Abstract
Autotransporters are secreted proteins produced by pathogenic Gram-negative bacteria. They consist of a membrane embedded β-domain and an extracellular passenger domain that is sometimes cleaved and released from the cell surface. We solved the structures of three non-cleavable mutants of the autotransporter EspP to examine how it promotes asparagine cyclization to cleave its passenger. We found that cyclization is facilitated by multiple factors. The active site asparagine is sterically constrained to conformations favorable for cyclization while electrostatic interactions correctly orient the carboxamide group for nucleophilic attack. During molecular dynamics simulations, water molecules were observed to enter the active site and form hydrogen bonds favorable for increasing the nucleophilicity of the active site asparagine. When the activated asparagine attacks its main chain carbonyl carbon the resulting oxyanion is stabilized by a protonated glutamate. Upon cleavage, this proton could be transferred to the leaving amine group helping overcome a significant energy barrier. Together these findings provide insight into factors important for asparagine cyclization, a broadly used mechanism for protein cleavage.
Keywords: EspP, autocleavage, outer membrane protein, crystal structure, asparagine cyclization
Introduction
Autotransporters are a large family of secreted proteins found in Gram-negative bacteria that are typically involved in virulence. They consist of two domains, an N-terminal passenger domain and a C-terminal β-domain. The β-domain is embedded in the outer membrane while the passenger domain is secreted to the extracellular space and contains the effector function 1. Passenger domains range in size but are usually large and can be over 3000 residues in length 2. Some of their virulence functions include promoting actin based motility 3; 4, biofilm formation 5, and cell vacuolation 6. There are two known subtypes of autotransporters, the monomeric autotransporters (also referred to as classical/conventional autotransporters) and the trimeric autotransporters.
For the monomeric autotransporters, the β-domain is comprised of a 12-stranded β-barrel that sits in the outer membrane and is formed by the C-terminal 250 – 300 residues 7; 8; 9; 10; 11; 12. A single polypeptide segment that passes through the barrel pore connects the surface exposed passenger to the first β-strand of the barrel. For the trimeric autotransporters, the β-domain is formed by ~70 residues located at the C-terminus. This domain contains a 4-stranded β-sheet that oligomerizes to form a trimeric 12-stranded β-barrel that is structurally very similar to the monomeric autotransporter barrels 8. For the trimeric autotransporters, the surface exposed passenger domain is also trimeric with each monomer connected to the β-domain by a polypeptide segment that passes through the barrel pore.
Passenger domain translocation is not well understood for either the monomeric or trimeric autotransporters. For the monomeric autotransporters, translocation models showing the passenger domain using the central pore of the β-domain or a pore created by the Bam complex have been proposed 9 (Supplementary Figure S1). The Bam complex (also known as the Omp85 or YaeT complex) assembles β-barrel outer membrane proteins into the outer membrane 13. In both models, a hairpin intermediate is present during translocation for the monomeric autotransporters because the C-terminus of the passenger is surface exposed while its N-terminus is still in the periplasm 14; 15. Folding above the cell surface, at the tip of the hairpin, would simultaneously prevent the passenger from slipping back through the translocation pore and provide energy to pull the N-terminal portion of the passenger from the periplasm to the cell surface.
After translocation, some monomeric autotransporters cleave their passenger domains allowing them to be released from the cell surface 16. For members of the SPATE (Serine Protease Autotransporters of Enterobacteriaceae) family of autotransporters, cleavage occurs by asparagine cyclization inside the β-domain pore 17 and can take less than a minute 16. In peptides, asparagine cyclization is slow (t1/2 = days) and an alternate cyclization pathway that results in deamidation of the asparagine rather than cleavage is favored 18. For this study we wanted to further examine the cleavage mechanism of the SPATE autotransporter EspP produced by E. coli O157:H7. Specifically, we wanted to visualize the active site prior to cleavage in order to determine how its conformation and the residues that surround it could facilitate asparagine cyclization. To do this we solved the pre-cleavage structures of three non-cleavable mutants of EspP. While our study was underway, the pre-cleavage structure of another SPATE, Hbp, was reported 10. The structures of Hbp and EspP are very similar and, since they are both SPATEs, they share the same cleavage mechanism. Comparison of the Hbp and three EspP pre-cleavage structures allowed us to see a conformational change that is likely an artifact that occurs when aspartate is substituted for the active site asparagine. This substitution was used by Tajima et al. to trap Hbp in its pre-cleavage state and it affects the proposed catalytic water molecule in their cleavage mechanism. We used molecular dynamics simulations to reveal potential alternate catalytic waters in EspP and Hbp that could increase the nucleophilicity of the active site asparagine to initiate cyclization. We report a modified version of the cleavage mechanism where a catalytic water molecule is positioned by one to three conserved acidic residues near the cleavage site. Additionally, further analysis of the EspP and Hbp pre-cleavage structures revealed how the active site asparagine side chain is sterically constrained to rotamers favorable for cyclization and its carboxamide group is correctly oriented over its main chain carbonyl carbon by electrostatics. Finally, we also identified a potential proton transfer event that would reduce the energy required for cleavage.
Results and Discussion
Construct design
Full length EspP consists of 1300 residues: residues 1–55 contain the signal peptide, residues 56-1023 contain the passenger domain, and residues 1024–1300 contain the β-domain. Asn1023 catalyzes cleavage of EspP between positions 1023 and 1024 by asparagine cyclization 19. To trap EspP in its pre-cleavage state we made substitutions at this residue to alanine (N1023A), serine (N1023S), or aspartate (N1023D). To facilitate purification and crystallization, we truncated the passenger domain near the β-domain at Ala999 and placed a 6XHIS tag at the N-terminus.
Crystals of the EspP pre-cleavage mutants were grown in the presence of the detergents lauryldimethylamine-N-oxide and tetraethylene glycol monooctyl ether. All three structures were solved at ~2.5 Å resolution by molecular replacement using the post cleavage structure of EspP 7 (PDB ID: 2QOM). Data collection and refinement statistics are shown in Table 1. Representative election density for each structure can be seen in Supplementary Figure S2.
Table 1.
Crystallographic data collection and refinement statistics
| EspP N1023A | EspP N1023D | EspP N1023S | |
|---|---|---|---|
| Data collection | |||
| Space group | P212121 | P212121 | P212121 |
| Cell dimensions | |||
| a, b, c (Å) | 31.1, 122, 124 | 31.1, 122, 123 | 31.2, 122, 122 |
| α, β, γ (°) | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 |
| Resolution (Å) | 50.0 - 2.48 | 50.0 - 2.52 | 50.0 - 2.47 |
| Rmerge | 0.087 (0.618) | 0.076 (0.570) | 0.079 (0.590) |
| I/σI | 16.7 (2.86) | 16.6 (1.97) | 18.6 (2.48) |
| Completeness (%) | 99.4 (100) | 98.6 (95.6) | 98.9 (97.9) |
| Redundancy | 4.8 (4.8) | 4.5 (3.9) | 5.4 (4.9) |
| Refinement | |||
| Resolution (Å) | 43.4 - 2.48 | 30.4 - 2.52 | 30.6–2.46 |
| No. of reflections | 17443 | 16442 | 17404 |
| Rwork/Rfree | 0.199/0.237 | 0.208/0.258 | 0.212/0.266 |
| Number of nonhydrogen atoms | |||
| Protein | 2318 | 2307 | 2322 |
| Water | 55 | 50 | 45 |
| B-factors | |||
| Protein | 44.9 | 39.6 | 38.5 |
| Water | 43.5 | 41.4 | 40.3 |
| r.m.s. deviations | |||
| Bond lengths (Å) | 0.007 | 0.019 | 0.0187 |
| Bond angles (°) | 1.063 | 1.82 | 1.82 |
| Ramachandran plot (%) a | |||
| outliers | 0.00 | 0.00 | 0.00 |
| favored | 98.0 | 96.7 | 97.7 |
Percentages were generated by MolProbity (http://molprobity.biochem.duke.edu/)
EspP pre-cleavage structures
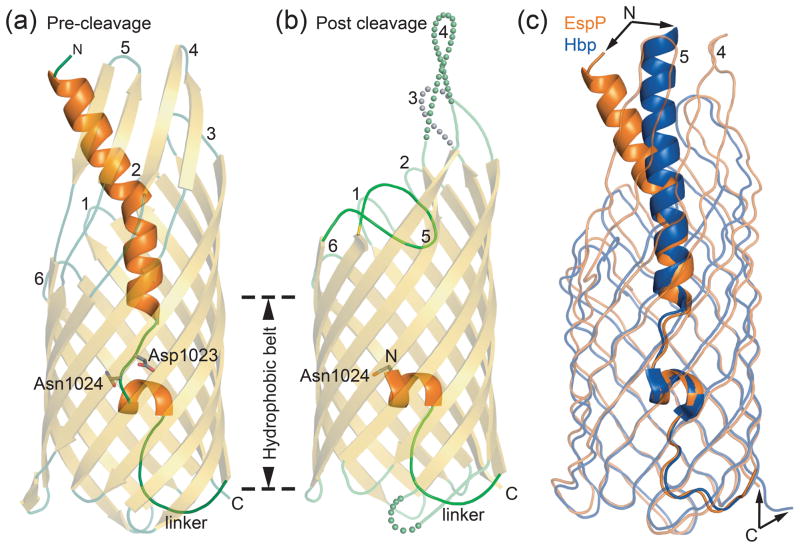
The structures of the three EspP pre-cleavage mutants are very similar. The N-terminus forms an α helix that protrudes from the extracellular side of the barrel pore (Fig. 1a). This helix is disrupted near the middle of the barrel and adopts an extended conformation for three residues (1022 to 1024). In wild-type EspP, this region contains an asparagine at position 1023 that catalyzes the cleavage reaction. Following this extended region, a single turn α helix that runs perpendicular to the barrel pore is connected by a linker to the first β-strand of the barrel. The barrel contains twelve antiparallel β-strands that are connected by extracellular loops of various lengths and short periplasmic turns. The first and last β-strands of the barrel pair to close it such that the C-terminus points towards the periplasm.
