Abstract
The interactions between glycosaminoglycans (GAGs), important components of the extracellular matrix, and proteins such as growth factors and chemokines play critical roles in cellular regulation processes. Therefore, the design of GAG derivatives for the development of innovative materials with bio-like properties in terms of their interaction with regulatory proteins is of great interest for tissue engineering and regenerative medicine. Previous work on the chemokine interleukin-8 (IL-8) has focused on its interaction with heparin and heparan sulfate, which regulate chemokine function. However, the extracellular matrix contains other GAGs, such as hyaluronic acid (HA), dermatan sulfate (DS) and chondroitin sulfate (CS), which have so far not been characterized in terms of their distinct molecular recognition properties towards IL-8 in relation to their length and sulfation patterns. NMR and molecular modeling have been in great part the methods of choice to study the structural and recognition properties of GAGs and their protein complexes. However, separately these methods have challenges to cope with the high degree of similarity and flexibility that GAGs exhibit. In this work, we combine fluorescence spectroscopy, NMR experiments, docking and molecular dynamics simulations to study the configurational and recognition properties of IL-8 towards a series of HA and CS derivatives and DS. We analyze the effects of GAG length and sulfation patterns in binding strength and specificity, and the influence of GAG binding on IL-8 dimer formation. Our results highlight the importance of combining experimental and theoretical approaches to obtain a better understanding of the molecular recognition properties of GAG–protein systems.
Keywords: GAG–protein interaction, glycosaminglycans (GAGs), interleukin-8 (IL-8), molecular modeling, NMR
Introduction
The extracellular matrix (ECM) consists of various charged and uncharged biopolymers and plays a crucial role in cell adhesion and proliferation. In addition, growth factors, cytokines and chemokines mediate numerous important cellular functions. Proper tissue function has to rely on the transport and availability of these regulatory proteins across the ECM, which is strongly mediated by the interactions between the proteins and the biopolymers of the ECM. Therefore, interactions between ECM and functional proteins play a decisive role in the regulation of processes such as hemostasis, anticoagulation, tumor progression or inflammation (Gandhi and Mancera 2008). Particularly important are the interactions between proteins and the negatively charged polysaccharides of the ECM, the glycosaminoglycans (GAGs). It has been suggested that GAGs may act as a storage site for highly diffusive functional proteins to aid their presentation to the respective receptor (Thiery and Boyer 1992). Thus, the ECM may provide a trap that sequesters secreted chemokines and prevents their diffusion from the inflammation site (Middleton et al. 1997).
Given the importance of the interaction between regulatory proteins and GAGs, it is astonishing that this topic represents a truly white spot in structural databases that contain tens of thousands of high-resolution protein structures obtained by X-ray crystallography or NMR, but fewer than 100 GAG–protein complexes (Imberty et al. 2007). Co-crystals of protein and oligosaccharides are difficult to obtain as GAGs may feature a high degree of conformational flexibility, especially regarding their glycosidic linkages. NMR assignments of oligosaccharides are complicated due to the highly repetitive nature of these molecules. Furthermore, chemical synthesis of pure oligosaccharides is extremely difficult due to their anomeric stereochemistry and the high complexity in terms of different possible structures that these molecules may offer (Gandhi and Mancera 2008). In nature, GAG conformational flexibility is further enhanced by variations in their sulfation patterns as well as by the contribution of metal ions and pH on their interactions with proteins (Gandhi and Mancera 2008). Although controlling the GAG sulfation pattern is experimentally extremely challenging, it can be addressed in computer simulations. Therefore, molecular modeling has also contributed largely to our understanding of GAG–protein interactions (Imberty et al. 2007; Gandhi and Mancera 2008; Sapay et al. 2011).
Interleukin-8 (IL-8) is a chemotactic cytokine that is able to trigger inflammation by recruiting and activating neutrophil granulocytes (Larsen et al. 1989). Leukocytes are typically activated by binding of a respective chemokine to a G protein-coupled receptor. However, IL-8 also strongly interacts with GAGs mediating the activation of leukocytes. Previous work has focused on the interaction of IL-8 with heparin and heparan sulfate, the most highly sulfated natural GAGs. One study has investigated the interaction of IL-8 with di- and trisulfated heparin-derived disaccharides using solution NMR spectroscopy (Kuschert et al. 1998). A general binding region in the C-terminal helix and a proximal loop was identified. Further, a horseshoe-like binding of heparan sulfate to IL-8 has been suggested from experimental work, where the GAG moiety runs parallel to the helix axis (Spillmann et al. 1998). Computational studies could reproduce the interaction sites of IL-8 and heparin and identify the residues that interact with the sugar moiety (Bitomsky and Wade 1999). Protein consensus sequences for the interaction of GAG with proteins have been identified (Cardin and Weintraub 1989), but binding of heparan sulfate to chemokines is rather unique (Lortat-Jacob et al. 2002). For IL-8, an orientation of the GAG molecule perpendicular to the helical axis bridging the two subunits in the IL-8 dimer has been proposed by computational work (Bitomsky and Wade 1999; Lortat-Jacob et al. 2002), which contrasts the above-mentioned horseshoe-like model (Bitomsky and Wade 1999; Krieger et al. 2004).
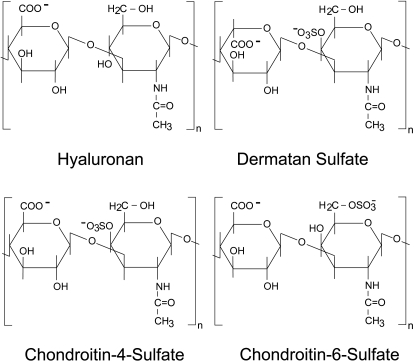
Although the interaction of IL-8 with heparin and heparan sulfate appears to be an important regulator of chemokine function, it should not be forgotten that the ECM also contains other GAGs such as hyaluronan (HA), chondroitin sulfate (CS) and dermatan sulfate (DS) (Figure 1) in much higher amounts (Gandhi and Mancera 2008). Some of these molecules are highly abundant as part of proteoglycans and undergo distinct dynamics in tissue (Schiller et al. 2001; Scheidt et al. 2010). While HA carries only one carboxylate group per disaccharide unit, the other GAGs feature an additional sulfate group.
Fig. 1.
Chemical structures of the different types of GAGs used in this study. The repeating disaccharide units of hyaluronan (HA), dermatan sulfate (DS), chondroitin-4-sulfate (CS4) and chondroitin-6-sulfate (CS6) are shown.
In our work, we have used fluorescence and NMR spectroscopy, docking and molecular dynamics (MD) simulations to study the molecular recognition properties of IL-8 with a variety of HA, DS, CS and their sulfated derivatives. These GAG derivatives hold an interesting potential for the development of innovative matrix materials with bio-like properties for tissue engineering applications and regenerative medicine. Such matrices are designed for the controlled delivery of bioactive proteins (Chung and Park 2007; Lee and Shin 2007; Dvir et al. 2011). The idea is to modify natural or synthetic matrix materials with GAGs to provide the scaffolding material with an environment that attracts regulatory proteins. The interaction strength between the matrix-associated GAGs and the regulatory proteins should be fine-tuned so that the matrix slowly releases these molecules to bind to the cellular receptors. Furthermore, an environment should be created that attracts specifically important regulatory proteins but is less attractive for unwanted pro-inflammatory regulators. For a rational design of such “intelligent” matrix materials, basic research has to address the details of the interaction of different GAGs with the respective regulatory proteins.
Results and discussion
Experimental binding
Experimental determination of GAG binding to IL-8 by fluorescence spectroscopy
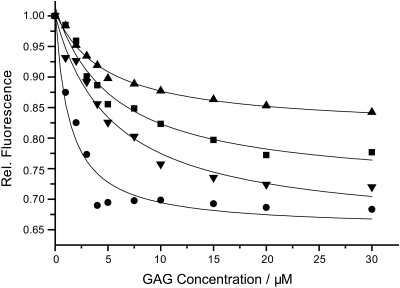
We analyzed the binding strength of negatively charged GAG hexasaccharides to IL-8. To this end, the tryptophan fluorescence intensity of IL-8 was recorded as a function of GAG concentration. GAG binding induces a decrease in Trp fluorescence and for chondroitin-6-sulfate (CS6) a slight shift in the fluorescence emission maximum. Experimental fluorescence titration curves are shown in Figure 2. Fluorescence intensity reaches a plateau value typically at around 20 µM GAG suggesting KD values in the low µM range. We determined experimental KD values using a binding model as described in the Methods section of 5.5 ± 1.3 µM for HA, 5.5 ± 1.0 µM for DS, 4.8 ± 0.7 µM for CS4 and 1.4 ± 0.4 µM for CS6 for the GAG hexasaccharides. Overall, these data show that all GAGs exhibit rather high affinity to IL-8. Differences in the overall strength of the decay in fluorescence intensity indicate slightly different binding geometry for the four GAGs. For CS6, the fluorescence intensity drops off quickly and reaches a plateau value already at ∼5 µM, indicating that the KD value might actually be smaller than the 1.4 µM determined by our model, which does not seem to describe the binding of CS6 to IL-8 correctly.
