Abstract
In E. coli, chemotactic behavior exhibits perfect adaptation that is robust to changes in the intracellular concentration of the chemotactic proteins, such as CheR and CheB. However, the robustness of the perfect adaptation does not explicitly imply a robust chemotactic response. Previous studies on the robustness of the chemotactic response relied on swarming assays, which can be confounded by processes besides chemotaxis, such as cellular growth and depletion of nutrients. Here, using a high-throughput capillary assay that eliminates the effects of growth, we experimentally studied how the chemotactic response depends on the relative concentration of the chemotactic proteins. We simultaneously measured both the chemotactic response of E. coli cells to L-aspartate and the concentrations of YFP-CheR and CheB-CFP fusion proteins. We found that the chemotactic response is fine-tuned to a specific ratio of [CheR]/[CheB] with a maximum response comparable to the chemotactic response of wild-type behavior. In contrast to adaptation in chemotaxis that is robust and exact, capillary assays revealed that the chemotactic response in swimming bacteria is fined-tuned to wild-type level of the [CheR]/[CheB] ratio.
Keywords: chemotaxis, Escherichia coli, capillary assay
Introduction
Chemotaxis in E. coli is a locomotion system that allows bacteria to move toward gradients of attractant chemicals, such as nutrients, or away from for toxic molecules that act as repellants. When bacteria are submerged in a liquid environment, they can swim using flagella that are powered by rotary motors. When all motors rotate counterclockwise (CCW), they form a bundle of flagella that acts like a propeller. Under this condition, swimming bacteria are described by long and smooth trajectories called runs. When most of the motors rotate clockwise (CW), the bundle breaks apart, and bacteria tumble. This alternating behavior of runs and tumbles gives rise to a random walk, which bacteria use to explore homogenous environments. By contrast, this system allows the bacteria to respond to sudden increases in nutrients in the environment by lowering the frequency of tumbles, which biases the random trajectory of bacteria towards the source of attractants. In this case, an abrupt drop in tumbling frequency is followed by an adaptive behavior in which the tumbling frequency returns exactly to its pre-stimulus level. Experimental and theoretical studies demonstrated that the property of exact adaptation is robust to variations of the chemotactic proteins levels [8,5]. Although perfect adaptation is an important characteristic of bacterial chemotaxis in E. coli, it is not clear if it implies that the chemotactic response itself is robust to variations in the concentrations of the chemotactic proteins.
To investigate how the chemotactic response depends on the concentrations of the chemotactic proteins, in an elegant study, Løvdok et al. used swarming assays on agar plates. They observed that the chemotactic efficiency varies weakly as a function of the chemotactic protein concentrations [16]. Although the swarming assay [1,27] is a convenient and standard approach to characterize chemotactic behaviors, it may often be difficult to accurately quantify the chemotactic response as a function of protein levels. The main difficulty lies in the fact that the swarming behavior of bacteria is accompanied with cell growth [9], which results in a population of cells with heterogeneous protein concentrations. These conditions make the swarming assay difficult to quantitatively interpret because we do not know whether the observed results are due to chemotaxis or the growth of bacteria or both.
An alternative assay commonly used is the capillary assay, initially developed by [3,19,2]. While this assay allows us to determine the chemotactic response associated with the swimming behavior decoupled from the growth of bacteria, it is also known to exhibit variable results from assay to assay. To circumvent this drawback, we quantitatively characterized the chemotactic behavior at the population level using a high-throughput capillary assay. This assay has been extended from [7] and is based on the parallel measurements of a large number of identical classic capillary assays [2]. This large number of measurements allows us to produce a robust average and to determine the chemotactic response in a reliable manner.
In E. coli, the bacterial chemotaxis pathway is one of the most well-characterized signal transduction systems [8,23,45,24]. When nutrients (or toxic molecules) bind to a specific receptor in the receptor-kinase complexes, the chemicals initiate a fast decrease (or increase) of activity of the kinase CheA [10]. After this fast initial response, the kinase activity adapts back to its pre-stimulus steady-state. The methylation/demythylation of the receptor-complexes by the methyltransferase CheR and the demethylatransferase CheBp (the phosphorylated form of CheB) govern this adaptive behavior [25]. Downstream in the pathway, the phosphorylation of the diffusible response regulator, CheY, relays the variations in activity of the kinase CheA [25]. The phosphorylated form CheYp binds preferentially to the basal part of the rotary motor (FliM) [28,29,26], and induces tumbles.
