Abstract
Traditionally molecular biology research has tended to reduce biological pathways to composite units studied as isolated parts of the cellular system. With the advent of high throughput methodologies that can capture thousands of data points, and powerful computational approaches, the reality of studying cellular processes at a systems level is upon us. As these approaches yield massive datasets, systems level analyses have drawn upon other fields such as engineering and mathematics, adapting computational and statistical approaches to decipher relationships between molecules. Guided by high quality datasets and analyses, one can begin the process of predictive modeling. The findings from such approaches are often surprising and beyond normal intuition. We discuss four classes of dynamical systems used to model genetic regulatory networks. The discussion is divided into continuous and discrete models, as well as deterministic and stochastic model classes. For each combination of these categories, a model is presented and discussed in the context of the yeast cell cycle, illustrating how different types of questions can be addressed by different model classes.
1. Introduction
Modern molecular biology technologies and the proliferation of Web-based resources containing information on various aspects of biomolecular networks in living cells have made it possible to mathematically model dynamical systems of molecular interactions that control various cellular functions and processes. Such models can then be used to predict the behavior of the system in response to different perturbations or stimuli and ultimately for developing rational control strategies intended to drive the cellular system toward a desired state or away from an undesired state that may be associated with disease. To this end, various dynamical models have been studied, most commonly in the context of genetic regulatory networks, for a variety of biological systems. Although there are a number of natural ways to categorize and classify dynamical models of genetic networks, this chapter presents a model class with an accompanying example in each combination of deterministic versus stochastic and continuous versus discrete model categories. The example used in each of the model classes is that of the yeast cell cycle, as this system has been extensively studied from a variety of different perspectives and with different model classes. It is not the intention of this chapter to go into an in-depth investigation of the cell cycle, but rather to use it as a running example to illustrate the kinds of questions that can be addressed by the different model classes considered.
A deterministic model of a genetic regulatory network may involve a number of different mechanisms that capture the collective behavior of the elements constituting the network. The models can differ in numerous ways, such as in the nature of the physical elements that are represented in the model (i.e., genes, proteins, and other factors); the resolution or scale at which the behavior of the network elements are captured (e.g., are genes discretized, such as being either on or off, or do they take on continuous values?); and how the network elements interact (e.g., interactions can either be present or absent or they may have a quantitative nature). The common aspect of deterministic models is the inherent lack of randomness or stochasticity in the model. This chapter presents Boolean networks and systems of differential equations as examples of discrete and continuous deterministic models of genetic networks, respectively.
Stochastic models of genetic regulatory networks differ from their deterministic counterparts by incorporating randomness or uncertainty. Most deterministic models can be generalized such that one associates probabilities with particular components or aspects of the model. Thus, stochastic models can also be categorized into discrete and continuous categories. The stochastic or probabilistic components in such models can either be associated with model structure, so that the interactions or rules of interaction are described by probability distributions, or by the incorporation of noise terms that capture intrinsic biological stochasticity or measurement uncertainty. Probabilistic Boolean networks (PBNs) and stochastic differential equations are presented as examples of discrete and continuous stochastic models of genetic networks, respectively.
2. Boolean Networks
Boolean networks are a class of discrete dynamical systems that can be characterized by the interactions over a set of Boolean variables. Random Boolean networks (RBN), which are ensembles of random network structures, were first introduced by Kauffman (1969a,b) as a simple model class for studying dynamical properties of gene regulatory networks at a time when the structure of such networks was largely unknown. The idea behind such an approach is to define an ensemble of Boolean networks such that it fulfills certain known features of biological networks and then study random instances of these networks to learn more about general properties of such networks (Kauffman, 1974, 1993, 2004). Boolean network modeling of genetic networks was further developed by Thomas (1973) and others.
The ensemble approach has been extraordinarily successful in shedding light on fundamental principles of complex living systems at all scales of organization, including adaptability and evolvability, robustness, coordination of complex behaviors, storage of information, and the relationships between the structure of such complex systems and their dynamical behavior. The reader is referred to several excellent review articles that cover the ensemble properties of Boolean networks (Aldana et al., 2002; Drossel, 2007). However, our focus here is on Boolean network models that can be used to capture the behavior of a specific gene regulatory network.
Consider a directed graph where the vertices represent genes and the directed edges represent the actions of genes, or rather their products, on other genes. For example, directed edges from genes A and B into gene C indicate that A and B jointly act on C. The specific mechanism of action is not represented in the graph structure itself, so an additional representation is necessary. One of the simplest representation of frameworks assumes that genes are binary-valued entities, meaning that they can be in one of two possible states of activity (e.g., ON or OFF) at any given point in time, and that they act on each other by means of rules represented by Boolean functions. For example, gene C may be determined by the output of a Boolean function whose inputs areAandB.The underlying directed graph merely represents the input–output relationships. We now present this idea more formally.
A Boolean network is defined by a set of nodes (genes) {x1, …, xn} and a list of Boolean functions {f1, f2, …, fn}. Each gene xi ∈{0, 1} (i = 1, …, n) is a binary variable whose value at time t + 1 is completely determined by the values of genes xj1, xj2, …, xjki at time t by means of a Boolean function fi : {0, 1}ki → {0, 1}. That is, there are ki regulatory genes assigned to gene xi that determine the “wiring” of that gene. Thus, one can write
| (13.1) |
In an RBN, the functions fi are selected randomly as are the genes that are used as their inputs. This is the basis of the ensemble approach mentioned above.
Each xi represents the state (expression) of gene i, where xi = 1 represents the fact that gene i is expressed and xi = 0 means it is not expressed. Such a seemingly crude simplification of gene expression has ample justification in the experimental literature (Bornholdt, 2008). Indeed, consider the fact that many organisms exhibit an amazing determinism of gene activity under specific experimental contexts or conditions, such as Escherichia coli under temperature change (Richmond et al., 1999). The determinism is apparent despite the prevalent molecular stochasticity and experimental noise inherent to measurement technologies such as microarrays. Furthermore, accurate mathematical models of gene regulation that capture kinetic level details of molecular reactions frequently operate with expressed molecular concentrations spanning several orders of magnitude, either in a saturation regime or in a regime of insignificantly small concentrations, with rapid switch-like transitions between such regimes (Davidich and Bornholdt, 2008a). Further, even higher organisms, which are necessarily more complex in terms of genetic regulation and heterogeneity, exhibit remarkable consistency when gene expression is quantized into two levels; for example, different subtypes of human tumors can be reliably discriminated in the binary domain (Shmulevich and Zhang, 2002).
In a Boolean network, a given gene transforms its inputs (regulatory factors that bind to it) into an output, which is the state or expression of the gene itself at the next time-point. All genes are assumed to update synchronously in accordance with the functions assigned to them and this process is then repeated. It is clear that the dynamics of a synchronous Boolean network are completely determined by Eq. (13.1). The artificial synchrony simplifies computation while preserving the qualitative, generic properties of global network dynamics. Synchronous updating has been applied in most analytical studies so far, as it is the only one that yields deterministic state transitions. Although the introduction of asynchronous updating, which typically involves a random update schedule, renders the system stochastic, asynchronous updating is not per se biologically more realistic and has to be motivated carefully in every case not to fall victim to artifacts (Chaves et al., 2005). Additionally, recent research indicates that some molecular control networks are so robustly designed that timing is not a critical factor (Braunewell and Bornholdt, 2006), that time ordering in the emergence of cell-fate patterns is not an artifact of synchronous updating in the Boolean model (Alvarez-Buylla et al., 2008), and that simplified synchronous models are able to reliably reproduce the sequence of states in biological systems. Nonetheless, PBNs, presented in Section 4, are able to model asynchronous updating as well as other stochastic generalizations of Boolean networks.
