Summary
Ebolavirus (EboV) is a highly pathogenic enveloped virus that causes outbreaks of zoonotic infection in Africa. The clinical symptoms are manifestations of the massive production of pro-inflammatory cytokines in response to infection1 and in many outbreaks, mortality exceeds 75%. The unpredictable onset, ease of transmission, rapid progression of disease, high mortality and lack of effective vaccine or therapy have created a high level of public concern about EboV2. Here we report the identification of a novel benzylpiperazine adamantane diamide-derived compound that inhibits EboV infection. Using mutant cell lines and informative derivatives of the lead compound, we show that the target of the inhibitor is the endosomal membrane protein Niemann-Pick C1 (NPC1). We find that NPC1 is essential for infection, that it binds to the virus glycoprotein (GP), and that the anti-viral compounds interfere with GP binding to NPC1. Combined with the results of previous studies of GP structure and function, our findings support a model of EboV infection in which cleavage of the GP1 subunit by endosomal cathepsin proteases removes heavily glycosylated domains to expose the N-terminal domain3–7, which is a ligand for NPC1 and regulates membrane fusion by the GP2 subunit8. Thus, NPC1 is essential for EboV entry and a target for anti-viral therapy.
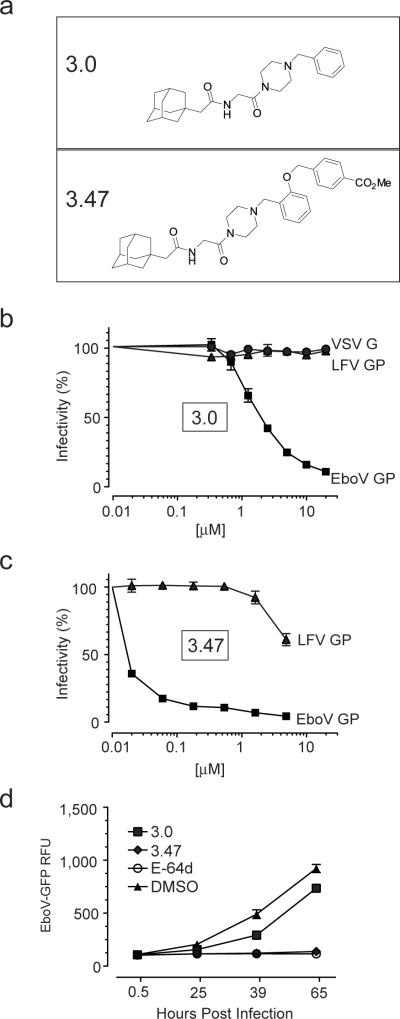
To identify chemical probes that target EboV host factors, we screened a library of small molecules and identified a novel benzylpiperazine adamantane diamide, 3.0, that inhibits infection of Vero cells by vesicular stomatitis virus particles (VSV) pseudotyped with EboV Zaire GP, but not with VSV G or Lassa fever virus (LFV) GP (Fig. 1a,b). To verify that 3.0 is a bona fide inhibitor, we measured EboV growth on Vero cells for 96 hours and found it was reduced by >99% in the presence of 3.0 (Supplementary Fig. 1a). We synthesized and tested >50 analogs of 3.0 and found that the addition of a (methoxycarbonyl) benzyl group at the ortho position of the benzene ring (compound 3.47) increased the potency as measured by a single cycle of EboV GP-dependent infection and efficacy as measured by growth of EboV on Vero cells (Fig 1a,c,d).
Figure 1. Structure and function of ebolavirus entry inhibitors.
a, Compounds 3.0 and 3.47.
b,c, Vero cells were cultured in media containing increasing concentrations of 3.0 (b) or 3.47 (c) for 90 minutes prior to the addition of VSV particles encoding luciferase (b) or GFP (c) and pseudotyped with either EboV GP, VSV G or Lassa fever virus GP (LFV GP). Virus infection is reported as percent of luminescence units (RLU) or GFP-positive cells relative to cells exposed to DMSO vehicle alone. Data is mean ± s.d. (n=4) and is representative of 3 experiments.
d, Vero cells were cultured in media containing 3.0 [40 μM], 3.47 [40 μM], vehicle (1% DMSO) or the cysteine cathepsin protease inhibitor E-64d [150μM] 90 minutes prior to the addition of replication competent ebolavirus Zaire-Mayinga encoding GFP (moi = 0.1). Results are mean relative fluorescence units ± s.e.m. (n=3).
Previous studies revealed that the endosomal protease cathepsin B is essential for EboV infection because it cleaves the GP1 subunit of GP3,4. To address the possibility that 3.0 and 3.47 target this step, we measured cathepsin B activity in the presence of these compounds and found no effect in vitro or in cells (data not shown). Moreover, 3.0 and 3.47 inhibited infection by VSV EboV particles treated with thermolysin, a metalloprotease that faithfully mimics cathepsin cleavage of the GP1 subunit of GP (Supplementary Fig. 1b)4,9. These findings demonstrate that cathepsin B is not the target of 3.0 and 3.47.
