Abstract
MicroRNAs (miRNAs) are important regulators of gene expression and play a role in many biological processes. More than 700 human miRNAs have been identified so far with each having up to hundreds of unique target mRNAs. Computational tools, expression and proteomics assays, and chromatin-immunoprecipitation-based techniques provide important clues for identifying mRNAs that are direct targets of a particular miRNA. In addition, 3'UTR-reporter assays have become an important component of thorough miRNA target studies because they provide functional evidence for and quantitate the effects of specific miRNA-3'UTR interactions in a cell-based system. To enable more researchers to leverage 3'UTR-reporter assays and to support the scale-up of such assays to high-throughput levels, we have created a genome-wide collection of human 3'UTR luciferase reporters in the highly-optimized LightSwitch Luciferase Assay System. The system also includes synthetic miRNA target reporter constructs for use as positive controls, various endogenous 3'UTR reporter constructs, and a series of standardized experimental protocols.
Here we describe a method for co-transfection of individual 3'UTR-reporter constructs along with a miRNA mimic that is efficient, reproducible, and amenable to high-throughput analysis.
Protocol
Introduction
MicroRNAs have emerged as important regulators of gene expression that modulate the activity of many important biological processes. With more than 700 human miRNAs identified so far1, it is not surprising that mutations and improper regulation of miRNAs have been linked to a variety of physiological disorders such as cancer and heart disease2,3. Recent studies have shown that the majority of miRNA regulatory binding sites are found in 3'UTRs4,5, and some researchers have estimated that there are hundreds of targets per miRNA in the cell6. Given the biological importance of miRNAs, the large number of miRNAs that exist and the large number of targets for each of them, it is critical that researchers are able to reliably identify endogenous targets of miRNAs, preferably on platforms amenable to high-throughput studies.
While utilizing computational tools like miRBase, TargetScan, and PicTar1,6,7 is a powerful way to generate lists of putative mRNA targets for any miRNA, the predictions are not yet accurate enough to be relied upon alone8. Therefore, researchers also leverage a number of experimental approaches to identify targets de novo or to validate predicted targets. Many teams have relied on high-throughput measures of steady-state mRNA or protein levels to identify genes for which expression changes when miRNA activity is perturbed4,9,10. Such changes are undoubtedly biologically meaningful and represent an important first step, however measuring changes in overall expression does not distinguish between direct effects of a miRNA on a transcript and indirect or downstream signaling effects. Other powerful tools that provide evidence for direct miRNA-mRNA interactions are chromatin-immunoprecipitation (ChIP) strategies that cross-link argonaute proteins (and thus the RISC complex) to miRNAs and mRNAs, though identifying the interacting partners does not immediately indicate the functional effects of such a binding event5,11. Thus, 3'UTR-reporter assays that provide functional evidence for a miRNA's effect on a particular UTR have become an important component of a thorough miRNA study. Such cell-based assay data taken together with computational target predictions or expression and proteomics data provides strong evidence that a particular 3'UTR is directly regulated by the miRNA of interest.
In a 3'UTR-reporter assay, the 3'UTR from a gene of interest is fused to the end of a luciferase reporter gene. If the 3'UTR is a target of a miRNA, the reporter transcript stability and/or its translation efficiency will change with variation in functional miRNA concentration. So unlike transcript-only based assays, reporter assays help identify effects on both steady state mRNA and protein levels. Reporter systems may also be used to study the functional effects of mutagenizing a putative miRNA binding site in a 3'UTR to confirm that it is indeed necessary for the interaction8. And while large-scale studies using expression arrays, proteomics techniques, and argonaute ChIP provide valuable genome-wide data for a small number of experimental conditions, cell-based reporter assays are the most amenable to scaling up for the study of miRNA-3'UTR interactions in hundreds or thousands of conditions as may become necessary for high-throughput screening of small molecules or other library-based effectors.
Many scientists have applied reporter assay technology to the study of miRNA-3'UTR interactions. However, with the idea that many miRNAs each target hundreds of different transcripts, it has been difficult for researchers to utilize this powerful approach on a high-throughput scale because the cloning, validation, and optimization steps for creating thousands of different 3'UTR-reporter vectors are often cost prohibitive. In an effort to make small- and large-scale 3'UTR-reporter studies accessible to any researcher, we have created a genome-wide collection of human 3'UTRs pre-cloned into the highly-optimized LightSwitch Luciferase Assay System. The reporter system also includes validated control vectors, a set of more than 700 synthetic miRNA target reporter constructs for use as positive controls, and a series of standardized experimental protocols.