Let us start with a simple example to illustrate the dynamics of Boolean networks and present the key idea of attractors. Consider a Boolean network consisting of five genes {x1, …, x5} with the corresponding Boolean functions given by the truth tables shown in Table 13.1. Note that x4(t + 1) = f4(x4(t)) is a function of only one variable and is an example of autoregulation. The maximum connectivity (i.e., maximal number of regulators) K = maxiki is equal to 3 in this case.
Table 13.1.
Truth tables of the functions in a Boolean network with five genes
| f1 | f2 | f3 | f4 | f5 | |
|---|---|---|---|---|---|
| 0 | 0 | 0 | 0 | 0 | |
| 1 | 1 | 1 | 1 | 0 | |
| 1 | 1 | 1 | – | 0 | |
| 1 | 0 | 0 | – | 0 | |
| 0 | 0 | 1 | – | 0 | |
| 1 | 1 | 1 | – | 0 | |
| 1 | 1 | 0 | – | 0 | |
| 1 | 1 | 1 | – | 1 | |
| j1 | 5 | 3 | 3 | 4 | 5 |
| j2 | 2 | 5 | 1 | – | 4 |
| j3 | 4 | 4 | 5 | – | 1 |
The indices j1, j2, and j3 indicate the input connections for each of the functions.
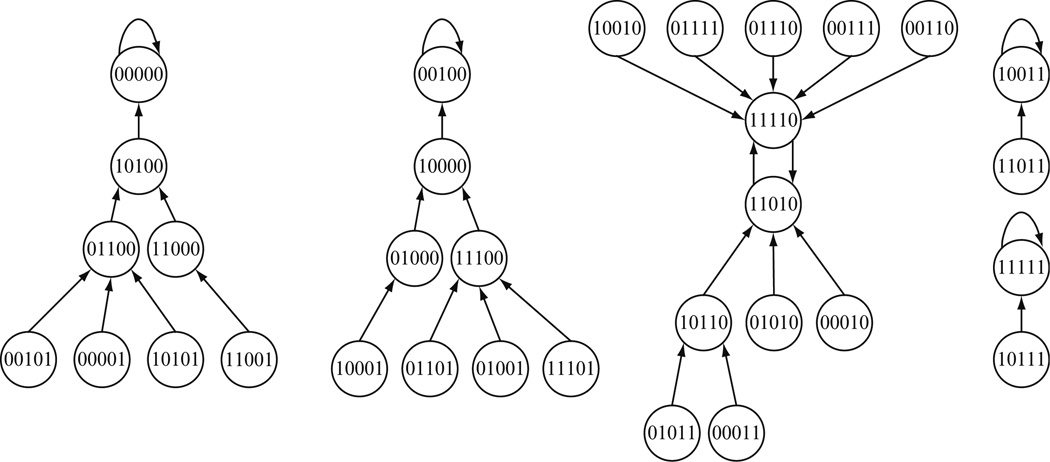
The dynamics of this Boolean network are shown in Fig. 13.1. Since there are five genes, there are 25 = 32 possible states that the network can be in. Each state is represented by a circle and the arrows between states show the transitions of the network according to the functions in Table 13.1. It is easy to see that because of the inherent deterministic directionality in Boolean networks as well as only a finite number of possible states, certain states will be revisited infinitely often if, depending on the initial starting state, the network happens to transition into them. Such states are called attractors and the states that lead into them, including the attractors themselves, comprise their basins of attraction. For example, in Fig. 13.1, the state (00000) is an attractor and together with the seven other (transient) states that eventually lead into it comprise its basin of attraction.
Figure 13.1.
The state-transition diagram for the Boolean network defined in Table 13.1 (Shmulevich et al., 2002c).
The attractors represent the fixed points of the dynamical system, thus capturing the system’s long-term behavior. The attractors are always cyclical and may consist of more than one state. Starting from any state on an attractor, the number of transitions necessary for the system to return to it is called the cycle length. For example, the attractor (00000) has cycle length 1 while the states (11010) and (11110) comprise an attractor of length 2.
Real genetic regulatory networks are highly stable in the presence of perturbations, since the cell must be able to maintain homeostasis in metabolism or its developmental program in the face of such external perturbations and variety of stimuli. Within the Boolean network formalism, this means that when a minimal number of genes transiently change value (say, by means of some external stimulus), the system typically transitions into states that reside in the same basin of attraction and the network eventually “flows” back to the same attractor. Generally speaking, large basins of attraction correspond to higher stability. Such stability of networks in living organisms allows the cells to maintain their functional state within their environment.
Although in developmental biology, epigenetic, heritable changes in cell determination have been well established, it is now becoming evident that the same type of mechanisms may also be responsible in carcinogenesis and that gene expression patterns can be inherited without the need for mutational changes in DNA (MacLeod, 1996). In the Boolean network framework, this can be explained by so-called hysteresis; that is, a change in the system’s state caused by a stimulus that does not change back when the stimulus is withdrawn (Huang, 1999). Thus, if the change of some particular gene does in fact cause a transition to a different attractor, the network will often remain in the new attractor even if that gene is switched off. Thus, the structure of the state space of a Boolean network, in which every state in a basin of attraction is associated with the corresponding attractor to which the system will ultimately flow, represents a type of associative memory.
2.1. Attractors as cell types and cellular functional states
Real gene regulatory networks exhibit spontaneous emergence of ordered collective behavior of gene activity, captured by the attractors. Indeed, recent findings provide experimental evidence for the existence of attractors in real regulatory networks (Chang et al., 2008; Huang and Ingber, 2000; Huang et al., 2005). At the same time, many studies have shown (e.g., Wolf and Eeckman, 1998) that dynamical system behavior and stability of equilibria can be largely determined from regulatory element organization. This suggests that there must exist certain generic features of regulatory networks that are responsible for their inherent robustness and stability. Since in multicellular organisms, the cellular “fate” is determined by which genes and proteins are expressed, the attractors in the Boolean networks should correspond to cell types, an idea originally due to Kauffman (2004). This interpretation is quite reasonable if cell types are characterized by stable recurrent patterns of gene expression (Jacob and Monod, 1961).
