Abstract
Small-molecule protein kinase inhibitors are central tools for elucidating cellular signaling pathways and are promising therapeutic agents. Due to evolutionary conservation of the ATP-binding site, most kinase inhibitors that target this site promiscuously inhibit multiple kinases. Interpretation of experiments utilizing these compounds is confounded by a lack of data on the comprehensive kinase selectivity of most inhibitors. Here we profiled the activity of 178 commercially available kinase inhibitors against a panel of 300 recombinant protein kinases using a functional assay. Quantitative analysis revealed complex and often unexpected kinase-inhibitor interactions, with a wide spectrum of promiscuity. Many off-target interactions occur with seemingly unrelated kinases, revealing how large-scale profiling can be used to identify multi-targeted inhibitors of specific, diverse kinases. The results have significant implications for drug development and provide a resource for selecting compounds to elucidate kinase function and for interpreting the results of experiments that use them.
Protein kinases are among the most important classes of therapeutic targets because of their central roles in cell signaling pathways and the presence of a highly conserved ATP-binding pocket that can be exploited by synthetic chemical compounds. However, achieving highly selective kinase inhibition is a significant challenge1–6. Knowledge of target selectivity for kinase inhibitors is critical for predicting and interpreting the effects of inhibitors in both the research and clinical settings. However, kinase inhibitor selectivity is generally not comprehensively known for most inhibitors. Recent technological advances have led to the development of methods to profile kinase target selectivity against significant fractions of the 518 human protein kinases7, 8. In many cases, however, these methods measure compound-kinase binding rather than functional inhibition of catalytic activity. The ability of these assays to predict functional inhibition is, therefore, an important outstanding question.
Traditionally, kinase inhibitors have been discovered in a target-centric manner in which inhibitors of interest are identified by high throughput screening using a particular kinase. The resulting compounds are then tested for selectivity against a panel of representative kinases. An alternative approach has been suggested in which libraries of compounds are screened in a target-blind manner against a comprehensive panel of recombinant protein kinases to reveal the selectivity of each compound9, 10. Compounds showing desired selectivity patterns are identified and then chemically optimized. This parallel approach is predicted to identify unexpected new inhibitors for kinases of interest and reveal “multi-targeted” inhibitors, whose inhibitory activity is focused toward a small number of specific kinase targets rather than toward a single primary target11, 12. Indeed, multi-targeted inhibitors are challenging to identify by conventional target-centric screens15.
We have conducted a large-scale parallel screen of 178 known kinase inhibitors against a panel of 300 protein kinases in duplicate using a high-throughput enzymatic assay. Our goals were to identify novel inhibitor chemotypes for specific kinase targets and to reveal the target specificities of a large panel of kinase inhibitors. The compounds tested represent widely used research compounds and clinical agents targeting all of the major kinase families. The resulting dataset, to our knowledge the largest of its type available in the public domain, comprises over 100,000 independent functional assays measuring pairwise inhibition of a single enzyme by a single compound. Systematic, quantitative analysis of the results revealed kinases that are commonly inhibited by many compounds, kinases that are resistant to small-molecule inhibition, and unexpected off-target activities of many commonly used kinase inhibitors. In addition we report potential leads for orphan kinases for which few inhibitors currently exist and starting points for the development of multi-targeted kinase inhibitors.
RESULTS
A kinase-inhibitor interaction map
To directly test the kinase selectivity of a large number of kinase inhibitors, we conducted low volume kinase assays using a panel of 300 recombinant human protein kinases. We utilized HotSpot, a radiometric assay based on conventional filter-binding assays, that directly measures kinase catalytic activity toward a specific substrate. This well-validated method is the standard against which more indirect assays for kinase inhibition are compared 7. Our collection of kinase inhibitors included FDA-approved drugs, compounds in clinical testing, and compounds primarily used as research tools. The library comprised 178 compounds known to inhibit kinases from all major protein kinase subfamilies (Fig. 1a). A complete listing of the inhibitors is included in Supplementary Table 1.
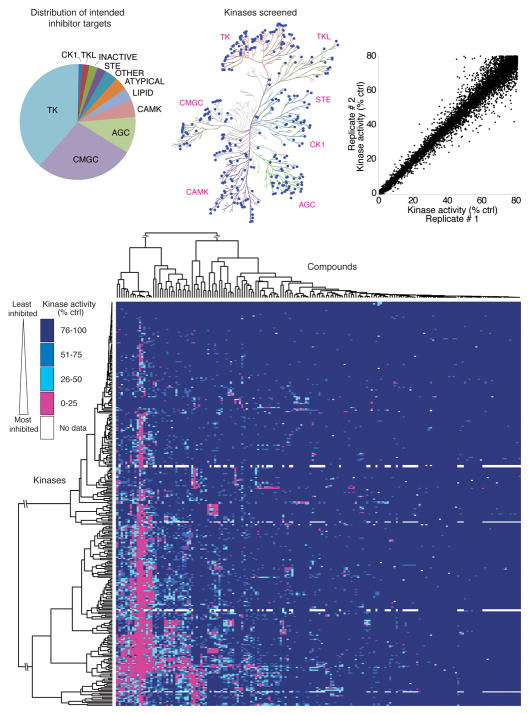
Figure 1.
Large-scale inhibitor-kinase interaction analysis. (a) Distribution of the intended targets of the inhibitor library, by kinase family. (b) The distribution of kinases in the screening panel is represented by blue dots on a dendrogram representing the human kinome (Kinome illustration was adapted and is reproduced courtesy of Cell Signaling Technology, Inc. (www.cellsignal.com) based on Manning et al.33). (c) Scatter plot of the kinase activity in replicate 1 versus replicate 2 for each kinase-inhibitor pair for which > 20% inhibition of kinase activity was observed. (d) Two-way hierarchical clustering analysis of the entire kinase-inhibitor interaction map presented as a heatmap of kinase activity. A fully labeled, high-resolution version of this heatmap is presented in Supplementary Figure 2, as a data table in Supplementary Table 3, and via the Kinase Inhibitor Resource (KIR) online tool (http://kir.fccc.edu).