Fig. 2.
Fluorescence titration curves of IL-8 (1 µM) with HA (squares), DS (inverted triangles), CS4 (triangles) and CS6 (circles). The lines represent fits to the theoretical binding model as described in the text.
Resonance assignment and 1H-15N HSQC NMR spectra of IL-8
To obtain additional structural insights into the GAG–IL-8 interaction, we carried out solution NMR experiments on 15N-labeled IL-8. NMR resonance assignment and the three-dimensional structure determination of IL-8 (residues 1–72) have been reported before (Clore et al. 1989, 1990). Here, we have confirmed the assignment by carrying out an nuclear overhauser effect spectroscopy (NOESY) and a total correlation spectroscopy (TOCSY) experiment and further assigned the additional five N-terminal residues of the IL-8 (1–77) variant. A 1H-15N heteronuclear single-quantum coherence (HSQC) spectrum of uniformly 15N-labeled IL-8 (1–77) with the full assignments is shown in Figure 3. All resonance lines are narrow and well-dispersed as reported in the literature.
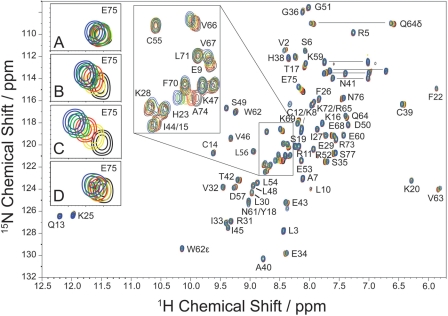
Fig. 3.
1H-15N HSQC NMR spectra of 15N-labeled IL-8 (1.0 mM, pH 7.0) in the presence of varying concentrations of CS6 hexasaccharide. Each color corresponds to a titration step: black, 0 µM CS6; yellow, 124 µM CS6; red, 244 µM CS6; green, 359 µM CS6; and blue, 471 µM CS6. (A–D) Enlargement of the response of IL-8 residue E75 on titration with various GAGs. (A) IL-8 (0.6 mM) titrated with HA hexasaccharides (black, 0 µM HA; yellow, 86 µM HA; red, 252 µM HA; green, 412 µM HA; blue, 566 µM HA); (B) IL-8 (0.8 mM) titrated with CS4 hexasaccharides (black, 0 µM CS4; yellow, 132 µM CS4; red, 261 µM CS4; green, 389 µM CS4; blue, 541 µM CS4) and (C) IL-8 (1.0 mM) titrated with CS6 hexasaccharides (black, 0 µM CS6; yellow, 124 µM CS6; red, 244 µM CS6; green, 359 µM CS6; blue, 471 µM CS6). (D) IL-8 (1.0 mM) titrated with DS hexasaccharide (black, 0 µM DS; yellow, 117 µM DS; red, 277 µM DS; green, 468 µM DS; blue 637 µM DS).
Interaction of IL-8 with GAGs
To probe the interaction of IL-8 with GAG hexamers, titration experiments were carried out. Figure 3 shows the superposition of 1H-15N HSQC spectra of IL-8 representing five titration steps with CS6 hexasaccharides. Clearly, several backbone sites showed pronounced chemical shift changes indicative of an interaction with the GAG. Chemical shifts change gradually as a function of GAG concentration, indicating a fast exchange process. However, not all GAGs investigated in this study showed a strong interaction with IL-8. Exemplary, the response of residue E75 of IL-8 to GAG binding is shown in the insets A–D. There is only a negligible chemical shift change upon titration of IL-8 with HA hexamers (A). Nevertheless, our binding measurements as well as literature data (David et al. 2008) show that HA interacts with IL-8, apparently without larger structural alterations in the protein. In contrast, much stronger effects were observed when IL-8 was titrated with CS4 (B), CS6 (C) and DS (D) indicating that the negatively charged sulfate group of CS or DS has a pronounced effect on the backbone structure of residue E75 of IL-8. We have also carried out HSQC experiments in the presence of higher salt concentration (500 mM NaCl), which showed essentially the same chemical shift changes upon GAG titration (data not shown).
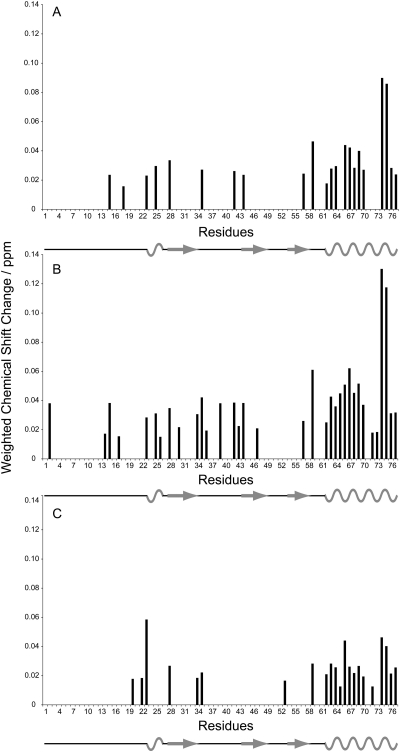
In order to map out the IL-8 residues that showed strongest interaction with the respective GAG, chemical shift perturbation charts were plotted (Figure 4). As inferred from Figure 3A, titration with HA did not result in any significant chemical shift perturbations in the HSQC spectra. In contrast, titration with both CS4 (Figure 4A) and CS6 (Figure 4B) as well as with DS (Figure 4C) showed significant chemical shift changes. Two protein regions experiencing strong changes in chemical shifts during titration were identified: residues 15–45 and 57–77. The largest chemical shift changes occurred for residues K59, V66, V67, K69, A74 and E75. Overall, the chemical shift changes observed are moderate but compare well with what has been observed for the interaction between heparin disaccharides and IL-8 (Kuschert et al. 1998).
Fig. 4.
Weighted chemical shift perturbation plots of IL-8 showing the chemical shift changes of each residue after titration with (A) 663 µM CS4, (B) 471 µM CS6 and (C) 637 µM DS.
In principle, the NMR titration experiments should also provide binding curves similar to those measured in the fluorescence experiments (Figure 2). However, the IL-8 concentration necessary for meaningful NMR experiments needs to be on the order of 1 mM, which is ∼3 orders of magnitude higher than the KD and therefore renders its determination by NMR very difficult. The experiment is further hampered by the fact that part of the protein is lost due to aggregation. This has also been observed in other studies of GAG–chemokine interaction (Zhao and Liwang 2010). Therefore, in NMR studies that typically require large protein concentration, just dimeric GAGs are used (Kuschert et al. 1998; Zhao and Liwang 2010). For the sake of higher biological relevance, we decided to carry out the GAG binding measurements with hexameric GAGs. Longer GAGs induce protein aggregation and the last titration steps showed the precipitation of IL-8–GAG complexes. As a result, no binding constants could be determined from the NMR titrations.
Previous work identified a BxxxBxxBB binding motif of heparin and heparan sulfate to the α-helix of IL-8, which involves the basic residues R65, K69, K72 and R73. In addition, H23 and K25 of the loop connecting the N-terminal region with the first strand of the β-sheet appear to be important binding regions (Kuschert et al. 1998; Bitomsky and Wade 1999; Lortat-Jacob et al. 2002). Although no significant changes in the HSQC spectra resulted from HA titration, both DS and CS binding caused these basic residues to shift their spectral position. This suggests a similar binding motif for the monosulfated GAGs studied here in comparison to heparin and heparan sulfate. However, residues R65, K72 and R73 did not show a very pronounced response to titration with CS4, whereas distinct effects are observed for CS6.