Another interpretation of attractors in Boolean networks is that they correspond to cellular states, such as proliferation (cell cycle), apoptosis (programmed cell death), and differentiation (execution of tissue-specific tasks) (Huang, 1999). Such an interpretation can provide new insights into cellular homeostasis and cancer progression, the latter being characterized by a disbalance between these cellular states. For instance, an occurrence of a structural mutation can result in a reduction of the probability of the network entering the apoptosis attractor(s), making the cells less likely to undergo apoptosis and exhibiting uncontrolled growth. Similarly, an enlargement of the basins of attraction for the proliferation attractor would hyperstabilize it, resulting in hyperproliferation, typical of tumorigenesis. Such an interpretation need not be at odds with the interpretation that attractors represent cellular types. To the contrary, these views are complementary to each other, since for a given cell type, different cellular functional states must exist and be determined by the collective behavior of gene activity. Thus, one cell type can comprise several “neighboring” attractors each corresponding to different cellular functional states.
Biological networks can often be modeled as logical circuits from well-known local interaction data in a straightforward way. This is clearly one of the advantages of the Boolean network approach. Though logical models may sometimes appear obvious and simplistic, compared to detailed kinetic models of biomolecular reactions, they may help to understand the dynamic key properties of a regulatory process. Further, a Boolean network model can be formulated as a coarse-grained limit of the more detailed differential equations model for a system (Davidich and Bornholdt, 2008a), discussed in Section 3. They may also lead the experimentalist to ask new questions and to test them first in silico.
Let us consider a Boolean network model of the cell cycle control network in the budding yeast Saccharomyces cerevisiae proposed in Li et al. (2004). The core regulatory network involving activations and inhibitions among cyclins, transcription factors, and check points, such as cell size, consists of 11 binary variables. The Boolean functions, Eq. (13.1), assigned to each variable are chosen from the subclass of threshold Boolean functions (Muroga, 1971), which sum up their inputs with weights and if the sum exceeds a threshold, then the output of the function is equal to 1, else it is equal to 0. This is equivalent to a perceptron and represents a hyperplane that cuts the Boolean hypercube into two halves, zeros on one side, and ones on the other. The model, shown in Fig. 1 in Li et al. (2004), also has self-degradation loops such that nodes that are not negatively regulated by others are degraded at the next time point. The dynamics of the model are described by
| (13.2) |
and the weights were all set to 1 or −1, depending on activation or inhibition, respectively (Li et al., 2004).
Since there are 11 nodes in the network, there are 2048 states in total and all the state transitions can be computed directly through Eq. (13.2). One of the attractors, among seven, is the most stable and attracts approximately 86% of all states. This stable (fixed point) attractor, in which the molecules Cdhl and Sicl are equal to 1 and all others (Cln3, MBF, SBF, Cln1/2, Swi5, Cdc20, Clb5/6, Clb1/2, Mcml) are equal to 0, represents the biological G1 stationary state (one of the four phases of the cell cycle process in which the cell grows and can commit to division), guaranteeing cellular stability in this state. It is further demonstrated in Li et al. (2004) that the dynamic state trajectories starting from each of the states in the basin of attraction of the G1 stationary state converge rapidly onto an attracting state trajectory that is highly stable, ensuring that starting from any point in the cell cycle process, the system does not deviate from this trajectory. It is also shown, by comparison with random networks, that the highly stable attractor is unlikely to arise by chance (Li et al., 2004). Additionally, the results were fairly insensitive to the values of the weights, justifying setting them both equal to 1. Other similar studies have been carried out with the cell cycle of the fission yeast Schizosaccharomyces pombe (Davidich and Bornholdt, 2008b) and the mammalian cell cycle (Fauré et al., 2006). Recently, a new more accurate Boolean network model, which can incorporate time delays, has been proposed as a model of the budding yeast cell cycle (Irons, 2009).
3. Differential Equation Models
A model of a genetic network based on a system of differential equations expresses the rates of change of an element, such as a gene product, in terms of the levels of other elements of the network and possibly external inputs. In general, a nonlinear time-dependent differential equation has the form
| (13.3) |
where x is a state vector denoting the values of the physical variables in the system, ẋ = dx/dt is the elementwise derivative of x, u is a vector of external inputs, and t is time.
If time is discretized and the functional dependency specified by f does not depend on time, then the system is said to be time-invariant. If f is linear and time-invariant, then it can be expressed as
| (13.4) |
where A and B are constant matrices (Weaver et al., 1999).
When ẋ = 0, the variables no longer change with time and thus define the steady state of the system, which is analogous to a fixed point attractor in a Boolean network. Consider the simple case of a gene product x (a scalar) whose rate of synthesis is proportional, with kinetic constant k1, to the abundance of another protein a that is sufficiently abundant such that the overall concentration of a is not significantly changed by the reaction. However, x is also subject to degradation, the rate of which is proportional, with constant k2, to the concentration of x itself. This situation can be expressed as
| (13.5) |
Let us analyze the behavior of this simple system. If initially x = 0, then the decay term is also 0 and ẋ = k1a. However, as x is produced, the decay term k2x will also increase thereby decreasing the rate ẋ toward 0 and stabilizing x at some steady-state value x̄. It is easy to determine this value, since setting ẋ = 0 and solving for x yields
| (13.6) |
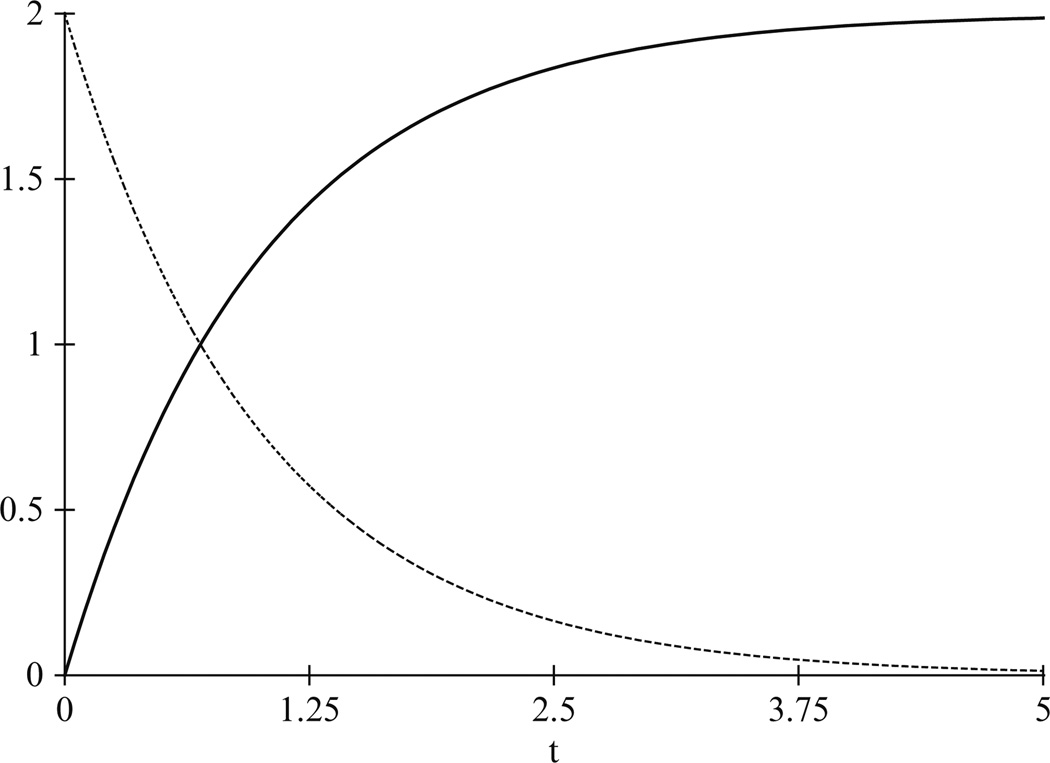
This behavior is shown in Fig. 13.2, where x starts off at x = 0 and approaches the value in Eq. (13.6). The exact form of the kinetics is
| (13.7) |
Figure 13.2.
