Abstract
African Americans are a genetically diverse population with a high burden of many, common heritable diseases. However, our understanding of genetic variation in African Americans is substandard because of a lack of published population-based genetic studies. We report the distribution of copy-number variation (CNV) in African Americans collected as part of the Hypertension Genetic Epidemiology Network (HyperGEN) using the Affymetrix 6.0 array and the CNV calling algorithms Birdsuite and PennCNV. We present population estimates of CNV from 446 unrelated African-American subjects randomly selected from the 451 families collected within HyperGEN. Although the majority of CNVs discovered were individually rare, we found the frequency of CNVs to be collectively high. We identified a total of 11 070 CNVs greater than 10 kb passing quality control criteria that were called by both algorithms – leading to an average of 24.8 CNVs per person covering 2214 kb (median). We identified 1541 unique copy-number variable regions, 309 of which did not overlap with the Database of Genomic Variants. These results provide further insight into the distribution of CNV in African Americans.
Keywords: DNA copy-number variation, African American, calling algorithm, Birdsuite, PennCNV, HyperGEN
Introduction
Genome-wide association studies based on single-nucleotide polymorphisms have been modestly successful in identifying genetic correlates of phenotypic variation for many complex disorders, including psychiatric, autoimmune, and cardiovascular traits.1 However, much of the genetic variance attributed to these diseases remains unexplained. Copy-number variation (CNV) has been cited as a potential source of genetic variation, contributing to the development of complex diseases, potentially through rare variation driven by purifying selection acting on exonic and intronic deletions.2, 3 Redon et al4 constructed a first-generation map of CNV in the human genome from 270 individuals in the HapMap collection.5 They found over 1400 copy-number variable regions (CNVRs) covering roughly 12% of the genome. These regions overlapped with over half of the known reference sequence genes. Other early studies examined CNV in different racial/ethnic populations – including unrelated healthy individuals from Northern Germany,6 Caucasian males from Northern France,7 and Koreans8 among others – many of which are included in the Database of Genomic Variants (DGVs).9
Yet there remains a lack of published genome-wide data on CNV in African Americans – a racial/ethnic group with a particularly high burden of many common, heritable diseases that occur within numerous biological pathways, including cardiovascular diseases such as hypertension and stroke,10 metabolic disorders such as type 2 diabetes11 and kidney disease,12 and autoimmune and neurological disorders such systemic lupus erythematosus13 and Alzheimer's disease.14 Only one such study has investigated CNVs exclusively in African Americans.15 This study identified 1362 copy-number variants in 385 African Americans using the Affymetrix Genome-Wide SNP Array 5.0 (Affymetrix, Santa Clara, CA, USA). However, the continued development of CNV genotyping platforms and statistical calling algorithms dictates the routine reevaluation of the distribution of CNV on a genome-wide level.
In this study we examined CNVs in an unrelated sample of African-American individuals derived from 451 families in the Hypertension Genetic Epidemiology Network (HyperGEN) who were genotyped on the Affymetrix Genome-Wide SNP Array 6.0. This platform was specifically designed in consideration of CNV and provides a more accurate assessment of CNV compared with its predecessor in terms of copy-number probe density, measurement reliability, and detection.16 CNVs were inferred using the normalization procedures and calling applications in Birdsuite16 and PennCNV.17 We present the genome-wide distribution of CNV. In particular, we have focused on the size and location of CNV, and distinguish between known and novel CNVRs. Our work provides further insight into specific structural variation in African Americans.
Methods
Data set
Study participants were obtained from the HyperGEN Study. HyperGEN is one of four Family Blood Pressure Program networks supported by the National Heart, Lung, and Blood Institute to identify genetic contributors to hypertension.18 In the first phase of the HyperGEN Study, hypertensive sibships eligible for recruitment consisted of probands, with onset of hypertension by age 60 years and one or more hypertensive siblings who were willing to participate in the study. In the second phase of the study, the offspring of the hypertensive siblings were recruited. Hypertension was defined as having an average systolic blood pressure ≥140 mm Hg or average diastolic blood pressure ≥90 mm Hg during at least two evaluations or receiving medical treatment for hypertension.
