Abstract
Functional assays have long played a key role in measuring of immunogenicity of a given vaccine. This is conventionally expressed as serum bactericidal titers. Studies of serum bactericidal titers in response to childhood vaccines have enabled us to develop and validate cut-off levels for protective immune responses and such cut-offs are in routine use. No such assays have been taken forward into the routine assessment of vaccines that induce primarily cell-mediated immunity in the form of effector T cell responses, such as TB vaccines. In the animal model, the performance of a given vaccine candidate is routinely evaluated in standardized bactericidal assays, and all current novel TB-vaccine candidates have been subjected to this step in their evaluation prior to phase 1 human trials. The assessment of immunogenicity and therefore likelihood of protective efficacy of novel anti-TB vaccines should ideally undergo a similar step-wise evaluation in the human models now, including measurements in bactericidal assays.
Bactericidal assays in the context of tuberculosis vaccine research are already well established in the animal models, where they are applied to screen potentially promising vaccine candidates. Reduction of bacterial load in various organs functions as the main read-out of immunogenicity. However, no such assays have been incorporated into clinical trials for novel anti-TB vaccines to date.
Although there is still uncertainty about the exact mechanisms that lead to killing of mycobacteria inside human macrophages, the interaction of macrophages and T cells with mycobacteria is clearly required. The assay described in this paper represents a novel generation of bactericidal assays that enables studies of such key cellular components with all other cellular and humoral factors present in whole blood without making assumptions about their relative individual contribution. The assay described by our group uses small volumes of whole blood and has already been employed in studies of adults and children in TB-endemic settings. We have shown immunogenicity of the BCG vaccine, increased growth of mycobacteria in HIV-positive patients, as well as the effect of anti-retroviral therapy and Vitamin D on mycobacterial survival in vitro. Here we summarise the methodology, and present our reproducibility data using this relatively simple, low-cost and field-friendly model.
Note: Definitions/Abbreviations
BCG lux = M. bovis BCG, Montreal strain, transformed with shuttle plasmid pSMT1 carrying the luxAB genes from Vibrio harveyi, under the control of the mycobacterial GroEL (hsp60) promoter. CFU = Colony Forming Unit (a measure of mycobacterial viability).
Protocol
1. Preparation of BCG lux reporter mycobacteria stock
Grow BCG lux (M. bovis BCG Montreal strain transformed with the shuttle plasmid pSMT1) with shaking at 200rpm to mid-logarithmic phase in Middlebrook 7H9 broth containing 0.2% glycerol, 0.05% tween 80 and 10% ADC enrichment.
Prepare 1ml of a 1:10 dilution of BCG lux culture in sterile PBS in a luminometer tube (900μl PBS + 100μl BCG lux culture) in duplicate.
Load tube containing N-decyl aldehyde (luciferase enzyme substrate) onto the back of the machine and ensure the lid is secured and tubes in place.
Prime luminometer injector with substrate via the prime program, set to inject 100 ucl x 3 into each of 3 empty priming tubes placed in the luminometer.
Load the 2 tubes into the luminometer and take readings via a program set at injecting 100 ucl of substrate into each tube and read for 20 sec in 1 sec intervals.
When the stock has reached 1x108 RLU/ml to 2x108 RLU/ml (this takes about 3-4 days of growth), add an equal volume of sterile 30% glycerol to the culture in a 50ml falcon tube and gently mix.
Aliquot 1.5ml volumes into labeled 2ml screw-cap microtube and store at -80°C.
2. Determining stock RLU/CFU correlation and contents of aliquots
Add 15ml of Middlebrook 7H9 culture medium with 10% ADC supplement to a 200ml Erlenmeyer flask with a vented cap.
Add 15μl of hygromycin and 30μl of 20% Tween.
Remove a vial of BCG lux from the freezer and defrost at room temperature (RT) in the safety cabinet. Add the contents of the vial to the medium and tighten the cap.
Incubate with shaking at 200rpm at 37°C for 4 days.
On each day, make up serial 10-fold dilutions (1:10, 1:100, 1:1000, 1:10,000 and 1:100,000) for CFU determination.