The behavior of the solution to ẋ = k1a − k2x, x(0) = 0, where k1 = 2, k2 = 1, and a = 1. As can be seen, the gene product x, shown with a solid plot, tends toward its steady-state value given in Eq. (13.6). The time derivative ẋ, which starts at initial value of k1a and tends toward 0, is shown with a dashed plot.
Similarly, the derivative ẋ, also shown in Fig. 13.2, starts off at the initial value of k1a and thereafter tends toward zero.
Now suppose that a is suddenly removed after the steady-state value x̄ is reached. Since a = 0, we have ẋ = −k2x and since the initial condition is x = k1a/k2, ẋ = −k1a initially. The solution of this equation is
| (13.8) |
and it can be seen that it will eventually approach zero.
This example describes a linear relationship between a and ẋ. However, most gene interactions are highly nonlinear. When the regulator is below some critical value, it has very little effect on the regulated gene. When it is above the critical value, it has virtually full effect that cannot be significantly amplified by increased concentrations of the regulator. This nonlinear behavior is typically described by sigmoid functions, which can be either monotonically increasing or decreasing. A common form is the so-called Hill functions given by
| (13.9) |
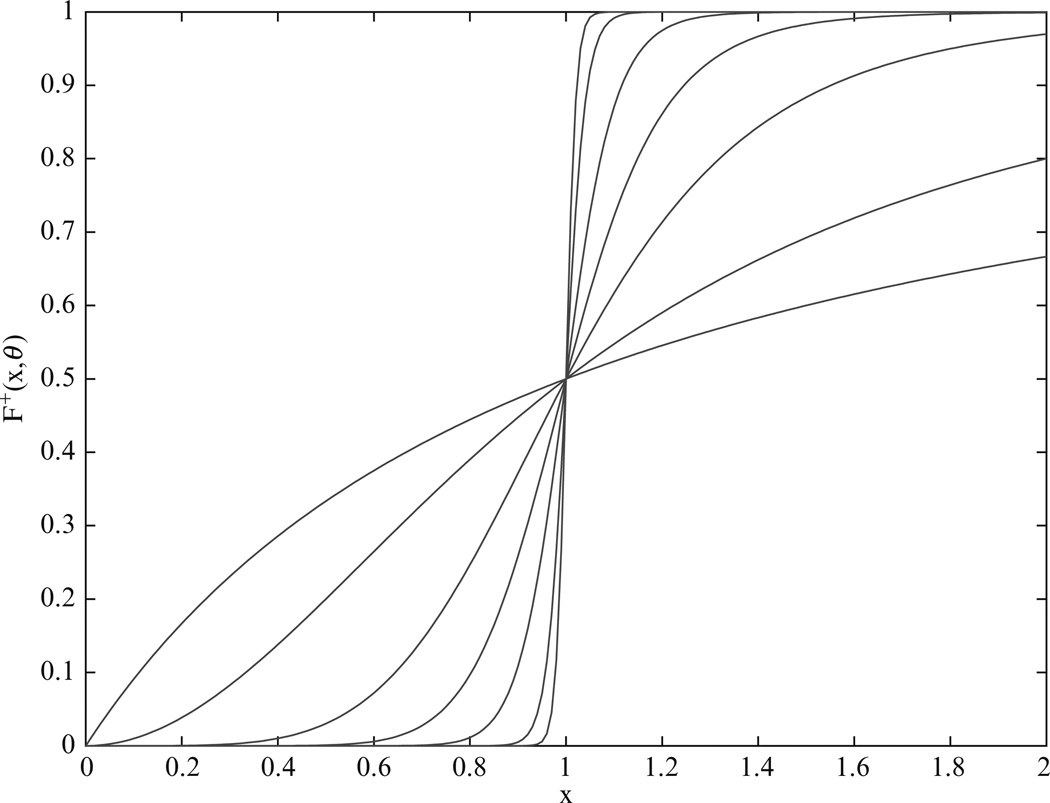
The function F+ (x, 1) is illustrated in Fig. 13.3 for n = 1, 2, 5, 10, 20, 50, and 100. It can be seen that it approaches an ideal step function with increasing n, thus approximating a Boolean switch. In fact, the parameter θ essentially plays the role of the threshold value. Glass (1975) used step functions in place of sigmoidal functions in differential equation models, resulting in so-called piecewise linear differential equations. Glass and Kauffman (1973) also showed that many systems exhibit the same qualitative behavior for a wide range of sigmoidal steepnesses, parameterized by n.
Figure 13.3.
The function F+ (x, θ) for θ = 1 and n = 1, 2, 5, 10, 20, 50, and 100. As n gets large, F+ (x, θ) approaches an ideal step function and thus functions as a Boolean switch.
Given that gene regulation is nonlinear, the differential equation models can incorporate the Hill functions into their synthesis and decay terms. There are many available computer tools for simulating and analyzing such dynamical systems using a variety of methods and algorithms (Lambert, 1991), including DBsolve (Goryanin et al., 1999), GEPASI (Mendes, 1993), and Dizzy (Ramsey et al., 2005). Additionally, there are toolboxes available for MATLAB® that can be used for modeling, simulating, and analyzing biological systems with ordinary differential equations (Schmidt and Jirstrand, 2006). MathWorks’ SimBiology® toolbox (http://www.mathworks.com/products/simbiology) also provides a graphical user interface for constructing models and entering reactions, parameters, and kinetic laws, which can be simulated deterministically or stochastically. A useful review of nonlinear ordinary differential equation modeling of the cell cycle is available in Sible and Tyson (2007).
3.1. Accurate description of cellular growth and division and prediction of mutant phenotypes
Let us return to the regulatory network controlling the cell cycle in budding yeast. If the goal of the modeling is to predict detailed quantitative phenomena, such as cell cycle duration in parent and daughter cells, the length of the different phases of the cell cycle, or ratios between certain regulatory proteins, then logical models such as Boolean networks are not appropriate, and systems of ordinary differential equations with detailed kinetic parameters must be used. Chen et al. (2004) constructed such a detailed model of the cell cycle regulatory network containing 36 equations with 148 constants, in addition to algebraic equations (available in Table 1 in that paper, with Table 2 containing parameter values). The model incorporates protein concentrations, cell mass, DNA mass, the state of the emerging bud, and of the mitotic spindle.
After manual fitting of some of the parameters, the dynamics generated by the model were able to accurately describe the growth and division of wild-type cells. Remarkably, the model also conformed to the phenotypes of more than 100 mutant strains, in terms of experimentally observed properties such as size at bud emergence or at onset of DNA synthesis, viability, or growth rate, relative to these properties in the wild type.