Fig. 1. EspP and Hbp structures.
(a) Pre-cleavage structure of the EspP N1023D mutant. The side chains of Asp1023 and Asn1024 are shown. (b) Post cleavage structure of EspP. The N-terminal residue, Asn1024, is shown. Disordered loops are depicted as spheres. (a and b) β-strands, yellow; α-helices, orange; loops, green. The location of the hydrophobic belt, N and C-termini, extracellular loops 1 through 6, and linker loops are labeled. (c) Structural alignment of pre-cleavage EspP (orange) and pre-cleavage Hbp (blue). For EspP, loops 4 and 5 are labeled. These loops are disordered in the Hbp structure. Figures 1, 2, 5, 6, and 7 were created using The PyMOL Molecular Graphics System (Schrödinger, LLC).
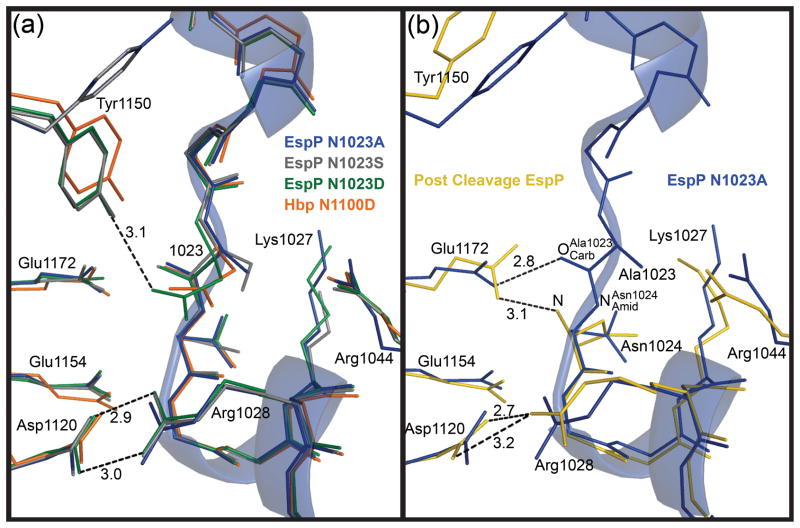
Superposing the N1023D or N1023S mutant on the N1023A mutant resulted in RMSDs of 0.3 and 0.2 Å for C∝ atoms, respectively. The extra negative charge introduced by the aspartate side chain in the N1023D mutant did not affect the conformation of the cleavage site significantly because the cleavage sites for all three structures superpose closely (Fig. 2a). One exception is Tyr1150, which occupies different conformations between the pre-cleavage mutants (Supplementary Figure S3). In the N1023A mutant, Tyr1150 points away from the cleavage site. For the N1023S mutant, it is present in two conformations, one that points toward the cleavage site and one that points away. In the N1023D mutant, Tyr1150 points towards the cleavage site and hydrogen bonds with the side chain oxygen of Asp1023. Thus, as more negative charge is introduced at position 1023, the side chain of Tyr1150 favors the conformation that point towards the cleavage site.
Fig. 2. Superposition of the EspP and Hbp cleavage sites.
The cleavage site region of the EspP N1023A mutant is shown as a transparent ribbon. For clarity, side chains are shown only for residues discussed in the text. These residues are labeled with EspP numbering. N1023A mutant, blue; N1023S mutant, gray; N1023D mutant, green; Hbp, orange; post cleavage EspP, yellow. Hydrogen bonds are shown as dashed lines and distances in angstroms are labeled. (a) Superposition of the pre-cleavage EspP mutants and Hbp. Tyr1150 is present in two different conformations. The salt bridge between Asp1120 and Arg1028 is shown for the EspP N1023D mutant. This salt bridge is present in all the EspP structures and Hbp. The hydrogen bond between Tyr1150 and Asp1023 is shown for the EspP N1023D mutant. (b) Superposition of the N1023A EspP pre-cleavage mutant and post cleavage EspP. The hydrogen bond between Glu1172 and is shown for the N1023A mutant. The hydrogen bond between Glu1172 and the N-terminus (N) and the salt bridge between Asp1120 and Arg1028 are shown for post cleavage EspP.
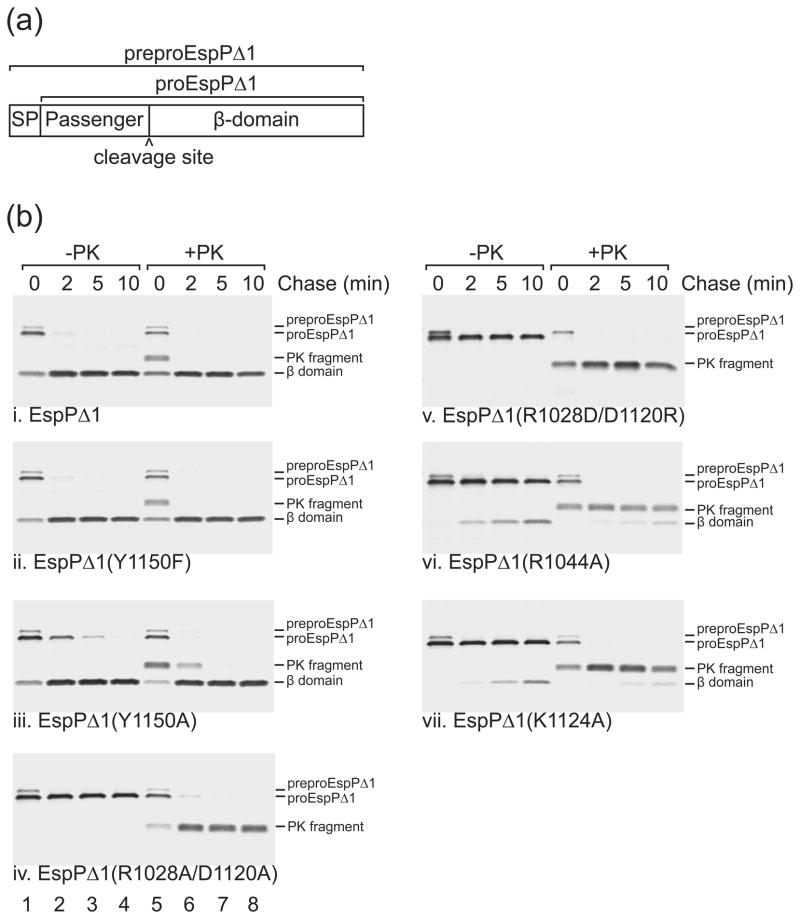
Pulse-chase experiments showed that the mutation of Tyr1150 to phenylalanine had no effect on cleavage. In these experiments, a truncated version of EspP that contains a 116 residue passenger domain (EspPΔ1; see Fig. 3a) was radiolabeled, and passenger domain cleavage was assessed by comparing the rate at which the free ~30 kD β-domain was released from the pro form of the protein (which contains covalently linked passenger and β-domains). EspPΔ1 was used because its passenger domain is secreted more rapidly and synchronously than the full-length passenger domain and it displays subtle defects in protein biogenesis more readily 16; 20. Whenever a fraction of the passenger domain was translocated across the outer membrane but not cleaved, treatment of cells with proteinase K (PK) removed most of the passenger domain from proEspPΔ1 and yielded a ~33 kD C-terminal fragment. Consistent with previous results 16, the wild-type EspPΔ1 passenger domain was cleaved very rapidly and a ~33 kD PK fragment could be observed only at the 0 min time point (Fig. 3b, gel i). EspPΔ1 containing the Y1150F mutation was processed at essentially the same rate (Fig. 3b, gel ii). Therefore, Tyr1150 does not appear to play a direct role in the cleavage reaction. However, if an alanine substitution was made at position 1150, cleavage was slightly delayed, indicating that an aromatic or bulky side chain is important (Fig. 3b, gel iii).
