Abstract
The molecular mechanisms conferring resistance to docetaxel in prostate cancer patients remain partially understood. We generated docetaxel resistant derivatives of the androgen independent prostate cancer cell lines PC-3 and DU-145. Docetaxel rapidly induces DU-145 cell death via apoptosis and the drug resistant cells were produced by periodically exposing proliferating DU-145 cultures to small doses of docetaxel. In PC-3 cells docetaxel induces delayed cell death via mitotic catastrophe evident by profound multinucleation and formation of giant cells. Mononucleated progeny of the giant PC-3 cells shows significant resistance to docetaxel. Gene expression profiling of these docetaxel resistant PC-3 cells revealed sets of docetaxel inducible and constitutively expressed genes associated with major cancer pathways. A contradictory overlap with DU-145 docetaxel resistant cells was also found. Analyses suggested significant changes associated with apoptotic function, DNA repair, cell growth, survival and proliferation, metabolism, maintenance of cytoskeleton and extracellular matrix formation. These cellular processes often contribute to drug resistance and our study identified a set of genes managing this phenotype. Additional analyses of the drug resistant PC-3 cells using shRNA constructs determined direct relevance of Cyclin G2 to docetaxel resistance as well as prevention of multinucleation, whereas the knockdown of upregulated CYP1B1 showed no effect on either of these processes. Downregulated GBP1 was explored by ectopic overexpression and even though GBP1 has a potential to mediate resistance to docetaxel, it was not utilized in PC-3 cells. The results suggest complex combination of gene expression pattern changes that enables resistance to docetaxel while preventing death via multinucleation.
Key words: multinucleation, giant cells, drug resistant phenotype, docetaxel, prostate, cancer, p53 negative
Introduction
Docetaxel, a member of taxane family of anti-cancer drugs, is widely used as a single agent or in combination with other drugs to treat advanced stages of prostate cancer. However emergence of the drug resistance is a frequent outcome and is often accompanied by an androgen independent phenotype. To generate a framework for studies of docetaxel activity against prostate cancer cells we have developed a series of docetaxel resistant derivatives of the androgen independent prostate cancer cell line PC-3.1 Using western blot and immunostaining techniques we determined that the resistant phenotype of these clones was not associated with gene products often associated with drug-resistance such as docetaxel target β-tubulin, drug efflux pump P-glycoprotein and anti-apoptotic BCL-2 and BCL-xL.1 To further elucidate the molecular mechanisms mediating resistance to docetaxel we performed a broad gene expression profiling using high throughput gene arrays in this study. Docetaxel induces mitotic catastrophe in PC-3 cells and the drug resistant cells emerge from the profoundly multinucleated cells. The process of asymmetric cell division when viable mononucleated descendants arise from drug- or radiation-induced multinucleated progenitors (neosis for short) was found to be quite common in various cancers and serves as a mechanism for tumor cell survival and a promoter of tumor progression.2 A parallel phenomenon occurring in normal tissues is also known and is important during tissue regeneration and differentiation.3 Because multinucleation might result in a severe genetic instability, we have reasoned that the comparative analysis of several docetaxel resistant PC-3 cell lines should reveal a more consistent pattern of gene expression alterations. Importantly, the high level of docetaxel resistance in these cells allows maintenance of the drug-resistant cultures in the presence of docetaxel at low concentrations without inducing multinucleation and cell death. We thus compared two selected docetaxel resistant PC-3 cell cultures showing high levels of resistance under two experimental conditions: when the cells were grown either in the absence or in the presence of docetaxel in culture media. These experimental conditions were sought to help in discriminating between the genes constitutively altered following neosis and the genes induced by low-dose docetaxel during routine maintenance in culture. Furthermore, we also extended our investigation to include the docetaxel sensitive and resistant cultures of androgen independent DU145 cells. These cells do not die via multinucleation but instead undergo apoptosis in response to docetaxel treatment. Our screening of RNA samples from docetaxel sensitive and resistant DU145 cultures provided a basis to extrapolate between the multinucleation vs. apoptosis mechanisms.
A number of genes potentially relevant to drug resistance were identified in our gene array screens and merit detailed investigation. We verified the gene array data by using qRT-PCR amplifications and then performed functional tests with several genes to determine their relevance to docetaxel resistance. We at first studied three relatively under-characterized genes identified in previous studies as potentially pertinent to docetaxel resistance. These included cytochrome P450 CYP1B1 that was shown to improve ability in cell survival in docetaxel resistant MCF-7 breast cancer cells4 and was strongly induced by docetaxel in resistant PC-3 cells. We also reasoned that the persistent upregulation of Cyclin G2, the only cell cycle related gene detected in the docetaxel resistant PC-3 derivatives, might be also relevant to docetaxel resistance. Yiwei Li and co-workers described an elevated expression of Cyclin G2 in naïve PC-3 cells treated with docetaxel which suggested this cyclin might be an important early indicator of an emerging drug resistance.5 Both upregulated CYP1B1 and Cyclin G2 were knocked down by small hairpin RNA (shRNA) silencing, but only Cyclin G2 silencing induced changes in drug sensitivity. Curiously, most of the genes previously attributed to multidrug and/or taxane resistance were downregulated in docetaxel resistant PC-3 cells. These included an interferon-inducible guanylate binding protein GBP1 established earlier as a prominent mediator of paclitaxel resistance.6 We found that the ectopic overexpression of GBP1 using cDNA plasmid constructs notably increases docetaxel sensitivity. Altogether, our studies suggested existence of different, complex and likely complementary molecular mechanisms mediating resistance to docetaxel and suggested new potential target leads for future investigations.
Results
Cytotoxicity of docetaxel in DU145 cells.
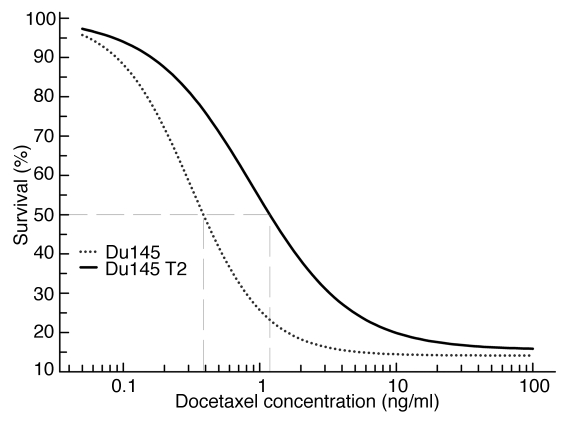
The level of relative resistance to docetaxel in DU145 T2 cells was established by comparing to the parental DU145 cells in the MTT cytotoxicity assay. It was determined that the IC50 value for the resistant cells was 1.18329 ng/ml compared to 0.38624 ng/ml in parental DU145 cells, thus showing ∼3 times higher levels of docetaxel resistance in the T2 derivative (Fig. 1).