It should be pointed out that parameter estimation of the model must be approached with care. First, the objective function, for example mean-squared error between the model predictions and the experimental data, may have multiple local optima in the parameter space. Thus, an apparently good model fit may nonetheless contain unrealistic sets of parameters that will ultimately fail to generalize. For example, as was found in Chen et al. (2004), changing parameters to “rescue” a model with respect to a mutant (i.e., make it agree with experimental observations) often exhibit unintended and unanticipated effects on other mutants. Second, model selection must be carefully considered, since a model that is overly complex, meaning that it has many degrees of freedom, is likely to “overfit” the data and thereby, sacrifice predictive accuracy. In other words, the model may appear to predict very well when tested against data on which it was trained, but when tested against data under new conditions, the model will predict very poorly. There are powerful tools, such as minimum description length, and indeed, entire frameworks based on algorithmic information theory and Bayesian inference, devoted to these fundamental issues (Rissanen, 2007).
4. Probabilistic Boolean Networks
PBNs are probabilistic or stochastic generalizations of Boolean networks. Essentially, the deterministic dynamics are replaced by probabilistic dynamics, which can be framed within the mature and well-established theory of Markov chains, for which many analytical and numerical tools have been developed. Recall that Markov chains are stochastic processes having the property that future states depend only on the present state, and not on the past states. The transitions from one state to another (possibly itself) are specified by state transition probabilities. Boolean networks are special cases of PBNs in which state transition probabilities are either 1 or 0, depending on whether Eq. (13.1) is satisfied for all i = 1,…,n. The probabilistic nature of this model class affords flexibility and power in terms of making inferences from data, which necessarily contain uncertainty, as well as in terms of understanding the dynamical behavior of biological networks, particularly in relation to their structure.
Once the state transition probabilities for a Markov chain corresponding to a PBN are determined, it becomes possible to study the steady-state (long-run) behavior of the stochastic system. This long-run behavior is analogous to attractors in Boolean networks or fixed points in systems of differential equations. Kim et al. (2002) investigated the Markov chain corresponding to a small network based on microarray data observations of human melanoma samples. The steady-state behavior (distribution) of the constructed Markov chain was then compared to the initial observations. If the Markov chain is ergodic, meaning that it is possible to reach any state from any other state after an arbitrary number of steps, then the steady-state probability corresponds to the fraction of the time that the system will spend in that particular state.
The remarkable finding was that only a small number of all possible states had significant steady-state probabilities and most of those states with high probability were observed in the data. Furthermore, it was found that more than 85% of those states with high steady-state probability that were not observed in the data were very close to the observed data in terms of Hamming distance, which is equal to the number of genes that “disagree” in their binary values. Based on the transition rules inferred from the data, the model produced localized stability, meaning that the system tended to flow back to the states with high steady-state probability mass if placed in their vicinity. Thus, the stochastic dynamics of the Markov chain were able to mimic biological regulation. It should be noted that Markov chains are commonly used to model gene expression dynamics using so-called dynamic Bayesian networks (Murphy and Mian, 1999; Yu et al., 2004; Zou and Conzen, 2005). Indeed, PBNs and dynamic Bayesian networks are able to represent the same joint probability distribution over their common variables (i.e., genes) (Lähdesmäki et al., 2006).
Except in very restricted circumstances, gene expression data refute the determinism inherent to the Boolean network model, there typically being a number of possible successor states to any given state. Consequently, if one continues to assume the state at time t + 1 is independent of the state values prior to time t, then, as stated above, the network dynamics are described by a Markov chain whose state transition matrix reflects the observed stochasticity. In terms of gene regulation, this stochasticity can be interpreted to mean that several regulator gene sets are associated with each gene and at any time point one of these “predictor” sets, along with a corresponding Boolean function, is randomly chosen to provide the value of the gene as a function of the values within the chosen predictor set. It is this reasoning that motivated the original definition of a PBN in which the definition of a Boolean network was adapted in such a way that, for each gene, at each time point, a Boolean function (and predictor gene set) is randomly chosen to determine the network transition (Shmulevich et al., 2002a,c).
Rather than simply randomly assigning Boolean functions at each time point, one can take the perspective that the data come from distinct sources, each representing a “context” of the cell. From this perspective, the data derive from a family of deterministic networks and, in principle, the data could be separated into separate samples according to the contexts from which they have been derived. Given the context, the overall network would function as a Boolean network, its transition matrix reflecting determinism (i.e., each row contains one 1, in the column that corresponds to the successor state, and the rest are 0s). If defined in this manner, a PBN is a collection of Boolean networks in which a constituent network governs gene activity for a random period of time before another randomly chosen constituent network takes over, possibly in response to some random event, such as an external stimulus or the action of a (latent) regulator that is outside the scope of the network. Since the latter is not part of the model, network switching is random. This model defines a “context-sensitive” PBN (Brun et al., 2005; Shmulevich et al., 2002c). The probabilistic nature of the constituent choice reflects the fact that the system is open, not closed, the idea being that changes between the constituent networks result from the genes responding to latent variables external to the model network.
We now formally define PBNs. Although we retain the terminology “Boolean” in the definition, this does not refer to the binary quantization assumed in standard Boolean networks, but rather to the logical character of the gene predictor functions. In the case of PBNs, quantization is assumed to be finite, but not necessarily binary. However, we restrict ourselves to the binary domain here for simplicity. Formally, a PBN consists of a sequence of n nodes, where xi ∈ {0, 1}, and a sequence of vector-valued functions, defining constituent networks. In the framework of gene regulation, each element xi represents the expression value of a gene. Each vector-valued function determines a constituent network, or context, of the PBN. The function is a predictor of gene i, whenever network l is selected. At each updating epoch, a decision is made whether to switch the constituent network. This decision depends on a binary random variable ξ: if ξ = 0, then the current context is maintained; if ξ = 1, then a constituent network is randomly selected from among all constituent networks according to the selection probability distribution
| (13.10) |
The switching probability q = P(ξ = 1) is a system parameter. If the current network is maintained, then the PBN behaves like a fixed network and synchronously updates the values of all the genes according to the current context. Note that, even if ξ = 1, a different constituent network is not necessarily selected because the “new” network is selected from among all contexts. In other words, the decision to switch is not equivalent to the decision to change the current network. If a switch is called for (ξ = 1), then, after selecting the predictor function fl, the values of genes are updated accordingly; that is, according to the network determined by fl. If q < 1, the PBN is said to be context-sensitive; if q = 1, the PBN is said to be instantaneously random, which corresponds to the original definition in Shmulevich et al. (2002a).
Whereas a network switch corresponds to a change in a latent variable causing a structural change in the functions governing the network, a random perturbation corresponds to a transient value change that leaves the network wiring unchanged, as in the case of activation or inactivation owing to external stimuli such as stress conditions, small molecule inhibitors, etc. In a PBN with perturbation, there is a small probability p that a gene may change its value at each epoch. Perturbation is characterized by a random perturbation vector γ = (γ1, γ2, …, γn), γi ∈ {0, 1}, and P(γi = 1) = p, the perturbation probability; γi is also known as a Bernoulli(p) random variable. If x(t) is the current state of the network, and γ (t + 1) = 0, then the next state of the network is given by x(t + 1) = fl(x(t)), as in Eq. (13.1); otherwise, x(t + 1) = x(t) ⊕ γ(t + 1), where ⊕ is componentwise exclusive OR. The probability of no perturbation, in which case the next state is determined according to the current network function fl, is (1 − p)n and the probability of a perturbation is 1 − (1 − p)n. The perturbation model captures the realistic situation where the activity of a gene undergoes a random alteration (Shmulevich et al., 2002b).