In addition to the response of the basic residues, we observe large chemical shift changes for V66 and V67, which are in between the first two basic residues of the consensus sequence. Interestingly, the largest shift changes were observed for A74 and E75, which are directly adjacent to the two basic residues at the end of the consensus sequence. It should be emphasized that the NMR shifts report the response of the protein backbone to GAG binding, which takes place at the end of the basic side chains. Apparently, GAG binding to IL-8 causes moderate structural alterations of the C-terminal α-helix of IL-8, which also affects residues that do not directly interact. In particular, the negatively charged E75 appears to undergo considerable structural changes. This is likely related to the electrostatic repulsion of the negatively charged carboxylate group of E75 and the sulfate groups of CS, which causes a structural change of this residue. Compared with CS, the response of IL-8 to DS binding is more moderate, which might be related to the orientation of the negatively charged groups on the GAG (vide infra). DS binding also caused residue H23 to shift significantly, which was also reported to be involved in heparin binding (Kuschert et al. 1998; Bitomsky and Wade 1999; Lortat-Jacob et al. 2002).
Computational binding
Although the NMR approach allows studying the interaction of IL-8 with various GAG site specifically, it is limited to those sugar molecules that are readily available. However, for biotechnological applications, GAGs with non-natural sulfation patterns, such as sulfated derivatives of HA and CS, may provide more favorable interaction properties than natural ones. Further, determining the exact structure of protein–GAG complexes by NMR is challenging due to the highly repetitive nature of the sugar moieties, which introduces a considerable ambiguity. Here, computer-derived models and simulation provide great complementarity as even a sparse set of experimental constraints can largely improve the predicting power of modeling and simulation techniques. Further, any given sugar molecule with a specific sulfation pattern can be studied computationally prior to laborious chemical synthesis. Therefore, in order to complement our experimental studies and to extend them with regard to other sulfated sugar molecules, we carried out extensive computational analysis on GAGs with natural (HA, CS4, CS6, heparin and DS) and non-natural sulfation patterns binding to IL-8. First, we have used GAG tetrasaccharides since nowadays available docking approaches are restricted to the number of degrees of ligand's freedom and, consequently, to the GAG length. In order to be more consistent with our NMR studies, we characterized the influence of GAG length on the binding to IL-8 and, in particular, we also analyzed the binding of those GAG hexasaccharides that were studied experimentally.
Docking of GAGs to IL-8
We have observed that docking heparin/heparan sulfate disaccharides to the IL-8 heparin-binding site (Kuschert et al. 1998) does not reveal binding specificity that could help suggesting a most probable binding pose for longer GAGs (data not shown). These results indicated the prevalence of unspecific electrostatic interactions between sulfate and carboxylic groups of GAGs with positively charged residues H23, K25, R65, K69, K72 and R73 of IL-8. When docking tetrameric GAGs, the results, however, clearly differ. In spite of the substantial differences in sulfation patterns of the 14 docked ligands, we observe one common and highly scored binding pose for all GAGs. Although not reflected in terms of root-mean-square deviation (RMSD)-based clustering (Table I), the conformational differences observed qualitatively speak for one common pose, which corresponds to an extended conformation located between the C-terminal α-helix of IL-8 (residues 60–77) and its N-terminal loop/310-helix (residues 14–25) and oriented from non-reducing to reducing terminus in a parallel manner to the IL-8 C-terminal α-helix. A similar pose was also observed in previous studies on hexameric heparin docking to IL-8 (Krieger et al. 2004). We find the same pose prevailing in the top 10 docking solutions also when docking is carried out with the 24 explicit water molecules in the binding site (Table I). There is a correlation (adjusted R2 = 0.47, P < 0.05) between the pose abundance for docking with and without water for different GAGs, supporting the idea that this pose does not appear randomly and is not purely electrostatically driven, since the abundance of the pose is not correlated with the charges of the GAGs. The second-most representative pose obtained by docking is an antiparallel pose to the above described (Supplementary data, Figure S1), confirming that the complexity of GAG docking can be partly explained by the high symmetry of GAGs (Forster and Mulloy 2006). To summarize, the combination of complementary experimental and theoretical approaches allowed us to discern a distinct, highly scoring and representative common binding pose to IL-8 for all studied GAGs, even though being of different length and containing different sulfation patterns. This observed binding pose is in agreement with previously proposed binding modes for heparin on IL-8.
Table I.
Docking results of tetrasaccharide derivatives of hyaluronan and CS, DS and heparin
| Ligand | Receptor without water molecules |
Receptor with 24 water molecules |
||
|---|---|---|---|---|
| Cluster number (members)a | Pose in top 10b | Cluster number (members)a | Pose in top 10b | |
| HA | 1 (14) | 7 | 2 (14) | 8 |
| HA4 | 1 (5) | 7 | 1 (6) | 6 |
| HA6 | 6 (6) | 6 | 6 (2) | 4 |
| HA46 | 1 (4) | 5 | 1 (3) | 7 |
| HA462′ | 2 (4) | 4 | 1 (2) | 8 |
| HA463′ | 6 (1) | 4 | 1 (5) | 6 |
| HA462′3′ | 1 (1) | 3 | 1 (2) | 5 |
| CS_de | 9 (1) | 2 | 7 (2) | 1 |
| CS4 | 2 (1) | 4 | 10 (1) | 2 |
| CS6 | 3 (3) | 4 | 2 (2) | 3 |
| CS46 | 5 (4) | 2 | 4 (2) | 2 |
| CS462′ | 8 (3) | 2 | 7 (2) | 1 |
| CS463′ | 2 (1) | 6 | 3 (3) | 6 |
| CS462′3′ | 3 (2) | 2 | 3 (3) | 2 |
| DSc | 2 (7) | 5 | 1 (6) | 8 |
| Heparinc | 5 (2) | 5 | 4 (1) | 4 |
aCluster rank with the number of cluster members, clustering is done for the RMSD value of 4 Å for GAGs heavy atoms.
bNumber of “correct” poses in the top 10 docking solutions by visual inspection.
cIn DS and heparin the iduronic acid was in 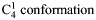 .
.
MD and energetic analysis of GAG–IL-8 complexes
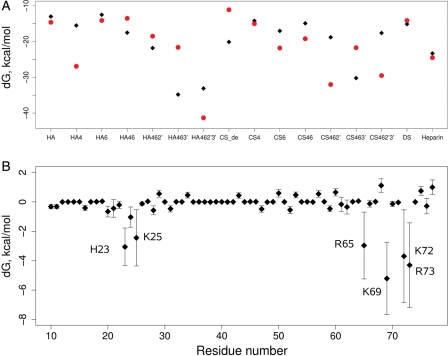
We used MD to simulate the GAG–IL-8 complexes obtained for the best scoring binding poses and to analyze their stability. We observed a clear improvement of binding with increasing GAG sulfation (Figure 5A) independently of the constraints applied to the IL-8 backbone. Calculated binding energies for other natural GAGs, DS and heparin, which have one and three sulfates per disaccharide, respectively, do not differ from binding energies of other GAGs with the same level of sulfation (Figure 5A). Despite this, the influence of individual sulfated positions is challenging to estimate. For example, a more favorable binding energy for HA4 in comparison to HA and HA6 cannot be unambiguously attributed to specificity or to inaccuracy of the method, which is especially sensitive to electrostatics and is dramatically affected by small changes of charged atoms positions. Moreover, statistical sampling, which is an issue for MD approaches in general, could strongly affect the molecular mechanics poisson-Boltzmann solvent accessible surface area (MM-PBSA) calculations as well. These considerations based on the obtained results lead to the conclusion that binding differences for similar GAGs observed with MM-PBSA calculations should be analyzed carefully and, in the best case, be complemented with additional experimental data in order to allow drawing conclusions on interactions specificity. For instance, our calculations show that both CS6 tetramer and hexamer bind stronger to IL-8 than the corresponding CS4 tetramer and hexamer, which is in agreement with our NMR experimental data (Figure 3). This is a good example on how the complementation of theoretical and experimental approaches represents a valuable approach for the analysis of the molecular recognition properties of GAG–protein systems.
Fig. 5.
(A) MM-PBSA binding free energies for monomeric IL-8 with different tetrameric GAGs in calculated binding pose. Black diamonds and red circles correspond to MD simulations carried out with harmonically constrained IL-8 backbone and free IL-8, respectively. (B) MM-GBSA binding free energies for monomeric IL-8 per residue obtained from 28 MD simulations with different tetrasaccharides of HA and CS derivatives.
To understand the impact of individual IL-8 residues to the binding of GAGs, we carried out generalized born poisson-Boltzmann solvent accessible surface area (MM-GBSA) energy decomposition, and we summed up the results for all complexes in the simulations with and without harmonic constraints on the IL-8 backbone to obtain reasonable statistical and conformational sampling. The data obtained from these simulations (Figure 5B) agree with the literature (Kuschert et al. 1998), demonstrating the crucial role of residues H23, K25, R65, K69, K72 and R73 for binding GAGs also in terms of energetics (six lowest points in the plot). These results implicitly confirm the reliability of the calculated binding pose. Observation of the unfavorable energy impact of some residues (i.e. negatively charged E75 close to the binding site) could serve as valuable information for protein engineering aimed to improve the binding mode of GAGs to IL-8.