The kinase panel tested includes members of all major human protein kinase families (Fig. 1b) and includes the intended targets of 87.6% of the compounds tested. A complete listing of the kinase constructs and substrates used is provided in Supplementary Table 2. For simplicity, all compounds were tested at a concentration of 0.5 μM in the presence of 10 μM ATP. 0.5 μM was chosen despite an average reported IC50 for these compounds toward their primary targets of 66 nM in order to capture weaker off-target inhibitory activity.
Each kinase-inhibitor pair was tested in duplicate and results were expressed as average substrate phosphorylation as a percentage of solvent control reactions (henceforth referred to as “remaining kinase activity”). Disparate replicates, representing only 0.18 % of the dataset, were identified and eliminated from the analysis (see Methods and Supplementary Fig. 1). Figure 1c illustrates the reproducibility of the resulting dataset as a scatter plot in which each point represents one kinase-inhibitor pair plotted as the remaining kinase activity in one replicate versus the second replicate, for all kinase-inhibitor pairs in which at least 20% kinase inhibition was observed.
Mean remaining kinase activity for each kinase-inhibitor pair is presented as a heatmap in Figure 1d (in high-resolution form in Supplementary Fig. 2) and as a downloadable Microsoft Excel spreadsheet in Supplementary Table 3. In addition, we created the Kinase Inhibitor Resource (KIR) database, an internet website that allows compound or kinase specific queries of the dataset to be downloaded or analyzed within a browser window (http://kir.fccc.edu). Two-way hierarchical clustering was performed to cluster both kinases and inhibitors based on the similarity of their activity patterns. As expected, structurally related compounds were generally grouped. Similarly, kinases closely related by sequence identity were often clustered and were inhibited by similar patterns of compounds. Exceptions included members of the clinically relevant Aurora, PDGFR, and FGFR family kinases (see Supplementary Fig. 2), suggesting the possibility that members of these families can be differentially targeted by small molecules. Consistent with this finding, isoform specific inhibitors of Aurora kinases have been reported and structural studies have revealed the structural basis for isoform-specific inhibition 13.
Comparison of data across multiple assay platforms
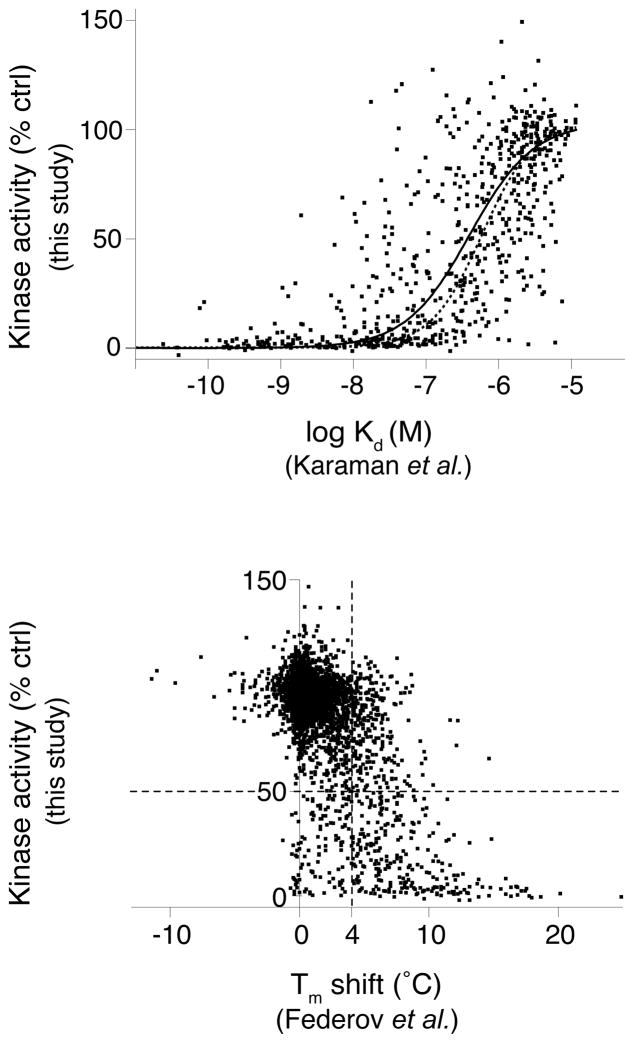
A variety of high-throughput screening approaches have been devised to detect compound-kinase interactions without directly measuring inhibition of kinase catalytic activity. Though convenient for screening, it remains an important question to what degree these binding assays predict inhibition of catalytic activity. To assess this, we compared our kinase inhibition data with previous large-scale studies of compound-kinase binding. Two recent studies utilized a competitive binding assay to derive affinities for a large number of kinase-inhibitor interactions1, 2. 654 kinase-inhibitor pairs overlapped with our study and their affinities showed generally good agreement with the expected kinase activity measured in our single dose study (Fig. 2a). 90.2% of kinase-inhibitor interactions with high affinity (stronger than 100 nM Kd) showed functional inhibition (>50%). Conversely, only 13.1% of the kinase-inhibitor pairs with low affinity (weaker than 1 μM Kd) showed significant inhibition, as expected.
Figure 2.
Comparison of this study’s functional inhibition data with previous kinase-inhibitor interaction profiling studies. Scatter plots compare our results with studies that examined interactions of overlapping kinase-inhibitor pairs by (a) a quantitative kinase-inhibitor binding assay1, 2, or (b) an assay measuring resistance to thermal denaturation by kinases in the presence of individual inhibitors3. In (a), remaining kinase activity is plotted as a function of kinase- compound binding affinity (Kd) for 654 kinase-inhibitor pairs. The resulting data was fit to a sigmoidal dose-response curve (solid line) and can be compared with a theoretical curve (dotted line) for expected remaining kinase activity for an inhibitor of the given affinity. The theoretical activity curve was calculated according to the equation: activity=(100-(100/(1+(IC50/0.5 μM))) and the Cheng-Prusoff equation34 relating Ki and IC50. This calculation assumes a Hill coefficient of 1 for the binding and a Km,ATP of 10 μM for all kinases. In (b) remaining kinase activity is plotted against the change in melting temperature (Tm), relative to solvent control, caused by compound binding for 3,926 kinase-inhibitor pairs. The dotted vertical line denotes the Tm shift threshold used by Federov et al.3. The dotted horizontal line highlights the 50% threshold for inhibition of catalytic activity. The resulting upper right quadrant includes compounds that showed significant thermal stabilization without inhibiting kinase activity whereas the lower left quadrant contains compounds which only marginally affect thermal stability yet show > 50% inhibition of catalytic activity.