Set up the same dilutions in parallel and duplicate to determine luminescence (see part 1.2-1.6).
Prepare two 3-compartment plates of Middlebrook 7H11 agar (1 l of liquid medium containing 0.5% glycerol, 10% OADC supplement, 1 ml of 20% Tween and 1 ml of hygromycin).
Plate out 100μl of each dilution onto a segment of each plate using separate spreaders, distributing the liquid equally across the individual chamber.
Seal each plate with parafilm, place in a sterile plastic bag, seal with autoclave tape, and incubate at 37°C for 2 weeks with lids facing down.
Inspect regularly until CFUs appear (2-3 weeks).
To count colonies, remove the plates from the incubator and place on a colony counter. Calculate the mean for each dilution from the duplicate plates.
Calculate the RLU/CFU ratio using equivalent RLU and CFU counts for each dilution. The ratio should be between 3 and 5 RLU/CFU.
3. Preparation of BCG lux culture for inoculation into whole blood
Add 15ml of Middlebrook 7H9 culture medium with 10% ADC supplement to a 200ml Erlenmeyer flask with a vented cap.
Add 15μl of hygromycin and 30μl of 20% Tween.
Remove a vial of BCG lux from the freezer and defrost at RT in the safety cabinet. Add the contents of the vial to the medium and tighten the cap.
Incubate with shaking at 200rpm at 37°C for 2 to 4 days.
Prepare 1ml of a 1:10 dilution of BCG lux culture in sterile PBS in a luminometer tube (900μl PBS + 100μl BCG lux culture) in duplicate.
Prime luminometer with substrate (as described above), gently vortex the 2 tubes and load into the luminometer and take readings (as described above).
Use this reading to dilute down the culture in PBS to give an equivalent of 7x106 RLU (this will give an inoculum of about 2x105 CFU/ml blood and a ratio of BCG to monocytes of about 1:1, assuming a monocyte count of about 2x105 to 4x105 monocytes per ml of blood).
4. Preparation of whole blood
Take 3 to 5ml of venous blood in a tube containing preservative-free heparin.
Transfer the blood to a 50ml falcon tube and dilute with an equal volume of RPMI 1640 containing glutamine and 25mM HEPES (no pen/strep).
Aliquot 900μl of diluted blood in triplicate into sterile bijou tubes for each time point (6 tubes in total: 3 for 0hrs and 3 for 96hrs. Set up additional tubes for additional timepoints, if necessary.
Aliquot 900μl of Middlebrooks 7H9 culture medium with 10% ADC supplement + 50μg/ml hygromycin (the same medium as used to culture the BCG lux) in duplicate for the growth controls.
Add 100μl of the diluted BCG lux to each tube of blood, and to the 2 growth control tubes.
Mix well and place the 3 bijous for the 96hr time-point and the 2 growth controls on their side in the rocking incubator at 37°C, 20 rev/min (CO2 not required).
5. Measuring luminescence (at 0hr and 96hrs)
Centrifuge the 3 bijous at 2000g for 10 min.
Carefully remove 300μl of supernatant without disturbing the pellet and place in 2ml screw-cap microtubes to freeze at -80°C for subsequent cytokine analysis.
Add 300μl of PBS to each bijou to replace the volume of supernatant removed.
Aspirate the contents of each bijou into the corresponding 50ml falcon tube and add 8ml distilled water to each tube. Begin a timer for 10 min. Nb: This is a time-sensitive step, and this incubation must not be any longer than 10 min, beginning from when the cells first come into contact with water.
Rinse each bijou with 2ml of distilled water and vortex for 5 seconds before tipping into the corresponding falcon tube.
At the end of the 10 minute incubation, centrifuge the falcon tubes at 2000g for 10 minutes.
Decant supernatant into Surfanios disinfectant.
Add a few glass beads to each pellet and vortex.
Add 1ml of sterile PBS to each tube and vortex.
In luminometer tubes, make 1:10 dilutions for each sample in duplicate with PBS (900μl PBS + 100μl sample) and at the 96hr time-point. Make the same dilutions for each of the growth controls.