The HyperGEN Study includes 1549 African-American subjects recruited from centers located in Birmingham, AL, USA, and Forsyth County, NC, USA. Cardiovascular-related phenotypes were measured on the subjects, such as laboratory measurements, blood pressure, body mass index, left ventricular wall thickness, and pulse pressure/stroke volume ratio among others. These phenotypic characteristics have been extensively studied elsewhere. A number of positive genetic associations with these phenotypes have been reported including the MYH9 gene and albuminuria,19 the KCNB1 gene and left ventricular mass,20 and the CRCP gene and aortic root diameter.21
Genotyping methods
Genetic data were obtained from 1224 HyperGEN African-American subjects from 451 families via the Affymetrix Genome-Wide Human SNP Array 6.0. DNA samples were processed in 34 batches, in which one batch consisted of all the samples that were processed on a particular day (average batch size=36). The Affymetrix genotyping protocol was followed.
CNV analysis
CNV calling algorithms can often produce discrepant results on the same data set. These inconsistencies can occur due to the process by which samples are normalized and compared with a reference genome, the type of calling algorithm used, and deviation in the parameters between similar types of algorithms.22 Recent CNV studies have supported a stringent discovery criterion of only reporting copy-number segments that are similarly identified by at least two different algorithms.23 This standard increases the confidence of identified CNVs. Therefore, copy-number calls were made using Birdsuite16 and PennCNV17 software. In general, confidence thresholds implemented in the algorithms were set as close to the developers' default values or references as possible. Genetic samples obtained from small batches (three batches with less than 10 samples each), or that did not meet quality control criteria in either algorithm, were removed from further analysis. Among the remaining samples, one participant from each family was randomly selected to be included in downstream analysis (N=446). The findings of this paper include only copy-number segments >10 kb in autosomes. Results using differing probe counts as an alternative to segment length were explored, but found them to be inferior in terms of agreement between the calling algorithms (Supplementary Table 1).
Birdsuite
Birdsuite version 1.5.5 was used for copy-number analysis.16 Samples were processed by batch to eliminate batch effects. This allows for better clustering of the data and improves sensitivity and specificity of the algorithm compared with combining data across batches. Samples with copy-number sample variances greater than two were removed from downstream analysis. Copy-number events were measured using the Hidden Markov Model (HMM)-based Birdseye application, using segments with LOD values >5.0 to indicate a positive call. Output files were processed with version 1.4 of the Birdsuite to PLINK Pipeline24 and summarized using the CNV applications in PLINK version 1.05 where appropriate.25
PennCNV
CNVs were also called using the PennCNV algorithm,17 which takes into consideration the total signal intensity and allelic intensity ratio at each SNP marker, the distance between neighboring SNPs, and the allele frequency of each SNP through a HMM. All samples were processed simultaneously. The PennCNV-Affymetrix protocol (http://www.openbioinformatics. org/penncnv/penncnv_tutorial_affy_gw6.html) was first applied to transform the intensities from the raw CEL files into log R ratios (LRRs) and B allele frequencies (BAFs) on all samples. We used the developer's default settings for the HMM, and also applied the model adjustment for genomic waves.26 We removed noisy samples from downstream analyses based on previously used criteria for quality control:27 standard deviation for autosomal LRR>0.28, a median BAF of >0.55 or <0.45 or a BAF drift of >0.002.