Figure 1.
Dose response curves showing 3 fold difference in relative level of resistance to docetaxel in drug-resistant DU145 T2 cells (IC50 = 1.18329 ng/ml, p = 1.0812 E-06) compared to parental DU145 cultures (IC50 = 0.38624 ng/ml, p = 2.9165 E-06).
Genes differentially expressed in the docetaxel resistant PC-3 cell lines.
We have compared the control replicate arrays of the docetaxel-untreated parental PC-3 cell line to the experimental replicate arrays of the docetaxel resistant PC-3 cell lines 8b2 and 1-INT under two conditions: without docetaxel in media and with docetaxel present at 0.1 ng/ml. Because the resistant PC-3 cells were clonally derived via neosis, e.g., following substantial genetic alterations in the parental cells, we anticipated a great variability in the gene expression pattern. However, the analyses indicated that at the selected cutoff level, less than 250 transcripts out of 47,000 tested were expressed differentially in the 8B2 and 1-INT cells compared to the parental PC-3 cells. Combined data of the two experimental conditions (docetaxel + or − in media) revealed 132 deregulated genes common to both 8B2 and 1-INT PC-3 clones. The level of 103 genes remained near constant after the drug treatment suggesting constitutive deregulation, while the expression of another 29 transcripts was induced by docetaxel. The latter group included CYP1B1, EGR1 and Serpin1 genes which were altered before the treatment and were further deregulated upon treatment with docetaxel. Functional annotation using DAVID search engine enabled to stratify most of these genes into distinct but biologically closely related gene clusters that included: regulation of cell death/apoptosis, cell cycle, cell growth/proliferation, extracellular matrix/adhesion, cytoskeleton, microtubules, response to stress/chemical stimulus, receptors, signal transduction and transcription, GTPase binding and regulator activity, transporters and metabolic processes, chromatin organization and a group of other genes without known associations (Table 1, see supplemental materials). Functional dependence of many of these genes and their respective pathways on availability of calcium ions was also evident. Analyses of the pathway associations established that numerous genes were related to major pathways involved in cancer. These included tightly associated signaling cascades of Homologous recombination (RAD51L1) and chromatin maintenance (histone cluster H1), Hedgehog and WNT signaling pathways (WNT5A, ZIC2, CDH11, PCDH7), Notch (DLL1, HES1) and MAPK (RELB, NFκB2, IL1, FGF1, FGF5, EGR1), p53 signaling pathway (CyclinG2, IGFBP3, SERPIN1), Toll receptor (TLR4, NFκB2, PTGS2, TNFAIP3, RELB), TGFbeta (ID2, FKBP1B), a closely associated coagulation and plasminogen activation cascades (F3, PLAUR, C3, SERPINE1, MMP13, SERPINB2) and steroid hormone biosynthesis (CYP1B1, ALDH3A2). A set of genes with an established role in multidrug and/or taxane specific resistance was also found and included GBP1,6 Serpin1 and SerpinB2, CXCL5, AKAP12,7 LAMC2,8 as well as TGFβI9 all of which were constitutively downregulated in PC-3 cell clones. Exception was a constitutively upregulated tubulin α1, a member of the tubulin family of microtubule proteins, the target for docetaxel binding and a well established taxane resistance mediator.
Gene array profiling of the drug treated PC-3 clones identified a set of docetaxel induced genes signifying the changes associated with chronic exposure to sub-lethal concentration of the drug. Annotation clustering scattered this gene group along most of the functional clusters and several pathways uncovered in the untreated cells. Notable was an upregulation of the genes carrying an anti-apoptotic function (DEPDC6, MAL, HOXA13, NEFL) and downregulation of the cell growth and proliferation associated chemokines CXCL3 and CCL26. An enhanced expression of the microtubule associated centrosomal protein CEP68 and interferon-induced IFIT2 associated well with an activation of the transcription (ELK3, ATXN, EGR1, STRN3) and translation (EIF2C4) regulatory factors involved into microtubule synthesis. These concomitant changes suggested coordinated effort to modify the processes maintaining microtubules in response to docetaxel. A similar paradigm was noted in the chromatin remodeling group of genes which was further enriched by upregulation of yet another histone 1H as well as latexin (LTN), a carboxypeptidase inhibitor involved into histone hyperacetylation pathway. Treatment also downregulated RASA2, a GTPase regulator of RAS p21 an important activator of many signaling cascades. Metabolism associated genes in this group included further upregulated CYP1B1 and an induced aldo-keto reductase AKR1C1 as well as downregulated SGMS2. Particularly interesting was identification of an induced upregulation of a microRNA 21 (MIR21), that was recently similarly identified by microarray screens of docetaxel resistant PC-3 cells and characterized as a contributing factor in mediating resistance to docetaxel.10
Genes differentially expressed in the docetaxel resistant DU145 T2 cells.
A total of 313 deregulated gene transcripts have met the cutoff criteria and were clustered according to their biological function (Table 2, see supplemental materials). Analyses in DAVID environment indicated that on the whole the pattern of gene deregulation in DU145 T2 cells was suggestive of an increased and coordinated activity of the negative and positive regulators of cell death and apoptosis as well as the factors promoting cell survival, growth and proliferation. Deregulation of the cell cycle related molecules suggested increased activity primarily during G2/M phase of the cycle. These changes are commonly recognized as consistent with an activation of numerous biologic pathways and processes in response to cytotoxic drug activity and ultimately aid in drug resistance. Many gene transcripts pointed toward profound modifications in extracellular matrices and related focal adhesion processes, and also activation of elements within cytochrome P450 metabolic cascade. Similarly to PC-3 cells, we found a set of genes previously attributed to multidrug and/or taxane drug resistance. However, unlike in PC-3 cells most of such genes were upregulated in DU145 T2 cells. These included chemokine CXCL5; an aldehyde dehydrogenase ALDH1A3; an ATP-binding cassette transporter ABCG2; guanylate binding protein1 GBP1; and also extracellular matrix components DNAJC15, DnaJ (HSP40) homolog and laminin gamma2, LAMC2. In contrast, GSTA4, glutathione S-transferase alpha 4, and an ATP-binding cassette transporter ABCC3 were downregulated. Intriguingly, this pattern of inverse associations was also found in a set of genes overlapping in both PC-3 and DU145 T2 cells which included the drug resistance related molecules CXCL5, GBP1 and LAMC2 (Table 3). A common trait with PC-3 cells was a great number of genes and associated pathways dependent on calcium ions to sustain their biological functions. For example, upregulation of RRAD, ras-related associated with diabetes, a member of small GTPase binding family of proteins was an important indicator of the increased activity of the L type Ca2+ channel regulation cascade.11
Table 3.