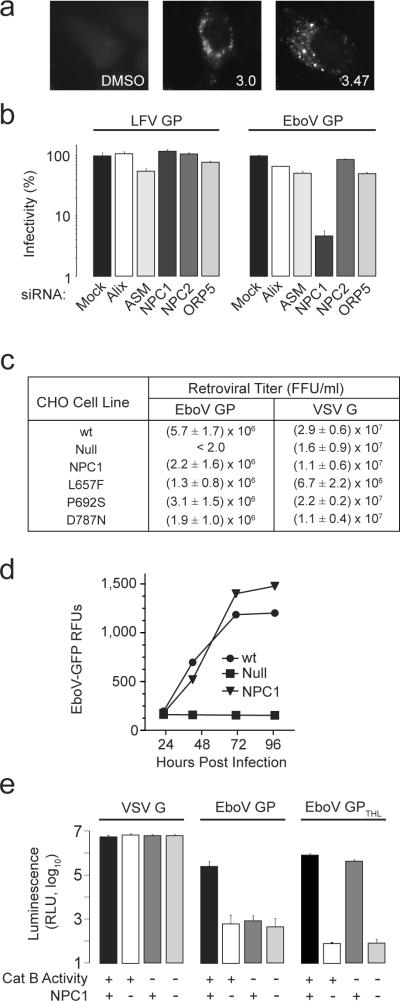
HeLa cells treated with 3.0 or 3.47 for more than 18 hours developed cytoplasmic vacuoles that were labeled by cholesterol-avid filipin (Fig. 2a). The induction of filipin-stained vacuoles by the compounds suggested that they target one or more proteins involved in regulation of cholesterol uptake in cells. To test this hypothesis, we used mutant cell lines and cells treated with siRNA to analyze proteins for which loss of activity had been previously associated with cholesterol accumulation in late endosomes10–12. We found that EboV GP infection is dependent on the expression of Niemann-Pick C1 (NPC1), but not Niemann-Pick C2 (NPC2), acid sphingomyelinase (ASM), ALG-2-interacting protein X (Alix), or oxysterol binding protein 5 (ORP5) (Fig 2b, Supplementary Fig. 2a–c). NPC1 is a polytopic protein that resides in the limiting membrane of late endosomes and lysosomes (LE/LY) and mediates distribution of lipoprotein-derived cholesterol in cells10,13. To analyze the role of NPC1 in infection, we studied Chinese hamster ovary (CHO)-derived cell lines that differ in expression of NPC1. We found that the titer of a murine leukemia virus (MLV) vector pseudotyped with EboV GP on wild type CHO cells (CHOwt) exceeded 106 infectious units/ml (Fig. 2c). Importantly, CHO cells lacking NPC1 (CHOnull) were completely resistant to infection by this virus and infection of these cells was fully restored when NPC1 was expressed (CHONPC1). Thus, NPC1 expression is essential for EboV infection.
Figure 2. NPC1 is essential for ebolavirus infection.
a, HeLa cells were treated with 3.0 (20 μM), 3.47 (1.25 μM) or vehicle for 18 hours, then fixed and incubated with the cholesterol-avid fluorophore filipin.
b, HeLa cells were transfected with siRNAs targeting ASM, Alix, NPC1, NPC2, and ORP5. After 72 hours, VSV EboV GP or LFV GP infection of these cells was measured as in Fig 1c. Data is mean ± s.d. (n=3) and is representative of 3 experiments.
c, CHOwt, CHOnull and CHOnull cells stably expressing mouse NPC1 (CHONPC1) or NPC1 mutants L657F, P692S, D787N were exposed to MLV particles encoding LacZ and pseudotyped with either EboV GP or VSV G. Results are the mean ± s.d. (n=4) and is representative of 3 experiments.
d, CHOwt, CHOnull, and CHONPC1 cells were infected with replication competent ebolavirus Zaire-Mayinga encoding GFP (moi = 1). Results are mean relative fluorescence units ± s.d. (n=3).
e, CHOwt and CHOnull cells were treated with the cathepsin B inhibitor CA074 (80 μM) or vehicle. These cells were challenged with VSV G particles or VSV EboV GP particles treated with thermolysin (EboV GPTHL) or untreated control (EboV GP). Infection was measured as in Fig 1b. Data is mean ± s.d. (n=9).
In CHOnull cells, LE/LY are enlarged and contain excess cholesterol (Supplementary Fig. 3)14. To determine if EboV infection is inhibited by endosome dysfunction secondary to the absence of NPC1, we studied a well-characterized NPC1 mutant P692S that is defective in cholesterol uptake and NPC1-dependent membrane trafficking13–15 and found that expression of NPC1 P692S fully supports infection of CHOnull cells (Fig. 2c). Conversely, gain-of-function mutants NPC1 L657F and NPC1 D787N14 did not enhance EboV GP infection. Thus, EboV entry is strictly dependent on NPC1 expression but not NPC1-dependent cholesterol transport activity. Consistent with the conclusion that NPC1 expression is essential for EboV GP-dependent entry, we found that ebolavirus did not grow on CHOnull cells (Fig. 2d). In addition, we tested a single round of infection by MLV particles bearing GPs from the filoviruses EboV Sudan, EboV Côte d'Ivoire, EboV Bundibugyo, EboV Reston and marburgvirus and found that all are strictly NPC1-dependent (Supplementary Fig. 4). Since these viruses are not closely related16, these findings suggest that the requirement for NPC1 as an entry factor is conserved among viruses in the Filoviridae family.
Since NPC1 and cathepsin B are both essential host factors, we analyzed their relationship during infection. In our initial experiment, we compared cathepsin B activity in CHOnull cells and found it was not significantly different from CHOwt cells (Supplementary Fig. 5). To determine if NPC1 is required for virus processing by cathepsin B, we tested whether thermolysin-cleaved particles are dependent on NPC1. As expected, we found that thermolysin-cleaved particles are infectious and resistant to inactivation of cathepsin B when NPC1 is present (Fig. 2e). However, thermolysin cleavage did not bypass the barrier to virus infection of NPC1 deficient cells. Taken together, these findings indicate that cathepsin B and NPC1 mediate distinct steps in infection.