An alternative approach to monitoring compound-kinase binding has been established based on protection of kinases from thermal denaturation by compound binding3. To assess this approach to predict kinase inhibition, we plotted the remaining kinase activity in our functional assay as a function of the change in reported melting temperature (Tm) of each kinase-inhibitor pair (Fig. 2b). Generally, compounds that increased the kinase melting temperature also showed inhibition of catalytic activity, as predicted. However a significant number of compounds showed Tm changes > 4 C, the hit threshold used by Federov et al3, without significantly affecting kinase activity (upper right dashed quadrant). Likewise, 117 out of 3926 inhibitor pairs showed > 50% inhibition of kinase activity without exhibiting Tm changes > 4 C (lower left dashed quadrant). The findings from these comparisons, taken together, suggest that inhibitor-kinase binding assays exhibit appreciable false positive and false negative rates with respect to their ability to predict compounds that functionally inhibit catalytic activity, though binding and inhibition are significantly correlated.
Analysis of kinase druggability
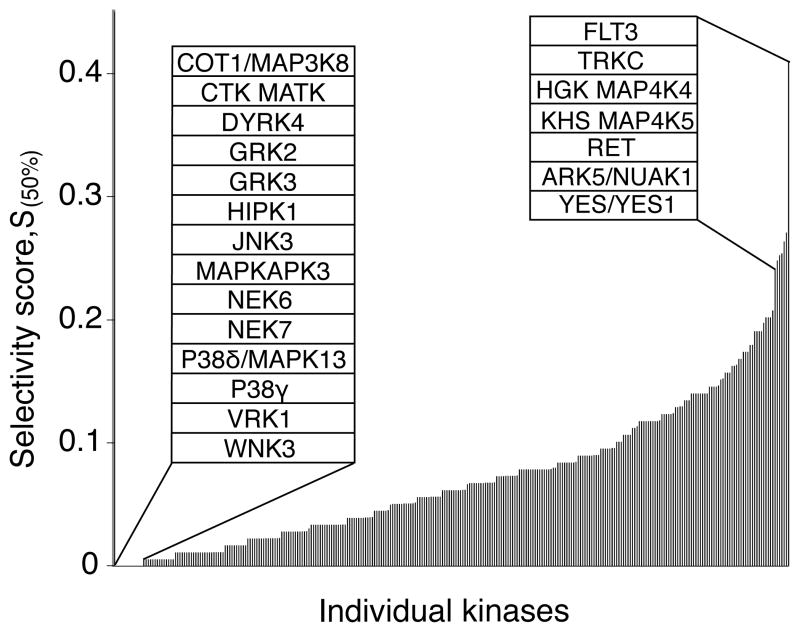
We next asked whether each kinase in the panel was equally likely to be inhibited by a given compound or whether certain kinases were more sensitive to small-molecule inhibition while others were more resistant. To do so, we ranked the kinases with respect to a Selectivity score (S(50%)) the fraction of all compounds tested that inhibited each kinase by >50% (Fig. 3; complete data in Supplementary Table 4). Only 14 kinases in the panel were not inhibited by any of the compounds tested (left inset), demonstrating good coverage of the kinome by this inhibitor set. The untargeted kinases, including COT1, NEK6/7, and p38δ, suggest a target list for which screens utilizing traditional ATP-mimetic scaffolds may be less successful. By contrast, a subset of kinases including FLT3, TRKC, and HGK/MAP4K4 were broadly inhibited by large numbers of compounds (right inset), potentially representing kinases highly susceptible to chemical inhibition. This broad range of kinase sensitivity to small molecules has significant implications for the assessment of kinase inhibitor selectivity with small kinase panels and suggests that screening panels should include these sensitive kinases. We cannot completely exclude the possibility, however, that the results could reflect hidden biases in our compound library.
Figure 3.
Kinase selectivity. A ranked bar chart of Selectivity scores (S(50%)) for all tested kinases. This score corresponds to the fraction of all tested inhibitors that inhibit catalytic activity by >50%. Each bar represents the Selectivity score of an individual kinase. Insets identify the fourteen kinases that were not inhibited by any compound (left) and the top seven most frequently inhibited kinases (right). The complete table is presented in Supplementary Table 4.
Kinase inhibitor selectivity
Kinase inhibitors are commonly used as research tools to reveal the biological consequence of acute inactivation of their kinase targets. Interpretation of the results of such experiments depends critically on knowing the inhibitor target(s). The selectivity of novel kinase inhibitors is frequently assessed by testing against a limited panel of closely related kinases based on the assumption that off-target interactions are more likely to be found with kinases most closely related by amino acid sequence. To test this quantitatively, we assessed the fraction of kinase targets that are within the same kinase subfamily versus outside the family of the primary target. Since highly promiscuous compounds would increase the apparent frequency of “out of family” targets, the top ten most promiscuous compounds were removed prior to the analysis. On average, 42% of the kinases inhibited by a given compound were from a different kinase subfamily than the subfamily of the intended kinase target (Supplementary Fig. 3). For inhibitors developed against tyrosine kinases, 24% of off-target hits were serine/threonine kinases. The within-family selectivity of tyrosine kinase-targeting compounds may be explained, in part, by the fact that these compounds include almost all of the clinical agents in our compound set and are, therefore, likely more optimized with regard to specificity than research tool compounds. These results highlight the importance of assessing the selectivity of kinase inhibitors against as broad a panel of kinases as possible.