Overlapping genes in PC-3 and DU145 docetaxel resistant cells
| Gene | Function | 8B2 | 8B2 DOX | 1-INT | 1-INT DOX | DU145 T2 |
| BCL6: B-cell CLL/lymphoma 6 | Regulation of cell death/apoptosis | 3.42 | 4.66 | 3.37 | 5.26 | −2.13 |
| CXCL5: chemokine (C-X-C motif) ligand 5 | Regulation of cell growth/proliferation | −3.49 | −4.30 | −2.59 | −2.87 | 7.53 |
| LAMC2: laminin, gamma2 | Extracellular matrix | −5.72 | −6.02 | −2.55 | −2.63 | 1.92 |
| HSPB8: heat shock 22 kDa protein 8 | Response to stress/chemical stimulus | −3.66 | −3.65 | −2.41 | −2.77 | 3.01 |
| F3: coagulation factor III (thromboplastin, tissue factor) | Response to stress/coagulation | −5.02 | −5.40 | −5.46 | −5.93 | −2.01 |
| PLAUR: plasminogen activator, urokinase receptor | Receptor, coagulation cascade | −2.86 | −3.15 | −2.19 | −2.27 | 1.87 |
| GBP1: guanylate binding protein 1, interferon-inducible | GTPase activity | −8.65 | −9.73 | −6.93 | −7.03 | 2.11 |
| AKR1C3: aldo-keto reductase family 1, member C3 | Lipid metabolic process | 2.55 | 6.04 | −9.62 | ||
| FAM83A: family with sequence similarity 83, member a | Unknown | −7.04 | −5.52 | −2.45 | −2.85 | −3.33 |
| FAM46C: family with sequence similarity 46, member C | Unknown | 2.53 | 2.57 | 2.67 | 3.08 | −4.25 |
Validation of the gene array results by quantitative real-time PCR (qRT-PCR).
To verify variations in gene expression detected by the microarrays we chose representative genes with varying expression profiles for real-time quantitative RT-PCR validation. The results of the qRT-PCR for these selected genes were in direct agreement with the microarray data (Table 4). Although the fold change in the expression levels was not always exactly the same between these two different analytical methods, the data was consistent and thus supported the findings obtained from the microarray experiments.
Table 4.
Validation of the gene expression profiling data by quantitative real-time PCR
| 8b2 | 8b2 + docetaxel | 1-INT | 1-INT + docetaxel | |||||
| Gene | Affy | QPCR | Affy | QPCR | Affy | QPCR | Affy | QPCR |
| SERPINE1 | −11.2 | −4.55 ± 0.3, p < 0.0001 | −15.62 | −18.89 ± 2.1, p < 0.0001 | −8.04 | −6.86 ± 1.25, p = 0.0005 | −12.75 | −12.53 ± 1.45, p = 0.0001 |
| CXCL5 | −4.04 | −3.0 ± 0.24, p = 0.0132 | −5.11 | −3.8 ± 0.15, p < 0.0001 | −3.11 | −2.47 ± 0.13, p = 0.0023 | −4.3 | −2.71 ± 0.44, p < 0.0001 |
| TGFBI | −7.38 | −6.9 ± 2.32, p = 0.0008 | −8.38 | −5.83 ± 0.02, p < 0.0001 | −2.25 | −2.84 ± 0.3, p = 0.0001 | −2.45 −3.01 | ± 0.5, p < 0.0001 |
| GBP1 | −9.93 | −5.27 ± 0.3, p = 0.0046 | −9.73 | −5.76 ± 0.73, p < 0.0001 | −7.03 | −7.22 ± 0.4, p = 0.0007 | −8.08 | −4.98 ± 0.52, p = 0.0018 |
| CYP1B1 | 12.43 | 6.62 ± 0.11, p = 0.0009 | 13.83 | 13.49 ± 2.42, p = 0.0004 | 5.99 | 3.4 ± 0.75, p = 0.0008 | 9.19 | 5.29 ± 0.56, p = 0.0002 |
| KCNMA1 | 7.09 | 4.7 ± 0.52, p < 0.0001 | 6.22 | 5.39 ± 0.8, p = 0.0004 | 7.7 | 4.62 ± 1.62, p = 0.0001 | 6.77 | 2.43 ± 0.2, p = 0.0004 |
| CCNG2 | 2.52 | 3.64 ± 0.48, p = 0.0017 | 2.98 | 4.33 ± 0.31, p < 0.0001 | 3.51 | 5.3 ± 2.03, p = 0.0006 | 4.01 | 8.95 ± 1.6, p = 0.0028 |
| HES1 | 2.80 | 5.81 ± 1.48, p = 0.0053 | 2.38 | 4.18 ± 0.19, p = 0.0006 | 2.67 | 4.88 ± 0.59, p = 0.0001 | 2.43 | 4.73 ± 0.62, p = 0.0078 |
Randomly selected genes showing differential pattern of expression by gene arrays (affy) were measured by quantitative real-time PCR (qRT-PCR) and showed fold change values consistent with gene arrays. QPCR values are mean ± s.e.m and the p values were calculated using two-tailed student's t-test.
Sensitivity to docetaxel in shCCNG2, shCYP1B1 and pCMVGBP1 derivatives of 8B2 cells.
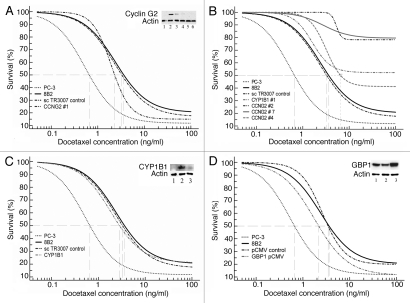
Transfection of 8B2 PC-3 cells with shCCNG2 construct was lethal to most cells. Transfected cultures displayed profound multinucleation in most cells although a fraction of mononucleated cells was also observed. To rescue subcultures showing sustained growth the transfected cells were cloned. Of the four clones derived, clone #1 showed an ability to proliferate steadily without multinucleating as the parental cultures. In contrast clone #4 also initially composed primarily of mono-nucleated cells was prone to accumulate the giant multinucleated cells during extended culturing. Clones #2 and #7 were mostly composed of multinucleated cells that could have been serially passaged and showed an ability to survive for over 3 months in culture although the rate of proliferation was low. Western blot analyses determined partial silencing of cyclin G2 in the clones #1 and #4, and complete inhibition in the other two clones. As expected, the MTT cytotoxicity assays of these individual shCCNG2 8B2 clones showed diverse response to the docetaxel treatment. The steadily proliferating shCCNG2 clone #1 displayed significant increase in drug sensitivity as evident by the decreased IC50 values (Fig. 2A). This suggested that the upregulation of CCNG2 contributes to the overall level of resistance to docetaxel. The MTT assay showed significantly lower differential between cell death and proliferation in clones 2, 4 and 7 due to an overwhelming presence of the slowly cycling giant cells. This discrepancy limited the utility of the MTT cell proliferation assay in determining true cytotoxic activity of the drug (Fig. 2B). Because the transfection with shCCNG2 construct had induced giant cells in most but not all 8B2 cells, and the shRNA inhibition was only partial in the mononucleated clones we have concluded that the cyclin G2 function is required for cell survival whereas its silencing effectively induces cell death via multinucleation.