Verification and comparison procedures
Both the Birdseye application in Birdsuite and PennCNV are five-state HMMs that produces results at a given probe by estimating the integer copy-number state as 0 copies, 1 copy, 2, 3, or 4 copies. This allows for greater accuracy in predicting SNP allele counts in CNVRs as opposed to more traditional CNV calling algorithms, which simply use ‘loss', ‘normal', or ‘gain'. However, in terms of comparing CNV calls between Birdsuite and PennCNV, we combined 0 and 1 copy calls and refer them as deleted segments, 3 and 4 copy calls as duplicated segments, and 2 copies as normal. This allowed the comparison between algorithms to not be overly conservative. For example, if Birdsuite assigned a copy number of 3 to a given locus and PennCNV assigned a copy number of 4 to that locus, then we concluded that both algorithms assigned ‘duplication' to this region. For CNVs in which both algorithms made similar calls and had at least one base pair overlap, we assumed they were calling the same segment.
A similar concern is the overlap of CNV calls across individuals, as there is often ambiguity in terms of the exact break points of a CNV. We defined CNVRs as the region starting at the first base pair of all overlapping deleted or duplicated segments across samples (at least one base pair) and ending at the last base pair of these segments. After localizing these CNVRs, we compared these with known copy-number variants in the DGVs.9 We considered a CNVR to be novel if it did not overlap with any of the CNVs included in this database at the time of preparation of this manuscript.
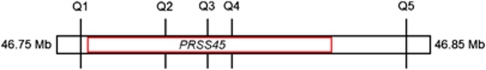
We validated one CNVR called by both algorithms using quantitative PCR (qPCR). This CNVR was a common duplicated region identified on chromosome 3: 46 738 146–46 855 144. This region overlaps portions of the PRSS50 and PRSS42 genes, and completely encompasses the PRSS45 gene. We selected premade and commercially available qPCR probes as provided by Applied Biosystems Inc. (Foster City, CA, USA) in this region (Figure 1). These probes have been tested by the vendor and performed according to their specifications for CNV testing. Samples were diluted to 5 ng/μl, and 2 μl of each sample was used for each reaction. A real-time PCR reaction was set up using Genotyping Master Mix (ABI Cat no.: 4371355), RNase P, the probe, and water. Each sample was set up in quadruplicate. The standard real-time PCR protocol on the ABI 7900 was used. A set of cases (n=50, defined as participants having more than two copies in this region identified by Birdsuite and PennCNV) and eight controls (two copies) were analyzed. The controls were selected from HyperGEN Cohort showing only two copies of the alleles for the probes covering PRSS45 gene in the initial array analyses. For each probe in this region, we verified that controls had two copies. Cases and controls were randomly placed on each plate. Sample analysis on the instrument was performed using manual baseline set at 0.2. Data were exported and analyzed with Applied Biosystem's copy-number macro to determine the copy-number state.
Figure 1.
Relative location of probes used for CNV validation via qPCR. Probes are labeled Q1–Q5 and represent locations in the genome spanning the PRSS45 gene, 46 755 803–46 848 989 bp (hg18) on chromosome 3. The red box indicates the gene boundaries and the black box represents a 100-kb genomic region containing these probes. The colour reproduction of this figure is available at the European Journal of Human Genetics online.
HyperGEN includes genetic information and a variety of phenotypic data on hypertensive siblings and their offspring. The long-term goals of this study are to determine which genes may be responsible for a multitude of cardiovascular-related traits. Knowing the extent to which CNV is present in known genes may influence the way future association testing in HyperGEN and other studies are designed when considering both SNPs and CNVs. To accomplish this, we constructed a record of known genes from the UCSC and Entrez gene databases based upon the Human Genome 18 reference map and compared our CNV calls with this list.28, 29
Results
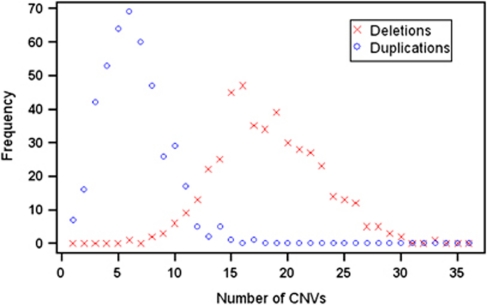
We identified 1541 unique CNVRs identified by both Birdsuite and PennCNV, of which 309 were novel. There were differences in the total number of deleted and duplicated segments called by each algorithm. We found a total of 14 522 and 10 529 deleted segments and 6014 and 5631 duplicated segments using Birdsuite and PennCNV, respectively. These values equate to 32.5 and 24.2 deleted segments, and 13.5 and 12.6 duplicated segments per person found in Birdsuite and PennCNV, respectively. We identified 11 070 CNVs that were called by both algorithms (Figure 2), including 8385 deletions and 2685 duplications – an agreement of 79.6% and 47.7% between algorithms, respectively. These segments covered a total genomic area of 2214 kb per person (median). Perhaps not surprisingly, duplicated segments tended on average to be larger than deleted segments. Among the CNVRs identified by both algorithms, 655 were present in more than one individual (see Supplementary Table 2 for complete CNVR results and a comparison with regions recognized in the DGVs9 and Conrad et al3).
