a transcription factory occupied by Igh. PLoS Biol. 2007;5:e192. doi: 10.1371/journal.pbio.0050192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel JH, McMahon SB. Targeting of Miz-1 is essential for Myc-mediated apoptosis. J Biol Chem. 2006;281:3283–3289. doi: 10.1074/jbc.M513038200. [DOI] [PubMed] [Google Scholar]
- Patel JH, McMahon SB. BCL2 is a downstream effector of MIZ-1 essential for blocking c-MYC-induced apoptosis. J Biol Chem. 2007;282:5–13. doi: 10.1074/jbc.M609138200. [DOI] [PubMed] [Google Scholar]
- Perini G, Diolaiti D, Porro A, Della Valle G. In vivo transcriptional regulation of N-Myc target genes is controlled by E-box methylation. Proc Natl Acad Sci USA. 2005;102:12117–12122. doi: 10.1073/pnas.0409097102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peukert K, Staller P, Schneider A, Carmichael G, Hanel F, Eilers M. An alternative pathway for gene regulation by Myc. EMBO J. 1997;16:5672–5686. doi: 10.1093/emboj/16.18.5672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reguart N, He B, Xu Z, You L, Lee AY, Mazieres J, et al. Cloning and characterization of the promoter of human Wnt inhibitory factor-1. Biochem Biophys Res Commun. 2004;323:229–234. doi: 10.1016/j.bbrc.2004.08.075. [DOI] [PubMed] [Google Scholar]
- Rhee I, Bachman KE, Park BH, Jair KW, Yen RW, Schuebel KE, et al. DNMT1 and DNMT3b cooperate to silence genes in human cancer cells. Nature. 2002;416:552–556. doi: 10.1038/416552a. [DOI] [PubMed] [Google Scholar]
- Rountree MR, Bachman KE, Baylin SB. DNMT1 binds HDAC2 and a new co-repressor, DMAP1, to form a complex at replication foci. Nat Genet. 2000;25:269–277. doi: 10.1038/77023. [DOI] [PubMed] [Google Scholar]
- Schuebel KE, Chen W, Cope L, Glockner SC, Suzuki H, Yi JM, et al. Comparing the DNA hypermethylome with gene mutations in human colorectal cancer. PLoS Genet. 2007;3:1709–1723. doi: 10.1371/journal.pgen.0030157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seoane J, Pouponnot C, Staller P, Schader M, Eilers M, Massague J. TGFbeta influences Myc, Miz-1 and Smad to control the CDK inhibitor p15INK4b. Nat Cell Biol. 2001;3:400–408. doi: 10.1038/35070086. [DOI] [PubMed] [Google Scholar]
- Seoane J, Le HV, Massague J. Myc suppression of the p21(Cip1) Cdk inhibitor influences the outcome of the p53 response to DNA damage. Nature. 2002;419:729–734. doi: 10.1038/nature01119. [DOI] [PubMed] [Google Scholar]
- Shah JN, Shao G, Hei TK, Zhao Y. Methylation screening of the TGFBI promoter in human lung and prostate cancer by methylation-specific PCR. BMC Cancer. 2008;8:284. doi: 10.1186/1471-2407-8-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smale ST, Baltimore D. The ‘initiator’ as a transcription control element. Cell. 1989;57:103–113. doi: 10.1016/0092-8674(89)90176-1. [DOI] [PubMed] [Google Scholar]
- Staller P, Peukert K, Kiermaier A, Seoane J, Lukas J, Karsunky H, et al. Repression of p15INK4b expression by Myc through association with Miz-1. Nat Cell Biol. 2001;3:392–399. doi: 10.1038/35070076. [DOI] [PubMed] [Google Scholar]
- Tang Y, Simoneau AR, Liao WX, Yi G, Hope C, Liu F, et al. WIF1, a Wnt pathway inhibitor, regulates SKP2 and c-myc expression leading to G1 arrest and growth inhibition of human invasive urinary bladder cancer cells. Mol Cancer Ther. 2009;8:458–468. doi: 10.1158/1535-7163.MCT-08-0885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi H, Yamamoto H, Hirata T, Miyamoto N, Oki M, Nosho K, et al. Frequent epigenetic inactivation of Wnt inhibitory factor-1 in human gastrointestinal cancers. Oncogene. 2005;24:7946–7952. doi: 10.1038/sj.onc.1208910. [DOI] [PubMed] [Google Scholar]
- Tiwari VK, McGarvey KM, Licchesi JD, Ohm JE, Herman JG, Schubeler D, et al. PcG proteins, DNA methylation, and gene repression by chromatin looping. PLoS Biol. 2008;6:2911–2927. doi: 10.1371/journal.pbio.0060306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urakami S, Shiina H, Enokida H, Kawakami T, Tokizane T, Ogishima T, et al. Epigenetic inactivation of Wnt inhibitory factor-1 plays an important role in bladder cancer through aberrant canonical Wnt/beta-catenin signaling pathway. Clin Cancer Res. 2006;12:383–391. doi: 10.1158/1078-0432.CCR-05-1344. [DOI] [PubMed] [Google Scholar]
- van de Wetering M, Sancho E, Verweij C, de Lau W, Oving I, Hurlstone A, et al. The beta-catenin/TCF-4 complex imposes a crypt progenitor phenotype on colorectal cancer cells. Cell. 2002;111:241–250. doi: 10.1016/s0092-8674(02)01014-0. [DOI] [PubMed] [Google Scholar]
- Villa R, Pasini D, Gutierrez A, Morey L, Occhionorelli M, Vire E, et al. Role of the polycomb repressive complex 2 in acute promyelocytic leukemia. Cancer Cell. 2007;11:513–525. doi: 10.1016/j.ccr.2007.04.009. [DOI] [PubMed] [Google Scholar]
- Wu S, Cetinkaya C, Munoz-Alonso MJ, von der Lehr N, Bahram F, Beuger V, et al. Myc represses differentiation-induced p21CIP1 expression via Miz-1-dependent interaction with the p21 core promoter. Oncogene. 2003;22:351–360. doi: 10.1038/sj.onc.1206145. [DOI] [PubMed] [Google Scholar]
- Yu L, Liu C, Vandeusen J, Becknell B, Dai Z, Wu YZ, et al. Global assessment of promoter methylation in a mouse model of cancer identifies ID4 as a putative tumor-suppressor gene in human leukemia. Nat Genet. 2005;37:265–274. doi: 10.1038/ng1521. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Wen G, Shao G, Wang C, Lin C, Fang H, et al. TGFBI deficiency predisposes mice to spontaneous tumor development. Cancer Res. 2009;69:37–44. doi: 10.1158/0008-5472.CAN-08-1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinn RL, Pruitt K, Eguchi S, Baylin SB, Herman JG. hTERT is expressed in cancer cell lines despite promoter DNA methylation by preservation of unmethylated DNA and active chromatin around the transcription start site. Cancer Res. 2007;67:194–201. doi: 10.1158/0008-5472.CAN-06-3396. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.