Figure 2.
(A) Dose response curves showing relative level of resistance to docetaxel in the shCCNG2 transfected 8B2 PC-3 cells (clone #1, IC50 = 2.30325 ng/ml, p = 3.061 E-06) compared to drug sensitive PC-3 cells (IC50 = 0.66182 ng/ml, p = 4.7827 E-015), resistant parental cells 8B2 PC-3 (IC50 = 3.62045 ng/ml, p = 1.5035 E-010) and scTR3007 vector control (IC50 = 3.27586 ng/ml, p = 3.6625 E-07). Drug sensitivity in shCCNG2 transfected cells increased by 1.57 fold compared to parental 8B2 cultures. Inset: Immunoblot confirming knockdown of cyclin G2. Lanes (1) parental PC-3 cells; (2) sc TR3007 vector control; (3) shCCNG2 clone #1; (4) shCCNG2 clone #4; (5) shCCNG2 clone #2; (6) shCCNG2 clone #7. (B) examples of limited utility of the MTT cell proliferation assay to determine cytotoxicity profile of the giant multinucleated cells derived from cyclin G2 and CYP1B1 shRNA transfectants. See details in the text. (C) Dose response curves showing relative level of resistance to docetaxel in the shCYP1B1 transfected 8B2 PC-3 cells (IC50 = 2.82571 ng/ml, p = 7.0043 E-016) compared to drug sensitive PC-3 cells (IC50 = 0.66182 ng/ml, p = 4.7827 E-015), resistant parental cells 8B2 PC-3 (IC50 = 3.62045 ng/ml, p = 1.5035 E-010) and scTR3007 vector control (IC50 = 3.27586 ng/ml, p = 3.6625 E-07). an 1.28 upward change in drug sensitivity upon shCYP1B1 transfection compared to 8B2 parental cells was considered significant but insufficient to fully explain robust drug resistance in 8B2 cells. Inset: Immunoblot confirming knockdown of CYP1B1 (lane 3) compared to vector control (lane 2) and parental PC-3 cells (lane 1). (D) Dose response curves showing relative level of resistance to docetaxel in the pCMV GBP1 transfected 8B2 PC-3 cells (IC50 = 2.17439 ng/ml, p = 4.9621 E-013) compared to drug sensitive PC-3 cells (IC50 = 0.66182 ng/ml, p = 4.7827 E-015), resistant parental cells 8B2 PC-3 (IC50 = 3.62045 ng/ml, p = 1.5035 E-010) and pCMV vector control (IC50 = 3.45328 ng/ml, p = 3.022 E-009). Drug sensitivity in pCMV GBP1 transfected cells increased by 1.67 fold compared to 8B2 parental cultures. Inset: Immunoblot confirming successful overexpression of GBP1 (lane 3) compared to the vector control (lane 2) and parental PC-3 cells (lane 1).
The incidence of giant cells in shCYP1B1 8B2 cultures was low and comparable to the 8B2 parental cells. Levels of resistance to docetaxel remained virtually unchanged in the shCYP1B1 8B2 cells as determined by the MTT cytotoxicity assay. To evaluate a possibility of clonal variations in these cultures we derived eight individual shCYP1B1 8B2 clones upon transfection. Seven of these clones were composed primarily of mononucleated cells and showed little or no growth differences. According to the MTT assays, the levels of sensitivity to docetaxel in these clones were nearly identical to the uncloned shCYP1B1 cultures. An insignificant downward shift in IC50 values was detected suggesting that silencing of CYP1B1 did not lead to a measurable reduction of drug resistance in these cultures. Figure 2C shows combined data obtained by the MTT assays in these clones. Cloning have also yielded one culture (#1) containing a large fraction of the multinucleated cells which were propagated for over 3 month similarly to the shCCNG2 giant cell clones. Because the overall occurrence of the giant multinucleated cells among shCYP1B1 8B2 cells was low, we have concluded that the randomly isolated clone #1 represents the cells predetermined to form giant cells rather than induced by silencing of CYP1B1. As with the shCCNG2 giant cells, the MTT assay of the shCYP1B1 clone #1 was not informative (Fig. 2B).
The GBP1 transfected cells were steadily proliferating, showed rare occurrence of multinucleated cells and therefore were not cloned. Following the antibiotic selection for 2 weeks, the cell lysate testing by the western blot technique indicated successful overexpression of GBP1 compared to the vector control. The MTT cytotoxicity assay showed notable increase in sensitivity to docetaxel in GBP1 8B2 cells (Fig. 2D) suggesting that GBP1 might be directly relevant to docetaxel resistance but was not utilized by 8B2 parental cells.
Discussion
Differential gene displays obtained by the gene array screens and validated by the qRT-PCR have surpassed our expectations. The results indicated striking resemblance between the studied PC-3 clones and were limited to a narrow set of genes of which a significant proportion was in common. To discriminate specific genes involved in mediating docetaxel resistance from those related to multinucleation we examined the docetaxel resistant PC-3 cells under two experimental conditions when the cells were treated or untreated with docetaxel. The screens of the treated cells allowed verifying the data from the untreated cells and also uncovered several genes showing specific docetaxel-induced pattern of expression. By treating the cells with sub-toxic drug concentrations to avert formation of the giant multinucleated cells we aimed to distinguish an early compensatory cellular response to the drug. Overall analysis of the gene array data suggested consistent changes associated with increased anti-apoptotic function; DNA repair; cell growth, survival, proliferation; metabolism and maintenance of cytoskeleton and extracellular matrix formation. These processes are often involved in drug resistance and their combination might explain substantial levels of docetaxel resistance observed in our model system. Examination of the gene expression patterns suggested that DU145 T2 cells might be following a more canonic pathway of resistance that was not obvious in PC-3 cells. These phenotype variations might reflect differences in the initial genetic background as well as experimental approaches used to derive docetaxel resistant cells. Disparities in the expression pattern of the commonly known drug resistance associated genes and other genes overlapping with DU145 T2 is quite arcane and needs further investigation.