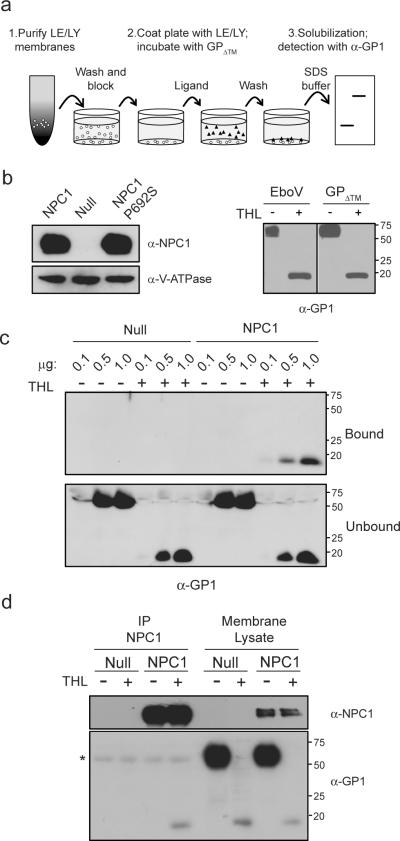
Previous studies suggest that the product of cathepsin B cleavage of the GP1 subunit of EboV GP is a ligand for a host factor6,17–20. To test this hypothesis, we performed a series of experiments measuring binding of EboV GP to LE/LY membranes from CHOnull, CHONPC1 and CHOP692S cells (Fig. 3a,b left panel). The source of EboV GP is a purified recombinant protein that is truncated just before the transmembrane domain (EboV GPΔ™). EboV GPΔ™ is a trimer that is faithfully cleaved by thermolysin (Fig 3b, right panel). We found that binding of EboV GPΔ™ to LE/LY membranes is concentration dependent, saturable, and strictly dependent on both thermolysin cleavage of GP1 and membrane expression of NPC1 or NPC1 P692S (Fig. 3c and Supplementary Fig. 6a,b). To determine if cleaved GP binds to NPC1, we performed a co-immunoprecipitation experiment. LE/LY membranes were incubated with EboV GPΔ™ and then solubilized in detergent. NPC1 was recovered from the lysate by immunoprecipitation and the immune complexes were analyzed for GP1. The findings indicate that cleaved EboV GPΔ™ binds to NPC1 and that uncleaved EboV GPΔ™ does not (Fig. 3d).
Figure 3. Protease-cleaved EboV GP binds to NPC1.
a, Schematic diagram of EboV GP1 binding assay used in panel c.
b, (left) LE/LY membranes from CHONPC1, CHOnull and CHO NPC1 P692S cells were analyzed by immunoblot using antibodies to NPC1 or V-ATPase B1/2. (right) VSV-EboV GP particles and EboV GPΔ™ protein were incubated in the presence or absence of thermolysin (THL) and analyzed by immunoblot for GP1.
c, EboV GPΔ™ or thermolysin-cleaved EboV GPΔ™ (0.1, 0.5, or 1.0 μg) was added to LE/LY membranes purified from CHOnull or CHONPC1 cells. Membrane bound and unbound GP1 were analyzed by immunoblot.
d,. LE/LY membranes from CHOnull or CHOhNPC1 cells were incubated with EboV GPΔ™ or thermolysin-cleaved EboV GPΔ™. Following binding, membranes were dissolved in CHAPSO, NPC1 was precipitated using an NPC1-specific antibody, and the immunoprecipitate and the input membrane lysate were analyzed by immunoblot for NPC1 (top) or GP1 (bottom). * IgG heavy chain.
Since the small molecules 3.0 and 3.47 inhibit infection of thermolysin-treated VSV EboV GP particles (Supplementary Fig. 1b) and inhibit cholesterol uptake from LE/LY into cells (Fig. 2a), both of which require NPC1, this suggests the possibility that these compounds directly target NPC1. To test this hypothesis, we synthesized the 3.47 derivative 3.98. This compound has anti-EboV activity and contains two additional functional moieties: an aryl-azide for photo-affinity labeling of target proteins and an alkyne for click conjugation with biotin21 (Supplementary Fig. 7). Compound 3.98 was incubated with LE/LY membranes, activated by UV light and coupled to biotin. NPC1 was then isolated by immunoprecipitation and analyzed using streptavidin-HRP. The findings show that NPC1 is cross-linked to 3.98 and that cross-linking is inhibited by the presence of 3.47 but not by the closely-related analog 3.18, which has weak anti-viral activity (Fig. 4a, Supplementary Fig. 7). In addition, we observed that overexpression of NPC1 conferred resistance to the anti-viral activity of 3.0 and 3.47 (Supplementary Fig. 8), thus providing additional functional evidence supporting the conclusion based on the results of the cross-linking experiment using 3.98 that NPC1 is a direct target of the anti-viral compounds.