Using the following protocol, you can ask whether a particular human 3'UTR is a target of your miRNA of interest. The critical steps in this workflow are experimental design, transfection of reporter constructs and miRNA mimics, luciferase reporter assays, and data analysis. For experimental design, it is important thoughtfully select a transfectable cell line, experimental and control reporter constructs, and miRNA mimics and non-targeting controls. Common highly-transfectable adherent cell lines like HT1080, HepG2, HCT-116, Hela, 293, and others often work well with this protocol.
Important notes on experimental design
The LightSwitch Luciferase Assay System is a fully optimized reporter system that includes GoClone constructs utilizing the RenSP luciferase gene and LightSwitch Luciferase Assay Reagents. Use all components of the LightSwitch System as recommended to obtain optimal reporter assay results.
Co-transfection of any oligos/plasmids with SwitchGear GoClone reporter constructs reduces overall luciferase signals in a non-specific manner. Therefore, it is important to perform the following transfection combinations: reporter only, reporter + non-targeting control miRNA, and reporter + specific miRNA.
Our empty_3UTR vector (strong promoter, no UTR) provides a very high level of luciferase signal and serves as a positive control for transfection efficiency, and a synthetic miRNA target reporter vector may serve as a positive control for mimic activity. In addition, we recommend using the GoClone 3'UTR housekeeping controls and random controls for accurately separating sequence-specific vs. non-specific effects.
Study your protocol and whenever possible make master mixes to decrease the variability in your data due to pipetting error or evaporation.
This protocol uses DharmaFect Duo transfection reagent (Dharmacon).
Day 1
IMPORTANT TIP: Choose your cell line wisely. For best results, use a cell line that is known to be transfectable. We have also found that the measurable response of a 3' UTR reporter to a miRNA mimic is attenuated in cell lines with high endogenous levels of that particular miRNA. If you don't know how much miRNA your cell line expresses, try SGG's synthetic microRNA target which indirectly measures the abundance of a particular miRNA in a cell as well as informing you as to the dynamic range of your assay.
1. Seed cells to yield ≥80% confluence at time of transfection.
Seed cells in a 96-well white TC plate in 100µl total volume.
In parallel, seed the appropriate number of cells in a clear 96-well plate for assessing confluence.
IMPORTANT TIP 1: Confluence is key to achieving good results in a mimic experiment. For optimal knockdown, cells should be 100% confluent at time of transfection.
IMPORTANT TIP 2: Use low passage cells for best results. Cells that have been passaged too many times tend to yield noisy transfection data.
Day 2
Prepare GoClonereporter constructs and miRNAs, then transfect into the confluent cells.
2. Prepare Constructs
Thaw GoClone constructs (plasmid DNA) at room temperature. Mix well.
Centrifuge to remove condensation from the cap.
3. Prepare Micro RNAs
Thaw microRNAs (specific mimics and non-targeting controls) at room temperature and centrifuge to remove condensation from the caps.
Dilute to a working concentration of 2 µM in RNase-free water.
In addition to an experimental miRNA mimic ALWAYS include a non-targeting miRNA control. Co-transfecting a plasmid and a small RNA oligo leads to lower signal than transfecting the plasmid alone.
Include one or more positive controls. Consider using one of the hundreds of synthetic miRNA target reporter vectors in SGG's catalog.
Include one or more negative or non-specific controls. SGG has a range of controls including 3' UTRs of housekeeping genes, random fragments and/or the empty (no UTR insert) control. The use of one or more of these controls alongside an experimental construct will allow you to normalize for any non-specific effects attributable to the overexpression of your specific miRNA or the non-targeting control.
4. Prepare Transfections
For each transfection combine the following reagents for a single assay:
Mixture 1
| Individual GoClone reporter: | 3.33 µL |
| microRNA: | Varies* |
| Serum-free media: | to 10 µL |
* Effective concentration of miRNA can vary from 10 nM-100 nM. Consult miRNA manufacturer for appropriate concentration.
IMPORTANT TIP: Do not add too much mimic. A reasonable range for mimic concentrations is 10-50 nM in the final 100uL reaction.
Perform at least three replicate transfections for each treatment/UTR combination and include additional volume to allow for pipetting error.
For example, multiply the single reaction volumes by 3.5 to get enough transfection mix for 3 replicate transfection wells including extra for pipetting error or evaporation.
For each reporter, set up replicates for the following: reporter only, reporter + non-targeting control miRNA and reporter + specific miRNA.
Set up transfections for the targeting microRNA at the same time as those for a non-targeting control.
Make up the DharmaFECT DUO (Dharmacon) mixture as follows for each transfection:
Mixture 2
| DharmaFect Duo: | 0.15 µL |
| Serum free media: | 9.85 µL |
The amount of DharmaFECT Duo to be added may be cell line dependent.