As with Boolean networks, attractors play a major role in the study of PBNs. By definition, the attractor cycles of a PBN consist of the attractor cycles of the constituent networks, and their basins are likewise defined. Whereas in a Boolean network two attractor cycles cannot intersect, attractor cycles from different contexts can intersect in a PBN. The presentation of the state transition probabilities of the Markov chain corresponding to the (context-sensitive) PBN is beyond the scope of this chapter, and the reader is referred to Brun et al. (2005). Suffice it to say that from the state transition matrix of the Markov chain, which is guaranteed to be ergodic under a gene perturbation model as described above, even for very small p, one can compute the steady-state distribution. A Markov chain is said to possess a steady-state distribution if there exists a probability distribution π = (π1, π2, …, πM) such that for all states i, j ∈{1, 2, …, M},
| (13.11) |
where is the r-step transition probability between states i and j. If there exists a steady-state distribution, then regardless of the initial state, the probability of the Markov chain being in state i in the long run can be estimated by sampling the observed states in the simulation (by simply counting the percentage of time the chain spends in that state). Such an approach was used to analyze the joint steady-state probabilities of several key molecules (NFκB; Tie-2 and TGFB3) in a 15-gene network derived from human glioma gene expression data (Shmulevich et al., 2003).
4.1. Steady-state analysis and stability under stochastic fluctuations
The Boolean network model of the cell cycle, discussed in Section 2.1, was generalized in Zhang et al. (2006) such that network dynamics are described by a Markov chain with transition probabilities:
| (13.12) |
and
| (13.13) |
The term T appears in Eq. (13.2). Note that this is essentially a way of introducing noise and therefore making the Markov chain ergodic, so that a steady-state distribution exists. The positive number β plays the role of temperature that characterizes the strength of the noise introduced into the system dynamics. The parameter α is used to characterize the stochasticity when the input to a node is zero and determines the probability for a protein to maintain its state when there is no input to it. It should be noted that when α, β → ∞, the stochastic model converges to the deterministic Boolean network model in Li et al. (2004).
The state transition probabilities allow the computation of the steady-state distribution in Eq. (13.11). In addition, the so-called net probability flux πi Pij —πj Pji from state i to state j can be determined, where Pij is the state transition probability. The steady-state probability of the stationary G1 phase of the cell cycle was studied relative to the noise level determined by β. It was found that this state is indeed the most probable state of the system and that it decreases with increasing noise strength, as expected, since random perturbations will tend to move the system away from the attractor (Zhang et al., 2006). Interestingly, a type of phase transition was found whereby at a critical value of the parameter β, the steady-state probability of the stationary G1 state virtually vanishes and the system becomes dominated by noise and cannot carry out coordinated behavior. Nonetheless, this critical temperature is quite high and the system is able to tolerate approximately 10% of its rules misbehaving, implying that the cell cycle network is robust against stochastic fluctuations (Zhang et al., 2006). Additionally, the probability flux from states other than those on the cell cycle trajectory from the excited G1 state is convergent onto this trajectory, implying homeostatic stability.
5. Stochastic Differential Equation Models
The stochastic generalization of Boolean networks, leading to Markovian dynamics, is intended to capture uncertainty in the data, whether due to measurement noise or biological variability, intrinsic or extrinsic, the latter being caused by latent variables external to the model. On the other hand, if the intention of the modeling is to capture quantitative molecular or physical details, as in systems of ordinary differential equations discussed in Section 3, then stochastic fluctuations on the molecular level can be incorporated explicitly into the model using stochastic differential equations. For example, as most regulatory molecules are produced at very low intracellular concentrations, the resulting reaction rates exhibit large variability. Such intrinsic molecular noise has been found to be important for many biological functions and processes (Ozbudak et al., 2002; Raser and O’Shea, 2005).
There exist powerful stochastic simulation methods for accurately simulating the dynamics of a system of chemically reacting molecules that can reflect the discrete and stochastic nature of such systems on a cellular scale. A recent review of such methods is available in Cao and Samuels (2009). However, there are undoubtedly other intrinsic and extrinsic contributions to variability in gene and protein expression, for example, due to spatial heterogeneity or fluctuations in cellular components (Swain et al., 2002). Stochastic differential equations allow for a very general incorporation of stochasticity into a model without the need to assume specific knowledge about the nature of such stochasticity.
Manninen et al. (2006) developed several approaches to incorporate stochasticity into deterministic differential equation models, obtaining socalled Itô stochastic differential equations, and applied them to neuronal protein kinase C signal transduction pathway modeling. By a comparative analysis it was shown that such approaches are preferred to the stochastic simulation algorithm methods, as the latter are considerably slower by several orders of magnitude when simulating systems with a large number of chemical species (Manninen et al., 2006). The stochastic differential equation framework additionally allows the incorporation of stochasticity into the reaction rates, rate constants, and concentrations.
The basic model can be written as a Langevin equation with multiplicative noise (Rao et al., 2002), so that for a single species
| (13.14) |
where fi(x, u, t) is the deterministic model and ξi(t) is zero mean unit variance Gaussian white noise. The function g(xi) represents the contribution of the fluctuations and it is commonly assumed to be proportional to the square root of the concentration, that is, . The solution to such stochastic differential equations can be obtained by numerical integration using standard techniques.
5.1. The influence of noise on system behavior
Let us turn to the cell cycle control network of the fission yeast S. pombe, for which a system of ordinary differential equations was proposed (Novak et al., 2001), consisting of eight deterministic differential equations and three algebraic equations. We mention in passing that a Boolean network model for this network is available in Davidich and Bornholdt (2008b). The differential equation model in Novak et al. (2001) was found to be in good agreement with wild-type cells as well as with several mutants. Steuer (2004)converted this model to a system of stochastic differential equations and compared the simulations with experimental data. It was found that the cycle time and division size distributions within a cell population were predicted well by the model; for example, the model predicted a negative correlation between cycle time and mass at birth, meaning that the cells that are large at birth have shorter cycle times, which ensures homeostasis in successive generations (Steuer, 2004). The stochastic model also accounted for a characteristic ratio of the coefficients of variation for the cycle time and division length.
The stochastic differential equation model was also applied to study a certain double mutant (wee1− cdc25 Δ) that exhibits quantized cycle lengths. A deterministic model of the mutants can be obtained by removing the corresponding parameters from the system of differential equations. However, the simulation of the deterministic differential equation model of the double mutant results in periodically alternating long and short cycle times, which are determined exclusively by cell mass at birth, meaning that small cells have long cycles and have large daughters, and large cells have short cycles and give rise to small daughters. The simulation of the stochastic differential equation model produces very different results: cell mass at birth no longer determines the length of the next cycle and the (nonintuitive) characteristic clusters (i.e., “quantization”) in a plot of cycle time versus mass at birth are in good agreement with experimental observations (Steuer, 2004). Additionally, in the stochastic model, the oscillation between long and short cycles disappears, which is consistent with experimental observations. Thus, the inclusion of stochastic fluctuations in the model was able to account for several features not accounted for by the deterministic model. The fact that noise is able to qualitatively alter macroscopic system behavior suggests that stochastic fluctuations play a key role in modulating cellular regulation. Stochastic differential equation models provide a powerful framework for gaining an understanding of these phenomena.