Figure 4. NPC1 is a target of the small molecule inhibitors.
a, LE/LY membranes from CHOnull or CHOhNPC1 cells were incubated at the indicated concentrations of 3.47, 3.18 or DMSO (5%) prior to the addition of the photoactivatable 3.98 (25 μM). After incubation, 3.98 was activated by UV light and then conjugated to biotin. NPC1 was immunoprecipitated and analyzed by immunoblot for conjugation of 3.98 to NPC1 using streptavidin-HRP (top) and recovery of NPC1 (bottom).
b, Thermolysin-cleaved EboV GPΔ™ protein (1 μg) was added to LE/LY membranes from CHOnull or CHONPC1 cells in the presence of DMSO (10%) or the indicated concentrations of 3.47, 3.0, or 3.18 (left panel), and 3.47 or U18666A (U18, right panel). Membrane bound and unbound GP1 were analyzed by immunoblot.
c, Proposed model of EboV entry. Following EboV uptake and trafficking to late endosomes24,25, EboV GP is cleaved by cathepsin protease to remove heavily glycosylated domains (CHO) and expose the putative receptor binding domain (RBD) of GP16,17–19. Binding of cleaved GP1 to NPC1 is necessary for infection and is blocked by the EboV inhibitor 3.47.
The evidence that NPC1 is the target of the 3.0-derived small molecules selected for anti-EboV suggested that these compounds interfere with binding of cleaved GP to NPC1. Consistent with this hypothesis, we found that 3.0 and 3.47 inhibited binding of cleaved EboV GPΔ™ to NPC1 membranes in a concentration-dependent manner (Fig. 4b). Importantly, we observed a direct correlation between the potency of 3.47, 3.0, and 3.18 in inhibiting binding (Fig. 4b, left panel) and in inhibiting EboV infection (Supplementary Fig. 7). We also tested U18666A, a small molecule inhibitor of LE/LY cholesterol transport and membrane trafficking22,23, and found that it does not inhibit binding of cleaved EboV GP to NPC1 membranes (Fig. 4b, right panel). These results support the conclusion that the 3.0-derived compounds inhibit EboV infection by interfering with binding of cleaved GP to NPC1.
Previous studies show that cleavage of GP by endosomal cathepsin proteases removes heavily-glycosylated domains in the GP1 subunit and exposes the N-terminal domain3–7. It has been proposed that binding of this domain to a host factor is essential for infection6,17–20. The most straight forward interpretation of the findings in this report is that NPC1 is this host factor. This conclusion is based on the observations that NPC1 is strictly required for infection, that cleaved GP1 binds to NPC1, and that small molecules that target NPC1 are potent inhibitors of binding and infection.
Analysis of the EboV GP structure reveals that the residues in the N-terminal domain of GP1 that mediate binding to NPC1 are interspersed with the residues that make stabilizing contacts with GP25. This structural feature is consistent with the possibility that binding of cleaved GP1 to NPC1 relieves the GP1-imposed constraints on GP2 and promotes virus fusion to the limiting membrane (Fig. 4c). The role of cathepsin proteases in cleavage of GP1 to expose the NPC1 binding site during EboV infection is analogous to the role of CD4 in inducing a conformational change in gp120 to expose the co-receptor binding site during human immunodeficiency virus infection8. An alternative possibility is that binding of protease-cleaved GP1 to NPC1 is an essential step in infection, but virus membrane fusion is not completed until an additional signal is received, possibly including further cleavage of GP by cathepsin proteases, as has been proposed3,4,9. These studies provide an example of how small molecules identified by screening and medicinal chemistry optimization can be used as molecular probes to analyze virus-host interactions.
Methods Summary
Screening of small molecules was performed at the New England Regional Centers of Excellence for Biodefense and Emerging Infectious Diseases at Harvard Medical School. Infection was assayed using VSV pseudotyped viruses encoding GFP or luciferase. Experiments with native ebolavirus were performed under BSL-4 conditions at the United States Army Medical Research Institute for Infectious Diseases. Cells were infected with EboV Zaire-Mayinga GFP and growth was measured by mean fluorescence. EboV GPΔ™ is a derivative of EboV GP in which the transmembrane domain has been replaced by a GCN4-derived trimerization domain followed by a His6 tag for purification. Late endosomes/lysosomes (LE/LY) were isolated by differential centrifugation and further purified by Percoll density gradient centrifugation. LE/LY were disrupted by incubation with methionine methyl ester and coated onto high binding ELISA plates. Following attachment, unbound LE/LY membranes were removed and plates were blocked. Bound membranes were incubated with the indicated amounts of native or thermolysin-cleaved EboV GPΔ™ protein. Unbound EboV GPΔ™ protein was removed, membranes were washed and bound EboV GPΔ™ protein was recovered in SDS loading buffer and analyzed by immunoblot using GP1 antiserum. Where applicable, membranes were pre-incubated with 3.0, 3.47, 3.18 or vehicle prior to the addition of EboV GPΔ™. To analyze EboV GPΔ™ binding to NPC1, LE/LY membranes were dissolved in 10mM CHAPSO and NPC1 was recovered by immunoprecipitation and the immune complexes were analyzed by immunoblot probed with EboV GP1 antiserum.
Methods
Cell lines
Vero, 293T, HeLa (ATCC) and human fibroblasts26 (Coriell) were maintained in DMEM (Invitrogen) supplemented with 5% FetalPlex, 5% FBS (Gemini) or 10% FBS (HeLa, human fibroblasts). All CHO derived cell lines were grown as previously described14,27. We have designated the CHO-K1 cell line as CHOwt, CHO-M12 as CHOnull, CHO-wt8 as CHONPC1, and the CHO-derived cell lines expressing NPC1 mutants as CHO NPC1 P692S, CHO NPC1 L657F, and CHO NPC1 D787N. CHO/NPC1-1, designated here as CHO hNPC1, expresses high levels of human NPC127.