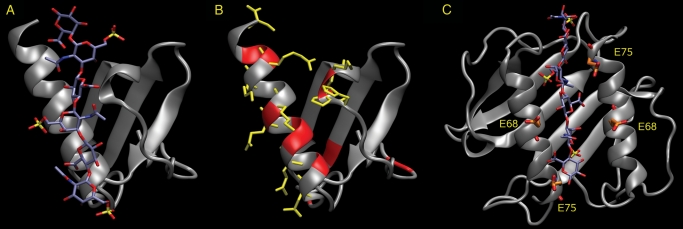
The data we obtain on energy decomposition per residue for hexasaccharide binding agree very well with our NMR data on CS6 binding (Figure 6A and B) and partially for CS4 binding, which corroborates the predicted common binding pose. Residues R65, K72 and R73 were not observed by NMR to be responding essentially to CS4 binding in comparison to CS6. MD simulations with no constraints on the protein showed significantly less favorable free energies of interaction for these residues with CS4. Thus, though increase in GAG sulfation in general improves binding, both experimental and computational methods also detect specific differences for binding GAGs with the same level of sulfation, which are not purely electrostatically driven.
Fig. 6.
(A) CS6 hexasaccharide (in sticks colored by atom type) in the highly scored binding pose with a monomeric IL-8 (in cartoon in gray). (B) The backbones of the 10 residues with the highest changes of backbone chemical shifts when binding a CS6 hexasaccharide (highlighted in red), and the side chains of the 10 residues with the highest absolute impact for binding according to MM-PBSA free energy decomposition (in sticks in yellow). (C) IL-8 dimer (in cartoon in gray) in complex with the hexasaccharide of CS6 in the “alternative” binding pose (in sticks colored by atom type). Glutamate residues located near the bound GAG are shown in sticks and labeled.
Comparison of bound GAG conformations with free GAGs
To explore the conformational space available to free GAGs in solution and to compare it with that in their bound state, we carried out MD simulations of hexameric GAGs in explicit solvent and plotted the sampled glycosidic linkage dihedrals. In general, both glycosidic linkages for HA and CS derivatives explore a larger area in conformational space with the increase in sulfation (Supplementary data, Figure S2). The reason for higher conformational flexibility for GAGs with higher sulfation could be explained by the increase in the number of possible local energy minima emerging after the introduction of additional groups due to the formation of stabilizing hydrogen bonds and salt-bridges. HA derivatives form ∼1.6 times more hydrogen bonds in comparison to CS derivatives because of the 4-OH group equatorial configuration participating more often in intramolecular interactions.
To explore the differences between the conformational space visited by glycosidic linkages in the simulated free GAGs and the experimentally available in the PDB, we analyzed the data for bound HA and CS4 (Supplementary data, Figure S3). Simulated GAG glycosidic linkage conformations overlap very well with PDB data except for two cases. The fact that one of these (for the type 1 of glycosidic linkage for HA, PDB ID: 1LXM) is not sampled in the free GAG conformational space could be explained by the presence of an unsaturated bond between C'4 and C'5 in the derivative of the glucuronic acid used to obtain this structure, which probably affects the geometry of the glycosidic linkage. The second (CS4_1, PDB ID: 3CE9), where ϕ ∼ 180°, can be reached by most free GAGs.
Comparison of the glycosidic linkages in the bound form of the predicted common binding pose (Supplementary data, Figure S2) also shows that most of the observed conformations for bound GAGs resemble free GAG conformations or converge to them in the simulation. Therefore, our results suggest that GAG glycosidic linkage conformations do not change significantly upon IL-8 binding compared with free GAGs. This has significant implications for the further development of theoretical and experimental approaches to study GAG–protein interactions, as it may assist in the process of discriminating accessible GAG glycosidic linkage conformations and, therefore, help simplifying the treatment of larger GAGs in computational studies and with the challenging interpretation of NMR data.
Elongation of GAGs bound to IL-8
In order to explore the effect of GAG elongation on binding to IL-8, we used 8-, 10- and 12-mers of HA463′ bound in the common binding pose predicted for tetrameric and hexameric GAGs to have high affinity. MM-PBSA calculations showed that HA463′ elongation did not lead to improvement of binding (Supplementary data, Figure S4). Moreover, energy decomposition per residue shows that there is a very high correlation between the energy values obtained for tetrameric and elongated GAGs: adjusted R2 values for all IL-8 residues are 0.94, 0.78, 0.91 and 0.85 for 6-, 8-, 10- and 12-mers of HA463′, respectively. These values are even higher than values for correlation between different GAGs in the same binding pose (e.g. adjusted R2 for HA and HA462′3′ is only 0.49), suggesting that tetrasaccharide is the minimal GAG unit required to achieve specific binding to IL-8. Therefore, conclusions made for tetrameric GAGs could be extrapolated to longer GAGs bound to IL-8. Nevertheless, these speculations analyzing the results obtained after GAG elongation should be taken with special care because of two aspects. First, GAG structure minimization after each step of elongation can lead the system to be trapped in a local minimum. Second, even if elongation of a bound GAG does not significantly affect the enthalpic component calculated by MM-PBSA, there is still an entropic component of binding, which is expected to vary significantly for GAGs of different length and that is not explicitly taken into account for MM-PBSA calculations.
Docking GAGs to dimeric IL-8
We performed docking calculations with dimeric IL-8 in order to explore if the binding pose found for different GAGs on monomeric IL-8 is also a plausible binding pose for GAGs on the IL-8 dimer. Indeed, the same binding pose turned out to be highly representative among the top scoring poses together with its antiparallel pose, as it was also observed for monomeric IL-8. In addition, we found another highly scoring and representative docking pose specific for dimeric IL-8 binding in which the GAG is in extended conformation and located in a groove between the two C-terminal α-helices (Figure 6C). Since the C-terminal α-helices of each IL-8 subunit in the dimer are antiparallel, both parallel and antiparallel docking poses of GAGs are symmetric, represented equally and can be considered as the same pose. Our MD simulations showed that this pose is energetically unfavorable (ΔG ≥ 0 kcal/mol), which could be explained in terms of the proximity of E68 and E75 residues to the bound GAG. On the contrary, in the case of monomeric IL-8, the side chains of these negatively charged residues are oriented away from the bound GAGs, which avoids the unfavorable interactions remaining very close to the GAG-binding site in the dimer. Free energy decomposition shows a very unfavorable impact on binding of these four glutamate residues (Supplementary data, Figure S5). In contrast to previous studies (Bitomsky and Wade 1999; Lortat-Jacob et al. 2002), we did not find the perpendicular binding pose to dimeric IL-8 to be well clustered and highly scored but rather a pose agreeing with the horseshoe-like model (Spillmann et al. 1998), where two GAGs in the calculated binding pose could be connected with a spacer, which does not interact with IL-8.
Dimerization of IL-8 vs binding to GAGs
According to the available data from literature, GAG binding affects the process of dimerization for chemokines (Jansma et al. 2010), in particular for IL-8, which in turn leads to changes in their receptor binding (Das et al. 2010). We performed MD simulations with the dimeric IL-8, where one or both IL-8 subunits were complexed to HA463′ of the length of 4, 6, 8, 10 and 12 monosaccharide units in the predicted common binding pose. Our results show that binding of one GAG of this length does not significantly influence dimerization, whereas simultaneous binding of two GAGs to IL-8 monomers within the dimer weakens monomers association (Supplementary data, Figure S6). This effect is due to a direct electrostatic repulsion between the two highly charged GAGs and to the reorientation upon binding of the charged residues at the C-terminal α-helix (R65, E59, K60, K63, R64 and E66) and is more pronounced for the longer GAGs. As a result, either one of the GAGs could dissociate from the IL-8 dimer or the IL-8 dimer could dissociate into two monomers, each binding a GAG. Therefore, GAG binding could regulate association equilibrium for IL-8 monomers, and GAG's sulfation degree could be used to attune this regulation. However, this model of IL-8 dimerization does not consider possible horseshoe-like binding of a longer GAG, which could assist the dimerization, whereas each IL-8 monomeric unit binds a GAG in the pose proposed in this study.