The steady-state of the bacterial motor is defined by its CW bias, which is the proportion of time spent in CW rotation. The CW bias of the bacterial motor has a one-to-one functional relationship to the steady concentration of the signaling protein CheYp [11]. The CW bias and its corresponding CheYp concentration change quickly, reflecting the changes in kinase activity in response to an external stimulus, but slowly adapt back to the pre-stimulus state through the methylation-driven adaptive mechanism. This perfect adaptation is robust to the variations of the chemotactic proteins CheR and CheB [5], whose ratio determines the steady-state of the kinase activity in the adaptive network [15].
With the high-throughput capillary assay, we determined if the chemotactic response was also robust to the relative concentration of CheR and CheB. To measure [CheR] and [CheB], we created two fluorescent reporter fusion constructs, yfp-cheR and cheB-cfp, under the control of distinct inducible promoters. As a proxy for protein concentration, we measured the fluorescent intensities of the fusion proteins YFP-CheR and CheB-CFP in vivo and correlated them with the chemotactic responses from the same cells. These experiments allowed us to establish the functional relationship between population chemotactic response and [CheR] and [CheB] relative to wild-type levels. To characterize how effectively bacteria can respond to a gradient of attractant, we used metrics that are specific either to the swarming assay, ‘chemotactic efficiency’, or to the capillary assay, ‘chemotactic response’.
Materials and Methods
Bacterial strains and plasmids
RP437 is a wild-type E. coli strain for chemotaxis [20]. RP4968 is derived from the RP437 strain and is ΔcheR [20]. RP4968 cells were complemented with a LacR-inducible plasmid, pZA1tac-YFP-CheR, encoding the yfp-cheR fusion gene. To construct pZA1tac-YFP-CheR, the tac promoter and yfp-cheR fusion gene were PCR amplified from the plasmid pVS127 (gift from V. Sourjik [6]) with the primers VS127-5’ 5’-ggc tcg agg cgc cga cat cat aac gg-3’ and VS127-3’ 5’-cgc caa aac agc caa gct tgc-3’. The PCR fragment was cloned into the XhoI and HindIII sites of a pZA1 series plasmid that contains an ampicillin resistance cassette [18]. The tac promoter allows the plasmid to be LacR inducible. RP4992 is derived from the RP437 strain and is ΔcheB (gift from J.S. Parkinson). RP4992 cells were complemented with a TetR-inducible plasmid, pZE21-CheB-CFP, expressing CheB-CFP fusion proteins. To construct the plasmid pZE21-CheB-CFP, we PCR amplified the cheB-cfp fusion gene from the plasmid pVS103 (gift from V. Sourjik, CheB-eCFP in pBAD33 [13]) with primers SCheB-5’: 5’-gcg gta ccg cat gag caa aat cag ggt gtt atc-3’ and SCfp-3’: 5’-gct cta gat tac ttg tac agc tcg tcc atg cc-3’. The gene fragment was inserted into the KpnI and XbaI sites in a pZE21 series plasmid that contains a kanamycin resistance cassette and a TetR inducible promoter [18]. The RP2867 strain was derived from RP437 with a deletion of cheR and cheB genes [22]. Cells were complemented with pZA1tac-YFP-CheR and pZE21-CheB-CFP. The plasmid pUHS-IntI [17] encoding constitutively expressed lacR and tetR, controlled the expression of cheR and cheB gene fusions from plasmids. This plasmid carries a spectinomycin resistance gene.
Swarming assay
We grew wild-type (RP437) and mutant cells overnight in 3 mL Tryptone broth at 35°C shaking at 200 rpm. We inoculated swarming plates (1% Bacto-tryptone, 1% NaCl and 0.3% agar in distilled water) with 1 µL of the overnight culture. To induce different concentrations of YFP-CheR and CheB-CFP, the swarming plates contained various concentrations of isopropyl-β-D-thiogalactoside (IPTG) ([IPTG]=1000/20, 1000/21, 1000/22, 1000/23, 1000/24, 1000/25, 1000/26, 1000/27, 1000/28, 1000/29 and 0 µM for single mutant experiments and [IPTG]=1000/20, 1000/21, 1000/22, 1000/23, 1000/24, 1000/25, 1000/26, 1000/27 and 0 µM for double mutant experiments) and anhydrotetracycline ([aTc]= 200/20, 200/21, 200/22, 200/23, 200/24, 200/25, 200/26, 200/27, 200/28, 200/29 and 0 ng/mL for single gene mutant experiments and [aTc]= 200/22, 200/23, 200/24, 200/25, 200/26, 200/27 and 0 ng/mL for double gene mutant experiments), respectively. We also added antibiotics (ampicillin: 100µg/mL, kanamycin: 40µg/mL, or spectinomycin: 50µg/mL) to the swarming plates depending on the plasmids used. We incubated the swarming plates at 30°C for 12 hours and measured the diameters of the swarming rings.