Fig. 3. Effects of mutations near the EspP cleavage site on passenger domain cleavage.
(a) Diagram showing EspPΔ1, a truncated form of EspP that contains the C-terminal 116 residues of the passenger domain followed by the β-domain. The construct that we used also contains the OmpA signal peptide (SP) to target EspPΔ1 to the Sec machinery. The signal peptide of preproEspPΔ1 is removed during its translocation across the inner membrane to generate proEspPΔ1. Following translocation of the passenger domain across the outer membrane, proEspP is cleaved into discrete passenger domain and β domain fragments (b) AD202 expressing espPΔ1 or the indicated mutant were subjected to pulse-chase labeling and proteinase K (PK) was added to half of each sample. Immunoprecipitations were then conducted with an antiserum directed against a C-terminal EspP peptide. The length of the chase is indicated. Lanes 1–4, no PK added (−PK); lanes 5–8, PK added (+PK).
Comparison of pre and post cleavage EspP structures
Structural alignment of the pre and post cleavage EspP 7 showed that the structures are very similar with a RMSD of 1.2 Å for C∝ atoms. The only significant backbone differences between the pre and post cleavage states can be seen in the extracellular loops 3, 4, and 5 (Fig. 1a and 1b). Loop 5 is folded into the barrel pore in the post cleavage structure while in the pre-cleavage state the uncleaved passenger occupies this region. This forces loop 5 outside the barrel pore in an orientation that projects into the extracellular space. We note that the one turn α helix just below the cleavage site is in the same orientation in both structures, indicating that this region does not undergo a conformational change after cleavage as was previously proposed 7.
Comparison of pre-cleavage EspP and Hbp structures
Recently, the pre-cleavage structure of Hbp was solved and its structure is very similar to the pre-cleavage structure of EspP (Fig. 1c and 2a) 10. Structural alignment of these proteins results in a RMSD of 2.1 Å for C∝ atoms. A difference between the structures is the orientation of the passenger α helix that protrudes from the barrel pore (Fig. 1c). In both structures, this region is involved in crystal contacts and, for EspP, this region also contains the histidine tag. Both of these factors likely affect the positioning of the passenger N-termini. As the passenger domains of EspP and Hbp enter the barrel pore and approach the cleavage site, they superpose closely. To prevent cleavage of Hbp, the active site asparagine was mutated to aspartate. Similar to the EspP N1023D mutant, Tyr1227 of Hbp (equivalent to Tyr1150 in EspP) points towards the cleavage site (Fig. 2a) and has no effect on cleavage if mutated to phenylalanine 10.
Asparagine cyclization and EspP cleavage
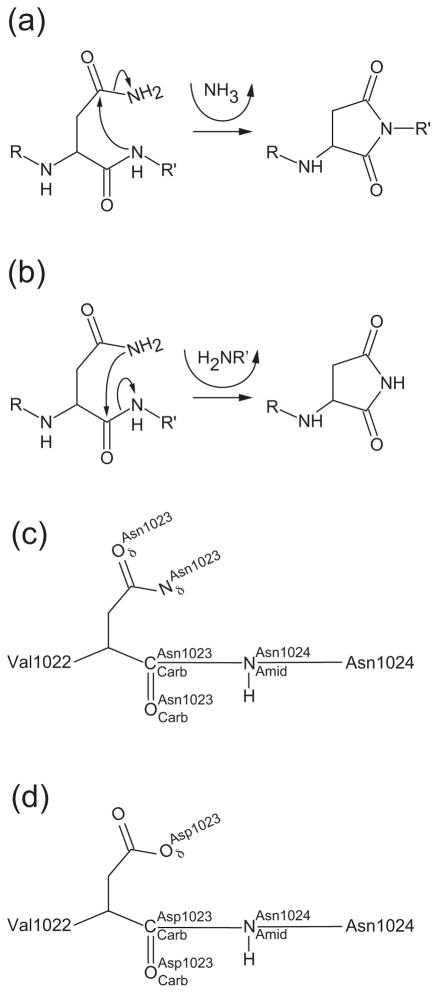
Asparagine cyclization is an intramolecular process that does not require external energy sources or accessory proteins. There are two pathways the cyclization reaction can follow: 1) the deamidation pathway where the peptide bond nitrogen performs a nucleophilic attack on the carboxamide carbon of the upstream asparagine side chain or 2) the cleavage pathway where the carboxamide nitrogen of the asparagine side chain performs a nucleophilic attack on its main chain carbonyl carbon (Fig. 4a and 4b). EspP uses the cleavage pathway to break the peptide bond between Asn1023 and Asn1024. From this point forward we will use the nomenclature shown in Fig. 4c and 4d when referring to the atoms involved in EspP cleavage. EspP initiates cleavage when attacks to form a tetrahedral intermediate. When the tetrahedral intermediate collapses the peptide bond is broken. This results in a succinimide ring at the C-terminus of the cleaved passenger domain and a primary amine at the newly formed N-terminus of the β-domain (Fig. 4b). Cleavage of EspP occurs within one to two minutes of translation 16 while uncatalyzed asparagine cyclization takes days in peptides and favors deamidation 18. However, there are several ways to catalyze the cleavage reaction such as: 1) optimizing the overall conformation of the cleavage site, 2) positioning Asn1023 for cyclization, 3) increasing the nucleophilicity of or the electrophilicity of , and 4) facilitating proton transfer. We will discuss how EspP could use each of these mechanisms to promote cleavage in the sections that follow. Since many of the residues near the cleavage site appear to promote cleavage by multiple means, we summarize their possible roles in Table 2.
Fig. 4. Consequences of asparagine cyclization.
(a) Deamidation pathway. The peptide bond nitrogen attacks the carboxamide group of the upstream asparagine residue resulting in the formation of a succinimide ring in the protein backbone and production of ammonia. (b) Cleavage pathway. The side chain nitrogen of asparagine attacks its own main chain carbonyl carbon resulting in cleavage of the peptide bond. A succinimide ring is formed at the new C-terminus. The new N-terminus begins with the residue directly downstream of the active site asparagine. (c and d) Nomenclature used in text for atoms involved in asparagine cyclization. (c) Wild-type asparagine shown at position 1023 or (d) substituted aspartate shown at position 1023.
Table 2.