Figure 2.
Number of CNVs per person identified by both Birdsuite and PennCNV. The x axis represents the number of CNVs called by both algorithms in a single study participant, and the y axis indicates the number of participants fitting each category.
One of our conditions for CNV detection was the inclusion of only copy-number segments that were larger than 10 kb. The smallest CNV we discovered that was identified by both algorithms was a deleted segment, 10 007 bp in length and located on chromosome 2. Meanwhile, the largest CNV identified by both algorithms was a deleted segment, 21.6 Mb in length and located on chromosome 1. We discovered more small CNVs than larger ones (Table 1). The median length deletions called by both algorithms was 36.6 kb and the median length of duplication was 94.8 kb. The median length of CNV regardless of deletion or duplication identified by both algorithms was 42.3 kb.
Table 1. Distribution of CNV sizes identified by both Birdsuite and PennCNV.
| Deletion | Duplication | |
|---|---|---|
| 10–20 kb | 2282 | 252 |
| 20–50 kb | 3140 | 663 |
| 50–100 kb | 1294 | 459 |
| 100–200 kb | 1009 | 594 |
| 200–500 kb | 475 | 459 |
| 500–1 Mb | 103 | 124 |
| 1–2 Mb | 81 | 117 |
| 2 Mb+ | 1 | 17 |
| Total | 8385 | 2685 |
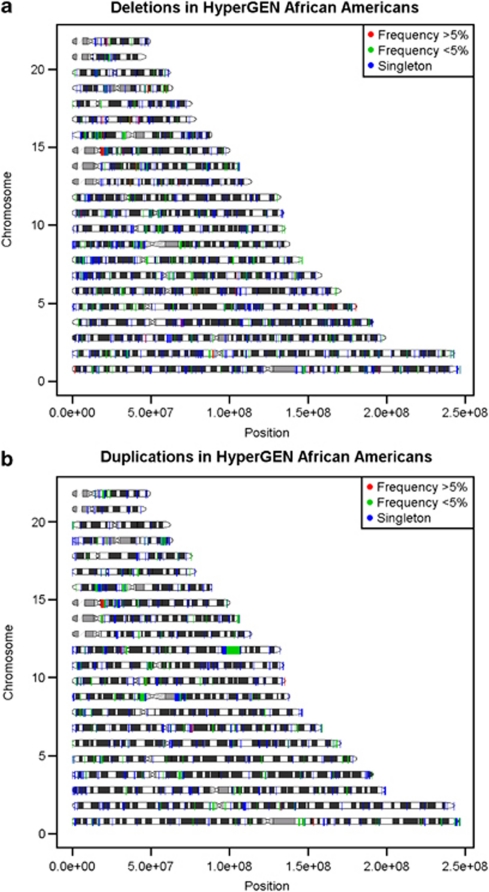
The majority of CNVRs that were called by both Birdsuite and PennCNV were rare (77.7%), occurring in <1% of the study population (four or less individuals). Many CNVRs were singleton (57.6%), only occurring in one individual (Table 2). The most common CNVR was a deletion present in 214 individuals and located on chromosome 1: 194 994 473–195 176 268 bp. This CNVR falls within the CFHR3 region that has previously been shown to be associated with age-related macular degeneration.30 Common CNVs were present in all the chromosomes, although the amount of regions with common CNVs did not appear to be uniform (Figure 3). The majority of novel CNVRs were only present in one person (87.7%), considerably higher than the frequency of singleton CNVRs intersecting known regions (49.9%). The most common, novel CNVR was a deletion on chromosome 8: 53 035 013–53 082 676 bp that overlaps the LOC286071 gene. Another equally common, novel CNVR was a duplication on chromosome 16: 5 456 063–5 468 535 bp. However, CNVs in both regions were only observed in five individuals. All novel CNVRs we detected in three or more individuals are listed in Table 3.