We estimated the expression levels of [YFP-CheR] and/or [CheB-CFP] from the relationship between the fluorescence intensity and the inducer concentrations ([IPTG]=0, 5, 10, 15, 30, 40, 80, 200 and 1000 µM; [aTc] =0, 2, 3, 5, 6, 7, 8, 9, 10, 12 and 15 ng/mL) (Fig. S1) in the capillary assay (see below). We measured the chemotactic efficiency, defined as the ratio of the diameters of the swarming rings of the mutant cells and wild-type (RP437) cells after 12 hours swarming at 30°C.
Capillary assay
The capillary assay is based on ref [2,7]. We grew cells overnight in 3 mL Tryptone broth at 35°C shaking at 200 rpm. We diluted the overnight culture 1:50 in 12 mL fresh Tryptone broth in a 250mL flask and grew the cells at 35°C shaking at 200 rpm until the optical density (O.D.) reached ~0.3 at 600 nm. To induce different concentrations of YFP-CheR and CheB-CFP, we varied the concentrations of IPTG and aTc ([IPTG] = 0, 5, 15, 30 and 1000 µM; [aTc] = 0, 3, 5, 7, 8 and 9 ng/mL) for single mutant experiments. For double mutant experiments, we used various combinations of the concentrations of IPTG and aTc ([IPTG] = 0, 10, 20, 30, 40, 50, 60, 100, 200, 500 and 1000 µM and [aTc] = 0, 5, 10, 20, 25, 30, 35, 38, 40, 50, 50, 100 and 200 ng/mL). We added antibiotics (ampicillin: 100µg/mL, kanamycin: 40µg/mL, or spectinomycin: 50µg/mL) specific to each plasmid. We washed the harvested cells (centrifuged twice at 3000 rpm for 5 min) and resuspended them in motility medium (0.1 mM EDTA, 0.1 mM L-methionine, 10mM potassium phosphate, pH 7.0). After the cells were washed, we measured [YFP-CheR] (excitation filter: 504/12 mm, emission filter: 540/10 nm) and/or [CheB-CFP] (excitation filter: 425/8 nm, emission filter: 486/10 nm) with a plate reader (Wallac 1420 Victor2, Perkin-Elmer).
We diluted the cells in motility medium to O.D.600 nm ~ 0.012 (corresponding to approximately ~7×106 cells/mL). We distributed 300 µL of this diluted cell solution into each well of a 96-well round bottom plate (Pro-bind™ U-Bottom, polystyrene, Falcon). In each plate, we divided the 12 columns into 6 adjacent column pairs. Each column pair contained cells grown at the same [IPTG] and/or [aTc].
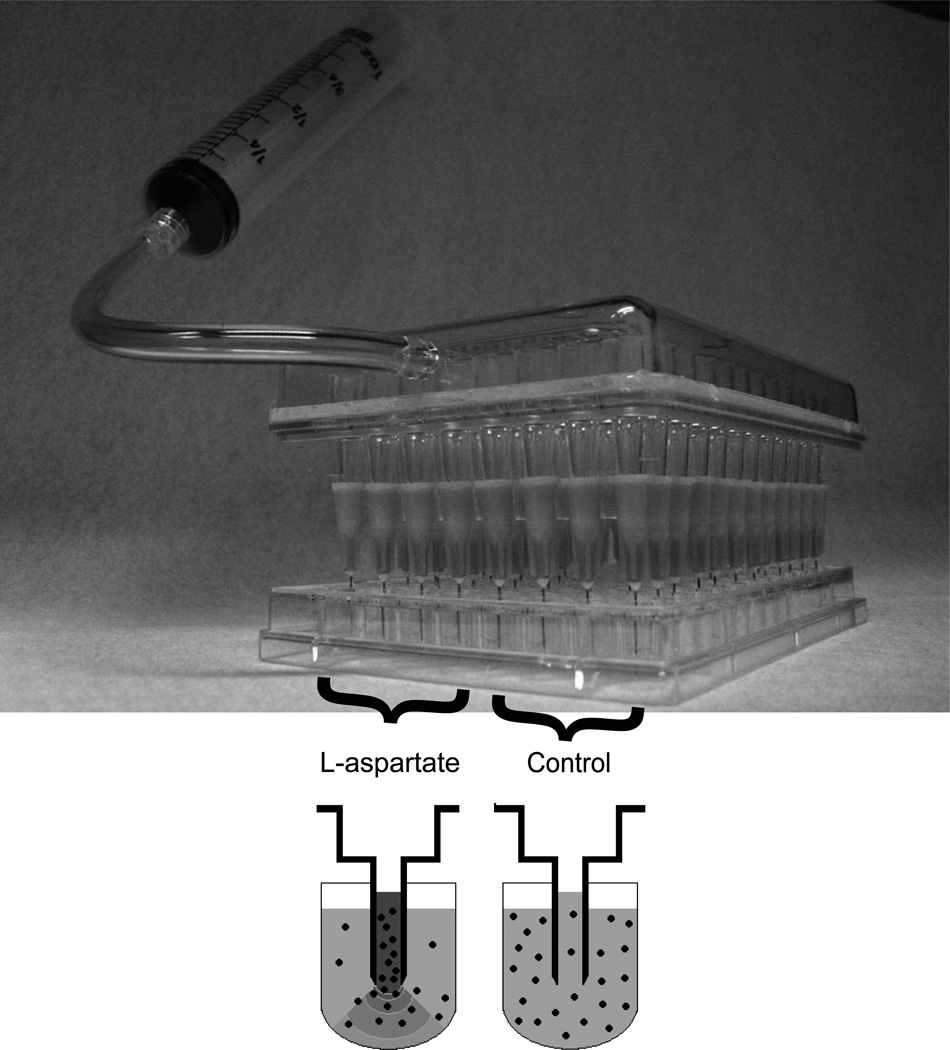
