Abstract
Cataracts are the commonest cause of blindness worldwide. Inherited cataract is a clinically and genetically heterogeneous disease that most often shows autosomal dominant inheritance. In this study, we report the identification of a novel locus for cerulean cataract type 5 (CCA5), also known as blue-dot cataract on chromosome 12q24. To date, four loci for autosomal dominant congenital cerulean cataract have been mapped on chromosomes, 17q24, 22q11.2–12.2, 2q33–35 and 16q23.1. To map this locus we performed genetic linkage analysis using microsatellite markers in a five-generation English family. After the exclusion of all known loci and several candidate genes we obtained significantly positive LOD score (Z) for marker D12S1611 (Zmax=3.60; at θ=0). Haplotype data indicated that CCA5 locus lies within a region of 14.3 Mb interval between the markers D12S1718 and D12S1723. Our data are strongly suggestive of a new locus for CCA5 on chromosome 12.
Keywords: cerulean cataract, linkage, heterogeneity
Introduction
Cataract, opacification of the eye lens, is the commonest cause of blindness in the world. During infancy and early childhood it frequently results in visual impairment and can lead to irreversible amblyopia. About one third of congenital cataracts are familial; the cataract may be isolated or be associated with other systemic abnormalities. Cataract is a key feature, of more than 200 genetic disorders.1 Inherited congenital cataract displays considerable genotypic and phenotypic heterogeneity. A number of distinct phenotypes have been described including: anterior polar, posterior polar, nuclear, lamellar, coralliform, blue-dot (cerulean), cortical, pulverulent, polymorphic and complete cataract.2, 3
Most inherited cataracts show autosomal dominant inheritance with complete penetrance, but expressivity is highly variable. Less frequently, autosomal recessive and X-linked inheritance patterns are seen. Recent research has paved the way for a better understanding of the underlying mechanisms of inherited forms of cataract. Progress has been made in elucidating cataract causing mutations in human genes encoding the transparent intracellular lens proteins (crystallins), membrane gap junction proteins (connexins), water channel proteins (aquaporins), solute carrier protein (SLC16A12), various cytoskeletal proteins (eg, phakinin, filensin and vimentin), transcription factors (FOXE3, EYA1, PAX6, MAF and PITX3) and transmembrane proteins (TMEM114, LIM2, CHMP-4B, EPHA2).4, 5 This insight also explains the basis for marked genotypic and phenotypic heterogeneity.
Cerulean cataract is an early onset, progressive cataract characterized by blue or white opacifications in the nucleus and cortex of the lens. This phenotype was first described by Vogt6 in 1922; it is not congenital but develops in childhood, progresses slowly and does not usually require surgery until adulthood. The opacities are distributed throughout the lens, with the outer nucleus containing blue-white dots that become more numerous in the cortex where they may form large cuneiform (wedgelike) shapes in the mid-periphery.
Previously ADCCCs (Autosomal Dominant Cerulean Congenital Cataracts) have been mapped to various chromosomes: 17q24 (CCA1, MIM 115660),7 22q11.2–q12.2 (CCA2, MIM 601547) close to CRYBB2 (MIM 123620) where Litt et al8 in 1997 identified a chain termination mutation in the CRYBB2 gene and showed that this mutation is associated with the CCA2 type. Recently, a Chinese family with cerulean cataract, has been identified with a point mutation in CRYBB2, causing a premature stop codon.9 Another large Moroccan pedigree with cerulean (blue dot) cataract (CCA3, MIM 608983) mapped to a region of chromosome 2q33–q35, and a missense mutation (P23T) in the CRYGD gene as the causative mutation.10 The same mutation has also been found in different Saudi families with cerulean cataract.11 A fourth cerulean locus in an Indian family has been mapped to 16q23.1, with a missense mutation in MAF gene;12 some affected members of this family also had microcornea. The causative gene at the CCA1 locus has not been identified. We have mapped a new locus for autosomal dominant congenital cerulean cataract to chromosome 12q24 in a five-generation family of English descent (Figure 1a).