Putative roles for residues that influence the cyclization of Asn1023
| EspP residue | Equivalent Hbp residue | Essential for cleavage site conformation | Sterically constrain Asn1023 | Electrostatically position the carboxamide group of Asn1023 | Position the putative catalytic water and accept a proton | Increase electrophilicity of . Stabilize the oxyanion intermediate. | Facilitate proton transfer to | Effect of alanine substitution on EspP cleavage |
|---|---|---|---|---|---|---|---|---|
| Lys1027 | Lys1104 | √ | Moderate delay 25 | |||||
| Arg1028 | Arg1105 | √ | √ | Abolishes cleavage 25 | ||||
| Arg1044 | Arg1121 | √ | √ | Severe delay 1 | ||||
| Asp1120 | Asp1197 | √ | √ | √ | Abolishes cleavage 23 | |||
| Lys1124 | Lys1201 | √ | Severe delay 1 | |||||
| Glu1154 | Glu1231 | √ | √ | Severe delay 23 | ||||
| Glu1172 | Glu1249 | √ | √ | √ | √ | Severe delay 23 |
This study
The extended conformation of the cleavage site facilitates cleavage
Based on secondary structure predictions and the NalP crystal structure 9 we previously predicted that an α-helix would transit the barrel pore prior to cleavage. However, this prediction is not entirely accurate. Asn1023 lies in a short extended region (residues 1022–1024) between two helices. This extended conformation facilitates Asn1023 cyclization compared to an α helical conformation because it allows to closely approach for nucleophilic attack.
For EspP, the only known alanine substitutions (with the exception of the N1023A mutation) that completely abolish cleavage disrupt the salt bridge between Arg1028 and Asp1120 19; 21; 22. This salt bridge is present in the pre and post cleavage structures of EspP and the pre-cleavage structure of Hbp (Fig. 2a and 2b). Alanine substitutions for EspP Arg1028 or Asp1120 completely abolish cleavage while conservative arginine to lysine or aspartate to glutamate substitutions allow cleavage 19; 22. To further examine the function of the salt bridge, we tested to see if changing both Asp1120 and Arg1028 to alanine might allow cleavage. The idea behind this double alanine mutant was that replacing both charged residues might have a milder effect than leaving one charged residue unpaired. However, the double alanine mutant abolished cleavage completely (Fig. 3b, gel iv) indicating that the salt bridge is necessary for cleavage and, since this mutation has a severe effect, suggests that the salt bridge is important for the overall conformation of the cleavage site. We also tried substituting aspartate for Arg1028 and arginine for Asp1120 to create a swapped mutant that could potentially still form the salt bridge. However, this mutant also abolished cleavage indicating that either the salt bridge cannot form in this mutant or that a reversed salt bridge cannot support cleavage (Fig. 3b, gel v). Importantly, double or single mutations at positions 1120 and 1028 had little to no effect on translocation of the passenger domain indicating that these mutations do not cause EspP to completely misfold (Fig. 3b, gel iv and v) 19; 22.
Asn1023 is positioned for nucleophilic attack
Another way to catalyze cleavage is to optimally orient to perform a nucleophilic attack on . For distances less than approximately 2.5 Å, the ideal angle of approach for a nucleophile attacking a carbonyl carbon is between 100° and 110° 23. We measured this distance and angle for Asp1023 in the N1023D mutant. This mutant was chosen because aspartate should most closely mimic the wild-type asparagine residue and, as mentioned above, it does not appear to affect the conformation of any nearby residues that are important for cleavage (Fig. 2a). For the N1023D mutant, Asp1023 is positioned over its main chain carbonyl carbon with placed 2.8 Å away from . The O···C=O angle between is 85° (Fig. 5a). Although this angle is outside the ideal trajectory, is in a reasonable position to approach at an angle of 100° –110° as the distance between the two atoms decreases. Furthermore, the wild-type asparagine side chain is likely positioned slightly differently than the mutant aspartate.
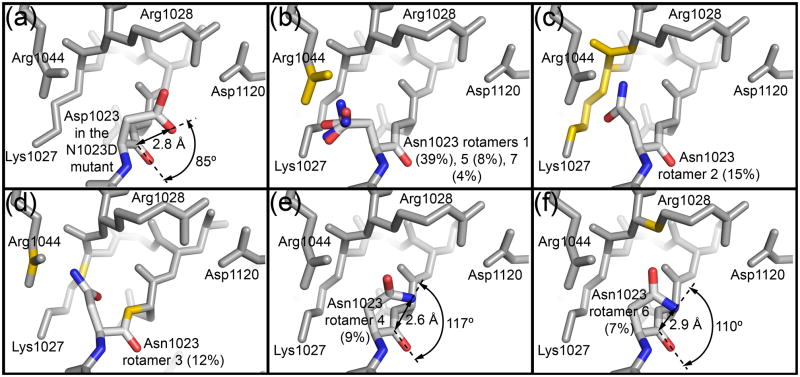
Fig. 5. Steric constraints limit the active site asparagine to rotamers favorable for cyclization.
(a) Burgi-Dunitz angle for Asp1023 in the EspP N1023D mutant. (b–f) The seven most common rotamers for asparagine from the Coot library were substituted at position 1023 in the EspP N1023A mutant to create the models. Each rotamer’s percentage abundance is shown in parentheses. Side chain and main chain atoms that clash with the asparagine rotamers are shown in yellow. For Panel (b), three similar rotamers (1,5,7) are shown. Rotamers 4 and 6 (Panels e and f) show the least clashes. When their carboxamide groups are rotated 180° such that is oriented towards , they are in a reasonable position for cyclization. The distance between and and the Burgi-Dunitz angle are shown for each of these rotamers.
We used molecular dynamics simulations to examine the average distance between and for wild-type and mutant forms of EspP and the corresponding atoms ( and ) for wild-type Hbp. To create the wild-type models, we modeled asparagine at position 1023 of the EspP N1023A and N1023D mutant structures and position 1100 of the Hbp N1100D structure. We named these wild-type models “A1023N”, “D1023N”, and “Hbp”, respectively. We made several more mutations in the EspP D1023N background to look at their effect on the average distance between and . We expected that mutations that had a more severe effect on cleavage would result in a greater distance between and . These mutations included R1028A, R1028K, R1044A, D1120N, E1154Q, E1172Q, and an E1154A/E1172A double mutant (Fig. 2a). We chose these mutations because their effects on cleavage range from mild to severe (Fig. 3b, gel vi, and Table 2) 7; 19; 22. For the simulations, each of these models was equilibrated for 5 ns with the active site asparagine restrained to a conformation ideal for cyclization. This was followed by 10 ns of unrestrained dynamics, a timescale similar to that used in previous molecular dynamics simulations of NalP 24. The 5 ns equilibration was included to ensure any motion of active site asparagine was not the result of a poor starting configuration; the root mean-square deviation of the protein backbone illustrates that equilibration is reached within 4–5 ns (Supplementary Figures 4 and 5). The four lowest average or distances ranged from 3.12 to 3.32 Å and were observed for the EspP wild-type models (A1023N and D1023N), the Hbp wild-type model, and the EspP cleavable mutant R1028K (Supplementary Figures S4a and S4b). These short distances are uncommon for asparagines; only 6.4% of 1.4 million structurally characterized asparagines have corresponding N-C distances less than 3.32 Å. In addition, these distances are comparable to the distances derived from quantum chemical calculations for an asparagine cyclization system comprised of an asparagine, two histidines, and two water molecules. For this system, the N-C distance was calculated to be ~3.7 Å during the first transition state where a proton was being removed from the active site asparagine side chain by a water molecule. After deprotonation of the asparagine and transfer of a proton to a nearby histidine, the N-C distance decreased to ~3.2 Å 25. For the EspP R1044A, D1120N, E1154Q, and E1172Q single mutants and the E1154/E1172A double mutant, the average distance ranged from 3.80 to 4.37 Å. All of these mutations affect cleavage in vivo as would be predicted by their longer distances. Only the EspP R1028A mutant did not give a clear result. This mutation abolishes cleavage in vivo, but the distance was intermediate (3.52 Å). To confirm reproducibility of the results, simulations of EspP A1023N, D1023N, and E1154Q were repeated. In these simulations, the average distances were 3.15, 3.23, and 3.86 Å, respectively, similar to those observed in the original simulations (Supplementary Figure S4b).