Table 2. Occurrences of CNVRs.
| Novel CNVRs | Known CNVRs | Total CNVRs | ||||
|---|---|---|---|---|---|---|
| Occurrence | Deletion | Duplication | Deletion | Duplication | Deletion | Duplication |
| 1 | 176 | 96 | 359 | 255 | 535 | 351 |
| 2–5 | 22 | 15 | 181 | 136 | 203 | 151 |
| 6–20 | 0 | 0 | 122 | 51 | 122 | 51 |
| 21+ | 0 | 0 | 97 | 31 | 97 | 31 |
| Total | 198 | 111 | 759 | 473 | 957 | 584 |
Abbreviation: DGV, Database of Genomic Variant.
Known CNVRs represent calls made by Birdsuite and PennCNV that overlap regions identified in the DGV. Novel CNVRs represent calls that have not been previously reported in the DGV.
Figure 3.
(a) Distribution of deleted segments throughout the genome. The x axis represents that genomic location in base pair (hg18) of deletions identified by Birdsuite and PennCNV. The y axis represents the chromosomes. (b) Distribution of duplicated segments throughout the genome. The x axis represents that genomic location in base pair (hg18) of duplications identified by Birdsuite and PennCNV. The y axis represents the chromosomes.
Table 3. Novel CNVRs.
| Chr | Start | Stop | Type | Genes |
|---|---|---|---|---|
| 2 | 50 343 433 | 50 368 605 | Deletion | NRXN1 |
| 5 | 32 948 750 | 32 964 072 | Deletion | AK022112 |
| 5 | 101 559 228 | 101 587 316 | Deletion | SLCO4C1 |
| 6 | 102 674 492 | 102 704 937 | Duplication | None |
| 8 | 53 035 013 | 53 082 676 | Deletion | LOC286071 |
| 16 | 5 456 063 | 5 468 535 | Duplication | None |
| 18 | 75 940 080 | 75 970 010 | Duplication | C18orf22 |
Regions included are those that were present in three or more individuals and not currently in the Database of Genomic Variants.
The duplication we validated by qPCR in the PRSS45 region on chromosome 3 was confirmed in all 50 cases. For most subjects, we were only able to confirm the location of one break point, as the amplification appeared to stretch beyond the Q5 probe (Figure 1). The break point that we were able to confirm occurred between the Q2 and Q3 probes in 38 of the 50 cases. Among the other 12 subjects, 10 had the break point occurring between Q3 and Q4, one between Q1 and Q2, and the remaining one missing. In 45 of the 50 cases, the amplification was witnessed in three copies. In four of the other five subjects, the amplification began as a three-copy duplication and increased to four copies at the Q5 probe. The remaining subject had a four-copy duplication, seen only at the Q4 probe.
Many of the CNVs called by PennCNV and Birdsuite overlapped known genes referenced in the Entrez and UCSC gene databases. Ignoring the type of CNV, there did not appear to be a considerable difference between the proportion of calls that covered known genes from the Birdsuite results (69.5%) and those that were also called by PennCNV (66.6%). However, duplicated segments generally appeared to overlap known genes more often than deleted segments – an observation that was more pronounced in copy-number segments that were also found by PennCNV. Among calls made by Birdsuite, 67.1% of deletions and 75.3% of duplications covered known genes. Meanwhile, among those calls that were also found by PennCNV, 61.8% of deletions and 81.7% of duplications demonstrated these findings.