Conclusions
In our work, we used fluorescence and NMR spectroscopy, docking and MD to study the molecular recognition properties of IL-8 with a variety of GAGs including HA and CS, their sulfated derivatives and DS. The combination of these complementary experimental and theoretical approaches allowed us to discern a distinct and common binding pose to IL-8 for all analyzed GAGs, even though being of different length and containing different sulfation patterns. This observed binding pose is in agreement with previously proposed binding modes for heparin on IL-8. Calculated free energy decomposition per residue and acquired backbone chemical shifts agree on the impact of individual IL-8 residues for GAG binding. Our results indicate that the sulfation pattern determines the strength of binding. Thus, increase in GAG sulfation in general improves binding as detected by both experimental and computational methods. In addition, we also observe binding specificity linked to the sulfate substitution positions in cases of equal sulfation degree. Our analysis on the elongation of the analyzed GAGs reveals the tetrasaccharide as the minimal GAG unit required to achieve specific binding to IL-8, which also agrees with experimental observations. We also studied the energetic influence of GAG binding in the process of IL-8 monomer association. We observed the same binding pose described for monomeric IL-8 highly scored and influencing energetically IL-8 dimer formation. Interestingly, we found that the GAG glycosidic linkage conformations do not change significantly upon IL-8 binding compared with free GAGs, which has significant implications for simplifying the treatment of longer GAGs in computational analysis and for the further development of theoretical and experimental approaches to study GAG–protein interactions. Our study demonstrates that the combination of experimental fluorescence and NMR spectroscopy with computational modeling and simulation can become very powerful for deriving structural and molecular recognition models for complicated systems such as GAGs and their complexes with proteins. In particular, these approaches may assist in constraining certain degrees of freedom associated with glycosidic linkages. This would translate into a decrease in high computational demands and avoid misleading interpretation of NMR data, which ultimately may allow handling of longer GAGs.
Materials and methods
Experimental studies
Materials
The pTXB1 vector encoding hIL-8 (1–77) was prepared as described in the literature (David et al. 2003). DS hexasaccharide was purchased from Iduron Ltd (Paterson Institute, Manchester, UK). HA hexasaccharide was purchased from Hyalose, L.L.C (Oklahoma) and used as supplied. High molecular weight CS4 from bovine trachea and CS6 from shark cartilage were purchased from Sigma-Aldrich (Taufkirchen, Germany), 15N-labeled ammonium salts from Eurisotop (Saarbrücken, Germany) and all other chemicals from Sigma-Aldrich.
GAG preparation
CS4 and CS6 polysaccharides were converted into a mixture of oligosaccharides by digestion with hyaluronate lyase from bovine testes (Schiller et al. 1999). Under these conditions, the GAG hexasaccharides are obtained in the highest yield. Hexasaccharides were separated from tetra- and larger oligomers by preparative high-performance thin-layer chromatography (HPTLC; Nimptsch et al. 2010). Afterwards, the lane corresponding to the respective hexasaccharide was re-eluted from the silica gel by three washing steps with distilled water. The aqueous solutions were combined and dried in vacuum; the dried powder was re-dissolved in buffer (20 mM PO43−, 50 mM NaCl, pH 7.0), yielding a concentration of 10 mg/mL. Purity of the preparation was checked by analytical HPTLC and negative ion matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) (Nimptsch et al. 2010). The concentration was verified by the established carbazole method (Bitter and Muir 1962) and glucuronic acid as reference.
According to the compositional data provided by the supplier, high-molecular CS from bovine trachea is characterized by a content of the 4-sulfate isomer higher than 65%, whereas CS from shark cartilage exhibits a 6-sulfate content of ∼90% (Torchia et al. 1977). As the isomeric composition may vary between individual preparations, the contribution of both isomers was checked by means of 1H NMR analysis subsequent to complete digestion with chondroitinase ABC. Evaluation of the resolved N-acetyl resonances of the 4- and 6-sulfate isomers of the unsaturated disaccharide resulted in 85% CS4 (bovine trachea) and ∼90% CS6 (shark cartilage). Although we did not obtain an isomerically pure product, it is obvious that significant differences in the sulfation sites can be obtained in dependence on the source of the used CS.
The CS hexasaccharide of interest was (subsequent to TLC purification) characterized by combined positive and negative ion MALDI-TOF MS. Comparisons with related electrospray (ESI) MS spectra showed that there was some loss of the sulfate residues under MALDI conditions but the mass of the intact molecules was also in all cases clearly detectable. We prefer the MALDI analysis due to the simplicity of performance (Nimptsch et al. 2009).
Protein expression
Escherichia coli strain ER2566 was transformed with a pTXB1 vector (New England BioLabs, Ipswich) including the gene for human IL-8 encoding the 77-amino acid form that was C-terminally linked to an Mxe intein/chitin-binding domain (David et al. 2003, 2004). A high-density feed-batch fermentation was carried out, cells were cultivated in 2 L minimal medium containing 15NH4Cl (2 g/L), (15NH4)2SO4 (9.84 g/L) and glucose (20 g/L) at 37°C. Protein expression was induced with 1 mM isopropyl-beta-D-thiogalactopyranoside (IPTG) at an OD600 of 10. Cells were harvested 4 h after induction by centrifugation. The cell pellet was resuspended in buffer A [20 mM Tris, 30 mM ethylenediaminetetraacetic acid (EDTA), 500 mM NaCl, pH 7.8] in the presence of lysozyme (15 µg/mL) over 1 h on ice and broken by three passages through a French press at 800 bar. Afterwards, the lysate was treated with 10 µg/mL of DNase I and 3 mM MgCl2 for 1 h at room temperature. After centrifugation at 38,000 × g for 50 min, IL-8 could be detected mainly in the supernatant. Expression and isolation of the target protein was monitored using sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS–PAGE) and the Bradford assay.
Protein purification and refolding
To isolate IL-8 from the protein extract, two purification steps were applied. In the first step, the IMPACT™ system was used. A column filled with chitin beads (New England BioLabs) was equilibrated in buffer B (20 mM Tris, 1 mM EDTA, 500 mM NaCl, pH 7.8) containing 1 M urea. Afterwards, the protein was loaded onto the column. After washing the column with 15 bed volumes of buffer B containing 1 M urea, the intein cleavage was induced by flushing with 3 bed volumes of dithiothreitol (DTT) (0.1 M) in buffer B. The cleavage proceeded at 4°C for 60 h on the column. IL-8 (1–77) was then eluted with buffer B and 3 mL fractions were collected, which were checked by SDS–PAGE. In the second step, DTT and other remaining cell proteins were removed by carrying out ion exchange chromatography, using a HiTrap SP FF Sepharose 1 mL column (GE Healthcare, Uppsala) with a flow of 1 mL/min. The elution was performed by a linear gradient of 8.3–100% 1.5 M NaCl in 20 mM Tris buffer containing 1 mM EDTA, pH 7.0. Fractions containing pure IL-8 were identified by SDS-PAGE. Finally, the protein was dialyzed twice against buffer B [containing 2.5 mM reduced gluthathione (GSH) and 0.5 mM oxidized gluthathione (GSSG), pH 9.3] and buffer C (20 mM PO43−, 50 mM NaCl, 1 mM EDTA, 1 mM arginine, pH 7.0). The folded protein was lyophilized or stored in aliquots at −80°C. Protein was characterized by SDS–PAGE and mass spectrometry. The concentration was determined spectrophotometrically using an extinction coefficient of 7240 M−1 cm−1 and by Bradford assay.
Fluorescence spectroscopy
Fluorescence binding measure ments were determined as described in the literature (Theisgen et al. 2011). Tryptophan fluorescence spectra were acquired on a Fluoromax 2 spectrometer (Jobin Yvon, Edison, NJ) using 290 nm as excitation wavelength. Emission was observed between 300 and 470 nm. Aliquots of the GAG hexamers in solution were titrated to the protein solution. Fluorescence intensities at the emission maximum of GAG bound protein were used for the calculation of GAG-bound fraction of IL-8 according to 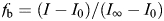 (Ladokhin et al. 2000), where fb denotes the GAG-bound fraction, I the fluorescence intensity, I0 the fluorescence intensity in the absence of GAG and I∞ the fluorescence intensity that corresponds to complete binding. This value for I∞ has to be determined from the fit to the recorded data. From the binding curve, the dissociation constant can be determined directly
(Ladokhin et al. 2000), where fb denotes the GAG-bound fraction, I the fluorescence intensity, I0 the fluorescence intensity in the absence of GAG and I∞ the fluorescence intensity that corresponds to complete binding. This value for I∞ has to be determined from the fit to the recorded data. From the binding curve, the dissociation constant can be determined directly 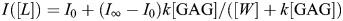 (Ladokhin et al. 2000), where [GAG] is the GAG concentration, k the ratio of GAG-associated protein and free protein and [W] the concentration of water (55.5 M). The respective KD value can be determined from the mole-fraction partition coefficient (KD = [W]/k).