Steric constraints and electrostatics position Asn1023 for cyclization
When the naturally occurring asparagine side chain was restored at position 1023 of the N1023A crystal structure, only two of its seven possible rotamers in the Coot library 26; 27 did not cause multiple clashes with surrounding side chains or backbone atoms (Fig. 5b–5f). These rotamers placed the asparagine over its main chain carbonyl carbon. Additionally, when their carboxamide groups were flipped 180° such that was oriented towards , they were in reasonable positions for cyclization (Fig. 5e and 5f). The side chains sterically constraining the asparagine include Lys1027, Arg1028, and Arg1044 while the backbone atoms include regions of the one turn α helix downstream of the cleavage site. All of these residues are conserved among the SPATEs. Consistent with these side chains preventing unfavorable conformations for cyclization, substituting Lys1027, Arg1028, and Arg1044 with alanine significantly delayed or abolished cleavage in pulse chase experiments (Fig. 3b, gel vi) 22. It could be argued that these side chains are mobile in vivo and would allow alternate conformations of Asn1023 not seen in the static crystal structure. However, Lys1027, Arg1028 and Arg1044 are in similar positions in the pre and post cleavage structures (Fig. 2b) suggesting that they are relatively stationary prior to and during cleavage. We also modeled the native asparagine at position 1100 for the Hbp N1100D mutant. We found that the same two rotamers of asparagine gave the fewest clashes and were in the best positions for cyclization while the remaining rotamers clashed with the equivalent residues (Lys1104, Arg1105, Arg1121) compared to EspP.
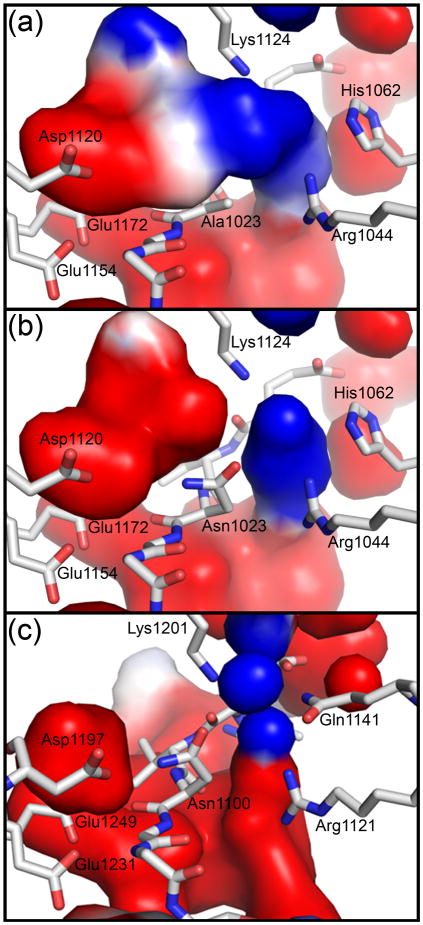
To examine the role of electrostatics in positioning Asn1023 for cleavage, we calculated the electrostatic surface of the cavities surrounding position 1023 using the N1023A crystal structure. A negatively charged surface connected to a positively charged surface was found above Ala1023 (Fig. 6a). For wild-type EspP, and would be expected to point towards the negatively and positively charged surfaces, respectively. To visualize this, we substituted Ala1023 with the non-clashing asparagine rotamer seen in Fig. 5e and recalculated the electrostatic surface. Indeed, this asparagine was placed between the positive and negative surfaces with near the negative surface and near the positive surface (Fig. 6b). The residues surrounding these surfaces include Asp1120, Glu1154, and Glu1172 for the negative surface and Arg1044, Lys1124, and His1062 for the positive surface. Consistent with these residues helping position Asn1023 for cleavage, alanine substitutions at each position except for His1062 severely delayed or abolished cleavage (Fig. 3b, gels vi and vii). His1062 was not tested because it is not conserved among the SPATEs. To see if similar electrostatic surfaces surround position 1100 in Hbp, we substituted Asp1100 with the asparagine rotamer seen in Fig. 5e and calculated the electrostatic surfaces of the surrounding cavities (Fig. 6c). Similar to EspP, we saw a positive and negative surface that would be expected to position Asn1100 for cyclization. The Hbp residues equivalent to those seen in EspP surrounded these surfaces with the exception of Hbp Gln1141, which occupies the space near EspP His1062 when the structures are superposed. Changing this glutamine to alanine was shown to have no discernable effect on cleavage 10.
Fig. 6. Electrostatic interactions position the carboxamide group of the active site asparagine for cyclization.
The electrostatic surfaces were calculated using APBS tools (red, negative; blue, positive; white, neutral) 41. (a) A region of negative charge and a region of positive charge are on either side above Ala1023 in the EspP N1023A mutant. (b) The asparagine rotamer seen in Fig. 5e was substituted at position 1023 of the EspP N1023A mutant and the electrostatic surfaces recalculated. The resulting surfaces would likely orient the carboxamide group of Asn1023 such that would be placed near and in position for cyclization. (c) The asparagine rotamer seen in Fig. 5e was substituted at position 1100 of the Hbp N1100D mutant. The electrostatic surfaces are similar to those seen for EspP and would be expected to correctly orient the carboxamide group of Hbp Asn1100 for cyclization.
Tajima et al. state that Hbp Arg1121 (Arg1044 in EspP) makes a long hydrogen bond (3.5 – 3.8 Å) with Asp1100 (Asn1023 in EspP). They conclude that this hydrogen bond would be present in wild-type Hbp and help position Asn1100 for nucleophilic attack on its main chain carbonyl carbon. However, Arg1121 is actually 4.21 Å (chain A) or 4.51 Å (chain B) from the nearest carboxyl group oxygen of Asp1100 in the Hbp structure. These distances are too long for Arg1121 to hydrogen bond with Asp1100. We think that a more plausible role of Arg1121 in Hbp (Arg1044 in EspP) is to help position the active site asparagine through electrostatic interactions and steric inhibition as discussed above.
A water molecule may catalyze cleavage by increasing the nucleophilicity of
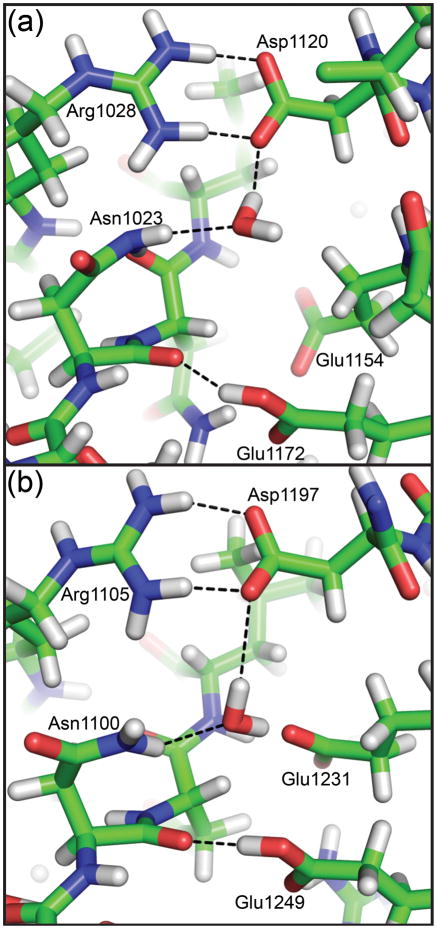
Other ways to catalyze the cleavage reaction are to increase the nucleophilicity of or the electrophilicity of . For EspP, there do not appear to be any side chains near position 1023 that could increase the nucleophilicity of the active site asparagine. However, in molecular dynamics simulations when the native asparagine was restored at position 1023 in the EspP N1023A structure, water molecules were observed to enter the active site and form hydrogen bonds favorable for catalysis. The oxygen atom for the water molecule shown in Fig. 7a hydrogen bonds with while one of its hydrogen atoms bonds with a carboxyl group oxygen of Asp1120. This interaction would increase the nucleophilicity of and promote cyclization. In previous studies, we predicted Asp1120 could directly interact with Asn1023 to catalyze cleavage, but these residues are too far apart for direct interaction in the pre-cleavage structures. Additionally, when the native asparagine was restored at position 1100 of the Hbp N1100D structure, similar water molecules were observed in molecular dynamics simulations. The water shown in Fig. 7b bridges Asp1197 and Asn1100 as seen for EspP and is also within hydrogen bonding distance of Glu1231 and Glu1249 (Fig. 7b). These additional hydrogen bonds should be possible in EspP by the equivalent glutamates at positions 1154 and 1172. Mutations in EspP Asp1120 abolish cleavage while mutations in Glu1154 and Glu1172 in EspP reduce cleavage 7; 19. The same results were seen for Hbp with the exception of Glu1249 (EspP Glu1172) where an alanine substitution abolished cleavage 28. However, the importance of the putative catalytic water is difficult to test by mutagenesis because the interacting residues appear to promote cleavage in multiple ways. EspP Asp1120 (or Asp1197 in Hbp) is also part of a salt bridge that appears to be essential for cleavage to occur. EspP Glu1154 hydrogen bonds with the main chain nitrogen of Leu1025 and could help stabilize the extended conformation of the cleavage site; the equivalent interaction is seen for Hbp Glu1231 and Leu1102. Possible additional roles of EspP Glu1172 and Hbp Glu1249 will be discussed in a following section.