Changes in metabolic processes were immediately apparent in both PC-3 and DU145 cells and included upregulation of the aldehyde dehydrogenase ALDH3A2 and ALDH1A3 respectively. Aldehyde dehydrogenases belong to the family of enzymes oxidizing aldehydes to carboxylic acids and are recognized drug resistance mediators. However, because the levels of ALDH3A2 remained virtually unchanged in PC-3 cells upon drug exposure, we concluded this enzymatic activity was likely related to processing of the therapeutically inactive and well known docetaxel metabolites containing aldehyde groups. In contrast, constitutively upregulated CYP1B1 was also strongly inducible and we reasoned it might have a role in conferring resistance to docetaxel. There is clinical evidence suggesting importance of CYP1B1 in prostate cancer patients treated with docetaxel.12 However CYP1B1 does not directly inactivate docetaxel and rather promotes cell survival.4 Other studies determined that docetaxel was not metabolized by recombinant human CYP1B1 but binds to CYP1B1 to act as an effector of this enzyme.13 Because shCYP1B1 silencing in 8B2 cells did not lead to measurable reduction in docetaxel resistance we concluded that CYP1B1 was not a key mediator of docetaxel resistance in our experimental model.
Among the drug-resistance associated genes downregulated in PC-3 clones were AKAP12,8 a putative tumor suppressor targeted at the control of cell proliferation and cytoskeletal remodeling,14 and CXCL5, a CXC-type chemokine—an inflammatory mediator and a powerful attractant for granulocytic immune cells which also promotes angiogenesis in cancer.15,16 It is believed that its overexpression stimulates cellular proliferation and gene transcription and also increases the capacity of aggressive prostate cancer cells to invade and metastasize.17 CXCL5 belongs to the same family of chemokines as S100, calcium binding proteins and known mediators of drug resistance including docetaxel. Similarly, an upregulation of GBP1 expression important in facilitating resistance to paclitaxel6 was also not observed. To investigate this trend we evaluated significance of the constitutive downregulation of GBP1 in PC-3 cells. Analysis of the relative levels of resistance in PC-3 8B2 and pCMV-GBP1 transfected 8B2 cells by the MTT cytotoxicity assay indicated that the ectopic overexpression of GBP1 have led to an increased docetaxel sensitivity. This finding suggested that even though GBP1 might function as a specific gene mediating drug resistance this function was not manifested in our experimental model. Although the importance of GBP1 downregulation in the PC-3 clones remains unclear, a related paradigm of GBP1 downregulation in radio-resistant lung cancer cells might suggest an alternative function and importance of this gene in recovery from multinucleation.18
CCNG2, cell cycle checkpoint cyclin G2, has potentially a dual role in our experimental system. On one hand, cyclin G2 was characterized as an inducer of G1/S phase cell cycle arrest that could result in nuclear aberrations resembling multinucleation and could be also significantly upregulated in response to DNA damage.19,20 However, this effect was only observed in the cells expressing functional p53 protein, which was not present in PC-3 cells. On the other hand, Cyclin G2 was shown to be active during other phases of the cell cycle. It is a centrosome-associated nucleo-cytoplasmic shuttling protein that influences microtubule stability and therefore could be directly relevant to docetaxel activity. Both possibilities are quite plausible because the levels of constitutively upregulated Cyclin G2 slightly increased upon treatment with docetaxel. The shRNA CCNG2 silencing indicated that the expression of cyclin G2 was important in preventing multinucleation. This effect could be interpreted such that upregulation of Cyclin G2 facilitates a delay in cell cycle transition from G1/S to G2/M phase thus allowing additional time for chromatin repairs and reorganization of microtubules deleteriously affected by docetaxel, thereby counteracting drug activity. Protraction of the cell cycle is also suggested by the overexpression of anti-apoptotic BCL-6.21 In this regard, Cyclin G2 also interacts with histones22 which play a central role in transcription regulation, DNA repair, DNA replication and chromosomal stability. Both constitutive and inducible upregulation of the 1H2 and 1H4 histone clusters suggested cytoprotective response to multinucleation23 and thus related to docetaxel resistance in PC-3 cells. Furthermore, an upregulation of HES1 in the absence of detectable changes in the affector molecule Notch1 might be indicative of increased chromosomal instability24 in aneuploid and p53 negative PC-3 cells. Intriguingly we also detected a concomitant downregulation of the signal transducer DLL1, another member of the Notch pathway, as well as an upregulation of transcription regulator NPAS2. These three genes are constituents of the circadian clock pathway which determines cyclical processes in the cells including the cell cycle. Deregulation of this pathway might be directly relevant to a mechanism by which the docetaxel resistant mono-nucleated PC-3 cells evade the cell cycle arrest and emerge from multinucleated progenitors.25 Additional studies of Cyclin G2 and associated genes in relationship to the cell cycle, giant cells and microtubule binding agents are warranted.
Calcium plays a central role in regulating contractility, gene expression, hypertrophy and apoptosis. Docetaxel binding to microtubules causes their stabilization and prevents depolymerisation from calcium ions thus changing tightly regulated calcium homeostasis. We detected a broad spectrum of deregulated genes dependant on calcium ions for proper function in both PC-3 and DU145 T2 cells. Alterations in calcium metabolism might be directly relevant to drug resistance and are also associated with cancer progression. For example, amplified expression of KCNMA1, potassium large conductance calcium-activated channel, was characterized as a promoter of prostate cancer cell proliferation and was also attributed to hormone-insensitive prostate cancer cells including PC-3.26 Similarly, over-expression of WNT5A is known to increase aggressive behavior of androgen independent prostate tumors and is also associated with anti-apoptotic function of WNT5A.27–29 A critical importance of calcium influx inhibition in reversing the docetaxel and multi-drug resistant phenotype was recently demonstrated in human lung cancer cell lines.30
Taken together, our efforts to identify specific genes and molecular cascades mediating resistance to docetaxel in prostate cancer cells have yielded a narrow set of genes and associated specific cellular processes that appear highly relevant. Data suggests that the resistance to docetaxel cannot be fully explained by alterations in any one specific gene, but represents a comprehensive cellular response enabling cell survival and neutralization of the drug activity. Undoubtedly, the tightly connected processes of the recovery from multinucleation while building resistance to the drug are complex and likely complement each other. These findings refine our understanding of the drug resistance phenotype in general and open a possibility to evaluate both individual genes and their combinations to further delineate these mechanisms. As our knowledge of the tumor stem cells is improving, it will be also important to relate the identified mechanisms of drug resistance to the stem/progenitor populations maintaining the tumor.31 In turn, this knowledge will aid in selecting new therapeutic agents helpful to enhance the efficacy of docetaxel treatment by preventing or nullifying the resistant phenotype.