To build the capillary system, we attached 96 narrow needles (inner diameter = 0.19mm, 27 G ½ needles, B-D) to the ends of the pipetting entrances of a 96-well pipetting device (Vaccu-Pette/96™, Scienceware). We positioned the capillaries approximately 3.5 mm above the center bottom of each well of the 96-well plate containing the 300 µL of motility medium with and without 10−2 M L-aspartate. By slowly withdrawing the syringe pump, we filled the capillaries with either motility medium (lower 48 wells) or motility medium containing 10−2 M of the attractant L-aspartate (upper 48 wells) (Fig. 1). We kept the media in the capillaries by locking the valve connected to the syringe pump. We placed the capillary device containing motility medium with or without L-aspartate approximately 3.5 mm above the bottom of each well in the 96-well round bottom plate containing the cell solutions.
Fig. 1. Schematic of the high-throughput capillary apparatus.
In the high-throughput capillary apparatus, the upper 48 capillaries (left) contained 10−2 M L-aspartate in motility medium and the lower 48 capillaries (right) contained only motility medium. We filled the capillaries with the medium by slowly pumping the syringe (Methods) and maintained the medium within the capillaries by locking a valve. When the capillary system was placed in a 96-well plate containing cells in motility medium, the L-aspartate solution in the upper capillaries produced a gradient in the wells, attracting the cells towards the capillary entrance (lower left). In the lower control wells, cells entered the capillaries exclusively by diffusion (lower right).
In the upper 48 capillaries, the L-aspartate solution in the capillaries created an L-aspartate concentration gradient in the wells. We incubated the capillary system at 30°C for 40 min and then emptied the capillaries into a new 96-well flat bottom plate (Clear Polystyrene flat bottom 96 wells, 300 µL, Whatman) by strongly pumping the capillary system with a 50 mL syringe. We added 200 µL LB to each well and covered it with 160 µL light mineral oil to prevent evaporation. We grew the cells overnight at 37°C with fast shaking inside the Wallac, measuring O.D.600nm of each well every 15 min. The chemotactic response (R) is then defined as the ratio of the number of cells in the capillary with the attractant and the number of cells in the capillary without the attractant. To count the number of cells in the capillaries, we used the method described in [7] with parameter b = 5277.2. We normalized the relative protein concentrations such that chemotactic responses in both single and double mutant experiments peaked at [YFP-CheR] = 1 and [CheB-CFP] = 1. All other protein concentrations are given relative to this wild-type level.