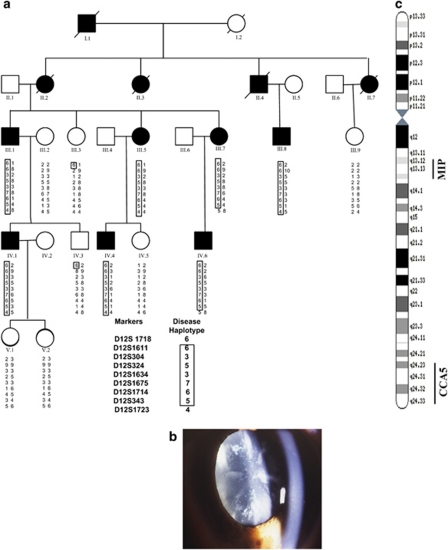
Figure 1.
(a) Abridged pedigree of the blue-dot cataract family used in this study showing the segregation of nine chromosome 12q markers listed in descending order. Squares and circles symbolize males and females, respectively. Open and filled symbols indicate unaffected and affected individuals. The disease haplotype is shown in the box. (b) Slit lamp view of a blue dot (cerulean) cataract in a 32-year-old male patient. (c) An idiogram of chromosome 12 showing locations of MIP and the new CCA5 locus.
Methods
Phenotyping
The family was drawn from the genetic clinic database at Moorfields Eye Hospital, London and had no other ocular or systemic abnormalities. Most of the patients underwent an eye examination with particular attention to the lens and cataract morphology. In this pedigree 7 of 13 patients were diagnosed as having cataract and of whom 5 were clinically examined. Cataract was diagnosed at a mean age of 17 years and patients underwent surgery at a mean age of 27 years (range 10–47). The cataract showed a typical cerulean phenotype with blue-white dots throughout the lens; they became more numerous in the cortex. Cuneiform (wedgelike) opacities were present in the mid-periphery of the lens (Figure 1b). The phenotype was consistent within the family. This pedigree has been excluded from the four known loci previously associated with cerulean cataract (data not shown).
Genotyping and linkage analysis
Genomic DNA was extracted from EDTA-sequestered blood samples using the Nucleon II DNA extraction kit (Scotlab Bioscience, Strathclyde, Scotland, UK). The local ethics committee approval was obtained for the studies and all individuals taking part in the study gave written informed consent. PCR-based genotyping was performed using ABI marker set HD-10 version 2.5 (Applied Biosystems, Warrington, UK). These markers are spaced at an average genetic distance of 10 cM. Analysis was performed using GeneMapper (version 4.0, Applied Biosystems) on ABI PRISM 3730 Genetic Analyzer (Applied Biosystems). We have developed a rapid analytical system that allows total genome genotyping using the 10 cM set within 3 days and is comparable to the speed of the SNP Chip analysis (protocol available on request). A full penetrance and a gene frequency of 0.0001 were used for the cataract locus. Two-point linkage analysis was carried out using the MLINK component of the LINKAGE program package version 5.10 (http://linkage.rockfelller.edu/soft/). The pedigree and haplotype data were managed by Cyrillic software (version 2.1.3, http://www.cyrillicsoftware.com). The region showing significant LOD score was refined using markers from Marshfield, GDB Human genome database (http://research.marshfieldclinic.org) and ENSEMBL genome data resources (http://www.ensembl.org).
Results
A five-generation family with cerulean cataract, comprising 14 members of the pedigree (Figure 1a) including 7 affected individuals, 6 unaffected individuals and 1 spouse were genotyped with microsatellite markers. We first excluded linkage from the four previously known loci for cerulean cataract (17q24 22q11.2–12.2, 2q33–35 and 16q23.1), and we then proceeded further to exclude all the other known cataract loci (data not shown). The genome-wide linkage analysis led us to obtain positive two-point LOD scores for markers D12S1611 (Z=3.62 at θ=0.00) and D12S1675, (Z=3.32 at θ=0.00). Seven other microsatellite markers in this region were genotyped in the family, and the two-point LOD scores of all nine marker loci are summarized in Table 1.