Gently vortex the tubes and load into the luminometer and take readings (as described above) Nb: Readings between triplicates of the same sample should be within 15% of one another.
6. Representative results:
It is important to use bacteria in logarithmic phase of growth for the whole blood lux assays, i.e. grown over 48-72 hours prior to inoculating the samples, as metabolic activity is suboptimal if either straight out of the freezer or in stationary phase. The prior growth should be standardized for a series of experiments to either 48 or 72 hours to avoid variability. Figure 1 shows the growth curve of a representative culture of BCG lux. The doubling time is about 24 hours until stationary phase is reached.
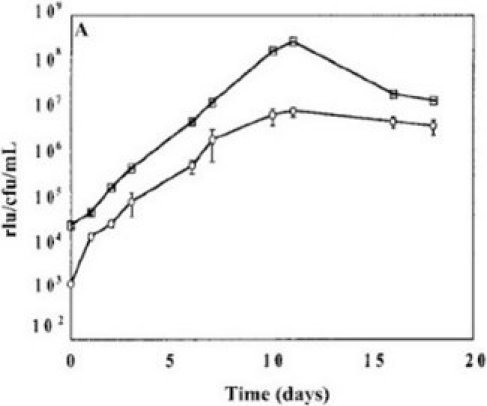 Figure 1. Representative growth curve of BCG lux over 96 hours in 7H9 medium and correlation of RLU and CFU.
Figure 1. Representative growth curve of BCG lux over 96 hours in 7H9 medium and correlation of RLU and CFU.
Although RLU values always correlate with CFU, the number of RLU corresponding to a single CFU can vary depending on the stock, which is why it is good practice to use the same frozen stock throughout a series of experiments to guarantee a consistent multiplicity of infection (MOI).
Table 1 shows an example of raw date at time of inoculation (T0) and at 96 hours. The growth ratios are calculated using the formula T96/T0, but other time intervals can of course also be measured. It is, however, advisable not to use the cultures beyond 96 hours, as significant cell death occurs.
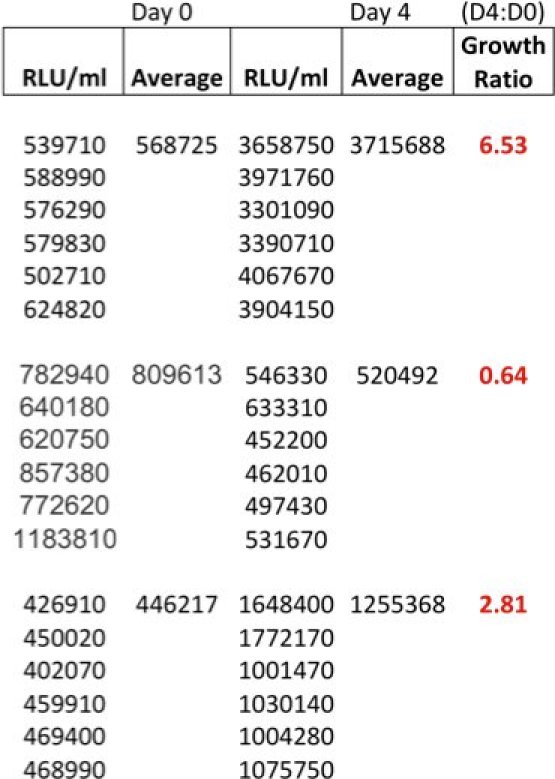 Table 1. Example of raw data at time of inoculation (T0) and at 96 hours (T96) and calculated growth ratios from 3 adult donors.
Table 1. Example of raw data at time of inoculation (T0) and at 96 hours (T96) and calculated growth ratios from 3 adult donors.
Depending on the available antigen-specific memory responses and possibly also neutrophil count, growth ratios vary among individuals, as shown here in Figure 2 in the blood of children with and without HIV-infection. On average, young children have higher growth ratios than adults, Tuberculin skin test (TST) +ve individuals have lower growth ratios than TST-ve individuals, and HIV-infected patients have high growth ratios due to the deficiency of CD4 T cell population, one of the key mediators of cellular immune responses to mycobacteria.
