Abstract
Class II photolyases ubiquitously occur in plants, animals, prokaryotes and some viruses. Like the distantly related microbial class I photolyases, these enzymes repair UV-induced cyclobutane pyrimidine dimer (CPD) lesions within duplex DNA using blue/near-UV light. Methanosarcina mazei Mm0852 is a class II photolyase of the archaeal order of Methanosarcinales, and is closely related to plant and metazoan counterparts. Mm0852 catalyses light-driven DNA repair and photoreduction, but in contrast to class I enzymes lacks a high degree of binding discrimination between UV-damaged and intact duplex DNA. We solved crystal structures of Mm0852, the first one for a class II photolyase, alone and in complex with CPD lesion-containing duplex DNA. The lesion-binding mode differs from other photolyases by a larger DNA-binding site, and an unrepaired CPD lesion is found flipped into the active site and recognized by a cluster of five water molecules next to the bound 3′-thymine base. Different from other members of the photolyase-cryptochrome family, class II photolyases appear to utilize an unusual, conserved tryptophane dyad as electron transfer pathway to the catalytic FAD cofactor.
Keywords: archaea, DNA repair, electron transfer, UV lesion, water cluster
Introduction
UV-induced lesions in B-type duplex DNA are either the pyrimidine-pyrimidone (6-4) photoproduct or the predominant cyclobutane pyrimidine dimer (CPD) in form of its cis-syn isomer (Heelis et al, 1993). These UV lesions are repaired by light-dependent DNA-repair enzymes called DNA photolyases, which are members of the structurally related photolyase-cryptochrome family that also comprises cryptochromes and DASH cryptochromes. Cryptochromes act as blue-light photoreceptors and exert various physiological functions like regulation of the circadian clock in animals and plants. Unlike photolyases they generally lack any kind of DNA-repair activity (Lin and Todo, 2005), whereas DNA photolyases can be specified according to their substrate specificity as (6-4) or CPD photolyases, respectively.
The photolyase-cryptochrome family is present in all three domains of life, that is, archaea, eubacteria and eukaryotes, and hence has arisen very early during evolution to protect genomes against the genotoxic effects of ultraviolet light originating from the sun. However, their evolution and the phylogenetic relationship of its members and subfamilies have been controversially discussed (Kanai et al, 1997; Falciatore and Bowler, 2005; Ozturk et al, 2008; Lucas-Lledo and Lynch, 2009). Based on sequence analyses, CPD photolyases have been initially subdivided into just two classes: class I enzymes occurring exclusively in microbes and class II photolyases mostly restricted to higher, multicellular eukaryotes. Only recently, cryptochromes of the DASH type have been recognized to catalyse light-driven CPD-repair activity in single-stranded and loop-structured duplex DNA as well (Selby and Sancar, 2006; Pokorny et al, 2008). Accordingly, other but distant subfamilies like class III photolyases present in some eubacteria (Ozturk et al, 2008) or a novel type of cryptochromes occurring in proteobacterial species (Hendrischk et al, 2009) have been discovered. Overall, this implies that the ancient photolyase-cryptochrome family is highly diversified and that the insights derived so far from the well-characterized class I CPD and (6-4) photolyases are not necessarily applicable to other subfamilies. For example, the cryptochrome subfamilies from plants and animals as well as the DASH cryptochromes have apparently branched off from class I CPD and (6-4) photolyases, respectively, but not from class II enzymes or other subfamilies.
Both photolyases and cryptochromes have a bilobal architecture consisting of two domains: an N-terminal domain that may contain a light-harvesting antenna chromophore to additionally broaden their activity spectra and a C-terminal α-helical catalytic domain comprising the light-sensitive FAD cofactor. This architecture is preserved in the structurally characterized class I photolyases (Park et al, 1995; Tamada et al, 1997; Komori et al, 2001; Fujihashi et al, 2007), (6-4) photolyases (Maul et al, 2008; Hitomi et al, 2009), plant cryptochromes (Brautigam et al, 2004) as well as DASH cryptochromes (Brudler et al, 2003; Klar et al, 2007), although some differences have been described. For example, the catalytic domain of the Thermus thermophilus CPD photolyase is C-terminally truncated by ∼20 residues as compared with other class I photolyases (Komori et al, 2001), whereas mature cry3 from Arabidopsis thaliana bears an N-terminal extension of 39 residues (Klar et al, 2007) that is present only in some plant but not in cyanobacterial orthologues (Brudler et al, 2003). Diverse classes of antenna chromophores like 5,10-methenyltetrahydrofolate (MTHF), 8-hydroxydeazaflavin, FMN or FAD have been identified in some photolyases/cryptochromes to broaden their activity spectra, whereas many others apparently lack any bound antenna chromophores.