Results and Discussion
Theoretical estimate
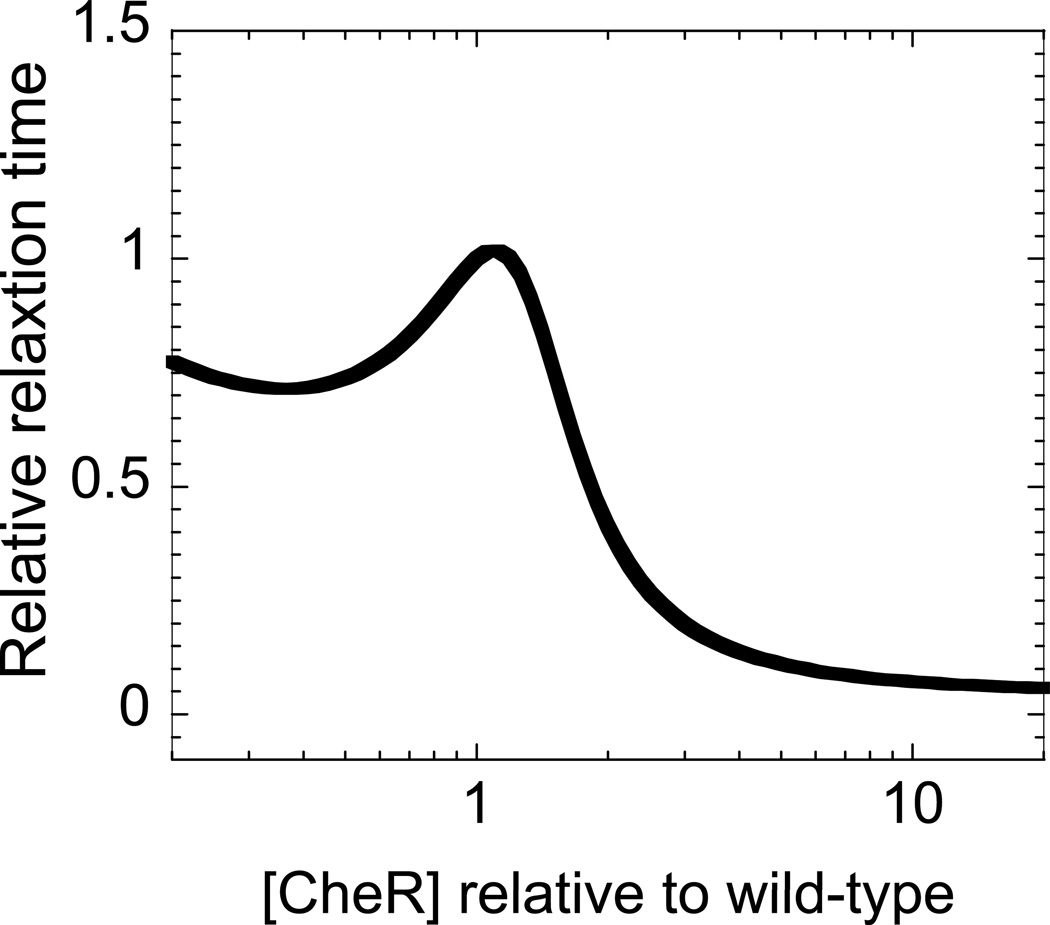
A recent kinetic model [12] of the bacterial chemotactic network showed that the relaxation time associated with the response to a small external stimulus (attractant increase) peaks at the wild-type [CheR]/[CheB] (Fig. 2). We observed that cells operating at the average wild-type level of [CheR] and [CheB] exhibit the longest relaxation time. This kinetic model also suggests that the relaxation time governs the chemotactic response and, therefore, cells with the largest relaxation time should also exhibit the largest response, that is, a longer drift toward the gradient of attractant. Another key result of this kinetic model is that the chemotactic response peaks for wild-type levels of CheR and CheB. We experimentally tested this prediction with swarming and capillary assays.
Fig. 2. Relative relaxation time as a function of [CheR].
Relative relaxation time is defined as the relaxation time associated with the total CheA kinase activity to a small external stimulus, normalized by the relaxation time of wild-type cells based on the model in ref. 12. [CheR] is calibrated relative to the wild-type level. The data is plotted from Figure 2D in ref. 12.