Antibodies
Rabbit polyclonal anti-serum was raised against a peptide corresponding to residues 83 to 98 of ebolavirus Zaire Mayinga GP1 (TKRWGFRSGVPPKVVC). Antibodies to NPC1 and V-ATPase B1/2 were obtained from Abcam and Santa Cruz, respectively.
Expression plasmids
Mucin domain-deleted EboV Zaire Mayinga GP (EboV GP) and VSV G were previously described3. Plasmids encoding Côte d'Ivoire-Ivory Coast GP, Sudan-Boniface GP, Reston-Penn. GP and Marburg-Musoke GP were obtained from Anthony Sanchez and the mucin domain-deleted (ΔMuc) derivatives were created: ZaireΔMuc GP (Δa.a. 309–489), Côte d'IvoireΔMuc GP (Δa.a. 310–489), SudanΔMuc GP (Δa.a. 309–490), and RestonΔMuc GP (Δa.a. 310–490). Bundibungyo-Uganda viral RNA was Trizol extracted and PCR used to generate a construct that expresses a mucin-deleted GP (Δa.a. 309–489). A plasmid encoding Lassa fever virus GP1 was kindly provided by Gary Nabel. A codon-optimized sequence encoding GP2 was generated and combined with the GP1 sequence in pCAGGS to complete a GP expression vector.
Production and purification of pseudotyped virions
VSV-ΔG pseudotyped viruses were created as described previously3. LacZ-encoding retroviral pseudotypes bearing the designated envelope glycoproteins were prepared as previously described28.
Thermolysin digestion of EboV GP Virus and EboV GPΔ™
Purified EboV GPΔ™ (50μg/mL) or VSV particles pseudotyped with EboV GP were incubated at 37°C for 1 hour with the metalloprotease thermolysin (Sigma, 0.2mg/ml) in NT buffer (10 mM Tris.Cl [pH 7.5], 135 mM NaCl). The reaction was stopped using 500 μM Phosphoramidon (Sigma) at 4°C. Cleaved EboV GPΔ™ was stored in phosphate buffered saline supplemented with 1 mM EDTA, 1 mM PMSF (Sigma) and 1× EDTA-Free Complete Protease Inhibitor Cocktail (Roche).
Infection assays with pseudotyped virus
VSV pseudotyped viruses expressing GFP were added to cells in serial 10-fold dilutions and assayed using fluorescence microscopy. An infectious unit (i.u.) is defined as one GFP-expressing cell within a range where the change in GFP-positive cells is directly proportional to the virus dilution. For VSV expressing the luciferase reporter, pseudotyped virus was added to cells and luciferase activity was assayed 6–20 hours post-infection using the firefly luciferase kit (Promega). Signal was measured in relative luminescence units (RLU) using an EnVison plate reader (Perkin Elmer). In experiments involving inhibitors, stock solutions of 3.0 (20mM) and 3.47 (10mM) in DMSO were diluted to a final concentration of 1% DMSO in media. Inhibitory activity was stable in the media of cultured cells for >72hours as assessed using a single cycle entry assay. Infection of target cells with LacZ-encoding retroviral pseudotypes was performed in the presence of 5 μg/ml polybrene (Sigma). Seventy-two hours post-infection, cells were stained for LacZ activity and titer was determined by counting positive foci and expressed as focus forming units (FFU) per ml of virus.
Ebolavirus infections under BSL-4 conditions
Vero cells or CHO cells were seeded to 96-well plates and exposed to EboV-GFP29. Vero cells were incubated with 3.0 (40 μM), 3.47 (40 μM), E-64-d (150 μM) or 1% DMSO 90 minutes prior to the addition of virus (moi=0.1). Virus was added to CHO cells at moi of 1 as measured on Vero cells. Virus-encoded GFP fluorescence was determined using a SpectraMax M5 plate reader (Molecular Devices) at Ex 485nm, Em 515nm, cutoff 495nm at 22.5, 42, 71 and 97 hours post-infection. An additional inhibitor experiment was performed using 3.0. Vero cells were treated with 3.0 (20μM) or 1% DMSO alone for 4 hours, and then infected with EBOV Zaire-1995 (moi=0.1). After 1 hour, the virus inoculum was removed, cells were washed, and fresh media containing 3.0 or DMSO was added. Cell supernatant was collected at 0, 24, 48, 72, or 91 hours post-infection. RNA was isolated from the supernatant using Virus RNA Extraction kits (Qiagen) and EboV NP RNA was measured using a real-time RT-PCR assay30. Virus titer was calculated using a standard curve obtained using a virus stock of known titer as determined by plaque assay.
Screen for ebolavirus entry inhibitors
Screening of small molecules was performed at the New England Regional Centers of Excellence for Biodefense and Emerging Infectious Diseases at Harvard Medical School. Vero cells were seeded in 384-well plates at a density of 5×104 cells per well using a Matrix WellMate (Thermo Scientific). The ChemBridge3, ChemDiv4, ChemDiv5 and Enamine2 compound libraries were transferred by robotics to the assay plates using stainless steel pin arrays. The compounds were screened at a constant dilution to achieve a final concentration between 10μM and 60μM. After incubation for 2 hours at 37 °C, viruses were dispensed into each well (moi =1) and incubated for an additional 6 hours to allow virus gene expression. Cells were lysed by addition of Steady-Glo (Promega) and after 10 minutes at room temperature luminescence was measured using an EnVision plate reader. Each compound was tested in duplicate. Candidate compounds that inhibited EboV GP infection by more than 80% were analyzed for potency, selectivity and absence of cytotoxicity (using Cyto-Tox assay, Promega) and 3.0 (2-((3r,5r,7r)-adamantan-1-yl)-N-(2-(4-benzylpiperazin-1-yl)-2-oxoethyl)acetamide) was identified. The antiviral activity of the inhibitors was verified on human cells (HeLa, A549, 293T), mouse embryonic fibroblasts, and Chinese hamster ovary cells.