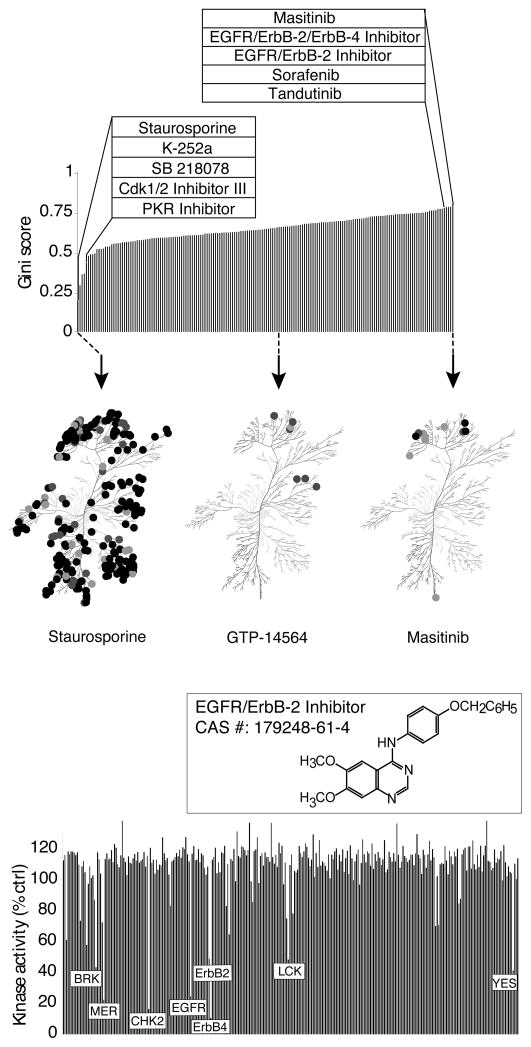
Inhibitors that exhibit selectivity for a very limited number of kinase targets are most valuable as research tools for probing kinase function. Various methods have been proposed to quantitatively assess kinase inhibitor selectivity. Karaman et al. defined a Selectivity score, S(x), where S is the number of kinases bound by an inhibitor (with an affinity greater than “x” μM) divided by the number of kinases tested2. A critical limitation of the Selectivity score is its dependence on an arbitrary hit threshold (“x” μM). For example, when we analyzed our data using an arbitrary percent inhibition as the hit criterion, several compounds scored favorably because they met the hit threshold with a limited number of kinases, despite significant inhibition of other kinases just below this threshold (not shown). Indeed, Selectivity scores generated from the same dataset but using different hit thresholds can produce different rank orders of compounds2. In addition, compounds that did not meet the hit threshold for any kinase could not be scored. We therefore calculated a previously described metric for kinase inhibitor selectivity based on the Gini coefficient14. Importantly, this method does not depend on defining an arbitrary hit threshold, although it is strongly influenced by the compound screening concentration. The Gini score reflects, on a scale of 0 to 1, the degree to which the aggregate inhibitory activity of a compound (calculated as the sum of inhibition for all kinases) is directed toward only a single target (a Gini score of 1) or is distributed equally across all tested kinases (a Gini score of 0). The results of this analysis were used to rank the compounds from most promiscuous to most selective (Fig. 4a; complete list in Supplementary Table 5). Not surprisingly, staurosporine and several of its structural analogues exhibited the lowest Gini scores (left inset), consistent with their known broad target spectrum. Among the most selective compounds (right inset) were several structurally distinct inhibitors of ErbB family kinases. The target spectra of the three compounds with the lowest, median, and highest Gini scores are shown in the bottom panels. Although a comparable number of kinases were targeted by the compounds with the median and highest Gini scores (middle and right dendrograms), Masitinib achieves a higher Gini score by more potently inhibiting its targets (darker spots).
Figure 4.
Kinase inhibitor selectivity. (a) A ranked list of kinase inhibitors sorted by Gini score14 as a measure of inhibitor selectivity. A Gini score of 0 indicates equal inhibition of all kinases (promiscuous inhibition) while a score of 1 indicates inhibition of only one kinase (selective inhibition). Left inset highlights the five compounds with the lowest Gini scores and the right inset, the five highest scoring compounds. The complete table is presented in Supplementary Table 5. Below, the selectivity of three representative compounds are presented on a dendrogram of all human kinases based on amino acid sequence similarity33. Spot color represents inhibitory potency: darkest, 0-10% remaining activity, lighter, 10–25% activity, lightest, 25–50% activity. The kinome dendrogram was adapted and is reproduced courtesy of Cell Signaling Technology, Inc. (www.cellsignal.com). (b) Target spectrum of 4-(4-benzyloxyanilino)-6,7-dimethoxyquinazoline, a multi-targeted inhibitor, highly selective for ErbB family members, a limited number of other tyrosine kinase targets, and the serine/threonine kinase CHK2. Each bar corresponds to the percent remaining activity for an individual kinase.
To understand the molecular features that contribute to inhibitor promiscuity, previous kinase/inhibitor profiling studies have identified correlations between compound physicochemical properties and promiscuity15, 16. We analyzed a variety of compound physicochemical properties with respect to either Gini score or Selectivity score but did not observe a consistent linear correlation with any single compound property (Supplementary Fig. 4). This finding and the discrepant findings of the previous studies suggest that compound promiscuity is unlikely to be strongly related to any one physical parameter in a simple, linear manner.
The clinical success of some kinase inhibitors that show poor kinase selectivity in vitro (e.g. dasatinib, sunitinib) has led to increasing interest in so-called “multi-targeted” kinase inhibitors12, 17. Such compounds, ideally, differ from promiscuous inhibitors in that they should show significant selectivity toward a limited number of clinically relevant targets with the goal of achieving greater therapeutic effect than targeting a single kinase18. Despite the promise of “polypharmacology”, it remains a significant technical challenge to rationally develop single compounds with a desired target spectrum18, 19. Parallel kinase profiling of large inhibitor libraries has been suggested as an approach to identify compound scaffolds that show promising activity against specific kinases of interest9, 19. We interrogated our data for examples of inhibitors with off-target activities against a limited number of cancer-relevant kinases. The ErbB family kinase inhibitor 4-(4-benzyloxyanilino)-6,7-dimethoxyquinazoline20 showed potent inhibition of a few tyrosine kinases beyond ErbB family members and, most surprisingly, potent inhibition of the serine/threonine kinase CHK2, a critical component of the DNA damage checkpoint (Fig. 4b). CHK2 inhibition has been proposed as a strategy to increase the therapeutic impact of DNA-damaging cancer therapies and inhibitors of CHK2 are in clinical trials21. This illustrates how kinase profiling can reveal unanticipated novel scaffolds that show activity against highly divergent kinases of therapeutic interest. Data mining of this and similar datasets can facilitate the identification of inhibitor scaffolds with activity towards multiple targets of interest.