Fig. 7. Representative snapshots of the cleavage sites of EspP and Hbp from molecular dynamics simulations.
Modeling asparagine at position 1023 of the EspP N1023A mutant (Panel (a)) or 1100 of the Hbp N1100D mutant (Panel (b)) created the wild-type models. A water molecule is shown for EspP and Hbp that forms hydrogen bonds with the active site asparagine and EspP Asp1120 or Hbp Asp1197. These hydrogen bonds would be favorable for increasing the nucleophilicity of the active site asparagine. All hydrogen bonds shown are less than 3.5 Å when measured between non-hydrogen atoms. The water molecule in Panel (a) is also within 3.5 Å of EspP Arg1028 and the water molecule in Panel (b) is within 3.5 Å of Hbp Glu1231 and Glu1249.
In their proposed cleavage model for Hbp, Tajima et al. show Tyr1227 (EspP Tyr1150) and Glu1249 (EspP Glu1172) positioning a water molecule that deprotonates the side chain nitrogen of the active site asparagine. We think that the aspartate substitution used by Tajima et al. and our group may artificially attract this tyrosine towards the cleavage site and would affect the water molecule shown in their proposed cleavage mechanism. Two pieces of evidence support this idea. First, changing Asn1023 in EspP to a non-polar alanine residue resulted in Tyr1150 pointing away from the cleavage site. Second, mutating EspP Tyr1150 or Hbp Tyr1227 to phenylalanine had no effect on cleavage.
Hydrogen bonding to Glu1172 increases the electrophilicity of
Glu1172 forms a hydrogen bond with in all of the EspP pre-cleavage structures. This bond is shown for the EspP N1023A mutant in Fig. 2b. In Hbp, Glu1249 makes the equivalent hydrogen bond with the main chain carbonyl oxygen of Asn1100. Normally, the glutamate side chain is present in its de-protonated form. However, the close distance (2.8 Å) between the carboxyl group oxygen of this glutamate residue and the main chain carbonyl oxygen of residue 1023 strongly suggests that a proton sits between these atoms. Consistent with this idea, the pKa of Glu1172 was calculated to be 8.39 29, indicating that it would be protonated at physiological pH. This hydrogen bond would increase the electrophilicity of the carbonyl group, promoting nucleophilic attack by . In addition, protonation of would also stabilize the negative charge of the oxyanion created during formation of the tetrahedral intermediate. In agreement with its potential role in catalysis, mutation of Glu1172 has been shown to delay cleavage 7; 19. As further support, when Glu1172 is left unprotonated in simulation, Asn1023 rapidly becomes distorted, and the distance rises to an average of 4.6 Å (Supplementary Figure 4b).
Glu1172 could facilitate proton transfer
During the cleavage reaction, protons are transferred between reaction intermediates and these transfers have been implicated as major energy barriers for cleavage 25. One of these transfers includes the protonation of during cleavage. It is possible that a proton could be transferred by Glu1172 to during cleavage and thereby help overcome a significant energy barrier. Two pieces of evidence support this idea. First, in the pre-cleavage structure the calculated pKa of Glu1172 is 8.39 while in the post cleavage structure its calculated pKa is −1.57 29. These pKa values indicate that Glu1172 loses a proton during or after the cleavage reaction. Second, Glu1172 is positioned in close proximity to throughout the cleavage reaction, making proton transfer between the two feasible (Fig. 2b). Glu1249 in Hbp could also mediate the same transfer.
EspP cleavage mechanism
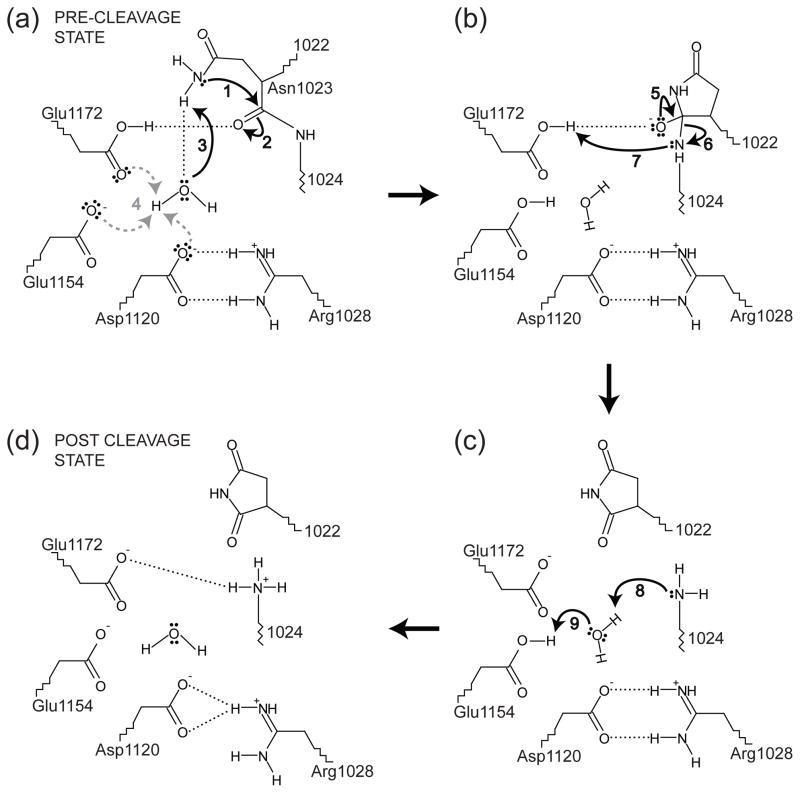
We propose the following cleavage mechanism for EspP, noting that Hbp would presumably use the same mechanism. First, the extended conformation of the cleavage site facilitates cleavage by allowing the Asn1023 side chain to easily be positioned over its main chain carbonyl oxygen while the residues surrounding the cleavage site sterically constrain Asn1023 to rotamers favorable for cyclization. Additionally, the carboxamide group of Asn1023 is oriented such that is correctly positioned over through electrostatic interactions. Next, a water molecule observed in molecular dynamics simulations but not in the crystal structures may initiate cyclization by hydrogen bonding with to increase its nucleophilicity (Fig. 8a). This water molecule could be positioned by hydrogen bonding to Asp1120, Glu1154, Glu1172, or a combination of these residues. Concurrently, Glu1172 also appears to facilitate cyclization by hydrogen bonding with to increase the electrophilicity of (Fig. 8a). During formation of the tetrahedral intermediate, the hydrogen bond between Glu1172 and could stabilize the negative charge of the oxyanion. Additionally, the putative catalytic water could absorb a proton from and then transfer a proton to either Asp1120, Glu1154, or Glu1172. All of these residues are reasonable candidates; for simplicity, we show Glu1154 accepting the proton in the cleavage model (Fig. 8a and 8b). When the tetrahedral intermediate collapses, the peptide bond between Asn1023 and Asn1024 is broken and the proton sitting between Glu1172 and could be transferred to to complete the cleavage reaction (Fig. 8b and 8c). In the post cleavage structure, is fully protonated with a pKa value of 8.55 29. This proton could be transferred to by a water molecule that absorbs a proton from Glu1154 (Fig. 8c). Again, Asp1120 or Glu1172 could perform this role if one of these residues is protonated instead of Glu1154. Finally, the hydrogen bond between Glu1172 and could form and the salt bridge between Asp1120 and Arg1028 could rearrange slightly to match the bonding pattern seen in the post cleavage EspP (Fig. 2b and 8d).
Fig. 8. Proposed mechanism for EspP passenger domain cleavage.