REFERENCES
- Aldana M, Coppersmith S, Kadanoff LP. Boolean dynamics with random couplings. In: Kaplan E, Marsden JE, Sreenivasan KR, editors. Perspectives and Problems in Nonlinear Science. New York: Springer; 2002. pp. 23–89. [Google Scholar]
- Alvarez-Buylla ER, Chaos A, Aldana M, Benítez M, Cortes-Poza Y, Espinosa-Soto C, Hartasánchez DA, Lotto RB, Malkin D, Escalera Santos GJ, Padilla-Longoria P. Floral morphogenesis: Stochastic explorations of a gene network epigenetic landscape. PLoS ONE. 2008;3(11):e3626. doi: 10.1371/journal.pone.0003626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bornholdt S. Boolean network models of cellular regulation: Prospects and limitations. J. R. Soc. Interface. 2008;5 Suppl. 1:S85–S94. doi: 10.1098/rsif.2008.0132.focus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braunewell S, Bornholdt S. Superstability of the yeast cell-cycle dynamics: Ensuring causality in the presence of biochemical stochasticity. J. Theor. Biol. 2006;245(4):638–643. doi: 10.1016/j.jtbi.2006.11.012. [DOI] [PubMed] [Google Scholar]
- Brun M, Dougherty ER, Shmulevich I. Steady-state probabilities for attractors in probabilistic Boolean networks. Signal Process. 2005;85(4):1993–2013. [Google Scholar]
- Cao Y, Samuels DC. Discrete stochastic simulation methods for chemically reacting systems. Methods Enzymol. 2009;454:115–140. doi: 10.1016/S0076-6879(08)03805-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang HH, Hemberg M, Barahona M, Ingber DE, Huang S. Transcriptome-wide noise controls lineage choice in mammalian progenitor cells. Nature. 2008;453(7194):544–547. doi: 10.1038/nature06965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaves M, Albert R, Sontag D. Robustness and fragility of Boolean models for genetic regulatory networks. J. Theor. Biol. 2005;235:431–449. doi: 10.1016/j.jtbi.2005.01.023. [DOI] [PubMed] [Google Scholar]
- Chen KC, Calzone L, Csikasz-Nagy A, Cross FR, Novak B, Tyson JJ. Integrative analysis of cell cycle control in budding yeast. Mol. Biol. Cell. 2004;15:3841–3862. doi: 10.1091/mbc.E03-11-0794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidich M, Bornholdt S. The transition from differential equations to Boolean networks: A case study in simplifying a regulatory network model. J. Theor. Biol. 2008a;255(3):269–277. doi: 10.1016/j.jtbi.2008.07.020. [DOI] [PubMed] [Google Scholar]
- Davidich MI, Bornholdt S. Boolean network model predicts cell cycle sequence of fission yeast. PLoS ONE. 2008b;3(2):e1672. doi: 10.1371/journal.pone.0001672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drossel B. Random Boolean networks. In: Schuster HG, editor. Annual Review of Nonlinear Dynamics and Complexity, Vol. 1. Wiley; 2007. [Google Scholar]
- Fauré A, Naldi A, Chaouiya C, Thieffry D. Dynamical analysis of a generic Boolean model for the control of the mammalian cell cycle. Bioinformatics. 2006;22(14):e124–e131. doi: 10.1093/bioinformatics/btl210. [DOI] [PubMed] [Google Scholar]
- Glass L. Classification of biological networks by their qualitative dynamics. J. Theor. Biol. 1975;54:85–107. doi: 10.1016/s0022-5193(75)80056-7. [DOI] [PubMed] [Google Scholar]
- Glass L, Kauffman SA. The logical analysis of continuous, nonlinear biochemical control networks. J. Theor. Biol. 1973;39:103–129. doi: 10.1016/0022-5193(73)90208-7. [DOI] [PubMed] [Google Scholar]
- Goryanin I, Hodgman TC, Selkov E. Mathematical simulation and analysis of cellular metabolism and regulation. Bioinformatics. 1999;15(9):749–758. doi: 10.1093/bioinformatics/15.9.749. [DOI] [PubMed] [Google Scholar]
- Huang S. Gene expression profiling, genetic networks, cellular states: An integrating concept for tumorigenesis and drug discovery. J. Mol. Med. 1999;77(6):469–480. doi: 10.1007/s001099900023. [DOI] [PubMed] [Google Scholar]
- Huang S, Ingber DE. Shape-dependent control of cell growth, differentiation, apoptosis: Switching between attractors in cell regulatory networks. Exp. Cell Res. 2000;261(1):91–103. doi: 10.1006/excr.2000.5044. [DOI] [PubMed] [Google Scholar]
- Huang S, Eichler G, Bar-Yam Y, Ingber DE. Cell fates as high-dimensional attractor states of a complex gene regulatory network. Phys. Rev. Lett. 2005;94(12):128701–128704. doi: 10.1103/PhysRevLett.94.128701. [DOI] [PubMed] [Google Scholar]
- Irons DJ. Logical analysis of the budding yeast cell cycle. J. Theor. Biol. 2009;257(4):543–559. doi: 10.1016/j.jtbi.2008.12.028. [DOI] [PubMed] [Google Scholar]
- Jacob F, Monod J. On the regulation of gene activity. Cold Spring Harb. Symp. Quant. Biol. 1961;26:193–211. doi: 10.1101/sqb.1961.026.01.048. [DOI] [PubMed] [Google Scholar]
- Kauffman SA. Metabolic stability and epigenesis in randomly constructed genetic nets. J. Theor. Biol. 1969a;22:437–467. doi: 10.1016/0022-5193(69)90015-0. [DOI] [PubMed] [Google Scholar]
- Kauffman SA. Homeostasis and differentiation in random genetic control networks. Nature. 1969b;224:177–178. doi: 10.1038/224177a0. [DOI] [PubMed] [Google Scholar]
- Kauffman SA. The large scale structure and dynamics of genetic control circuits: An ensemble approach. J. Theor. Biol. 1974;44:167–190. doi: 10.1016/s0022-5193(74)80037-8. [DOI] [PubMed] [Google Scholar]
- Kauffman SA. The Origins of Order: Self-Organization and Selection in Evolution. New York: Oxford University Press; 1993. [Google Scholar]
- Kauffman S. A proposal for using the ensemble approach to understand genetic regulatory networks. J. Theor. Biol. 2004;230(4):581–590. doi: 10.1016/j.jtbi.2003.12.017. [DOI] [PubMed] [Google Scholar]
- Kim S, Li H, Dougherty ER, Cao N, Chen Y, Bittner ML, Suh EB. Can Markov chain models mimic biological regulation? J. Biol. Syst. 2002;10(4):431–445. [Google Scholar]
- Lähdesmäki H, Hautaniemi S, Shmulevich I, Yli-Harja O. Relationships between probabilistic Boolean networks and dynamic Bayesian networks as models of gene regulatory networks. Signal Process. 2006;86(4):814–834. doi: 10.1016/j.sigpro.2005.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert JD. Numerical Methods for Ordinary Differential Equations. Chichester: Wiley; 1991. [Google Scholar]