Novel targets of uni-specific kinase inhibitors
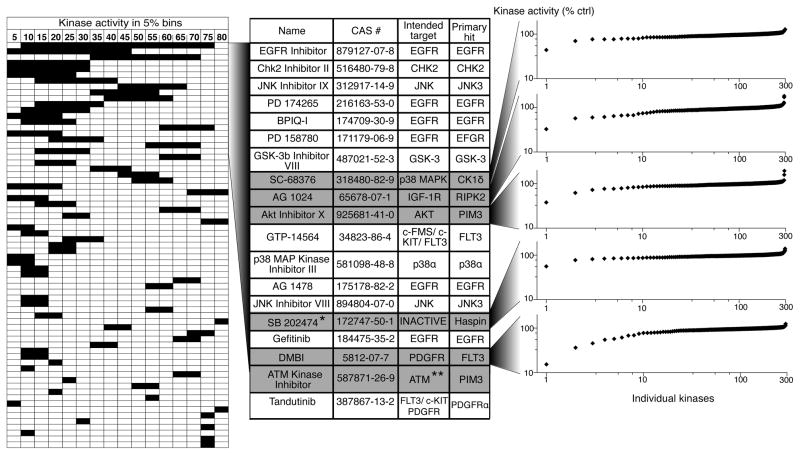
Even among the most selective inhibitors identified by the screen, most still targeted multiple kinases with similar potency (Fig. 4a, rightmost dendrogram), therefore confounding their use as research tools to elucidate the function of a single kinase. We therefore asked whether any compounds inhibited a single kinase significantly more potently than any other in our panel, a characteristic we termed “uni-specificity”. Importantly, this stringent criterion excludes compounds that target more than one kinase with similar potency, even if those kinases are closely related isoforms from the same subfamily. In addition it biases for kinase targets without close homologs in the screening panel. A uni-specificity score was calculated for each compound by subtracting the remaining kinase activity of the most potently inhibited kinase from the activity of next most potently inhibited kinase. Compounds were then ranked from most uni-specific (highest numerical score) to least. We plotted the results as a horizontal bar graph in which the leftmost edge of the bar denotes the remaining kinase activity for the most potently inhibited kinase and the rightmost edge indicates the remaining kinase activity of the second most potently inhibited target (Fig. 5, leftmost panel). The length of each bar, therefore, denotes the differential potency of inhibition of these two most sensitive kinase targets, and the left-right positioning of this bar indicates the absolute potency against these targets.
Figure 5.
“Uni-specific” kinase inhibitors. The left panel presents a graphical table of compounds ranked based on the compound’s ability to inhibit a single kinase significantly more potently than any other kinase tested. The left boundary of each horizontal bar depicts the potency with which the compound inhibits its most sensitive target and the right boundary reflects the potency with which the next most sensitive kinase is inhibited (% remaining kinase activity is shown in bins of 5%). Thus, the horizontal length of each bar reflects the differential activity of the corresponding inhibitor against its two most potently inhibited targets. Only compounds with a differential potency of at least 5% are shown. The central table identifies the compounds that showed at least 20% differential potency, their intended targets, and their most sensitive targets. Six compounds for which the most sensitive target is not the intended target are shown in grey. In the right panel, the effect of the individual compounds on each kinase in the panel is shown in a ranked plot. * SB 202474 is a negative control compound for the p38 MAP kinase inhibitor SB 202190. ** Note that ATM kinase was not included in our test panel.
Few compounds in the panel showed any degree of uni-specificity and most of these showed only slight potency differences between their primary and secondary targets (short bars in Fig. 5, leftmost panel). This finding highlights the challenge of achieving differential inhibition of closely related kinases. 19 compounds inhibited their primary target at least 20% more potently than any other kinase in the panel (Fig. 5, middle). Among these most uni-specific kinases are several inhibitors intended to target the epidermal growth factor receptor (EGFR). In fact, the most uni-specific inhibitor, a 4,6-dianilinopyrimidine EGFR inhibitor (CAS# 879127-07-8) with a reported IC50 of 21 nM for EGFR22, inhibited EGFR catalytic activity by >94% but inhibited its next most potent target, MRCKα, by only 22%. In contrast to other EGFR inhibitors tested, this compound also highlights the ability to achieve isoform selective inhibition among the closely related ErbB family kinases22. The significant selectivity of this and other uni-specific EGFR inhibitors identified here could reflect unique features of EGFR or, more likely, the unequal attention devoted to the development of inhibitors of this important therapeutic target.
Strikingly, 6 of the top 17 uni-specific compounds inhibited other kinases more potently than the kinases they were intended to target (Fig. 5, center, gray rows). The rightmost panel of Figure 5 shows the activity of 5 of these 6 compounds against all kinases in the panel as a sorted plot. The ATM kinase inhibitor was not included because ATM was not a part of the screening panel. In all cases these more potent “off-target” hits represent hitherto unknown kinase targets of these compounds. Remarkably, in all but one case, the compound DMBI, the most potent off-target hit falls outside of the kinase subfamily of the intended target. For example, we identified the serine/threonine kinase RIPK2 as a much more sensitive target of the IGF1R tyrosine kinase inhibitor AG1024, one of the most uni-specific compounds identified.
To validate the use of our single dose screening data to rank the sensitivity of different kinases to the same compound, we determined the dose-response relationship for five uni-specific compounds against both their intended and novel targets. In all cases the greater potency against the novel targets were confirmed (Supplementary Fig. 5). These findings confirm the accuracy of our single dose data and reveal potently inhibited new targets for these compounds. For example, the results revealed the weak platelet-derived growth factor receptor inhibitor, DMBI to be a highly potent inhibitor of FLT3 and TrkC. Additionally, SB202474, an inactive analog of the p38 MAP kinase inhibitor SB20219023, showed significant inhibition of only one kinase, the haploid germ cell-specific nuclear protein kinase Haspin (Fig. 5). This atypical family kinase phosphorylates histone H3 and contributes to chromosomal organization and has been suggested as an anti-cancer target, though few inhibitors have been reported24–26. Thus, the uni-specific compounds described here, provide new and selective inhibitors for their novel targets and in some cases starting points for multi-targeted kinase inhibition.