Table 1. Two-point LOD scores for linkage between the CCA5 locus and 12q24 markers.
| Distance | LOD scores at recombination fraction (θ) | |||||||
|---|---|---|---|---|---|---|---|---|
| MARKER | Mb | 0.0 | 0.01 | 0.05 | 0.1 | 0.2 | 0.3 | 0.4 |
| D12S1718 | 117.6 | 0.20 | 0.23 | 0.28 | 0.29 | 0.23 | 0.14 | 0.07 |
| D12S1611 | 124.9 | 3.62 | 3.56 | 3.29 | 2.95 | 2.22 | 1.44 | 0.65 |
| D12S304 | 125.8 | 2.62 | 2.57 | 2.38 | 2.14 | 1.60 | 1.03 | 0.44 |
| D12S324 | 126.6 | 2.40 | 2.36 | 2.17 | 1.93 | 1.44 | 0.91 | 0.39 |
| D12S1634 | 127.1 | 2.10 | 2.05 | 1.86 | 1.61 | 1.10 | 0.60 | 0.19 |
| D12S1675 | 128.1 | 3.32 | 3.26 | 3.02 | 2.70 | 2.03 | 1.31 | 0.57 |
| D12S1714 | 129.8 | 3.20 | 3.14 | 2.89 | 2.56 | 1.88 | 1.16 | 0.47 |
| D12S343 | 130.8 | 3.09 | 3.03 | 2.80 | 2.49 | 1.85 | 1.16 | 0.49 |
| D12S1723 | 131.9 | −3.36 | −3.36 | −1.22 | −0.33 | 0.03 | 0.26 | 0.26 |
Analysis of genotype data showed that all affected family members shared a common haplotype involving seven microsatellite markers (D12S1611, D12S304, D12S324, D12S1634, D12S1675, D12S1714 and D12S343). Critical recombination events were detected in affected individuals III 7 and IV 6 with marker D12S1723 and in unaffected individuals III 3 and IV 3 with marker D12S1718. These recombination events allowed us to narrow down the disease interval to approximately 14.3 Mb between markers D12S1718 and D12S1723 indicating that the CCA5 (cataract congenital cerulean type, 5) locus lies in a region flanked by these markers on the long arm of chromosome 12. There is no obvious candidate gene for Cerulean cataract currently localized on chromosome 12q in human and no murine cataract has been mapped to chromosome 5, the region of conserved synteny in mouse. With regards to the conserved synteny, it was noted that the human region contains an approximately 2 Mb of extra DNA sequence. Majority of this sequence (approximately 1.3 Mb) lies in the position 125.81–128.90 Mb on chromosome 12. The remaining approximately 0.7 Mb of extra sequence appears to be spread between 129.28–130.88. (http://www.ncbi.nlm.nih.gov/projects/homology/maps/mouse/chr5). This is a highly gene-rich area and a number of interesting genes map to this region. We have screened the following genes as candidates in our pedigree that might be relevant for lens biology, for example: transmembrane proteins (TMEM132C, TMEM132D) on the basis of involvement of TMEM114 in cataractogenesis13 and CAMKK2 (calcium/calmodulin-dependent protein kinase kinase 2), SFRS9 (serine/arginine-rich splicing factor 9) due to its expression pattern in the brain and in the eye (from data base http://www.genepaint.org).
Discussion
In this report, after excluding the known loci corresponding to ADCCC, we identified a novel locus (CCA5) on 12q23–24, associated with the cerulean congenital cataract in a five-generation English family. The first locus for blue-dot cataract was mapped on 17q24 in 1995 and so far no gene has been identified. Mutations in the CRYBB2 and CRYGD crystalline genes and the MAF transcription factor gene have been reported in association with cerulean cataract. There is thus considerable genetic heterogeneity for this distinctive cataract phenotype. The lens phenotype is the same in all reported families, but affected individuals with a mutation in MAF (CCA4) have in addition microcornea. To date the proteins encoded by the causative genes have a well-defined role in lens transparency, but it is not clear why the mutations result in a cerulean phenotype. The identification of the genes on chromosome 17q and 12q may identify new biological pathways associated with lens structure and function.