Materials and Methods
Cell culture, cytotoxicity assay and RNA preparation.
Prostate cancer cell lines PC-3 and DU145 were obtained from the ATCC collection. The development and characterization of the docetaxel resistant PC-3 cell lines has been previously described.1 In the current study we have used PC-3 8B2 and 1-int cell lines showing high levels of resistance to docetaxel of the 11 cell lines selected initially. In the transfection experiments preference was given to 8B2 clone since it demonstrated very stable levels of docetaxel resistance over extended time in culture. The docetaxel resistant DU145 T2 cell line (unpublished: Makarovskiy A, Siryaporn E) was produced by periodically exposing the actively proliferating DU145 cultures to low doses of docetaxel up to 0.1 ng/ml (once weekly for 8 hours). DU145 T2 cells cannot proliferate in the presence of the drug in media and die via apoptosis unlike the resistant PC-3 cells which can be cultured in media with docetaxel at concentrations up to IC25 value (∼1 ng/ml) without profound multinucleation and cell death. To determine early cellular response to docetaxel in the resistant PC-3 clones we reduced the drug concentration in culture media down to 0.1 ng/ml for the gene array experiments. Lowered drug concentration was used to reduce the stress on the cells, minimize induction of the giant cells and was also clinically relevant. The cell culture conditions and the MTT based cytotoxicity assay to determine relative levels of sensitivity to docetaxel have been carried out as previously described.1 Dose response curve fitting and determination of IC50 values (XLfit 5, IDBS) was based on the four parameter analyses of mean values from 6 replicate samples in 3 separate experiments conducted on consecutive cell passages (t test p < 0.005). Total RNA was isolated from the cells using RNeasy kit (Qiagen) following the manufacturer's instructions. Total RNA was quantified using NanoDrop spectrophotometer (NanoDrop) and RNA quality was assessed with a Bioanalyzer (Agilent Technologies).
Experimental design of gene expression profiling experiments.
Cell lines included in gene expression profiling experiments were grown in duplicates:
control biological duplicate of the PC-3 parental cell line sensitive to docetaxel.
experimental biological duplicate of the docetaxel resistant PC-3 clones 8B2 and 1-INT grown in the absence of docetaxel in media.
experimental biological duplicate of these docetaxel resistant clones cultured in media with docetaxel 0.1 ng/ml for 7 days prior to total RNA isolation.
control biological duplicate of the DU145 cells sensitive to docetaxel.
experimental biological duplicate of the DU145 T2 cell line resistant to docetaxel.
Gene expression profiling.
Gene expression profiling experiments were carried out according to the Affymetrix technology using Human Genome HG-U133 Plus 2.0 GeneChip arrays with more than 54,000 probe sets permitting analysis of over 47,000 transcripts. Because the major source of variations in microarray experiments comes from biological variations, these have been limited by using biological duplicates of the RNA samples to run each array. The two arrays representing a biological duplicate were considered and processed as replicate arrays. RNA labeling and hybridization on HG-U133 Plus 2.0 GeneChip arrays were carried out following the manufacturer's instructions. Briefly, reverse transcription of 7 micrograms of total RNA was carried out using SuperScript II reverse transcriptase (Invitrogen) and an oligo (dT) primer containing the T7 promoter. Second strand synthesis was performed using one-cycle cDNA synthesis protocol. The cDNA was purified and served as a template in the subsequent in vitro transcription carried out in the presence of T7 RNA polymerase and a biotinylated nucleotide analog/ribonucleotide mix for complementary RNA (cRNA) amplification and biotin labeling. Biotinylated cRNA was cleaned up and 20 micrograms of cRNA from each sample of biological duplicates was fragmented and hybridized to one HG-U133 Plus 2.0 GeneChip oligonucleotide array for 16 hrs at 45°C in a hybridization oven (Affymetrix). The arrays were automatically washed and stained with streptavidin-phycoerythrin conjugate in an Affymetrix GeneChip Fluidic Station 450 and fluorescence intensities were scanned with GeneArray Scanner (Affymetrix). The scanned images were inspected and analyzed with the Affymetrix Expression Console software using the RMA algorithm according to the established quality measures. An additional quality control was performed using the BioConductor simpleaffy package. All arrays were found to be easily within the acceptable quality range recommended by Affymetrix.
Gene expression profiling data mining.
GeneChip array analyses and identification of differentially expressed genes were performed using the DNA-Chip analyzer (dChip32). Normalization was carried out using the invariant-set normalization method.33 An array with median overall brightness was automatically selected as the baseline array against which the other arrays were normalized at probe intensity level. In our study the baseline array was the replicate array corresponding to the biological duplicate 8b2 treated with docetaxel. Normalization was carried out by an iterative procedure that identifies invariant sets of probes representing non-differentially expressed genes between the baseline array and the array to be normalized. The invariant sets of probes were used to fit a non-linear relationship between the baseline array and the arrays to be normalized. The non-linear relationship was then used to carry out normalization. All arrays were normalized one by one against the baseline array using this method. After normalization the arrays had similar overall brightness. Expression values were computed using the algorithm “PM-only model” that uses only Perfect Match probes. When comparing expression values of a transcript across the control and experimental biological replicates, a transcript was considered as differentially expressed when it satisfied two conditions: an increased or decreased fold change ≥2 and the paired t-test p value <0.05.
A second supplemental analysis method was then performed in BioConductor. Following RMA quantile normalization the data were processed using the RankProd package,34 which is based on the Rank Products algorithm.35 This permutation-based non-parametric test is more sensitive to variation between samples and is thus more suitable for experiments with few biological replicates to sample from. RankProd differential expression analyses were performed with the cutoff criteria defined as false discovery rate ≤0.05 and the paired t-test p value ≤0.05. After applying these cutoffs the resulting lists of deregulated genes were compared uncovering a high degree of concordance between the dChip and RankProd analysis methods. The gene ontology analysis, gene functional clustering and pathway analyses were conducted using David Bioinformatics Resources 6.7 (http://david.abcc.ncifcrf.gov/).36,37
Quantitative real-time PCR (qRT-PCR) analysis.
Several differentially expressed genes were selected for qRT-PCR analysis, with beta-actin as a control to verify the reliability of the oligonucleotide array data. qRT-PCR was performed with the iCycler IQ real-time detection system (Bio-Rad). 1 microgram of total RNA isolated to perform gene expression profiling experiments was used to generate cDNA using Superscript II reverse transcriptase according to the manufacturer's protocol (Invitrogen). PCR primers were obtained from the Real-time Primer Database (http://medgen.ugent.be/rtprimerdb/). qRT-PCR was performed by monitoring in real-time the increase in fluorescence of SYBR green dye from IQ™ SYBR green master mix (Bio-Rad). All assays were performed in triplicate. Two-tailed student t-test was used to compare threshold concentrations (Ct) values obtained between the resistant cell lines and parental cell line. Fold changes of expression were calculated following the Pfaffl method.38
Generation of stable shRNA 8b2 cells and stable GBP1 expressing 8b2 cells.