DISCUSSION
Previous kinase inhibitor profiling studies have revealed an unexpected number of off-target kinase interactions, even for highly characterized kinase inhibitors1, 2. These findings have emphasized the importance of broad kinase profiling of these compounds and are supported by our data. Quantitative assessment of inhibitor selectivity is increasingly important as ever larger kinase profiling datasets are reported. While strong kinase selectivity may not be essential for efficacy of therapeutic agents27, it is critical for tool compounds used to elucidate kinase biology. We therefore applied the Gini coefficient as a measure of kinase inhibitor selectivity14, thus avoiding the necessity for arbitrary hit thresholds used by previous methods2. Comparison of Gini scores across multiple inhibitors targeting a specific kinase of interest should provide a powerful basis for choosing the most selective inhibitor for investigating kinase function. For example, the compound collection contains four well-established inhibitors of the AGC subfamily kinase ROCK (Rho associated kinase): Rockout, glycyl-H-1152 (Rho Kinase Inhibitor IV), Y-27632 and the clinical agent Fasudil (HA-1077)28, 29. Gini score analysis revealed greatest selectivity for glycyl-H-1152 (0.738) and, indeed, this compound inhibited both ROCK I and II significantly more potently than any other kinase (not shown). By contrast Fasudil showed more potent inhibition of PRKX and KHS than ROCK. Strikingly, hierarchical clustering based on target spectrum clustered Rockout, Inhibitor IV and Y27632 together (Supplementary Fig. 2), despite no clear structural similarity in the compounds. In fact, the secondary targets shared by these compounds are almost all other members of the AGC kinase subfamily, demonstrating that a variety of distinct chemotypes can be employed to selectively inhibit AGC kinases, perhaps due to greater sequence divergence of this subfamily from other subfamilies. These findings illustrate the utility of the present dataset in guiding both tool compound selection and the development of new inhibitors selective for particular kinase subfamilies.
We also introduce the concept of uni-specificity as a way of quantitatively assessing the differential activity of an inhibitor toward its most sensitive and its next most sensitive kinase targets. Compounds exhibiting the greatest degree of uni-specificity are expected to provide the widest dosing window within which only a single kinase target is inhibited. We used this metric to prioritize the characterization of new inhibitor targets. Six uni-specific compounds were found that inhibit other kinases more potently than their intended targets. In all cases, these compounds represent previously unknown targets for these compounds.
While the high-throughput assay used here to systematically measure kinase activity is economical, rapid, and robust, extrapolation of these in vitro results to predict cellular efficacy must be made with caution. First our screen was carried out in the presence of 10 μM ATP regardless of the affinity of individual kinases for ATP. Potency of ATP-competitive kinase inhibitors in the cellular context is dictated not only by the intrinsic affinity of the inhibitor for the kinase, but also by the Michaelis-Menten constant for ATP binding30 and the cellular concentration of ATP. Thus, the relative rank order of inhibited kinases determined here may differ in the cellular context. Second, many kinases in the panel are represented by truncated constructs whose interactions with compound could differ in the context of the full-length kinase or in the cellular milieu. In addition, many kinases can adopt multiple conformational states and only one such state was assayed for each kinase. Third, though the kinase panel tested here is among the largest available for biochemical measurements of kinase catalytic activity, a minority of kinases are not included in the panel. Thus, additional off-target activities against untested kinases can be reasonably expected. Nevertheless, the data presented here provide a rich resource of information concerning kinase-inhibitor interactions, and biochemical analysis of kinase-inhibitor interactions generally correlates with cellular efficacy30.
Protein kinase research has been predominantly focused on a small subset of the kinome31. The identification of selective inhibitors targeting poorly understood kinases would greatly facilitate elucidation of their function. Our identification of a uni-specific inhibitor of Haspin provides one example of how large-scale kinase profiling can identify new tool compounds to stimulate new research. Crystallographic studies may also benefit from the present study. Protein kinases exhibit significant conformational plasticity that can make it difficult to obtain diffracting crystals of unliganded kinases32. ATP-competitive kinase inhibitors can be used to stabilize kinases for crystallographic structure determination3. The dataset presented here provides a library of candidates, on average nine per kinase, to support such studies. In addition, we illustrate how the present dataset can be mined to reveal new opportunities for multi-targeted kinase inhibition (Fig. 4b). Indeed, new statistical methods have been recently developed15 to facilitate analysis of potential drug polypharmacology using robust kinase-inhibitor interaction maps such as this. Finally, we expect that the inhibitor collection characterized here, with activity against the majority of human protein kinases, will be a powerful tool to elucidate kinase functions in cell models.
METHODS
Materials
Kinase inhibitors (detailed in Supplementary Table 1) were obtained either from EMD Biosciences or LC Laboratories with an average purity of >98%. A complete description of recombinant kinases used is provided in Supplementary Table 2.
Kinase assays
In vitro profiling of the 300 member kinase panel was performed at Reaction Biology Corporation (www.reactionbiology.com, Malvern, PA) using the “HotSpot” assay platform. Briefly, specific kinase / substrate pairs along with required cofactors were prepared in reaction buffer; 20 mM Hepes pH 7.5, 10 mM MgCl2, 1 mM EGTA, 0.02% Brij35, 0.02 mg/ml BSA, 0.1 mM Na3VO4, 2 mM DTT, 1% DMSO (for specific details of individual kinase reaction components see Supplementary Table 2). Compounds were delivered into the reaction, followed ~ 20 minutes later by addition of a mixture of ATP (Sigma, St. Louis MO) and 33P ATP (Perkin Elmer, Waltham MA) to a final concentration of 10 μM. Reactions were carried out at room temperature for 120 min, followed by spotting of the reactions onto P81 ion exchange filter paper (Whatman Inc., Piscataway, NJ). Unbound phosphate was removed by extensive washing of filters in 0.75% phosphoric acid. After subtraction of background derived from control reactions containing inactive enzyme, kinase activity data was expressed as the percent remaining kinase activity in test samples compared to vehicle (dimethyl sulfoxide) reactions. IC50 values and curve fits were obtained using Prism (GraphPad Software). Kinome tree representations were prepared using Kinome Mapper (http://www.reactionbiology.com/apps/kinome/mapper/LaunchKinome.htm).