Synthesis of 3.0 derivatives
Compound 3.47 (methyl 4-((2-((4-(2-(2-((3r,5r,7r)-adamantan-1-yl)acetamido)acetyl)piperazin-1-yl)methyl)phenoxy)methyl)benzoate) was prepared via a multi-step synthesis starting from N-Cbz-piperazine. Thus, coupling of N-Cbz-piperazine with N-Bocglycine followed by removal of the Boc group under acidic conditions yielded 4-Cbz-piperazine glycinamide. After acylation of the terminal amine with adamantan-1-acetyl chloride, the Cbz group was removed by hydrogenolysis to give (1-(adamantan-1-yl)acetamido)acetyl)piperazine. The piperazine was then benzylated via reductive amination with 2-(4-methoxycarbonyl)benzyloxybenzaldehyde using sodium triacetoxyborohydride to provide 3.47. Compound 3.18 was synthesized in a similar fashion. Compound 3.98 was prepared via a multi-step synthesis as follows. First, 2-hydroxy-5-nitrobenzaldehyde was alkylated by 4-ethynylbenzyl bromide in the presence of potassium carbonate in DMF. Resulting benzyloxy aldehyde underwent reductive amination with 2-((3r,5r,7r)-adamantan-1-yl)-N-(2-oxo-2-(piperazin-1-yl)ethyl)acetamide using sodium triacetoxyborohydride. The nitro group was then reduced to aniline (SnCl2), diazotized (NaNO2), and the diazonium finally converted to azide to yield 3.98. See Supplementary Information for detailed experimental procedures and characterization data.
Protease inhibitors and protease activity assays
The measurement of cathepsin B activity and the use of the inhibitor CA074 (Sigma) have been previously described3.
Detection of intracellular cholesterol
Cells were stained with filipin (50 μg/ml, Cayman Chemical) as previously described14. Images of stained cells were obtained using epifluorescence microscopy (Nikon Eclipse TE2000U). The images in the supplementary figures were processed using ImageJ software.
Production and purification of EboV GPΔ™ soluble protein
EboV GPΔ™ is a derivative of the mucin-deleted EboV Zaire-Mayinga GP in which the transmembrane domain and C-terminus (a.a. 657–676) has been replaced by a GCN4-derived trimerization domain (MKQIEDKIEEILSKIYHIENEIARIKKLIGEV) and a His6 tag. The expression plasmid encoding EboV GPΔ™ was transfected into 293T cells using lipofectamine2000. Eighteen to twenty-four hours later the culture medium was replaced with 293SFMII (Invitrogen) supplemented with 1× non-essential amino acids and 2 mM CaCl2 and harvested daily for four days. Media containing soluble EboV GPΔ™ was filtered and PMSF (1 mM)/ 1× EDTA-Free Complete Protease Inhibitor Cocktail was added. EboV GPΔ™ was purified by affinity chromatography using Ni-NTA agarose beads (Qiagen), dialyzed against PBS using a 3kDa dialysis cartridge (Pierce) and stored at −80°C. Purity and integrity of EboV GPΔ™ were analyzed by SDS-PAGE.
Membrane Binding Assay
Indicated cells were washed with PBS ×2, scraped in homogenization (HM) buffer (0.25 M sucrose, 1 mM EDTA, 10 mM Hepes pH7.0), and disrupted with a Dounce homogenizer. Nuclei and debris were pelleted by centrifugation at 1000 × g for 10 min. The post-nuclear supernatant was centrifuged at 15000 × g for 30 min at 4°C and the pellet, containing the LE/LY, was resuspended in a total volume of 0.9ml composed of 20% Percoll (Sigma) and 0.4% BSA (Sigma) in HM and centrifuged at 36000 × g for 30 min at 4°C. Fractions (0.150 ml) were collected from the bottom to the top of the tube and those containing the highest β-N-acetylglucosamidase activity, as assessed by release of 4-methylumbelliferone from 4-methylumbellifferyl-N-acetyl-β-D-glucosaminide (Sigma), were pooled and incubated in 20mM methionine methyl-ester (Sigma) for 1 hour at room temperature. Following LE/LY disruption, 1× EDTA-Free Complete Protease Inhibitor Cocktail and 1 mM PMSF was added. The amount of purified LE/LY membranes used for the binding assay was normalized using the activity of the marker β-N-acetylglucosamidase and validated by immunoblot using V-ATPase B1/2 antibody (Supplemental Fig. 5).
Disrupted LE/LY membranes were coated on high-binding ELISA plates (Corning) overnight at 4°C. Unbound membranes were removed and wells containing bound membranes were blocked for 2 hours at room temperature with binding buffer (PBS, 5% FBS, 1 mM PMSF, 1mM EDTA, 1× Complete Protease Inhibitor Cocktail). The indicated amount of purified EboV GPΔ™, pretreated or not with thermolysin, in binding buffer was added to each well and incubated for 1 hour at room temperature. Unbound proteins were removed and wells were washed 3 times with PBS. Membrane bound EboV GPΔ™ was solubilized in SDS-loading buffer. Bound and unbound EboV GPΔ™ were detected by immunoblot using the EboV GP1 anti-serum. For binding assays in the presence of inhibitors, the immobilized membranes were pre-incubated at room temperature with the inhibitor or vehicle (10% DMSO) in binding buffer. After 30 minutes, thermolysin cleaved EboV GPΔ™ was added in the continuous presence of compound and bound and unbound GP was measured as described above.