Predesigned shRNA constructs to silence the expression of CYP1B1 and CCNG2 genes were obtained from Origene (Rockville, Maryland, USA). For each gene, four 29-mer shRNAs were expressed under the control of U6 promoter into the pGFP-V-RS plasmid (Origene). Pre-confluent 8b2 cells were transfected in a 12-well culture plate with 1 microgram of shRNA construct using Lipofectamine LTX reagent and Opti-MEM I Reduced Serum Medium according to the manufacturer's instructions (Invitrogen). After 14–18 hours of incubation, the transfection solution was removed and replaced with fresh complete growth medium. At 48 hours post transfection, 8b2 cells stably expressing shRNA were selected with growth medium containing puromycin at 0.5 mg/ml concentration. Individual cell clones were picked, grown and screened by western blot. Alternatively, cell clones corresponding to the same shRNA construct were pooled and grown as a single culture. Vector pGFP-V-RS (TR3007) was transfected as a negative control.
Overexpression of GBP1 in 8b2 cells was achieved using the RSPD001 construct (Origene) in which GBP1 cDNA was inserted in the pCMV6-NEO plasmid. PC-3 8b2 cells were transfected as described above and stable GBP1 expressing 8b2 cells were selected with growth medium containing G418 at 100 mg/ml concentration. An increase of the antibiotic concentration to 500 mg/ml did not result in additional selection. Vector pCMV6-NEO as a negative control was transfected and selected similarly. The positive effect of gene silencing or overexpression was assessed by determining the levels of corresponding proteins in experimental cells compared to vector controls in a western blot assay with beta-actin as a normalization control. The western blot protocols were previously published.1 Monoclonal antibodies to CYP1B1 were purchased from BD Biosciences; Cyclin G2 from ABCAM Inc. and GBP1 was obtained from MBL International and used at a 1:500 dilution.
Acknowledgements
Funding for this research project was provided by Sanofi-Aventis (IST 10257, PI Makarovskiy) and Tufts Medical Center/Tufts University School of Medicine. The authors gratefully acknowledge the assistance of Tufts Center for Neuroscience Research P30 NS047243 (PI Jackson).
Conflict of Interest and Financial Disclosure
The authors declare no conflict of interest and no financial interest in Sanofi-Aventis who in part sponsored this study.
Supplementary Material
References
- 1.Makarovskiy AN, Siryaporn E, Hixson DC, Akerley W. Survival of docetaxel-resistant prostate cancer cells in vitro depends on phenotype alterations and continuity of drug exposure. Cell Mol Life Sci. 2002;59:1198–1211. doi: 10.1007/s00018-002-8498-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sundaram M, Guernsey DL, Rajaraman MM, Rajaraman R. Neosis: a novel type of cell division in cancer. Cancer Biol Ther. 2004;3:207–218. doi: 10.4161/cbt.3.2.663. [DOI] [PubMed] [Google Scholar]
- 3.Lo DC, Allen F, Brockes JP. Reversal of muscle differentiation during urodele limb regeneration. Proc Nat Acad Sci USA. 1993;90:7230–7234. doi: 10.1073/pnas.90.15.7230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Martinez VG, O'Connor R, Liang Y, Clynes M. CYP1B1 expression is induced by docetaxel: effect on cell viability and drug resistance. Br J Cancer. 2008;98:564–570. doi: 10.1038/sj.bjc.6604195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li Y, Hussain M, Sarkar SH, Eliason J, Li R, Sarkar FH. Gene expression profiling revealed novel mechanism of action of Taxotere and Furtulon in prostate cancer cells. BMC Cancer. 2005;5:7. doi: 10.1186/1471-2407-5-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Duan Z, Foster R, Brakora KA, Yusuf RZ, Seiden MV. GBP1 overexpression is associated with a paclitaxel resistance phenotype. Cancer Chemother and Pharmacol. 2006;57:25–33. doi: 10.1007/s00280-005-0026-3. [DOI] [PubMed] [Google Scholar]
- 7.Gelman IH. Metastasis suppression by SSeCKS/Gravin/AKA-P12 through the inhibition of distal angiogenesis. In: Jackson P, editor. New developments in metastasis suppressor research. 2007. pp. 273–288. [Google Scholar]
- 8.Gyorffy B, Surowiak P, Kiesslich O, Denkert C, Schafer R, Dietel M, Lage H. Gene expression profiling of 30 cancer cell lines predicts resistance towards 11 anticancer drugs at clinically achieved concentrations. Int J Cancer. 2006;118:1699–1712. doi: 10.1002/ijc.21570. [DOI] [PubMed] [Google Scholar]
- 9.Ahmed AA, Mills AD, Ibrahim AE, Temple J, Blenkiron C, Vias M, et al. The extracellular matrix protein TGFBI induces microtubule stabilization and sensitizes ovarian cancers to paclitaxel. Cancer Cell. 2007;12:514–527. doi: 10.1016/j.ccr.2007.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shi GH, Ye DW, Yao XD, Zhang SL, Dai B, Zhang HL, et al. Involvement of microRNA-21 in mediating chemo-resistance to docetaxel in androgen-independent prostate cancer PC3 cells. Acta Pharmacol Sinica. 31:867–873. doi: 10.1038/aps.2010.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yada H, Murata M, Shimoda K, Yuasa S, Kawaguchi H, Ieda M, et al. Dominant negative suppression of Rad leads to QT prolongation and causes ventricular arrhythmias via modulation of L-type Ca2+ channels in the heart. Circ Res. 2007;101:69–77. doi: 10.1161/CIRCRESAHA.106.146399. [DOI] [PubMed] [Google Scholar]
- 12.Sissung TM, Danesi R, Price DK, Steinberg SM, de Wit R, Zahid M, et al. Association of the CYP1B1*3 allele with survival in patients with prostate cancer receiving docetaxel. Mol Cancer Ther. 2008;7:19–26. doi: 10.1158/1535-7163.MCT-07-0557. [DOI] [PubMed] [Google Scholar]
- 13.Bournique B, Lemarie A. Docetaxel (Taxotere) is not metabolized by recombinant human CYP1B1 in vitro, but acts as an effector of this isozyme. Drug Metab Dispos. 2002;30:1149–1152. doi: 10.1124/dmd.30.11.1149. [DOI] [PubMed] [Google Scholar]