The residues shown are labeled using EspP numbering and the mechanism is described in the main text. In Panel (a), the water molecule hydrogen bonding to could be positioned by hydrogen bonding to Asp1120, Glu1154, Glu1172, or a combination these residues. Additionally, the proton from that is absorbed by the putative catalytic water in panel (a) could result in a proton being transferred to Asp1120, Glu1154 or Glu1172 (dashed, gray arrows). For simplicity, this proton is shown being transferred to Glu1154 in panels (b) and (c). In Panel (d) the hydrogen bonding pattern matches the post cleavage structure of EspP. The water shown in Panel (d) is not seen in the post cleavage structure. However, only six waters were included in the post-cleavage structure due to the resolution of the data.
Concluding Remarks
Although the pre-cleavage structures of EspP and Hbp are nearly identical we were able to gain several new insights into their shared cleavage mechanism by solving the structures of three non-cleavable mutants of EspP, comparing them with Hbp, and performing molecular dynamics simulations. First, analysis of the residues and electrostatic surfaces surrounding the active site asparagine revealed how its side chain is constrained to rotamers favorable for cleavage and its carboxamide group is correctly positioned over its main chain carbonyl carbon. Second, these structures show that an aspartate substitution for the active site asparagine likely artificially attracts EspP Tyr1150 and Hbp Tyr1227 towards the cleavage site. In Hbp this tyrosine and Glu1249 position a water molecule that could deprotonate the active site asparagine to initiate cleavage. However, mutation of this tyrosine in EspP or Hbp has no discernable effect on cleavage. The observation from the EspP structures that this tyrosine can point away from the cleavage site provides a plausible explanation for the mutagenesis results. Third, we used molecular dynamics simulations to look for alternate water molecules that could interact with the active site asparagine to increase its nucleophilicity. We observed water molecules in both EspP and Hbp that could be positioned by three conserved acidic residues to perform this role. Additionally, the position of these water molecules would allow them to act as relay stations for proton transfer between the active site asparagine and one of the three nearby acidic residues (Asp1120, Glu1154, and Glu1172 in EspP or the corresponding residues in Hbp). Finally, by comparing pre and post cleavage EspP, it is apparent that Glu1172 loses a proton during or after cleavage. This proton could be transferred to the new N-terminus during cleavage, helping overcome a significant energy barrier.
Materials and Methods
Data collection and structure determination
Data were collected at 100 K at the Southeast Regional Collaborative Access Team (SER-CAT) 22-ID beamline at the Advanced Photon Source, Argonne National Laboratory. All data were integrated and scaled with HKL2000 30. The structures were solved by molecular replacement using Phaser 31 and the post cleavage structure of EspP (PDB ID: 2QOM) as the search model. All of the structures had one molecule per asymmetric unit. Iterative manual model building and refinement were performed using COOT 27, REFMAC 32 from the CCP4 software suite 33, and PHENIX 34. Statistics for data collection and refinement are listed in Table 1.
Pulse chase experiments
E. coli strain AD202 (MC4100 ompT::kan) 35 was transformed with pJH62, a plasmid that encodes espPΔ1 20, or a derivative containing a mutation in the EspP β-domain. Cells were grown overnight in M9 medium containing 0.2% glycerol, all of the L-amino acids except methionine and cysteine, and 100 μg/ml ampicillin. Overnight cultures were washed and diluted into fresh medium at OD550=0.02. When the cultures reached OD550=0.2 the expression of espPΔ1 was induced by the addition of 10 μM IPTG. Cells were subjected to pulse-chase labeling, treated with PK, and processed as described 19. Immunoprecipitations were then conducted using an antiserum generated against a C-terminal EspP peptide 20 and proteins were resolved by SDS-PAGE on 8–16% Tris-glycine minigels (Invitrogen).
Molecular dynamics simulations
Construction of each system that was simulated began by restoring the wild-type Asn at position 1023 for the EspP N1023A and N1023D mutants and at position 1100 for Hbp (PDB code 3AEH) 10. Protonation states were generally chosen to represent standard values at pH 7.0; the only exception was Glu1172, which was protonated based on its measured pKa of 8.39 29. The protein was placed in a DMPE lipid bilayer, chosen to match the hydrophobic thickness of the EspP barrel, as done previously for BtuB 36. Similarly, DMPC was used in a simulation study of EspP 37 and NalP 24. The protein-membrane system was then solvated with water above and below, and ionized with NaCl to a concentration of 100 mM, resulting in a system size of approximately 55,000 atoms. All simulations were run using the program NAMD 38 with the CHARMM force field 39 and periodic boundary conditions. A constant temperature of 310 K was enforced using Langevin dynamics with a damping constant of 1 ps−1 and constant pressure of 1 atm using a Nosé-Hoover Langevin piston 38 A multiple-time-stepping algorithm was employed with bonded interactions evaluated every 1 fs, short-range every 2 fs (cutoff of 12 Å), and long-range every 4 fs. Long-range electrostatics were calculated using the particle-mesh Ewald method.
The wild-type EspP/Hbp systems were equilibrated in stages for 2 ns, the result of which were used as a branching points for subsequent simulations of point mutants. Mutations were made in silico and the system then subjected to another 3 ns of equilibration with N1023 restrained in a conformation optimal for cyclization. Finally, each system was simulated for 10 ns with no restraints. All analysis of the resulting trajectories was carried out using the visualization and analysis program VMD 40.
Supplementary Material
Acknowledgments
We would like to thank Jim Fairman, Lothar Esser, and Dan Appella for helpful discussions concerning refinement of the EspP structures and reaction mechanism. This work was supported by the Intramural Research Program of the National Institutes of Health, National Institute of Diabetes and Digestive and Kidney Diseases. Supporting institutions of SER-CAT may be found at http://www.ser-cat.org/members.html. J. G. is supported by a Director’s Postdoctoral Fellowship from Argonne National Laboratory. ET acknowledges support from National Institutes of Health grants R01-GM086749, R01-GM067887, P41-RR05969, and U54-GM087519. MD simulations were performed using supercomputing resources provided through TeraGrid grant MCA06N060.