Statistical methods
Outlier Detection
Raw data was measured as percentage of compound activity for each kinase-inhibitor pair in duplicate. All negative values were truncated to zero and kinase-inhibitor pairs with either missing observations or identical values across duplicates were removed from further analysis and the coefficient of variation (CV) and the difference (D) from duplicate observations were computed for each kinase-inhibitor pair. Using kernel density estimation and quantile-quantile plots, the difference D was determined to be double exponentially distributed (Supplementary Fig. 1a,b). Its location and scale parameters (and hence the mean and standard deviation) were estimated using maximum likelihood methods35. A scatter plot of CV versus D is displayed in Supplementary Figure 1e for all pairs of data points. In order to account for the inherent noise in the assay measurements, observations within 1 standard deviation of the mean of the distribution of differences D (as determined by the grey vertical lines in the double exponential density plot for D, Supplementary Fig. 1a) were retained for further analyses of compound activity. The region enclosed by these vertical lines contains 75.6% of the observations based on the estimated mean and standard deviation of this distribution. The red vertical lines in Supplementary Figure 1e also represent these limits while the green and black circles within this region represent these observations. These observations were excluded from the current set of data and the CV re-computed for the remaining kinase-inhibitor pairs.
The distribution based outlier detection method outlined by van der Loo36 was then applied to the CV based on this reduced set of data points. First, the distribution of CV was determined and its parameters estimated using methods described earlier for D35. The log-normal distribution provided the best fit for this data (Supplementary Fig. 1c,d). For outlier detection, the data (excluding the top and bottom 1%) was fit to the quantile-quantile plot positions for the log-normal distribution and its parameters were robustly estimated. A test was then performed to determine whether extreme observations are outliers by computing the threshold beyond which a certain pre-specified number of observations are expected. The pink horizontal line in Supplementary Figure 1e represents this threshold and corresponds to a CV cut-off of approximately 0.5. Based on this two-fold approach, the remainder of the observations that were located above the CV cut-off of 0.5 and outside this band, represented by blue circles, were identified as outlying observations and excluded from further analysis. The outliers (black data points) are shown within the context of the complete dataset in Supplementary Figure 1f.
Hierarchical clustering
Negative values for remaining kinase activity were truncated to zero and values in excess of 100 were truncated at that value. A re-ordered heat map of compound activity was obtained using two-way hierarchical clustering based on 1 - Spearman rank correlation as the distance metric and average linkage. No scaling was applied to the data.
Computations were carried out in the R statistical language and environment using libraries VGAM and extremevalues.
Supplementary Material
Supplementary Tables 1,2,4,5 and Supplementary Figures 1–5
Excel table of the complete pairwise kinase-compound activity dataset
Acknowledgments
We gratefully acknowledge B. Turk, A. Andrews and members of the Peterson laboratory for comments on the manuscript and R. Hartman of Reaction Biology Corp. for developing the Kinase Inhibitor Resource (KIR) web application tool. This work was supported by a W.W. Smith Foundation Award, funding from the Keystone Program in Head and Neck Cancer of Fox Chase Cancer Center and by US National Institutes of Health awards RO1 GM083025 to J.R.P. and P30 CA006927 to Fox Chase Cancer Center. HotSpot technology development was partially supported by the US National Institutes of Health (RO1 HG003818 and R44 CA114995 to H.M.).
Footnotes
Competing Financial Interests: S.W. Deacon and H. Ma are current employees of Reaction Biology Corporation.
AUTHOR CONTRIBUTIONS
The study was conceived by J.R.P, S.D., and H.M., experimental data was collected by S.D., statistical analysis was performed by K.D., data was analyzed by T.A. and J.R.P. with input from S.D. and H.M., the manuscript was written by J.R.P with input from the other authors.
References
- 1.Fabian MA, et al. A small molecule-kinase interaction map for clinical kinase inhibitors. Nat Biotechnol. 2005;23:329–336. doi: 10.1038/nbt1068. [DOI] [PubMed] [Google Scholar]
- 2.Karaman MW, et al. A quantitative analysis of kinase inhibitor selectivity. Nat Biotechnol. 2008;26:127–132. doi: 10.1038/nbt1358. [DOI] [PubMed] [Google Scholar]
- 3.Fedorov O, et al. A systematic interaction map of validated kinase inhibitors with Ser/Thr kinases. Proc Natl Acad Sci U S A. 2007;104:20523–20528. doi: 10.1073/pnas.0708800104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bain J, et al. The selectivity of protein kinase inhibitors: a further update. Biochem J. 2007;408:297–315. doi: 10.1042/BJ20070797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davies SP, Reddy H, Caivano M, Cohen P. Specificity and mechanism of action of some commonly used protein kinase inhibitors. Biochem J. 2000;351:95–105. doi: 10.1042/0264-6021:3510095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bain J, McLauchlan H, Elliott M, Cohen P. The specificities of protein kinase inhibitors: an update. Biochem J. 2003;371:199–204. doi: 10.1042/BJ20021535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ma H, Deacon S, Horiuchi K. The challenge of selecting protein kinase assays for lead discovery optimization. Exp Opin Drug Discov. 2008;3:607–621. doi: 10.1517/17460441.3.6.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smyth LA, Collins I. Measuring and interpreting the selectivity of protein kinase inhibitors. J Chem Biol. 2009;2:131–151. doi: 10.1007/s12154-009-0023-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goldstein DM, Gray NS, Zarrinkar PP. High-throughput kinase profiling as a platform for drug discovery. Nat Rev Drug Discov. 2008;7:391–397. doi: 10.1038/nrd2541. [DOI] [PubMed] [Google Scholar]