(Ladokhin et al. 2000), where [GAG] is the GAG concentration, k the ratio of GAG-associated protein and free protein and [W] the concentration of water (55.5 M). The respective KD value can be determined from the mole-fraction partition coefficient (KD = [W]/k).
NMR spectroscopy
NMR experiments were recorded on a Bruker DRX600 spectrometer (Bruker, Rheinstetten, Germany) using a 5 mm triple-inverse probe at 30°C. NMR samples contained 0.6–1 mM IL-8 in 90% H2O/10% D2O containing 20 mM PO43− buffer at pH 7.0. 1H NMR spectra were acquired using a WATERGATE W5 pulse sequence (Liu et al. 1998) for water suppression and 15N globally optimized alternating phase rectangular pulse (GARP) decoupling. NOESY (Jeener et al. 1979; mixing time 120 ms) and TOCSY (Bax and Davis 1985; mixing time 80 ms) spectra were collected for resonance assignment. For the 1H-15N HSQC experiment (Mori et al. 1995), 16 scans were recorded for each increment with spectral widths of 9.6 kHz for 1H and 1.6 kHz for 15N as well as 128 data points in the 15N dimension.
Titration of IL-8 with hexasaccharides were performed by adding 5 µL aliquots of concentrated hexasaccharide solutions. After each titration step, the sample was mixed and a HSQC spectrum was recorded. The pH value was checked after finishing the titration studies, only negligible pH shifts occurred. For each titration step, the significant chemical shift changes in the HSQC representing the peptide backbone for each residue were calculated according to 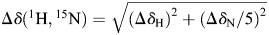 (Seo et al. 2010), where ΔδH and ΔδN represent the changes of the chemical shifts in the 1H and 15N dimension, respectively.
(Seo et al. 2010), where ΔδH and ΔδN represent the changes of the chemical shifts in the 1H and 15N dimension, respectively.
Computational studies
IL-8 and GAG structures
The X-ray structure of monomeric IL-8 (PDB ID 3IL8, 2.00 Å) was used for docking and MD simulations. For consistency between our results for monomeric and dimeric IL-8, we used this structure for building the dimeric IL-8 by superimposing it to each of the IL-8 subunits in the NMR IL-8 dimer structure (PDB ID 1IL8). The RMSD between the X-ray and the NMR structures is 1.57 Å for backbone atoms. The GAG structures were modeled based on the structures available in the PDB for octasaccharides of HA (PDB ID: 2BVK, NMR), hexameric CS4 (PDB ID: 1CS4, fiber diffraction) tetrasaccharides of DS (PDB ID: 1HM2, X-ray 2.00 Å), dodecasaccharide of heparin with iduronic acid in C14 ring conformation (PDB ID: 1HPN, NMR. Their geometries were optimized with the AMBER99 force field implemented in MOE (Chemical Computing Group, Inc., Montreal, CA). Fourteen HA and CS derivatives were used in our docking and MD calculations (Table II).
Table II.
GAG nomenclature and abbreviations for hyaluronan and CS derivatives
| Hyaluronan derivatives |
CS derivatives |
||
|---|---|---|---|
| HA | (-GlcUAβ1-3GlcNAcβ1-)n | CSde | (-GlcUAβ1-3GalNAcβ1-)n |
| HA4 | (-GlcUAβ1-3GlcNAc(4S)β1-)n | CS4 | (-GlcUAβ1-3GalNAc(4S)β1-)n |
| HA6 | (-GlcUAβ1-3GlcNAc(6S)β1-)n | CS6 | (-GlcUAβ1-3GalNAc(6S)β1-)n |
| HA46 | (-GlcUAβ1-3GlcNAc(4S,6S)β1-)n | CS46 | (-GlcUAβ1-3GalNAc(4S,6S)β1-)n |
| HA462′ | (-GlcUA(2′S)β1-3GlcNAc(4S,6S)β1-)n | CS462′ | (-GlcUA(2′S)β1-3GalNAc(4S,6S)β1-)n |
| HA463′ | (-GlcUA(3′S)β1-3GlcNAc(4S,6S)β1-)n | CS463′ | (-GlcUA(3′S)β1-3GalNAc(4S,6S)β1-)n |
| HA462′3′ | (-GlcUA(2′S,3′S)β1-3GlcNAc(4S,6S)β1-)n | CS462′3′ | (-GlcUA(2′S,3′S)β1-3GalNAc(4S,6S)β1-)n |
Water molecules localization
Crystallographic water molecules were taken into account for docking calculations. In addition, energetically favorable positions for additional water molecules in the IL-8 heparin-binding site (Kuschert et al. 1998) were calculated using a water probe with GRID (Goodford 1985). Grid points with negative energy values were chosen for placing water oxygens. In cases of two water oxygens closer than 2.8 Å, the water molecule with the weakest interaction energy was discarded. A total of 24 crystallographic and GRID-calculated water molecules were added.
Docking
Docking was performed with and without explicit water molecules in the binding site. Autodock3 (Morris et al. 1998) was used (150 runs) with the genetic algorithm and default parameters. A grid box, large enough to avoid biases of the docking results to a certain ligand orientation, was centered on the previously described heparin-binding site (H23, K25, R65, K69, K72 and R73; Kuschert et al. 1998). All torsion angles of the tetrameric ligands were left flexible. Hexasaccharides were optimized with the AMBER99 force field, and then rigidly docked. Obtained binding poses were clustered by RMSD (4 Å for heavy atoms). Docking poses for different ligands were considered similar by visual inspection when their orientations and placements were the same relative to the IL-8 contacting residues.
Molecular dynamics
MD simulations in a periodic box of TIP3P waters were performed as described previously (Samsonov et al. 2008). The AMBER 10.0 package (Case et al. 2008) was used with ff03 parameters (Duan et al. 2003) for protein and the GLYCAM06 for GAGs. Sulfate charges for HA and CS derivatives libraries were obtained from literature (Huige and Altona 1995). The scaling parameters SCEE and SCNB were set to 1 as required for the use of the GLYCAM06 force field within AMBER (Kirschner et al. 2008). Harmonic constraints (10 kcal/mol Å) for IL-8 backbone atoms were applied to retain the secondary structure, as it has been observed that the C-terminal α-helix, when unrestrained, experiences severe distortions (final backbone RMSD ∼ 4 Å). It is not clear if these distortions occur because of the force field instability for this small fold [similar challenge has been previously discussed (Kadirvelraj et al. 2008)]. To simulate longer GAGs, we elongated the docked structures to 8-, 10- and 12-mers by adding a monosaccharide residue stepwise to the GAGs-termini and minimized them using the AMBER99 force field in MOE.
Free GAG MD simulations
For free hexameric GAGs, simulations of the aforementioned derivatives of HA and CS were used, since hexasaccharide is the shortest GAG that exhibits all properties of a polymer (Sattelle et al. 2010). Simulations of 20 ns were carried out using the same protocol as described in the previous section. GAG glycosidic linkage conformations were analyzed in terms of the dihedral angles ϕ and ψ. Glycosidic linkage 1 (GlcUA–Glc/GalNAc) and glycosidic linkage 2 (Glc/GalNAc–GlcUA) are defined as the angles between the planes (H1-C1-O3-C3) and (C1-O3-C3-H3), and (H1-C1-O4-C4) and (C1-O4-C4-H4), respectively. The obtained results for all glycosidic linkages of the same type within the same molecule were taken together to create the Ramachandran-like plots. The structures of bound GAGs from the PDB were compared with the data on glycosidic linkages in MD simulations of free GAGs. To compare conformations of the modeled GAGs bound to IL-8 and free GAGs, MD frames were extracted each 200 ps and plotted together with the Ramachandran-like plot obtained for free GAGs.
MM-PBSA free energy calculations
Energetic post-processing of the trajectories and per residue energy decomposition were done in a continuous solvent model using MM-PBSA and MM-GBSA of AMBER 10. The statistically representative snapshots were chosen as described by Lafont et al. (2007).
Data analysis and its graphical representation were carried out with the R-package (R Development Core Team 2006).
Supplementary data
Supplementary data for this article is available online at http://glycob.oxfordjournals.org.
Funding
This work was supported by the German Research Council (SFB-TRR67; A2, A4, A6, A7).
Conflict of interest
None declared.