Swarming experiments
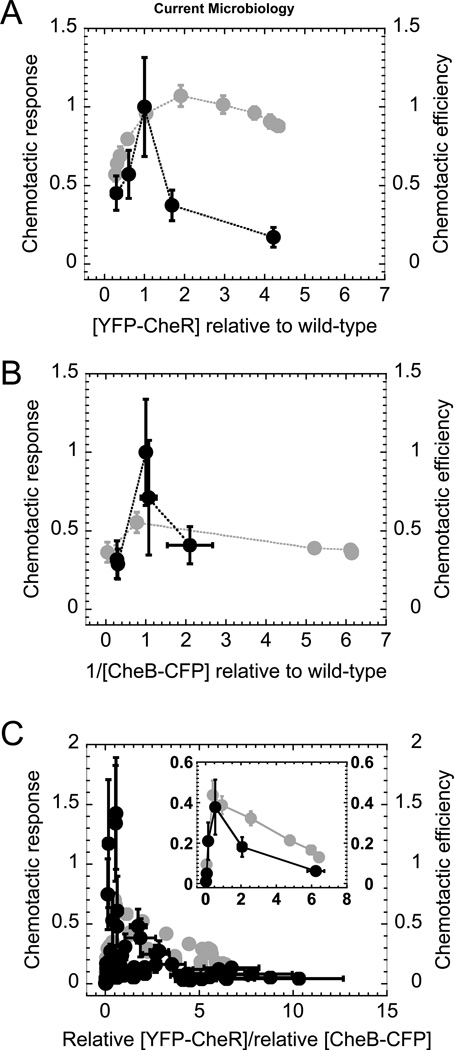
To characterize how the chemotactic efficiency is controlled by various levels of CheR and CheB, we first performed swarming assays by varying the expression of either a single gene (cheR or cheB) (Figs. 3A and 3B) or simultaneously two genes (Fig. 3C). We used ΔcheR and ΔcheB cells complemented with different YFP-CheR (for ΔcheR and ΔcheRΔcheB cells) and CheB-CFP (for ΔcheB and ΔcheRΔcheB cells) concentrations. We calibrated [YFP-CheR] and [CheB-CFP] relative to wild-type levels that re-establish the wild-type chemotaxis response from capillary experiments (methods) in the complemented mutants. We then used the relationship between inducer concentration and the [YFP-CheR] and [CheB-CFP] to estimate their concentration in swarming experiments (Fig. S1). To characterize how effectively bacteria can respond to a gradient of attractant, we defined (Methods) metrics that are specific either to swarming, called ‘chemotaxis efficiency’, or to capillary assays, called ‘chemotaxis response’. For relative [YFP-CheR] less than wild-type levels in the ΔcheR mutant cells, the chemotactic efficiency decreases sharply with [YFP-CheR]. By contrast, when [YFP-CheR] is greater than the wild-type level, we observed no significant change in the chemotactic efficiency compared to that of wild-type level. The highest [YFP-CheR], fivefold higher than wild-type levels, resulted in a slight decrease in chemotactic efficiency from that of wild-type. In the ΔcheB mutant cells, the maximum chemotactic efficiency at wild type CheB-CFP levels was roughly half of the wild-type value. In the double mutant cells, the chemotactic efficiency peaked near the ratio of relative concentrations [YFP-CheR]/[CheB-CFP] ~1, where the chemotactic efficiency was roughly half of that observed in wild-type cells. At lower [YFP-CheR]/[CheB-CFP] ratios, the chemotactic efficiency decreases steeply, but at higher ratios, it decreases weakly as a function of the ratio.
Fig. 3. Effect of relative chemotactic protein concentration on chemotactic behavior.
We measured the effects on chemotaxis of varying the relative concentrations of (A) YFP-CheR in ΔcheR cells (RP4968), (B) CheB-CFP in ΔcheB mutant cells (RP4992), and (C) both YFP-CheR and CheB-CFP in ΔcheR-ΔcheB cells (RP2867). In the capillary assay (black), we measured chemotactic behavior as the chemotactic response for various inducer concentrations (Methods). We normalized the chemotactic response such that it peaked at chemotactic response = 1. Protein concentrations were normalized such that chemotactic responses peaked at (A) [YFP-CheR] = 1, (B) [CheB-CFP] = 1. For (C) we kept the same normalizations for [YFP-CheR] and [CheB-CFP] as in (A) and (B). In the swarming assay (grey), we measured chemotactic behavior as chemotactic efficiency (Methods). For chemotactic efficiency, we estimated the relative [YFP-CheR] and 1/[CheB-CFP] values from the inductions curves for the single gene (Fig. S1) and double gene (Figs. S2 and S3) mutants. (B) Low [CheB-CFP] (high 1/[CheB-CFP]) have large error bars because these expression levels are close to the background level, and we do not show data for 1/[CheB-CFP] more than threefold higher than the wild-type level. (C, inset) We binned the data by the relative [YFP-CheR]/[CheB-CFP] (chemotactic response: 15 data points per bin. Chemotactic efficiency: 9 data points per bin) and averaged the functional relationship between the chemotactic response or the chemotactic efficiency and the relative [YFP-CheR]/[CheB-CFP] for each bin. Error bars are the standard error.