- 14.Potti A, Dressman HK, Bild A, Riedel RF, Chan G, Sayer R, et al. Genomic signatures to guide the use of chemotherapeutics. Nature Med. 2006;12:1294–1300. doi: 10.1038/nm1491. [DOI] [PubMed] [Google Scholar]
- 15.Strieter RM, Burdick MD, Mestas J, Gomperts B, Keane MP, Belperio JA. Cancer CXC chemokine networks and tumour angiogenesis. Eur J Cancer. 2006;42:768–778. doi: 10.1016/j.ejca.2006.01.006. [DOI] [PubMed] [Google Scholar]
- 16.Arenberg DA, Keane MP, DiGiovine B, Kunkel SL, Morris SB, Xue YY, et al. Epithelial-neutrophil activating peptide (ENA-78) is an important angiogenic factor in non-small cell lung cancer. J Clin Investigat. 1998;102:465–472. doi: 10.1172/JCI3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Begley LA, Kasina S, Mehra R, Adsule S, Admon AJ, Lonigro RJ, et al. CXCL5 promotes prostate cancer progression. Neoplasia. 2008;10:244–254. doi: 10.1593/neo.07976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu QY, Gao Y, Liu Y, Yang WZ, Xu XY. Identification of differential gene expression profiles of radioresistant lung cancer cell line established by fractionated ionizing radiation in vitro. Chinese Med J. 2008;121:1830–1837. [PubMed] [Google Scholar]
- 19.Arachchige Don AS, Dallapiazza RF, Bennin DA, Brake T, Cowan CE, Horne MC. Cyclin G2 is a centrosome-associated nucleocytoplasmic shuttling protein that influences microtubule stability and induces a p53-dependent cell cycle arrest. Exp Cell Res. 2006;312:4181–4204. doi: 10.1016/j.yexcr.2006.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bennin DA, Don AS, Brake T, McKenzie JL, Rosenbaum H, Ortiz L, et al. Cyclin G2 associates with protein phosphatase 2A catalytic and regulatory B' subunits in active complexes and induces nuclear aberrations and a G1/S phase cell cycle arrest. J Biol Chem. 2002;277:27449–27467. doi: 10.1074/jbc.M111693200. [DOI] [PubMed] [Google Scholar]
- 21.Alenzi FQ. BCL-6 prevents mammary epithelial apoptosis and promotes cell survival. Jpma. 2008;58:494–497. [PubMed] [Google Scholar]
- 22.Stossi F, Likhite VS, Katzenellenbogen JA, Katzenellenbogen BS. Estrogen-occupied estrogen receptor represses cyclin G2 gene expression and recruits a repressor complex at the cyclin G2 promoter. J Biol Chem. 2006;281:16272–16278. doi: 10.1074/jbc.M513405200. [DOI] [PubMed] [Google Scholar]
- 23.Illidge TM, Cragg MS, Fringes B, Olive P, Erenpreisa JA. Polyploid giant cells provide a survival mechanism for p53 mutant cells after DNA damage. Cell Biol Int. 2000;24:621–633. doi: 10.1006/cbir.2000.0557. [DOI] [PubMed] [Google Scholar]
- 24.Baia GS, Stifani S, Kimura ET, McDermott MW, Pieper RO, Lal A. Notch activation is associated with tetraploidy and enhanced chromosomal instability in meningiomas. Neoplasia (New York, NY) 2008;10:604–612. doi: 10.1593/neo.08356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sang L, Coller HA, Roberts JM. Control of the reversibility of cellular quiescence by the transcriptional repressor HES1. Science (New York, NY) 2008;321:1095–1100. doi: 10.1126/science.1155998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bloch M, Ousingsawat J, Simon R, Schraml P, Gasser TC, Mihatsch MJ, et al. KCNMA1 gene amplification promotes tumor cell proliferation in human prostate cancer. Oncogene. 2007;26:2525–2534. doi: 10.1038/sj.onc.1210036. [DOI] [PubMed] [Google Scholar]
- 27.Iozzo RV, Eichstetter I, Danielson KG. Aberrant expression of the growth factor Wnt-5A in human malignancy. Cancer Res. 1995;55:3495–3499. [PubMed] [Google Scholar]
- 28.Wang Q, Williamson M, Bott S, Brookman-Amissah N, Freeman A, Nariculam J, et al. Hypomethylation of WNT5A, CRIP1 and S100P in prostate cancer. Oncogene. 2007;26:6560–6565. doi: 10.1038/sj.onc.1210472. [DOI] [PubMed] [Google Scholar]
- 29.Torii K, Nishizawa K, Kawasaki A, Yamashita Y, Katada M, Ito M, et al. Anti-apoptotic action of Wnt5a in dermal fibroblasts is mediated by the PKA signaling pathways. Cell Signal. 2008;20:1256–1266. doi: 10.1016/j.cellsig.2008.02.013. [DOI] [PubMed] [Google Scholar]
- 30.Chiu LY, Ko JL, Lee YJ, Yang TY, Tee YT, Sheu GT. L-type calcium channel blockers reverse docetaxel and vincristine-induced multidrug resistance independent of ABCB1 expression in human lung cancer cell lines. Tox Letters. 192:408–418. doi: 10.1016/j.toxlet.2009.11.018. [DOI] [PubMed] [Google Scholar]
- 31.Mimeault M, Batra SK. Recent advances in the development of novel anti-cancer drugs targeting cancer stem/progenitor cells. Drug Dev Res. 2008;69:415–430. [Google Scholar]
- 32.Li C, Wong WH. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc Nat Acad Sci USA. 2001;98:31–36. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li C, Hung Wong W. Model-based analysis of oligonucleotide arrays: model validation, design issues and standard error application. Genome Biol. 2001;2:32. doi: 10.1186/gb-2001-2-8-research0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hong F, Breitling R, McEntee CW, Wittner BS, Nemhauser JL, Chory J. RankProd: a bioconductor package for detecting differentially expressed genes in meta-analysis. Bioinform (Oxford, England) 2006;22:2825–2827. doi: 10.1093/bioinformatics/btl476. [DOI] [PubMed] [Google Scholar]
- 35.Breitling R, Armengaud P, Amtmann A, Herzyk P. Rank products: a simple, yet powerful, new method to detect differentially regulated genes in replicated microarray experiments. FEBS letters. 2004;573:83–92. doi: 10.1016/j.febslet.2004.07.055. [DOI] [PubMed] [Google Scholar]
- 36.Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: database for annotation, visualization and integrated discovery. Genome Biol. 2003;4:3. [PubMed] [Google Scholar]
- 37.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature Prot. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 38.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nuc Acids Res. 2001;29:45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.