The amounts indicated above are appropriate for HT1080 cells.
Consult the manufacturer's documentation to determine the appropriate amount for your cell line.
Tip: Make a large transfection mixture by multiplying the volumes above by the number of reporters, the number of replicates and the number of miRNAs to be tested. When calculating the amount of transfection mix to prepare, be sure to include a small amount extra to account for pipetting errors and evaporation.
Allow the DharmaFECT Duo mixture to incubate at room temperature for 5 minutes.
After 5 minutes, add 10µL of the DharmaFECT Duo mixture (Mixture 2) to each prepared tube of 10uL plasmid and/or miRNA (Mixture 1). Each tube should then contain a volume of 20µL per replicate transfection.
Incubate each mixture for 20 minutes at room temperature.
After 20 minutes, add 80 µL of pre-warmed (37°C), antibiotic-free, complete media per replicate to each tube for a total of 100µL per replicate transfection. A deep-well block may be used for preparation of many replicates and samples simultaneously. Mix the solution by pipetting up and down.
Remove the seeded plate from the incubator. Verify that cells are at least 80% confluent.
Carefully pipet off the media from each well.
Add 100µL of the transfection mixture (20uL DNA/reagent mix + 80uL media) to each well.
Place plate in incubator overnight.
Don't over-incubate. Most mimics will produce significant results on bona fide targets within 24 hours. The level of repression measurable at 48 hours may be greater, but so will the well to well variability.
Mimic experiments are easier than inhibitor experiments. While most mimics will significantly repress true targets within 24 hours, significant de-repression attributable to the inhibitor can take 48 hours to be measurable and will almost always be less pronounced than the effect of the mimic.
Day 3
Note: Luciferase assays may be conducted immediately or the plates may be frozen at -80°C (freezing generally increases cell lysis and luciferase signal). Freeze and thaw your cells before assaying for best results.
REMEMBER to let frozen cells thaw to room temperature before adding LightSwitch Assay Reagent.
5. Measure luciferase activity with LightSwitch AssayReagents
Remove plate from incubator and bring to room temperature.
- Prepare LightSwitch Assay Reagents (for LS010 kit, protocols of other kit sizes may be found online):
- Reconstitute 100X Substrate by adding 100µL of Substrate Solvent to tube of lyophilized Assay Substrate. Protect from light and minimize time at room temperature. 100X Substrate may be stored at -20C for 2-3weeks. For best results, use freshly reconstituted substrate.
- Prepare Assay Solution by thawing 10mL bottle of Assay Buffer in room temperature water bath and add 100µL of reconstituted 100X Substrate prior to use. Mix well.
Use a multi-channel pipettor to add 100µL LightSwitch Assay Solution (buffer+substrate) directly to each sample well (containing 100uL cells + media) in a white 96-well plate. If cells were grown in another plate or flask format, transfer samples to a white 96-well plate in 100µL total volume (media or PBS).
Cover plate, protect from light, and incubate for 30 minutes at room temperature. If assaying more than one plate, stagger addition of assay solution so that each plate incubates for 30 minutes before reading.
Read each well for 2 seconds in a plate luminometer (SpectraMax L or equivalent).
Calculate the knockdown by calculating luciferase signal ratio for each construct for specific miRNA over the non-targeting control (luminescence = specific miRNA/non-targeting control). Use the data from housekeeping, random, and empty constructs to control for non-UTR specific treatment effects.
Representative Results
3'UTR targets in the presence of a targeting miRNA show luciferase knockdown.
The knockdown of 3'UTR-luciferase reporter activity by let-7a was measured in HT1080 cells by co-transfecting 100ng of each reporter construct with 20nM let-7a mimic or non-targeting control, incubating for 24 hours, then reading the luminescent reporter signal using LightSwitch Luciferase Assay Reagents.
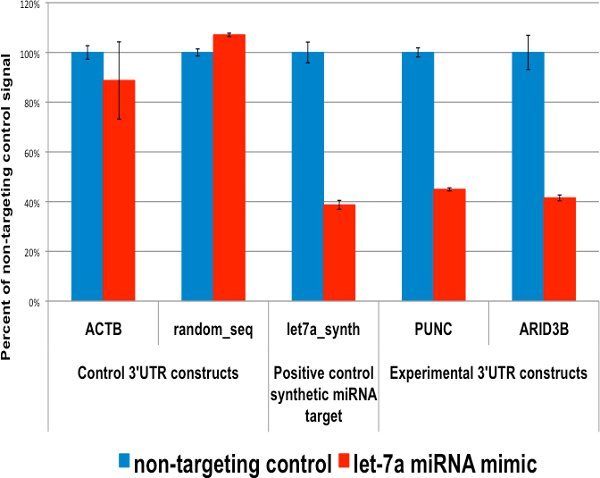 Figure 1. PUNC and ARID3B human 3'UTRs are targets of the let-7a microRNA. Signals from human 3'UTR reporters for the PUNC and ARID3B genes are significantly knocked down in the presence of the let-7a microRNA mimic while signals for housekeeping and random sequence UTRs are not.