Discussion
CNV presents a significant source of genetic variation. However, the genomic distribution of CNV in African Americans has only briefly been explored. The study by McElroy et al15 is the only other study to have investigated CNV architecture within this population. There are several similarities between their study and ours, as well as notable differences. Our results suggest that their estimates of CNV in African Americans were far too low. This is not particularly surprising, given the greater density of probes directed at CNVs in the Affymetrix 6.0 used in our study. Using the Affymetrix 5.0 array, McElroy et al found 3.5 copy-number events per person. We discovered more than seven times as many events per person.
Our results should be interpreted within the context of a number of limitations. Given the sample ascertainment scheme based on hypertensive status, it is possible that our sample may overestimate the population distribution of CNVs assuming a role of CNV in the development of hypertension. Also, the HyperGEN Cohort consists of Caucasian participants in addition to the African-American participants used in the present study. Unfortunately, Caucasian participants were genotyped before the African-American participants and before the Affymetrix 6.0 array became available. As a result, we were unable to compare CNV differences across racial group without confounding the results by array differences. In addition, although we chose to report CNVs on only unrelated subjects, the use of family data can provide interesting results particularly in identifying de novo CNVs. However, because of the ascertainment scheme of HyperGEN, data were generally only gathered from one parent. Thus we were unable to distinguish between inherited and de novo events. Finally, although the assessment of CNV derived from the Affymetrix 6.0 is a vast improvement over its predecessor, the results presented here are by no means a complete CNV map. Particularly, small CNVs tend to have very high false negative rates and, as was seen in the qPCR experiment we performed, CNV boundaries can vary amongst individuals.
Nevertheless, we found that on a genomic level CNV appeared to be quite common based upon the sheer number of events we witnessed. Among the 1541 unique CNVRs that we identified with both calling algorithms, 655 were present in more than one individual. These regions may include polymorphic CNVs, which can be more directly measured and lend themselves to traditional disease–gene association testing. Furthermore, about a fifth of all the CNVRs that we discovered had not previously been recognized in the DGVs9 – a few of which were present in multiple subjects. A total of 37 CNVRs that we discovered were present in more than one individual, but were not included in the DGV. This is in contrast to the report by McElroy et al, who found only three CNVs present in two or more individuals, but were not in the DGV. Similar to McElroy et al, we found a high frequency of a duplication on chromosome 15, but did not replicate their observed high frequency of a duplication on chromosome 9. The reason for this lack of replication is unclear, but would not appear to be due to the size restriction imposed by our study.
Many of the CNVRs that we discovered overlapped with known genes. Although potential pathological implications of these CNVRs cannot be determined from this study, the genes STK39 and SLCO4C1 have been implicated in hypertension31, 32 and DNAH5 with various forms of congenital heart disease.33 Other genes have been linked to asthma34 and to various forms of cancer.35, 36 Future studies would be warranted to determine the association of these novel CNVRs with specific disease states.
Although we were not able to include data from other populations to make a direct inference about relative occurrences of CNV in African Americans, we discovered 309 novel CNVs that did not overlap with CNVRs in the DGV.9 This provides some evidence that the distribution of CNV in African Americans is at least partially different from that of other populations. Our work provides further insight into the copy-number architecture of African Americans.