Abbreviations
CS, chondroitin sulfate; CS4, chondroitin-4-sulfate; CS6, chondroitin-6-sulfate; DS, dermatan sulfate; DTT, dithiothreitol; ECM, extracellular matrix; EDTA, ethylenediaminetetraacetic acid; GAG, glycosaminoglycan; GalNAc, β-d-N-acetylgalactosamine; GARP, globally optimized alternating phase rectangular pulse; GlcUA, β-d-glucuronic acid; GSH, reduced gluthathione; GSSG, oxidized gluthathione; HA, hyaluronan; HPTLC, high-performance thin-layer chromatography; HSQC, heteronuclear single-quantum coherence; IL-8, Interleukin-8; IPTG, isopropyl-beta-D-thiogalactopyranoside; MALDI-TOF, matrix-assisted laser desorption/ionization time-of-flight; MD, molecular dynamics; MM-GBSA, generalized born poisson-Boltzmann solvent accessible surface area; MM-PBSA, molecular mechanics poisson-Boltzmann solvent accessible surface area; MS, mass spectrometry; NOESY, nuclear overhauser effect spectroscopy; RMSD, Root-Mean-Square Deviation; SDS—PAGE, sodium dodecyl sulfate polyacrylamide gel electrophoresis; TOCSY, total correlation spectroscopy.
Supplementary Material
Acknowledgements
We are grateful to Ralf Gey for his invaluable technical support and to Clemens Horn for help with the assignment of the NOESY and TOCSY spectra of IL-8. We would like to thank the ZIH at TU Dresden for providing high-performance computational resources and assistance.
References
- Bax A, Davis DG. Mlev-17-Based two-dimensional homonuclear magnetization transfer spectroscopy. J Magn Reson. 1985;65:355–360. [Google Scholar]
- Bitomsky W, Wade RC. Docking of glycosaminoglycans to heparin-binding proteins: Validation for aFGF, bFGF, and antithrombin and application to IL-8. J Am Chem Soc. 1999;121:3004–3013. doi:10.1021/ja983319g. [Google Scholar]
- Bitter T, Muir HM. A modified uronic acid carbazole reaction. Anal Biochem. 1962;4:330–334. doi: 10.1016/0003-2697(62)90095-7. doi:10.1016/0003-2697(62)90095-7. [DOI] [PubMed] [Google Scholar]
- Cardin AD, Weintraub HJR. Molecular modeling of protein-glycosaminoglycan interactions. Arteriosclerosis. 1989;9:21–32. doi: 10.1161/01.atv.9.1.21. doi:10.1161/01.ATV.9.1.21. [DOI] [PubMed] [Google Scholar]
- Case DA, Darden TA, Cheatham T, Simmerling CL, Wang J, Duke RE, Luo R, Crowley M, Walker RC, Zhang W, et al. AMBER 10. San Francisco: University of California; 2008. [Google Scholar]
- Chung HJ, Park TG. Surface engineered and drug releasing pre-fabricated scaffolds for tissue engineering. Adv Drug Deliv Rev. 2007;59:249–262. doi: 10.1016/j.addr.2007.03.015. doi:10.1016/j.addr.2007.03.015. [DOI] [PubMed] [Google Scholar]
- Clore GM, Appella E, Yamada M, Matsushima K, Gronenborn AM. Determination of the secondary structure of interleukin-8 by nuclear magnetic resonance spectroscopy. J Biol Chem. 1989;264:18907–18911. [PubMed] [Google Scholar]
- Clore GM, Appella E, Yamada M, Matsushima K, Gronenborn AM. Three-dimensional structure of interleukin 8 in solution. Biochemistry. 1990;29:1689–1696. doi: 10.1021/bi00459a004. doi:10.1021/bi00459a004. [DOI] [PubMed] [Google Scholar]
- Das ST, Rajagopalan L, Guerrero-Plata A, Sai J, Richmond A, Garofalo RP, Rajarathnam K. Monomeric and dimeric CXCL8 are both essential for in vivo neutrophil recruitment. PLoS One. 2010;5:e11754. doi: 10.1371/journal.pone.0011754. doi:10.1371/journal.pone.0011754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David R, Gunther R, Baumann L, Luhmann T, Seebach D, Hofmann HJ, Beck-Sickinger AG. Artificial chemokines: Combining chemistry and molecular biology for the elucidation of interleukin-8 functionality. J Am Chem Soc. 2008;130:15311–15317. doi: 10.1021/ja802453x. doi:10.1021/ja802453x. [DOI] [PubMed] [Google Scholar]
- David R, Machova Z, Beck-Sickinger AG. Semisynthesis and application of carboxyfluorescein-labelled biologically active human interleukin-8. Biol Chem. 2003;384:1619–1630. doi: 10.1515/BC.2003.180. doi:10.1515/BC.2003.180. [DOI] [PubMed] [Google Scholar]
- David R, Richter MP, Beck-Sickinger AG. Expressed protein ligation. Method and applications. Eur J Biochem. 2004;271:663–677. doi: 10.1111/j.1432-1033.2004.03978.x. doi:10.1111/j.1432-1033.2004.03978.x. [DOI] [PubMed] [Google Scholar]
- Duan Y, Wu C, Chowdhury S, Lee MC, Xiong G, Zhang W, Yang R, Cieplak P, Luo R, Lee T, et al. A point-charge force field for molecular mechanics simulations of proteins based on condensed-phase quantum mechanical calculations. J Comput Chem. 2003;24:1999–2012. doi: 10.1002/jcc.10349. doi:10.1002/jcc.10349. [DOI] [PubMed] [Google Scholar]
- Dvir T, Timko BP, Kohane DS, Langer R. Nanotechnological strategies for engineering complex tissues. Nat Nanotechnol. 2011;6:13–22. doi: 10.1038/nnano.2010.246. doi:10.1038/nnano.2010.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forster M, Mulloy B. Computational approaches to the identification of heparin-binding sites on the surfaces of proteins. Biochem Soc Trans. 2006;34:431–434. doi: 10.1042/BST0340431. doi:10.1042/BST0340431. [DOI] [PubMed] [Google Scholar]
- Gandhi NS, Mancera RL. The structure of glycosaminoglycans and their interaction with proteins. Chem Biol Drug Des. 2008;72:455–482. doi: 10.1111/j.1747-0285.2008.00741.x. doi:10.1111/j.1747-0285.2008.00741.x. [DOI] [PubMed] [Google Scholar]
- Goodford PJ. A computational procedure for determining energetically favorable binding sites on biologically important macromolecules. J Med Chem. 1985;28:849–857. doi: 10.1021/jm00145a002. doi:10.1021/jm00145a002. [DOI] [PubMed] [Google Scholar]
- Huige CJM, Altona C. Force-field parameters for sulfates and sulfamates based on ab initio calculations: extensions of AMBER and CHARMm fields. J Comput Chem. 1995;16:56–79. doi:10.1002/jcc.540160106. [Google Scholar]
- Imberty A, Lortat-Jacob H, Perez S. Structural view of glycosaminoglycan–protein interactions. Carbohydr Res. 2007;342:430–439. doi: 10.1016/j.carres.2006.12.019. doi:10.1016/j.carres.2006.12.019. [DOI] [PubMed] [Google Scholar]
- Jansma AL, Kirkpatrick JP, Hsu AR, Handel TM, Nietlispach D. NMR analysis of the structure, dynamics, and unique oligomerization properties of the chemokine CCL27. J Biol Chem. 2010;285:14424–14437. doi: 10.1074/jbc.M109.091108. doi:10.1074/jbc.M109.091108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeener J, Meier BH, Bachmann P, Ernst RR. Investigation of exchange processes by two-dimensional NMR spectroscopy. J Chem Phys. 1979;71:4546–4553. doi:10.1063/1.438208. [Google Scholar]
- Kadirvelraj R, Foley BL, Dyekjaer JD, Woods RJ. Involvement of water in carbohydrate–protein binding: Concanavalin A revisited. J Am Chem Soc. 2008;130:16933–16942. doi: 10.1021/ja8039663. doi:10.1021/ja8039663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirschner KN, Yongye AB, Tschampel SM, Gonzalez-Outeirino J, Daniels CR, Foley BL, Woods RJ. GLYCAM06: A generalizable biomolecular force field. Carbohydrates. J Comput Chem. 2008;29:622–655. doi: 10.1002/jcc.20820. doi:10.1002/jcc.20820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieger E, Geretti E, Brandner B, Goger B, Wells TN, Kungl AJ. A structural and dynamic model for the interaction of interleukin-8 and glycosaminoglycans: Support from isothermal fluorescence titrations. Proteins. 2004;54:768–775. doi: 10.1002/prot.10590. doi:10.1002/prot.10590. [DOI] [PubMed] [Google Scholar]