Co-Immunoprecipitation
CHOnull and CHOhNPC1 cells were homogenized as described above. The 15000 × g membrane pellet was resuspended in HM buffer and protein content was measured using the BCA assay (Pierce). The LE/LY membranes contained in the 15000 × g resuspended pellet were disrupted by incubation with 20 mM methionine methyl-ester for 1 hour at RT. Membranes of equal protein content were incubated with indicated amounts of EboV GPΔ™, pre-treated or not with thermolysin, for 1 hour at RT in the presence of Complete Protease Inhibitor Cocktail (Roche) and incubated for an additional hour on ice before the addition of membrane lysis buffer (12.5mM CHAPSO, 150mM NaCl, 1mM EDTA, 10mM Tris/HCl pH7.4) for a final concentration of 10mM CHAPSO. Proteins were solubilized on ice for 20 min and debris was removed by centrifugation at 12000 × g for 10 min at 4°C. The soluble membrane lysates were incubated with anti-NPC1 antibody for 1 hour at 4°C and then incubated with Protein A-agarose beads (Sigma) for an additional 4 hours at 4°C. Beads were then washed 3 times with 8mM CHAPSO, 150 mM NaCl, 1mM EDTA, 10 mM Tris/HCl pH7.4 and immunoprecipitated product was eluted by incubation in 0.1M glycine pH 3.5 for 5 min at RT. The eluted complex was then neutralized and analyzed by immunoblot using the indicated antibody.
Photoactivation and click chemistry
Photoactivation and click chemistry were performed as described previously with some modifications21. Briefly, the 15000× g pellets from homogenized CHOhNPC1 or CHONull cells were resuspended in PBS and incubated with the indicated concentrations of 3.47, 3.18 or DMSO for 10 minutes at room temperature. Membranes were then incubated with 25μM of 3.98 for an additional 10 minutes and exposed to ultraviolet light (365 nm) for 1 min on ice. Proteins were solubilized in lysis buffer (1% Triton X-100, 0.1% NP-40, 20mM HEPES pH7.4) containing protease inhibitors and 150 μM of biotin-azide (Invitrogen) was added, followed by 5 mM L-ascorbic acid. The cycloaddition reaction (click chemistry) was initiated by the addition of 1 mM CuSO4 and samples were incubated for 3 hours at room temperature. NPC1 was immunoprecipitated and the product was resolved by SDS-PAGE, transferred to PVDF membrane, and analyzed for conjugation of 3.98 to NPC1 using streptavidin-HRP (Sigma).
Supplementary Material
Acknowledgements
We thank Bryden Considine, Anna Nilsson, and Sean Wilkes for assistance, Su Chiang for critical reading of the manuscript, Gerald Beltz, Nathanael Gray, Sergio Grinstein, Yiannis Iannou, Rodney Infante, Johannes Kornhuber, Francis Sharom and Sean Whelan for discussion. This work was supported by grants from U54 AI057159, R01 CA104266 to JC, PIDS-Sanofi-Pasteur Fellowship, K12-HD052896 and 5K08AI079381 to JM, 5-T32- HL007623 to AB, and fellowship from Fonds de la Recherche en Santé du Québec to MC. CF was supported by the Postgraduate Research Participation Program at the U.S. Army Medical Research and Material Command administered by the Oak Ridge Institute for Science and Education through an interagency agreement between the U.S. Department of Energy and USAMRMC.
Footnotes
Additional experimental procedures are available online at www.nature.com/nature.
Supplementary Information is linked to the online version of the paper at www.nature.com/nature
Author Information: Reprints and permission information is available at www.nature.com/reprints.
Competing financial interests: None.
References
- 1.Zampieri CA, Sullivan NJ, Nabel GJ. Immunopathology of highly virulent pathogens: insights from ebola virus. Nat. Immunol. 2007;8:1–6. doi: 10.1038/ni1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Geisbert TW, Jahrling PB. Exotic emerging viral diseases: progress and challenges. Nat. Med. 2004;10:S110–121. doi: 10.1038/nm1142. [DOI] [PubMed] [Google Scholar]
- 3.Chandran K, Sullivan NJ, Felbor U, Whelan SP, Cunningham JM. Endosomal proteolysis of the Ebola virus glycoprotein is necessary for infection. Science. 2005;308:1643–1645. doi: 10.1126/science.1110656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schornberg K, et al. Role of endosomal cathepsins in entry mediated by the Ebola virus glycoprotein. J. Virol. 2006;80:4174–4178. doi: 10.1128/JVI.80.8.4174-4178.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee JE, et al. Structure of the Ebola virus glycoprotein bound to an antibody from a human survivor. Nature. 2008;454:177–182. doi: 10.1038/nature07082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dube D, et al. The primed ebolavirus glycoprotein (19-kilodalton GP1,2): Sequence and residues critical for host cell binding. J. Virol. 2009;83:2883–2891. doi: 10.1128/JVI.01956-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hood CL, et al. Biochemical and structural characterization of cathepsin L-processed Ebola virus glycoprotein: Implications for viral entry and immunogenicity. J. Virol. 2010;84:2972–2982. doi: 10.1128/JVI.02151-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harrison SC. Viral membrane fusion. Nat. Struct. Mol. Biol. 2008;15:690–698. doi: 10.1038/nsmb.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wong A, Sandesara R, Mulherkar N, Whelan S, Chandran K. A forward genetic strategy reveals destabilizing mutations in the ebolavirus glycoprotein that alter its protease dependence during cell entry. J. Virol. 2010;84:163–175. doi: 10.1128/JVI.01832-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kolter T, Sandhoff K. Lysosomal degradation of membrane lipids. FEBS Lett. 2010;584:1700–1712. doi: 10.1016/j.febslet.2009.10.021. [DOI] [PubMed] [Google Scholar]