Abbreviations
- SPATE
Serine Protease Autotransporters of Enterobacteriaceae
- PK
proteinase k
- SP
signal peptide
Footnotes
Protein Data Bank accession numbers
EspP N1023A, PDB ID: 3SLJ; EspP N1023D, PDB ID: 3SLO; EspP N1023S, PDB ID: 3SLT.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Dautin N, Bernstein HD. Protein secretion in gram-negative bacteria via the autotransporter pathway. Annu Rev Microbiol. 2007;61:89–112. doi: 10.1146/annurev.micro.61.080706.093233. [DOI] [PubMed] [Google Scholar]
- 2.Yen MR, Peabody CR, Partovi SM, Zhai Y, Tseng YH, Saier MH. Protein-translocating outer membrane porins of Gram-negative bacteria. Biochim Biophys Acta. 2002;1562:6–31. doi: 10.1016/s0005-2736(02)00359-0. [DOI] [PubMed] [Google Scholar]
- 3.Goldberg MB, Theriot JA. Shigella flexneri surface protein IcsA is sufficient to direct actin-based motility. Proc Natl Acad Sci U S A. 1995;92:6572–6. doi: 10.1073/pnas.92.14.6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kocks C, Marchand JB, Gouin E, d’Hauteville H, Sansonetti PJ, Carlier MF, Cossart P. The unrelated surface proteins ActA of Listeria monocytogenes and IcsA of Shigella flexneri are sufficient to confer actin-based motility on Listeria innocua and Escherichia coli respectively. Mol Microbiol. 1995;18:413–23. doi: 10.1111/j.1365-2958.1995.mmi_18030413.x. [DOI] [PubMed] [Google Scholar]
- 5.Danese PN, Pratt LA, Dove SL, Kolter R. The outer membrane protein, antigen 43, mediates cell-to-cell interactions within Escherichia coli biofilms. Mol Microbiol. 2000;37:424–32. doi: 10.1046/j.1365-2958.2000.02008.x. [DOI] [PubMed] [Google Scholar]
- 6.Telford JL, Covacci A, Ghiara P, Montecucco C, Rappuoli R. Unravelling the pathogenic role of Helicobacter pylori in peptic ulcer: potential new therapies and vaccines. Trends Biotechnol. 1994;12:420–6. doi: 10.1016/0167-7799(94)90031-0. [DOI] [PubMed] [Google Scholar]
- 7.Barnard TJ, Dautin N, Lukacik P, Bernstein HD, Buchanan SK. Autotransporter structure reveals intra-barrel cleavage followed by conformational changes. Nat Struct Mol Biol. 2007;14:1214–20. doi: 10.1038/nsmb1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meng G, Surana NK, St Geme JW, 3rd, Waksman G. Structure of the outer membrane translocator domain of the Haemophilus influenzae Hia trimeric autotransporter. EMBO J. 2006;25:2297–304. doi: 10.1038/sj.emboj.7601132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oomen CJ, van Ulsen P, van Gelder P, Feijen M, Tommassen J, Gros P. Structure of the translocator domain of a bacterial autotransporter. EMBO J. 2004;23:1257–66. doi: 10.1038/sj.emboj.7600148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tajima N, Kawai F, Park SY, Tame JR. A novel intein-like autoproteolytic mechanism in autotransporter proteins. J Mol Biol. 2010;402:645–56. doi: 10.1016/j.jmb.2010.06.068. [DOI] [PubMed] [Google Scholar]
- 11.van den Berg B. Crystal structure of a full-length autotransporter. J Mol Biol. 2010;396:627–33. doi: 10.1016/j.jmb.2009.12.061. [DOI] [PubMed] [Google Scholar]
- 12.Zhai Y, Zhang K, Huo Y, Zhu Y, Zhou Q, Lu J, Black I, Pang X, Roszak AW, Zhang X, Isaacs NW, Sun F. Autotransporter passenger domain secretion requires a hydrophobic cavity at the extracellular entrance of the beta-domain pore. Biochem J. 2011;435:577–87. doi: 10.1042/BJ20101548. [DOI] [PubMed] [Google Scholar]
- 13.Tommassen J. Assembly of outer-membrane proteins in bacteria and mitochondria. Microbiology. 2010;156:2587–96. doi: 10.1099/mic.0.042689-0. [DOI] [PubMed] [Google Scholar]
- 14.Ieva R, Bernstein HD. Interaction of an autotransporter passenger domain with BamA during its translocation across the bacterial outer membrane. Proc Natl Acad Sci USA. 2009;106:19120–5. doi: 10.1073/pnas.0907912106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Junker M, Besingi RN, Clark PL. Vectorial transport and folding of an autotransporter virulence protein during outer membrane secretion. Mol Microbiol. 2009;71:1323–32. doi: 10.1111/j.1365-2958.2009.06607.x. [DOI] [PubMed] [Google Scholar]
- 16.Skillman KM, Barnard TJ, Peterson JH, Ghirlando R, Bernstein HD. Efficient secretion of a folded protein domain by a monomeric bacterial autotransporter. Mol Microbiol. 2005;58:945–58. doi: 10.1111/j.1365-2958.2005.04885.x. [DOI] [PubMed] [Google Scholar]
- 17.Dautin N. Serine protease autotransporters of Enterobacteriaceae (SPATEs): Biogenesis and function. Toxins. 2010;2:1179–1206. doi: 10.3390/toxins2061179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Geiger T, Clarke S. Deamidation, isomerization, and racemization at asparaginyl and aspartyl residues in peptides. Succinimide-linked reactions that contribute to protein degradation. J Biol Chem. 1987;262:785–94. [PubMed] [Google Scholar]
- 19.Dautin N, Barnard TJ, Anderson DE, Bernstein HD. Cleavage of a bacterial autotransporter by an evolutionarily convergent autocatalytic mechanism. EMBO J. 2007;26:1942–52. doi: 10.1038/sj.emboj.7601638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Szabady RL, Peterson JH, Skillman KM, Bernstein HD. An unusual signal peptide facilitates late steps in the biogenesis of a bacterial autotransporter. Proc Natl Acad Sci U S A. 2005;102:221–6. doi: 10.1073/pnas.0406055102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Velarde JJ, Nataro JP. Hydrophobic residues of the autotransporter EspP linker domain are important for outer membrane translocation of its passenger. J Biol Chem. 2004;279:31495–504. doi: 10.1074/jbc.M404424200. [DOI] [PubMed] [Google Scholar]
- 22.Dautin N, Bernstein HD. Residues in a conserved {alpha}-helical segment are required for cleavage but not secretion of an E. coli serine protease autotransporter passenger domain. J Bacteriol. 2011 doi: 10.1128/JB.05070-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Burgi H, Dunitz JD, Lehn JM, Wipff G. Stereochemistry of reaction paths at carbonyl centres. Tetrahedron. 1974;30:1563–1572. [Google Scholar]
- 24.Khalid S, Sansom MS. Molecular dynamics simulations of a bacterial autotransporter: NalP from Neisseria meningitidis. Mol Membr Biol. 2006;23:499–508. doi: 10.1080/09687860600849531. [DOI] [PubMed] [Google Scholar]
- 25.Mujika JI, Lopez X, Mulholland AJ. Modeling protein splicing: reaction pathway for C-terminal splice and intein scission. The journal of physical chemistry B. 2009;113:5607–16. doi: 10.1021/jp808911p. [DOI] [PubMed] [Google Scholar]
- 26.Lovell SC, Davis IW, Arendall WB, 3rd, de Bakker PI, Word JM, Prisant MG, Richardson JS, Richardson DC. Structure validation by Calpha geometry: phi, psi and Cbeta deviation. Proteins. 2003;50:437–50. doi: 10.1002/prot.10286. [DOI] [PubMed] [Google Scholar]
- 27.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yen YT, Tsang C, Cameron TA, Ankrah DO, Rodou A, Stathopoulos C. Importance of conserved residues of the serine protease autotransporter beta-domain in passenger domain processing and beta-barrel assembly. Infect Immun. 2010;78:3516–28. doi: 10.1128/IAI.00390-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rostkowski M, Olsson MH, Sondergaard CR, Jensen JH. Graphical analysis of pH-dependent properties of proteins predicted using PROPKA. BMC Struct Biol. 2011;11:6. doi: 10.1186/1472-6807-11-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods in Enzymology. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 31.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Murshudov GN, Skubak P, Lebedev AA, Pannu NS, Steiner RA, Nicholls RA, Winn MD, Long F, Vagin AA. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr D Biol Crystallogr. 2011;67:355–67. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AG, McCoy A, McNicholas SJ, Murshudov GN, Pannu NS, Potterton EA, Powell HR, Read RJ, Vagin A, Wilson KS. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 2011;67:235–42. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–21. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Akiyama Y, Ito K. SecY protein, a membrane-embedded secretion factor of E. coli, is cleaved by the ompT protease in vitro. Biochem Biophys Res Commun. 1990;167:711–5. doi: 10.1016/0006-291x(90)92083-c. [DOI] [PubMed] [Google Scholar]
- 36.Gumbart J, Wiener MC, Tajkhorshid E. Coupling of calcium and substrate binding through loop alignment in the outer-membrane transporter BtuB. J Mol Biol. 2009;393:1129–42. doi: 10.1016/j.jmb.2009.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tian P, Bernstein HD. Molecular basis for the structural stability of an enclosed beta-barrel loop. J Mol Biol. 2010;402:475–89. doi: 10.1016/j.jmb.2010.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kale L, Schulten K. Scalable molecular dynamics with NAMD. J Comput Chem. 2005;26:1781–802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.MacKerell AD, Bashford D, Bellott, Dunbrack RL, Evanseck JD, Field MJ, Fischer S, Gao J, Guo H, Ha S, Joseph-McCarthy D, Kuchnir L, Kuczera K, Lau FTK, Mattos C, Michnick S, Ngo T, Nguyen DT, Prodhom B, Reiher WE, Roux B, Schlenkrich M, Smith JC, Stote R, Straub J, Watanabe M, Wi√rkiewicz-Kuczera J, Yin D, Karplus M. All-Atom Empirical Potential for Molecular Modeling and Dynamics Studies of Proteins A†. The journal of physical chemistry B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 40.Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J Mol Graph. 1996;14:33–8. 27–8. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 41.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci U S A. 2001;98:10037–41. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.