- Kuschert GS, Hoogewerf AJ, Proudfoot AE, Chung CW, Cooke RM, Hubbard RE, Wells TN, Sanderson PN. Identification of a glycosaminoglycan binding surface on human interleukin-8. Biochemistry. 1998;37:11193–11201. doi: 10.1021/bi972867o. doi:10.1021/bi972867o. [DOI] [PubMed] [Google Scholar]
- Ladokhin AS, Jayasinghe S, White SH. How to measure and analyze tryptophan fluorescence in membranes properly, and why bother? Anal Biochem. 2000;285:235–245. doi: 10.1006/abio.2000.4773. doi:10.1006/abio.2000.4773. [DOI] [PubMed] [Google Scholar]
- Lafont V, Schaefer M, Stote RH, Altschuh D, Dejaegere A. Protein–protein recognition and interaction hot spots in an antigen–antibody complex: Free energy decomposition identifies “efficient amino acids”. Proteins. 2007;67:418–434. doi: 10.1002/prot.21259. doi:10.1002/prot.21259. [DOI] [PubMed] [Google Scholar]
- Larsen CG, Anderson AO, Appella E, Oppenheim JJ, Matsushima K. The neutrophil-activating protein (NAP-1) is also chemotactic for T lymphocytes. Science. 1989;243:1464–1466. doi: 10.1126/science.2648569. doi:10.1126/science.2648569. [DOI] [PubMed] [Google Scholar]
- Lee SH, Shin H. Matrices and scaffolds for delivery of bioactive molecules in bone and cartilage tissue engineering. Adv Drug Deliv Rev. 2007;59:339–359. doi: 10.1016/j.addr.2007.03.016. doi:10.1016/j.addr.2007.03.016. [DOI] [PubMed] [Google Scholar]
- Liu ML, Mao XA, Ye CH, Huang H, Nicholson JK, Lindon JC. Improved WATERGATE pulse sequences for solvent suppression in NMR spectroscopy. J Magn Reson. 1998;132:125–129. doi:10.1006/jmre.1998.1405. [Google Scholar]
- Lortat-Jacob H, Grosdidier A, Imberty A. Structural diversity of heparan sulfate binding domains in chemokines. Proc Natl Acad Sci USA. 2002;99:1229–1234. doi: 10.1073/pnas.032497699. doi:10.1073/pnas.032497699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middleton J, Neil S, Wintle J, Clark-Lewis I, Moore H, Lam C, Auer M, Hub E, Rot A. Transcytosis and surface presentation of IL-8 by venular endothelial cells. Cell. 1997;91:385–395. doi: 10.1016/s0092-8674(00)80422-5. doi:10.1016/S0092-8674(00)80422-5. [DOI] [PubMed] [Google Scholar]
- Mori S, Abeygunawardana C, Johnson MO, Vanzijl PCM. Improved sensitivity of HSQC spectra of exchanging protons at short Interscan delays using a new fast HSQC (FHSQC) detection scheme that avoids water saturation. J Magn Reson B. 1995;108:94–98. doi: 10.1006/jmrb.1995.1109. doi:10.1006/jmrb.1995.1109. [DOI] [PubMed] [Google Scholar]
- Morris GM, Goodsell DS, Halliday RS, Huey R, Hart WE, Belew RK, Olson AJ. Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J Comput Chem. 1998;19:1639–1662. doi:10.1002/(SICI)1096-987X(19981115)19:14<1639::AID-JCC10>3.0.CO;2-B. [Google Scholar]
- Nimptsch A, Schibur S, Schnabelrauch M, Fuchs B, Huster D, Schiller J. Characterization of the quantitative relationship between signal-to-noise (S/N) ratio and sample amount on-target by MALDI-TOF MS: Determination of chondroitin sulfate subsequent to enzymatic digestion. Anal Chim Acta. 2009;635:175–182. doi: 10.1016/j.aca.2009.01.025. doi:10.1016/j.aca.2009.01.025. [DOI] [PubMed] [Google Scholar]
- Nimptsch K, Suss R, Riemer T, Nimptsch A, Schnabelrauch M, Schiller J. Differently complex oligosaccharides can be easily identified by matrix-assisted laser desorption and ionization time-of-flight mass spectrometry directly from a standard thin-layer chromatography plate. J Chromatogr A. 2010;1217:3711–3715. doi: 10.1016/j.chroma.2010.04.005. doi:10.1016/j.chroma.2010.04.005. [DOI] [PubMed] [Google Scholar]
- R Development Core Team. R. a language and environment for statistical computing. 2006. http://www.r-project.org .
- Samsonov S, Teyra J, Pisabarro MT. A molecular dynamics approach to study the importance of solvent in protein interactions. Proteins. 2008;73:515–525. doi: 10.1002/prot.22076. doi:10.1002/prot.22076. [DOI] [PubMed] [Google Scholar]
- Sapay N, Cabannes E, Petitou M, Imberty A. Molecular modeling of the interaction between heparan sulfate and cellular growth factors: Bringing pieces together. Glycobiology. 2011;21:1181–1193. doi: 10.1093/glycob/cwr052. [DOI] [PubMed] [Google Scholar]
- Sattelle BM, Shakeri J, Roberts IS, Almond A. A 3D-structural model of unsulfated chondroitin from high-field NMR: 4-sulfation has little effect on backbone conformation. Carbohydr Res. 2010;345:291–302. doi: 10.1016/j.carres.2009.11.013. doi:10.1016/j.carres.2009.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheidt HA, Schibur S, Magalhaes A, de Azevedo ER, Bonagamba TJ, Pascui O, Schulz R, Reichert D, Huster D. The mobility of chondroitin sulfate in articular and artificial cartilage characterized by (13)C magic-angle spinning NMR spectroscopy. Biopolymers. 2010;93:520–532. doi: 10.1002/bip.21386. [DOI] [PubMed] [Google Scholar]
- Schiller J, Arnhold J, Benard S, Reichl S, Arnold K. Cartilage degradation by hyaluronate lyase and chondroitin ABC lyase: A MALDI-TOF mass spectrometric study. Carbohydr Res. 1999;318:116–122. doi: 10.1016/s0008-6215(99)00063-4. doi:10.1016/S0008-6215(99)00063-4. [DOI] [PubMed] [Google Scholar]
- Schiller J, Naji L, Huster D, Kaufmann J, Arnold K. 1H and 13C HR-MAS NMR investigations on native and enzymatically-digested bovine cartilage. MAGMA. 2001;13:19–27. doi: 10.1007/BF02668647. doi:10.1007/BF02668647. [DOI] [PubMed] [Google Scholar]
- Seo ES, Blaum BS, Vargues T, De CM, Deakin JA, Lyon M, Barran PE, Campopiano DJ, Uhrin D. Interaction of Human beta-Defensin 2 (HBD2) with glycosaminoglycans. Biochemistry. 2010;49:10486–10495. doi: 10.1021/bi1011749. doi:10.1021/bi1011749. [DOI] [PubMed] [Google Scholar]
- Spillmann D, Witt D, Lindahl U. Defining the interleukin-8-binding domain of heparan sulfate. J Biol Chem. 1998;273:15487–15493. doi: 10.1074/jbc.273.25.15487. doi:10.1074/jbc.273.25.15487. [DOI] [PubMed] [Google Scholar]
- Theisgen S, Thomas L, Schroder T, Lange C, Kovermann M, Balbach J, Huster D. The presence of membranes or micelles induces structural changes of the myristoylated guanylate-cyclase activating protein-2. Eur Biophys J. 2011;40:565–576. doi: 10.1007/s00249-011-0680-9. doi:10.1007/s00249-011-0680-9. [DOI] [PubMed] [Google Scholar]
- Thiery JP, Boyer B. The junction between cytokines and cell adhesion. Curr Opin Cell Biol. 1992;4:782–792. doi: 10.1016/0955-0674(92)90101-h. doi:10.1016/0955-0674(92)90101-H. [DOI] [PubMed] [Google Scholar]
- Torchia DA, Hasson MA, Hascall VC. Investigation of molecular motion of proteoglycans in cartilage by 13C magnetic resonance. J Biol Chem. 1977;252:3617–3625. [PubMed] [Google Scholar]
- Zhao B, Liwang PJ. Characterization of the interactions of vMIP-II, and a dimeric variant of vMIP-II, with glycosaminoglycans. Biochemistry. 2010;49:7012–7022. doi: 10.1021/bi100549y. doi:10.1021/bi100549y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.