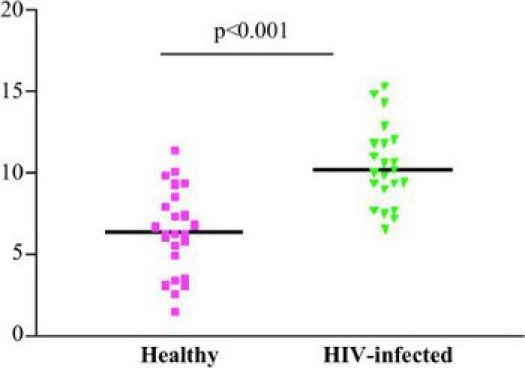 Figure 2. Inter-donor variability: Growth ratios of T0 versus 96 hours for a set of patients, depending on underlying HIV status.
Figure 2. Inter-donor variability: Growth ratios of T0 versus 96 hours for a set of patients, depending on underlying HIV status.
Reproducibility of growth ratios over a period of time is shown in Figure 3, which summarises results from 64 donors bled twice over a period of 12 months and from a single donor bled repeatedly for control experiments. Potential causes of variability could be changes in mycobacterial sensitization or variability within levels of host cytokines, as observed in many bioassays.
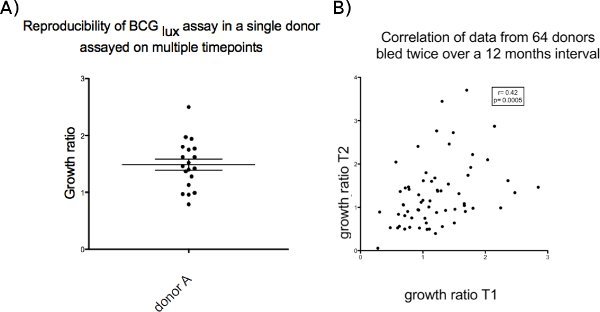 Figure 3. (a) Single donor on 12 occasions over 12 months. (b) multiple (64) donors on 2 occasions over 12 months.
Figure 3. (a) Single donor on 12 occasions over 12 months. (b) multiple (64) donors on 2 occasions over 12 months.
Discussion
Bactericidal assays in the context of tuberculosis vaccine research are well established in animal models, where they are applied to screen potentially promising vaccine candidates. Reduction of bacterial load in various organs functions as the main read-out of immunogenicity.
The first generation of human bactericidal assays were designed using macrophages alone, infected with M. tuberculosis. CFU served as the main read-out of mycobacterial survival after a minimum time of 3 weeks. These assays relied on preparation of PBMC, adherent monocytes/macrophages and lysis at given time points1,2.
As our understanding of the more complex interactions between macrophages and T cells to contain M. tuberculosis evolved, and the separation of various cell populations using magnetic beads was now possible, these systems were modified by adding T cells back into the assays and measuring CFU3,4,5. Some of these assays have used live M. tuberculosis, but one of the main impediments of work with M. tuberculosis in bactericidal assays is the need for containment facilities and the fact that CFU are only available after 3 weeks of culture. In addition, PBMC-based assays require a large amount of blood and still rely on CFU as read-out, which takes 3 weeks.
To facilitate more rapid read-outs, based on metabolic activity, new assays were designed. In these assays, metabolic activity is measured as a correlate of viable mycobacterial and three different systems have been designed in the last 10 years using either uracil incorporation4,5, reporter-gene tagged mycobacteria6-11 or a radiometric detection system via BacTec/MDGIT bottles7,8.
The whole blood lux assay is the only assay to date that has been successfully transferred to TB-endemic settings and used in children9,10. Furthermore, no other assay has published data of intra-donor variability over a period of time.