- 11.Du X, et al. A role for oxysterol-binding protein-related protein 5 in endosomal cholesterol trafficking. J. Cell Biol. 2011;192:121–35. doi: 10.1083/jcb.201004142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chevallier J, et al. Lysobisphosphatidic acid controls endosomal cholesterol levels. J. Biol. Chem. 2008;283:27871–27880. doi: 10.1074/jbc.M801463200. [DOI] [PubMed] [Google Scholar]
- 13.Ko DC, Gordon MD, Jin JY, Scott MP. Dynamic movements of organelles containing Niemann-Pick C1 protein: NPC1 involvement in late endocytic events. Mol. Biol. Cell. 2001;12:601–614. doi: 10.1091/mbc.12.3.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Millard EE, et al. The sterol-sensing domain of the Niemann-Pick C1 (NPC1) protein regulates trafficking of low density lipoprotein cholesterol. J. Biol. Chem. 2005;280:28581–28590. doi: 10.1074/jbc.M414024200. [DOI] [PubMed] [Google Scholar]
- 15.Ohgami N, et al. Binding between the Niemann-Pick C1 protein and a photoactivatable cholesterol analog requires a functional sterol-sensing domain. Proc. Natl. Acad. Sci. USA. 2004;101:12473–12478. doi: 10.1073/pnas.0405255101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Towner JS, et al. Newly discovered ebola virus associated with hemorrhagic fever outbreak in Uganda. PLoS Pathog. 2008;4:e1000212. doi: 10.1371/journal.ppat.1000212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuhn JH, et al. Conserved receptor-binding domains of Lake Victoria marburgvirus and Zaire ebolavirus bind a common receptor. J. Biol. Chem. 2006;281:15951–15958. doi: 10.1074/jbc.M601796200. [DOI] [PubMed] [Google Scholar]
- 18.Kaletsky RL, Simmons G, Bates P. Proteolysis of the Ebola virus glycoproteins enhances virus binding and infectivity. J. Virol. 2007;81:13378–13384. doi: 10.1128/JVI.01170-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brindley MA, et al. Ebola virus glycoprotein 1: Identification of residues important for binding and postbinding events. J. Virol. 2007;81:7702–7709. doi: 10.1128/JVI.02433-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dube D, et al. Cell adhesion-dependent membrane trafficking of a binding partner for the ebolavirus glycoprotein is a determinant of viral entry. Proc. Natl. Acad. Sci. USA. 2010;107:16637–16642. doi: 10.1073/pnas.1008509107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ban HS, et al. Identification of HSP60 as a primary target of ocarboranylphenylphenoxyacetanilide, an HIF-1α inhibitor. J. Am. Chem. Soc. 2010;132:11870–11871. doi: 10.1021/ja104739t. [DOI] [PubMed] [Google Scholar]
- 22.Sobo K, et al. Late endosomal cholesterol accumulation leads to impaired intraendosomal trafficking. PLoS ONE. 2007;2:e851. doi: 10.1371/journal.pone.0000851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huynh KK, Gershenzon E, Grinstein S. Cholesterol accumulation by macrophages impairs phagosome maturation. J. Biol. Chem. 2008;283:35745–35755. doi: 10.1074/jbc.M806232200. [DOI] [PubMed] [Google Scholar]
- 24.Saeed MF, Kolokoltsov AA, Albrecht T, Davey RA. Cellular entry of ebola virus involves uptake by a macropinocytosis-like mechanism and subsequent trafficking through early and late endosomes. PLoS Pathogen. 2010;6:e1001110. doi: 10.1371/journal.ppat.1001110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nanbo A, et al. Ebolavirus is internalized into host cells via macropinocytosis in a viral glycoprotein-dependent manner. PLoS Pathogen. 2010;6:e1001121. doi: 10.1371/journal.ppat.1001121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gelsthorpe ME, et al. Niemann-Pick type C1 I1061T mutant encodes a functional protein that is selected for endoplasmic reticulum-associated degradation due to protein misfolding. J Biol Chem. 2008;283:8229–8236. doi: 10.1074/jbc.M708735200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Millard EE, Srivastava K, Traub LM, Schaffer JE, Ory DS. Niemann-pick type C1 (NPC1) overexpression alters cellular cholesterol homeostasis. J Biol Chem. 2000;275:38445–38451. doi: 10.1074/jbc.M003180200. [DOI] [PubMed] [Google Scholar]
- 28.Soneoka Y, et al. A transient three-plasmid expression system for the production of high titer retroviral vectors. Nucleic Acids Res. 1995;23:628–633. doi: 10.1093/nar/23.4.628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Towner JS, et al. Generation of eGFP expressing recombinant Zaire ebolavirus for analysis of early pathogenesis events and high-throughput antiviral drug screening. Virology. 2005;332:20–27. doi: 10.1016/j.virol.2004.10.048. [DOI] [PubMed] [Google Scholar]
- 30.Weidmann M, Mühlberger E, Hufert FT. Rapid detection protocol for filoviruses. J Clin Virol. 2004;30:94–99. doi: 10.1016/j.jcv.2003.09.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.