Figure 1. PUNC and ARID3B human 3'UTRs are targets of the let-7a microRNA. Signals from human 3'UTR reporters for the PUNC and ARID3B genes are significantly knocked down in the presence of the let-7a microRNA mimic while signals for housekeeping and random sequence UTRs are not.
Discussion
We have created a genome-wide collection of pre-cloned human 3'UTR-reporter constructs to enable studies of miRNA-3'UTR interactions in living cells. Here we have shown a reproducible and high-throughput method for co-transfecting miRNA mimics with 3'UTR-reporters using a luciferase reporter system.
This method relies on researchers using a highly-transfectable cell line and perturbing miRNA activity with an overexpression technique. Researchers using poorly-transfectable cell lines may consider using our companion lentiviral 3'UTR-reporter collection created in conjunction with Sigma (Mission 3'UTR Lenti GoClones). Treating the cells with miRNA mimics results in an overexpression-type experiment, and with any such study, researchers must take into account that this may or may not represent actual physiological conditions. To address overexpression concerns, researchers may use miRNA inhibitors in addition to mimics to study the interaction with two different types of perturbation12-14.
Once likely miRNA targets are identified, researchers may wish to mutagenize the putative miRNA binding sites to determine whether the sites are necessary for the functional interaction. They may also wish to scale up to testing hundreds of different potential target UTRs (Boutz). In addition, if researchers are interested as to whether the miRNA is affecting transcript stability or translation efficiency, luciferase reporter data may be compared to measures of steady-state transcript levels. Any change in luciferase signal that is not associated with a proportional change in transcript levels will point to an effect on translation efficiency. Finally, researchers may use reporter vectors create a dose response curve to compare the relative sensitivity of different UTRs to changes in miRNA concentration14.
Disclosures
The authors are employees of SwitchGear Genomics who make tools and reagents appearing in this article. Production of this article was sponsored by SwitchGear Genomics.
References
- Griffiths-Jones S, Grocock RJ, van Dongen S. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–D144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calin GA, Croce CM. MicroRNA cancer connection: the beginning of a new tale. Cancer Res. 2006;66:7390–7394. doi: 10.1158/0008-5472.CAN-06-0800. [DOI] [PubMed] [Google Scholar]
- Chen JF, Murchison EP, Tang R. Targeted deletion of Dicer in the heart leads to dilated cardiomyopathy and heart failure. Proc. Natl. Acad. Sci. U.S.A. 2008;105:6–6. doi: 10.1073/pnas.0710228105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek D, Villen J, Shin C. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickson DG, Hogan DJ, Herschlag D. Systematic identification of mRNAs recruited to argonaute 2 by specific microRNAs and corresponding changes in transcript abundance. PLoS One. 2008;3:e2126–e2126. doi: 10.1371/journal.pone.0002126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lall S, Krek A, Chen K. A genome-wide map of conserved microRNA targets in. 2006;16:460–471. doi: 10.1016/j.cub.2006.01.050. [DOI] [PubMed] [Google Scholar]
- Boutz DR, Collins PJ, Suresh U. Two-tiered approach identifies a network of cancer and liver disease-related genes regulated by miR-122. J. Biol. Chem. 2011;286:18066–18078. doi: 10.1074/jbc.M110.196451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim LP, Lau NC, Garrett-Engele P. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- Selbach M, Schwanhausser B, Thierfeldner N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- Hutvagner G, Simard MJ, Mello CC, Zamore PD. Sequence-specific inhibition of small RNA function. PLoS Biol. 2004;2:e98–e98. doi: 10.1371/journal.pbio.0020098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister G, Landthaler M, Dorsett Y, Tuschl T. Sequence-specific inhibition of microRNA- and siRNA-induced RNA silencing. RNA. 2004;10:544–5450. doi: 10.1261/rna.5235104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao JM, Lu H. Auto-regulatory suppression of c-Myc by miR-185-3p. J. Biol. Chem. Epub. 2011 doi: 10.1074/jbc.M111.262030. Forthcoming. [DOI] [PMC free article] [PubMed] [Google Scholar]