A limitation of the lux assay is its reliance on genetically modified organisms and the need to culture the organism prior to inoculation. However, due to the exact quantification of organisms, it is possible to calculate a very accurate multiplicity of infection for each series of experiments and each batch, which aids comparability and reproducibility. Instructions need to be followed closely in the preparation of the strains to achieve good viability of stocks, which is essential for successful implementation of the assay. Since the assay is carried out on fresh blood, it is time-sensitive and storage of samples in the field is not possible. Ideally, the blood needs to reach the laboratory within 4 hours. It might be possible in the future to further reduce the volume of blood required, and our laboratory is currently working on this.
Given the low cost and applicability to small samples, it is technically feasible to integrate this assay into clinical trials of novel TB vaccines10 or anti-TB interventions11. This would provide an additional opportunity to assess not just the host response to the vaccine, as is the current practice using an array of immunological and molecular techniques (such as flow cytometry, ELISPOT and microarray), but to add the functional read out of mycobacterial survival following host-pathogen interaction. This is common practice for vaccine assessments in pre-clinical studies in the animal model or by using bactericidal assays based on serum inhibition for other infections not relying on cellular immune responses.
It is timely to apply this approach to novel TB vaccines.
Disclosures
No conflicts of interest declared.
Acknowledgments
This work was funded by personal fellowships from the Wellcome Trust to Beate Kampmann (056608, GRO77273).
References
- Cheng SH, Walker L. Demonstration of increased anti-mycobacterial activity in peripheral blood monocytes after BCG vaccination in British school children. Clin Exp Immunol. 1988;74:20–205. [PMC free article] [PubMed] [Google Scholar]
- Cheng SH, Walker KB. Monocyte antimycobacterial activity before and after Mycobacterium bovis BCG vaccination in Chingleput, India, and London, United Kingdom. Infect Immun. 1993;61:4501–4503. doi: 10.1128/iai.61.10.4501-4503.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver RF, Li Q. Expression of virulence of Mycobacterium tuberculosis within human monocytes: virulence correlates with intracellular growth and induction of tumor necrosis factor alpha but not with evasion of lymphocyte-dependent monocyte effector functions. Infect Immun. 1998;66:1190–1199. doi: 10.1128/iai.66.3.1190-1199.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoft DF, Worku S. Investigation of the relationships between immune-mediated inhibition of mycobacterial growth and other potential surrogate markers of protective Mycobacterium tuberculosis immunity. J Infect Dis. 2002;186:1448–1457. doi: 10.1086/344359. [DOI] [PubMed] [Google Scholar]
- Worku S, Hoft DF. Differential effects of control and antigen-specific T cells on intracellular mycobacterial growth. Infect Immun. 2003;71:1763–1773. doi: 10.1128/IAI.71.4.1763-1773.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kampmann B, Gaora PO. Evaluation of human antimycobacterial immunity using recombinant reporter mycobacteria. J Infect Dis. 2000;182:895–901. doi: 10.1086/315766. [DOI] [PubMed] [Google Scholar]
- Cheon SH, Kampmann B. Bactericidal activity in whole blood as a potential surrogate marker of immunity after vaccination against tuberculosis. Clin Diagn Lab Immunol. 2002;9:901–907. doi: 10.1128/CDLI.9.4.901-907.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janulionis E, Sofer C. Lack of activity of orally administered clofazimine against intracellular Mycobacterium tuberculosis in whole-blood culture. Antimicrob Agents Chemother. 2004;48:3133–3135. doi: 10.1128/AAC.48.8.3133-3135.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kampmann B, Tena GN. Novel human in vitro system for evaluating antimycobacterial vaccines. Infect Immun. 2004;72:6401–6407. doi: 10.1128/IAI.72.11.6401-6407.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kampmann B, Tena-Coki GN. Reconstitution of antimycobacterial immune responses in HIV-infected children receiving HAART. Aids. 20:1011–1018. doi: 10.1097/01.aids.0000222073.45372.ce. [DOI] [PubMed] [Google Scholar]
- Martineau AR, Wilkinson RJ. A single dose of vitamin D enhances immunity to mycobacteria. Am J Respir Crit Care Med. 2007;176:208–213. doi: 10.1164/rccm.200701-007OC. [DOI] [PubMed] [Google Scholar]