- 10.Miduturu CV, et al. High-Throughput Kinase Profiling: A More Efficient Approach toward the Discovery of New Kinase Inhibitors. Chem Biol. 2011;18:868–879. doi: 10.1016/j.chembiol.2011.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Daub H, Specht K, Ullrich A. Strategies to overcome resistance to targeted protein kinase inhibitors. Nat Rev Drug Discov. 2004;3:1001–1010. doi: 10.1038/nrd1579. [DOI] [PubMed] [Google Scholar]
- 12.Morphy R, Kay C, Rankovic Z. From magic bullets to designed multiple ligands. Drug Discov Today. 2004;9:641–651. doi: 10.1016/S1359-6446(04)03163-0. [DOI] [PubMed] [Google Scholar]
- 13.Dodson CA, et al. Crystal structure of an Aurora-A mutant that mimics Aurora-B bound to MLN8054: insights into selectivity and drug design. Biochem J. 2010;427:19–28. doi: 10.1042/BJ20091530. [DOI] [PubMed] [Google Scholar]
- 14.Graczyk PP. Gini coefficient: a new way to express selectivity of kinase inhibitors against a family of kinases. J Med Chem. 2007;50:5773–5779. doi: 10.1021/jm070562u. [DOI] [PubMed] [Google Scholar]
- 15.Metz JT, et al. Navigating the kinome. Nat Chem Biol. 2011 doi: 10.1038/nchembio.530. [DOI] [PubMed] [Google Scholar]
- 16.Bamborough P, Drewry D, Harper G, Smith GK, Schneider K. Assessment of chemical coverage of kinome space and its implications for kinase drug discovery. J Med Chem. 2008;51:7898–7914. doi: 10.1021/jm8011036. [DOI] [PubMed] [Google Scholar]
- 17.Faivre S, Djelloul S, Raymond E. New paradigms in anticancer therapy: targeting multiple signaling pathways with kinase inhibitors. Semin Oncol. 2006;33:407–420. doi: 10.1053/j.seminoncol.2006.04.005. [DOI] [PubMed] [Google Scholar]
- 18.Morphy R. Selectively nonselective kinase inhibition: striking the right balance. J Med Chem. 2010;53:1413–1437. doi: 10.1021/jm901132v. [DOI] [PubMed] [Google Scholar]
- 19.Knight ZA, Lin H, Shokat KM. Targeting the cancer kinome through polypharmacology. Nat Rev Cancer. 2010;10:130–137. doi: 10.1038/nrc2787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cockerill S, et al. Indazolylamino quinazolines and pyridopyrimidines as inhibitors of the EGFr and C-erbB-2. Bioorg Med Chem Lett. 2001;11:1401–1405. doi: 10.1016/s0960-894x(01)00219-0. [DOI] [PubMed] [Google Scholar]
- 21.Lapenna S, Giordano A. Cell cycle kinases as therapeutic targets for cancer. Nat Rev Drug Discov. 2009;8:547–566. doi: 10.1038/nrd2907. [DOI] [PubMed] [Google Scholar]
- 22.Zhang Q, et al. Discovery of EGFR selective 4,6-disubstituted pyrimidines from a combinatorial kinase-directed heterocycle library. J Am Chem Soc. 2006;128:2182–2183. doi: 10.1021/ja0567485. [DOI] [PubMed] [Google Scholar]
- 23.Lee JC, et al. A protein kinase involved in the regulation of inflammatory cytokine biosynthesis. Nature. 1994;372:739–746. doi: 10.1038/372739a0. [DOI] [PubMed] [Google Scholar]
- 24.Cuny GD, et al. Structure-activity relationship study of acridine analogs as haspin and DYRK2 kinase inhibitors. Bioorg Med Chem Lett. 2010;20:3491–3494. [PMC free article] [PubMed] [Google Scholar]
- 25.Huertas D, et al. Antitumor activity of a small-molecule inhibitor of the histone kinase Haspin. Oncogene. 2011 doi: 10.1038/onc.2011.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Patnaik D, et al. Identification of small molecule inhibitors of the mitotic kinase haspin by high-throughput screening using a homogeneous time-resolved fluorescence resonance energy transfer assay. J Biomol Screen. 2008;13:1025–1034. doi: 10.1177/1087057108326081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mencher SK, Wang LG. Promiscuous drugs compared to selective drugs (promiscuity can be a virtue) BMC Clin Pharmacol. 2005;5:3. doi: 10.1186/1472-6904-5-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liao JK, Seto M, Noma K. Rho kinase (ROCK) inhibitors. J Cardiovasc Pharmacol. 2007;50:17–24. doi: 10.1097/FJC.0b013e318070d1bd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yarrow JC, Totsukawa G, Charras GT, Mitchison TJ. Screening for cell migration inhibitors via automated microscopy reveals a Rho-kinase inhibitor. Chem Biol. 2005;12:385–395. doi: 10.1016/j.chembiol.2005.01.015. [DOI] [PubMed] [Google Scholar]
- 30.Knight ZA, Shokat KM. Features of selective kinase inhibitors. Chem Biol. 2005;12:621–637. doi: 10.1016/j.chembiol.2005.04.011. [DOI] [PubMed] [Google Scholar]
- 31.Fedorov O, Muller S, Knapp S. The (un)targeted cancer kinome. Nat Chem Biol. 2010;6:166–169. doi: 10.1038/nchembio.297. [DOI] [PubMed] [Google Scholar]
- 32.Huse M, Kuriyan J. The conformational plasticity of protein kinases. Cell. 2002;109:275–282. doi: 10.1016/s0092-8674(02)00741-9. [DOI] [PubMed] [Google Scholar]
- 33.Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 34.Cheng Y, Prusoff WH. Relationship between the inhibition constant (K1) and the concentration of inhibitor which causes 50 per cent inhibition (I50) of an enzymatic reaction. Biochem Pharmacol. 1973;22:3099–3108. doi: 10.1016/0006-2952(73)90196-2. [DOI] [PubMed] [Google Scholar]
- 35.Yee TW, Hastie TJ. Reduced-rank vector generalized linear models. Statistical Modelling. 2003;3:15–41. [Google Scholar]
- 36.van der Loo MPJ. Distribution based outlier detection for univariate data, Discussion paper 10003. Statistics Netherlands; The Hague: 2010. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Tables 1,2,4,5 and Supplementary Figures 1–5
Excel table of the complete pairwise kinase-compound activity dataset