Acknowledgments
This study was supported in part by the Marie and Emmett Carmichael Scholarship and by NIH grants R01HL055673, P01AR049084, T32HL079888, and T32HL072757. The opinions expressed herein are those of the authors and not necessarily those of the NIH or any organization with which the authors are affiliated.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- Donnelly P. Progress and challenges in genome-wide association studies in humans. Nature. 2008;456:728–731. doi: 10.1038/nature07631. [DOI] [PubMed] [Google Scholar]
- Manolio TA, Collins FS, Cox NJ, et al. Finding the missing heritability of complex diseases. Nature. 2009;461:747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad DF, Pinto D, Redon R, et al. Origins and functional impact of copy number variation in the human genome. Nature. 2010;464:704–712. doi: 10.1038/nature08516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redon R, Ishikawa S, Fitch KR, et al. Global variation in copy number in the human genome. Nature. 2006;444:444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International HapMap Consortium A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto D, Marshall C, Feuk L, Scherer SW.Copy-number variation in control population cohorts Hum Mol Genet 200716(Spec No. 2R168–R173.Erratum in: Hum Mol Genet. 2008;17(3):166–167. [DOI] [PubMed] [Google Scholar]
- de Smith AJ, Tsalenko A, Sampas N, et al. Array CGH analysis of copy number variation identifies 1284 new genes variant in healthy white males: implications for association studies of complex diseases. Hum Mol Genet. 2007;16:2783–2794. doi: 10.1093/hmg/ddm208. [DOI] [PubMed] [Google Scholar]
- Kang TW, Jeon YJ, Jang E, et al. Copy number variations (CNVs) identified in Korean individuals. BMC Genomics. 2008;9:492. doi: 10.1186/1471-2164-9-492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iafrate AJ, Feuk L, Rivera MN, et al. Detection of large-scale variation in the human genome. Nat Genet. 2004;36:949–951. doi: 10.1038/ng1416. [DOI] [PubMed] [Google Scholar]
- American Heart Association Heart Disease and Stroke Statistics – 2009Update ( ( http://www.americanheart.org )
- Center for Disease Control 2011 National Diabetes Fact Sheet ( http://www.cdc.gov )
- Ramirez S. Race and kidney disease outcomes: genes or environment. J Am Soc Nephrol. 2005;16:3461–3463. doi: 10.1681/ASN.2005101037. [DOI] [PubMed] [Google Scholar]
- Crosslin KL, Wiginton KL. The impact of race and ethnicity on disease severity in systemic lupus erythematosus. Ethn Dis. 2009;19:301–307. [PubMed] [Google Scholar]
- Gurland BJ, Wilder DE, Lantigua R, et al. Rates of dementia in three ethnoracial groups. Int J Ger Psy. 1999;14:481–493. [PubMed] [Google Scholar]
- McElroy JP, Nelson MR, Cailler SJ, Oksenber JR. Copy number variation in African Americans. BMC Genet. 2009;10:15. doi: 10.1186/1471-2156-10-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korn JM, Kuruvilla FG, McCarroll SA, et al. Integrated genotype calling and association analysis of SNPs, common copy number polymorphisms and rare CNVs. Nat Genet. 2008;40:1253–1260. doi: 10.1038/ng.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Li M, Hadley D, et al. PennCNV: an integrated hidden Markov model designed for high-resolution copy number variation detection in whole-genome SNP genotyping data. Genome Res. 2007;17:1665–1674. doi: 10.1101/gr.6861907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams RR, Rao DC, Ellison RC, et al. NHLBI family blood pressure program: methodology and recruitment in the HyperGEN network. Hypertension genetic epidemiology network. Ann Epidemiol. 2000;10:389–400. doi: 10.1016/s1047-2797(00)00063-6. [DOI] [PubMed] [Google Scholar]
- Freedman BI, Koop JB, Winkler CA, et al. Polymorphisms in the nonmuscle myosin heavy chain 9 gene (MYH9) are associated with albuminuria in hypertensive African Americans: the HyperGEN study. Am J Nephrol. 2009;29:626–632. doi: 10.1159/000194791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnett DK, Li N, Tang W, et al. Genome-wide association study identifies single-nucleotide polymorphism in KCNB1 associated with left ventricular mass in humans: the HyperGEN Study. BMC Med Genet. 2009;10:43. doi: 10.1186/1471-2350-10-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wineinger NE, Patki A, Meyers KJ, et al. Genome-wide joint SNP and CNV analysis of aortic root diameter in African Americans: the HyperGEN study. BMC Med Genomics. 2011;4:4. doi: 10.1186/1755-8794-4-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wineinger N, Kennedy R, Erickson S, Wojcynski M, Bruder C, Tiwari HK. Statistical issues in the analysis of DNA copy number variations data. Int J Comput Biol Drug Des. 2008;1:368–395. doi: 10.1504/IJCBDD.2008.022208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang TW, Jeon YJ, Jang E, et al. Copy number variations (CNVs) identified in Korean individuals. BMC Genomics. 2008;9:492. doi: 10.1186/1471-2164-9-492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitworth J, Voight B, Korn J, Nemesh J.‘CNV Analysis with Birdsuite and PLINK.'Broad Institute. 2009 〈 http://www.broadinstitute.org/ ftp/pub/mpg/birdsuite/Birdsuite_Pipeline.pdf 〉.