Capillary experiments
We performed capillary experiments in media that does not support cell growth, making the results easier to interpret than those of the swarming assay. We used the same strains and plasmid constructs as in the swarming assays. When we varied CheR and CheB independently, we found that the maximal chemotactic responses were approximately comparable to that of wild-type (Fig. 3A and 3B). In addition to the single gene mutants, we also performed the capillary experiments for the CheR and CheB double deletion strain (ΔcheR-ΔcheB, RP2867) transformed with the same plasmids and induced at various YFP-CheR and CheB-CFP levels (Figs. S2 and S3). In the double mutant cells, the chemotactic response peaked near the relative ratio [YFP-CheR]/[CheB-CFP] ~1 (Fig. 3C) to a value that approximately corresponded to the wild-type chemotactic response. We hypothesize that the lower peak in Fig. 3C (half of the peak height in Fig. 3B) is due to gene expression noise, which is larger when cheR and cheB are expressed from plasmids than when they are expressed directly from the chromosome [21]. When both genes are expressed from plasmids, it is expected that the noise smoothes out the chemotactic response by averaging the responses of single cells that have very different CheR and CheB expression levels in the same population. According to this hypothesis, wild-type cells should have an even higher peak in chemotactic response than that observed in Fig 3B. Moreover, when cheR and cheB are both expressed from distinct plasmids, the resulting noise is even larger than when only a single gene is expressed from a plasmid and smoothes out the average response in such a way that it is smaller than that of wild-type cells.
Because the chemotactic response in capillary assays is governed solely by the swimming behavior of bacteria and not by their growth, we can directly interpret the chemotactic response as a function of protein concentration of chemotaxis proteins. We found that the chemotactic response from the capillary assays peaked more sharply and, therefore, was more fine-tuned to the wild-type [CheR]/[CheB] compared to the swarming assay results. The chemotactic response collapsed to near zero at high and low [YFP-CheR], whereas the chemotactic efficiency from swarming assays remained nonzero for large concentrations of CheR and CheB.
Using high-throughput capillary assays allowed us to establish that the chemotactic response of a population of bacteria is fine-tuned and depends sharply on a specific ratio of CheR and CheB. This experimental result is in agreement with the kinetic model [12] that predicted how the relaxation time in response to a small stimulus varies as a function of [CheR]/[CheB]. In this model, the relaxation time was used as a proxy to estimate how the chemotactic response would vary with [CheR]/[CheB] and was also found to peak sharply in wild-type cells. Altogether, these results suggest that the ratio [CheR]/[CheB] is fined-tuned to a specific value that optimizes chemotaxis. For example, cheR and cheB are contiguous genes in meche operon, an arrangement that was found to reduce expression noise between the two genes [14,12]. Additionally, the negative-feedback loop on CheB is known to maintain a specific [CheR]/[CheB-P] value [14]. Our results suggest that the selection of such mechanisms may be the result of a strong evolutionary pressure to maintain bacteria with efficient chemotactic behavior.
Supplementary Material
Acknowledgments
We thank V. Sourjik (Heidelberg) for plasmids pVS103 and pVS127 and J.S. Parkinson (U. of Utah) for the RP4992 strain. Wendy Grus provided editorial assistance.
References
- 1.Adler J. Chemotaxis in bacteria. Science. 1966;153:708–716. doi: 10.1126/science.153.3737.708. [DOI] [PubMed] [Google Scholar]
- 2.Adler J. A method for measuring chemotaxis and use of the method to determine optimum conditions for chemotaxis by Escherichia coli. J Gen Microbiol. 1973;74:77–91. doi: 10.1099/00221287-74-1-77. [DOI] [PubMed] [Google Scholar]
- 3.Adler J, Dahl MM. A Method for Measuring Motility of Bacteria and for Comparing Random and Non-Random Motility. J Gen Microbiol. 1967;46:161. doi: 10.1099/00221287-46-2-161. [DOI] [PubMed] [Google Scholar]
- 4.Alon U, Camarena L, Surette MG, et al. Response regulator output in bacterial chemotaxis. EMBO J. 1998;17:4238–4248. doi: 10.1093/emboj/17.15.4238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alon U, Surette MG, Barkai N, et al. Robustness in bacterial chemotaxis. Nature. 1999;397:168–171. doi: 10.1038/16483. [DOI] [PubMed] [Google Scholar]
- 6.Amann E, Ochs B, Abel KJ. Tightly regulated tac promoter vectors useful for the expression of unfused and fused proteins in Escherichia coli. Gene. 1988;69:301–315. doi: 10.1016/0378-1119(88)90440-4. [DOI] [PubMed] [Google Scholar]