Acknowledgments
We would like to acknowledge funding from the Wellcome Trust project Grant 063969/Z/01, EU project ‘PYTHIA' (FP7-ICT2-224030), and NIHR (Moorfields Eye Hospital Biomedical Research Centre). We would like to thank the members of the family for taking part in this study.
The authors declare no conflict of interest.
References
- Krumpaszky HG, Klauss V. Epidemiology of blindness and eye disease. Ophthalmologica. 1996;210:1–84. doi: 10.1159/000310663. [DOI] [PubMed] [Google Scholar]
- Ionides A, Francis P, Berry V, et al. Clinical and genetic heterogeneity in autosomal dominant congenital cataract. Br J Ophthalmol. 1999;83:802–808. doi: 10.1136/bjo.83.7.802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis PJ, Berry V, Bhattacharya SS, Moore AT. The genetics of childhood cataract. J Med Genet. 2000a;37:481–488. doi: 10.1136/jmg.37.7.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graw J.The genetic and molecular basis of congenital eye defects Nat Rev Genet 20034876–888.Review. [DOI] [PubMed] [Google Scholar]
- Shiels A, Bennett TM, Hejtmancik JF. Cat-Map: putting cataract on the map. Mol Vis. 2010;16:2007–2015. [PMC free article] [PubMed] [Google Scholar]
- Vogt A. Lens and Zonule. Bonn: JP Weyenborgh; 1979. [Google Scholar]
- Armitage MM, Kivlin JD, Ferell R. A progressive early onset cataract gene maps to human chromosome 17q24. Nature Genet. 1995;9:37–40. doi: 10.1038/ng0195-37. [DOI] [PubMed] [Google Scholar]
- Litt M, Carrero-Valenzuela R, LaMorticella DM, et al. Autosomal dominant cerulean cataract is associated with a chain termination mutation in the human beta-crystallin gene CRYBB2. Hum Mol Genet. 1997;6:665–668. doi: 10.1093/hmg/6.5.665. [DOI] [PubMed] [Google Scholar]
- Wang L, Lin H, Gu J, Su H, Huang S, Qi Y. Autosomal-dominant cerulean cataract in a Chinese family associated with gene conversion mutation in beta-B2-crystallin. Ophthalmic Res. 2009;41:148–153. doi: 10.1159/000209668. [DOI] [PubMed] [Google Scholar]
- Rogaev EI, Rogaeva EA, Korovaitseva GI, et al. Linkage of polymorphic congenital cataract to the gamma-crystallin gene locus on human chromosome 2q33–35. Hum Mol Genet. 1996;5:699–703. doi: 10.1093/hmg/5.5.699. [DOI] [PubMed] [Google Scholar]
- Khan AO, Aldahmesh MA, Ghadhfan FE, Mesfer SA, Alkuraya FS. Founder heterozygous P23T CRYGD mutation associated with cerulean (and coralliform) cataract in 2 Saudi families. Mol Vis. 2009;15:1407–1411. [PMC free article] [PubMed] [Google Scholar]
- Vanita V, Singh D, Robinson PN, Sperling K, Singh JR. A novel mutation in the DNA-binding domain of MAF at 16q23.1 associated with autosomal dominant ‘cerulean cataract' in an Indian family. Am J Med Genet A. 2006;140:558–566. doi: 10.1002/ajmg.a.31126. [DOI] [PubMed] [Google Scholar]
- Jamieson RV, Farrar N, Stewart K, et al. Characterization of a familial t(16;22) balanced translocation associated with congenital cataract leads to identification of a novel gene, TMEM114, expressed in the lens and disrupted by the translocation. Hum Mutat. 2007;28:968–977. doi: 10.1002/humu.20545. [DOI] [PubMed] [Google Scholar]