- Li F, Long T, Lu Y, Quyang Q, Tang C. The yeast cell-cycle network is robustly designed. Proc. Natl. Acad. Sci. USA. 2004;101(14):4781–4786. doi: 10.1073/pnas.0305937101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLeod MC. A possible role in chemical carcinogenesis for epigenetic, heritable changes in gene expression. Mol. Carcinog. 1996;15(4):241–250. doi: 10.1002/(SICI)1098-2744(199604)15:4<241::AID-MC1>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- Manninen T, Linne ML, Ruohonen K. Developing Itô stochastic differential equation models for neuronal signal transduction pathways. Comput. Biol. Chem. 2006;30(4):280–291. doi: 10.1016/j.compbiolchem.2006.04.002. [DOI] [PubMed] [Google Scholar]
- Mendes P. GEPASI: A software package for modelling the dynamics, steady states and control of biochemical and other systems. Comput. Appl. Biosci. 1993;9(5):563–571. doi: 10.1093/bioinformatics/9.5.563. [DOI] [PubMed] [Google Scholar]
- Muroga S. Threshold Logic and its Applications. Wiley-Interscience; 1971. [Google Scholar]
- Murphy K, Mian S. Modelling Gene Expression Data using Dynamic Bayesian Networks. Technical Report. Berkeley: University of California; 1999. [Google Scholar]
- Novak B, Pataki Z, Ciliberto A, Tyson JJ. Mathematical model of the cell division cycle of fission yeast. Chaos. 2001;11(1):277–286. doi: 10.1063/1.1345725. [DOI] [PubMed] [Google Scholar]
- Ozbudak EM, Thattai M, Kurtser I, Grossman AD, van Oudenaarden A. Regulation of noise in the expression of a single gene. Nat. Genet. 2002;31(1):69–73. doi: 10.1038/ng869. [DOI] [PubMed] [Google Scholar]
- Ramsey S, Orrell D, Bolouri H. Dizzy: Stochastic simulations of large-scale genetic regulatory networks. J. Bioinform. Comput. Biol. 2005;3(2):1–21. doi: 10.1142/s0219720005001132. [DOI] [PubMed] [Google Scholar]
- Rao CV, Wolf DM, Arkin AP. Control, exploitation and tolerance of intracellular noise. Nature. 2002;420(6912):231–237. doi: 10.1038/nature01258. [DOI] [PubMed] [Google Scholar]
- Raser JM, O’Shea EK. Noise in gene expression: Origins, consequences, control. Science. 2005;309(5743):2010–2013. doi: 10.1126/science.1105891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richmond CS, Glasner JD, Mau R, Jin H, Blattner FR. Genome-wide expression profiling in Escherichia coli K-12. Nucleic Acids Res. 1999;27:3821–3835. doi: 10.1093/nar/27.19.3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rissanen J. Information and Complexity in Statistical Modeling. Springer; 2007. [Google Scholar]
- Schmidt H, Jirstrand M. Systems biology toolbox for MATLAB: A computational platform for research in systems biology. Bioinformatics. 2006;22(4):514–515. doi: 10.1093/bioinformatics/bti799. [DOI] [PubMed] [Google Scholar]
- Shmulevich I, Zhang W. Binary analysis and optimization-based normalization of gene expression data. Bioinformatics. 2002;18(4):555–565. doi: 10.1093/bioinformatics/18.4.555. [DOI] [PubMed] [Google Scholar]
- Shmulevich I, Dougherty ER, Kim S, Zhang W. Probabilistic Boolean networks: A rule-based uncertainty model for gene regulatory networks. Bioinformatics. 2002a;18(2):261–274. doi: 10.1093/bioinformatics/18.2.261. [DOI] [PubMed] [Google Scholar]
- Shmulevich I, Dougherty ER, Zhang W. Gene perturbation and intervention in probabilistic Boolean networks. Bioinformatics. 2002b;18(10):1319–1331. doi: 10.1093/bioinformatics/18.10.1319. [DOI] [PubMed] [Google Scholar]
- Shmulevich I, Dougherty ER, Zhang W. From Boolean to probabilistic Boolean networks as models of genetic regulatory networks. Proc. IEEE. 2002c;90(11):1778–1792. [Google Scholar]
- Shmulevich I, Gluhovsky I, Hashimoto R, Dougherty ER, Zhang W. Steady-state analysis of probabilistic Boolean networks. Comp. Funct. Genom. 2003;4(6):601–608. doi: 10.1002/cfg.342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sible JC, Tyson JJ. Mathematical modeling as a tool for investigating cell cycle control networks. Methods. 2007;41(2):238–247. doi: 10.1016/j.ymeth.2006.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SimBiology 3.0 Toolbox. http://www.mathworks.com/products/simbiology.
- Steuer R. Effects of stochasticity in models of the cell cycle: From quantized cycle times to noise-induced oscillations. J. Theor. Biol. 2004;228(3):293–301. doi: 10.1016/j.jtbi.2004.01.012. [DOI] [PubMed] [Google Scholar]
- Swain PS, Elowitz MB, Siggia ED. Intrinsic and extrinsic contributions to stochasticity in gene expression. Proc. Natl. Acad. Sci. USA. 2002;99(20):12795–12800. doi: 10.1073/pnas.162041399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas R. Boolean formalization of genetic control circuits. J. Theor. Biol. 1973;42:563–585. doi: 10.1016/0022-5193(73)90247-6. [DOI] [PubMed] [Google Scholar]
- Weaver DC, Workman CT, Stormo GD. Modeling regulatory networks with weight matrices. Pac. Symp. Biocomput. 1999;4:112–123. doi: 10.1142/9789814447300_0011. [DOI] [PubMed] [Google Scholar]
- Wolf DM, Eeckman FH. On the relationship between genomic regulatory element organization and gene regulatory dynamics. J. Theor. Biol. 1998;195(2):167–186. doi: 10.1006/jtbi.1998.0790. [DOI] [PubMed] [Google Scholar]
- Yu J, Smith VA, Wang PP, Hartemink AJ, Jarvis ED. Advances to Bayesian network inference for generating causal networks from observational biological data. Bioinformatics. 2004;20(18):3594–3603. doi: 10.1093/bioinformatics/bth448. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Qian M, Ouyang Q, Deng M, Li F, Tang C. Stochastic model of yeast cell-cycle network. Physica D. 2006;219(1):35–39. [Google Scholar]
- Zou M, Conzen SD. A new dynamic Bayesian network (DBN) approach for identifying gene regulatory networks from time course microarray data. Bioinformatics. 2005;21(1):71–79. doi: 10.1093/bioinformatics/bth463. [DOI] [PubMed] [Google Scholar]