- 7.Bainer R, Park H, Cluzel P. A high-throughput capillary assay for bacterial chemotaxis. J Microbiol Methods. 2003;55:315–319. doi: 10.1016/s0167-7012(03)00112-x. [DOI] [PubMed] [Google Scholar]
- 8.Barkai N, Leibler S. Robustness in simple biochemical networks. Nature. 1997;387:913–917. doi: 10.1038/43199. [DOI] [PubMed] [Google Scholar]
- 9.Belas R, Erskine D, Flaherty D. Proteus mirabilis mutants defective in swarmer cell differentiation and multicellular behavior. J Bacteriol. 1991;173:6279–6288. doi: 10.1128/jb.173.19.6279-6288.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bilwes AM, Alex LA, Crane BR, et al. Structure of CheA, a signal-transducing histidine kinase. Cell. 1999;96:131–141. doi: 10.1016/s0092-8674(00)80966-6. [DOI] [PubMed] [Google Scholar]
- 11.Cluzel P, Surette M, Leibler S. An ultrasensitive bacterial motor revealed by monitoring signaling proteins in single cells. Science. 2000;287:1652–1655. doi: 10.1126/science.287.5458.1652. [DOI] [PubMed] [Google Scholar]
- 12.Emonet T, Cluzel P. Relationship between cellular response and behavioral variability in bacterial chemotaxis. Proc Natl Acad Sci U S A. 2008;105:3304–3309. doi: 10.1073/pnas.0705463105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guzman LM, Belin D, Carson MJ, et al. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kollmann M, Lovdok L, Bartholome K, et al. Design principles of a bacterial signalling network. Nature. 2005;438:504–507. doi: 10.1038/nature04228. [DOI] [PubMed] [Google Scholar]
- 15.Li M, Hazelbauer GL. Cellular stoichiometry of the components of the chemotaxis signaling complex. J Bacteriol. 2004;186:3687–3694. doi: 10.1128/JB.186.12.3687-3694.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lovdok L, Kollmann M, Sourjik V. Co-expression of signaling proteins improves robustness of the bacterial chemotaxis pathway. J Biotechnol. 2007;129:173–180. doi: 10.1016/j.jbiotec.2007.01.024. [DOI] [PubMed] [Google Scholar]
- 17.Lutz R. Ph.D Thesis. Heidelberg; 1996. [Google Scholar]
- 18.Lutz R, Bujard H. Independent and tight regulation of transcriptional units in Escherichia coli via the LacR/O, the TetR/O and AraC/I1-I2 regulatory elements. Nucleic Acids Res. 1997;25:1203–1210. doi: 10.1093/nar/25.6.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mesibov R, Adler J. Chemotaxis toward Amino-Acids in Escherichia-Coli. J Bacteriol. 1972;112:315. doi: 10.1128/jb.112.1.315-326.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Parkinson JS, Houts SE. Isolation and behavior of Escherichia coli deletion mutants lacking chemotaxis functions. J Bacteriol. 1982;151:106–113. doi: 10.1128/jb.151.1.106-113.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Paulsson J. Summing up the noise in gene networks. Nature. 2004;427:415–418. doi: 10.1038/nature02257. [DOI] [PubMed] [Google Scholar]
- 22.Sherris D, Parkinson JS. Posttranslational processing of methyl-accepting chemotaxis proteins in Escherichia coli. Proc Natl Acad Sci U S A. 1981;78:6051–6055. doi: 10.1073/pnas.78.10.6051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Spiro PA, Parkinson JS, Othmer HG. A model of excitation and adaptation in bacterial chemotaxis. Proc Natl Acad Sci U S A. 1997;94:7263–7268. doi: 10.1073/pnas.94.14.7263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stock AM, Robinson VL, Goudreau PN. Two-component signal transduction. Annu Rev Biochem. 2000;69:183–215. doi: 10.1146/annurev.biochem.69.1.183. [DOI] [PubMed] [Google Scholar]
- 25.Stock JB, Lukat GS, Stock AM. Bacterial chemotaxis and the molecular logic of intracellular signal transduction networks. Annu Rev Biophys Biophys Chem. 1991;20:109–136. doi: 10.1146/annurev.bb.20.060191.000545. [DOI] [PubMed] [Google Scholar]
- 26.Toker AS, Macnab RM. Distinct regions of bacterial flagellar switch protein FliM interact with FliG, FliN and CheY. J Mol Biol. 1997;273:623–634. doi: 10.1006/jmbi.1997.1335. [DOI] [PubMed] [Google Scholar]
- 27.Tso WW, Adler J. Negative chemotaxis in Escherichia coli. J Bacteriol. 1974;118:560–576. doi: 10.1128/jb.118.2.560-576.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Welch M, Oosawa K, Aizawa S, et al. Phosphorylation-dependent binding of a signal molecule to the flagellar switch of bacteria. Proc Natl Acad Sci U S A. 1993;90:8787–8791. doi: 10.1073/pnas.90.19.8787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Welch M, Oosawa K, Aizawa SI, et al. Effects of phosphorylation, Mg2+, and conformation of the chemotaxis protein CheY on its binding to the flagellar switch protein FliM. Biochemistry. 1994;33:10470–10476. doi: 10.1021/bi00200a031. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.