- Purcell S, Neale B, Todd-Brown K, et al. PLINK: a toolset for whole-genome association and population-based linkage analysis. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diskin SJ, Li M, Hou C, et al. Adjustment of genomic waves in signal intensities from whole-genome SNP genotyping platforms. Nucleic Acids Res. 2008;36:e126. doi: 10.1093/nar/gkn556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucan M, Abrahams BS, Wang K, et al. Genome-wide analyses of exonic copy number variants in a family-based study point to novel autism susceptibility genes. PLoS Genetics. 2009;5:e1000536. doi: 10.1371/journal.pgen.1000536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, et al. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maglott D, Ostell J, Pruitt KD, Tatusova T. Entrez gene: gene-centered information at NCBI. Nucleic Acids Res. 2005;33 (Database Issue:D54–D58. doi: 10.1093/nar/gki031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes AE, Orr N, Esfandiary H, Diaz-Torres M, Goodship T, Chakravarthy U. A common CFH haplotypes, with deletion of CFHR1 and CRHR3, is associated with lower risk of age-related macular degeneration. Nat Genet. 2007;39:567. doi: 10.1038/ng1890. [DOI] [PubMed] [Google Scholar]
- Toyohara T, Suzuki T, Morimoto R, et al. SLCO4C1 transporter eliminates uremic toxins and attenuates hypertension and renal inflammation. J Am Soc Nephrol. 2009;20:2546–2555. doi: 10.1681/ASN.2009070696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, O'Connell JR, McArdle PF, et al. From the cover: whole-genome association study identifies STK39 as a hypertension susceptibility gene. Proc Natl Acad Sci USA. 2009;106:226–231. doi: 10.1073/pnas.0808358106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy MP, Omran H, Leigh MW, et al. Congenital heart disease and other heterotaxic defects in a large cohort of patients with primary ciliary dyskinesia. Circulation. 2007;115:2814–2821. doi: 10.1161/CIRCULATIONAHA.106.649038. [DOI] [PubMed] [Google Scholar]
- Laing IA, de Klerk NH, Turner SW, et al. Cross-sectional and longitudinal association of the secretoglobin 1A1 gene A38G polymorphism with asthma phenotype in the Perth Infant Asthma Follow-up cohort. Clin Exp Allergy. 2009;39:62–71. doi: 10.1111/j.1365-2222.2008.03102.x. [DOI] [PubMed] [Google Scholar]
- Hamamoto R, Furukawa Y, Morita M, et al. SMYD3 encodes a histone methyltransferase involved in the proliferation of cancer cells. Nat Cell Biol. 2004;6:731–740. doi: 10.1038/ncb1151. [DOI] [PubMed] [Google Scholar]
- Oppezzo P, Vasconcelos Y, Settegrana C, et al. The LPL/ADAM29 expression ratio is a novel prognosis indicator in chronic lymphocytic leukemia. Blood. 2005;106:650–657. doi: 10.1182/blood-2004-08-3344. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.