In DNA photolyases, binding and repair of UV-damaged DNA is the defining function of the catalytic domain. The low intrinsic affinity of class I CPD photolyases to intact DNA in the micromolar range is mediated by the basic nature of the protein's surface surrounding the active site with its catalytic FAD cofactor. The specific recognition and repair of the UV lesion occurs within the active site and its rectangular-shaped entrance. The pyrimidine dimer is flipped out of the double-stranded DNA (dsDNA) into the active site, forming numerous hydrophobic interactions at its bottom as well as hydrogen bonds between the C4-carbonyls of the 5′- and 3′-pyrimidine bases and the N6-amino group of the FAD's adenine moiety. This flavin cofactor participates in the light-mediated reactivation of UV-damaged DNA in its reduced and photoexcited FADH−* state. Within nanoseconds, injection of an electron into the CPD lesion causes the breakage of the C5-C5 and C6-C6 bonds and electron back-transfer to the FADH• intermediate (Sancar, 2003; Essen, 2006). Cocrystal structures of the class I photolyase from Anacystis nidulans (Mees et al, 2004) and the (6-4) photolyase from Drosophila melanogaster (Maul et al, 2008) with UV-damaged duplex DNA demonstrated that sequence-independent recognition of the UV lesion depended on salt bridges and hydrogen bonds mainly formed between the enzyme and the P−1, P+1, P+2 and P+3 phosphates of the DNA strand comprising the lesion. There is a distinct lack of interactions with the counter strand rationalizing why DNA photolyases catalyze very efficiently the repair of UV-B damaged ssDNA as well.
With sequence identities of <16%, class II photolyases are only distantly related to other photolyase/cryptochrome subfamilies (Yasui et al, 1994) suggesting that they have arisen early in evolution (Kanai et al, 1997) and may differ in their mode of FAD binding and catalysis, although photoreduction of their FAD cofactor occurs similarly to class I via a semireduced FADH• intermediate to the catalytically active FADH− (Okafuji et al, 2010). Class II photolyases occur ubiquitously in multicellular organisms, that is, plants and animals, but are missing in placental animals like humans and mice. Despite their functional importance, overall knowledge about class II photolyases lags behind their microbial class I relatives. Here, a combined structural, spectroscopic and functional analysis of the class II photolyase from the methanogenic archaeon Methanosarcina mazei Go1, the gene product Mm0852, is reported. Its close relationship to homologous enzymes from plants (Iwamatsu et al, 2008), and animals like the mammalian infraclass of marsupials, for example Potorous tridactylus (Yasui et al, 1994), makes the described structural insights highly transferable to all other metazoan photolyases.
Results and discussion
An archaeal orthologue as a representative of metazoan class II photolyases
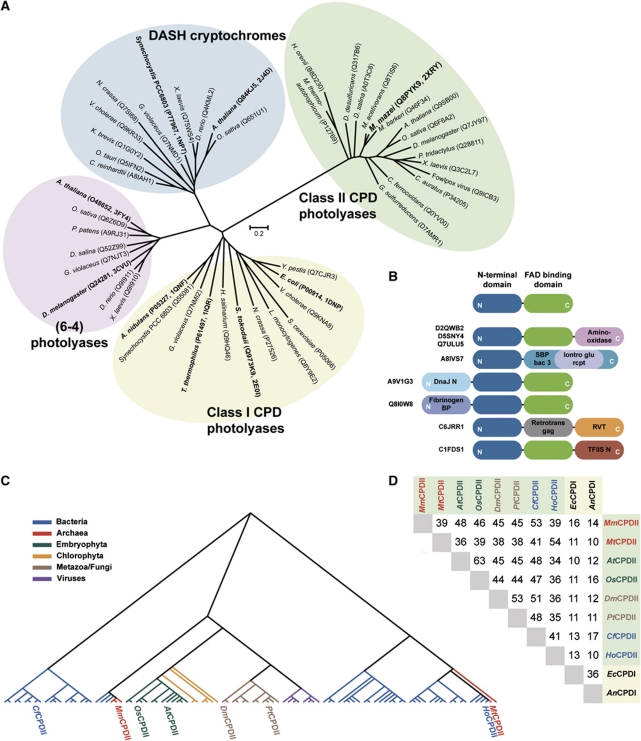
A phylogenetic sequence analysis of light-dependent DNA-repair enzymes was performed to identify orthologues of class II photolyases from plants which might be better suited for structural and biophysical analysis than the available recombinant plant enzymes (Kleiner et al, 1999; Kaiser et al, 2009). The unrooted phylogenetic tree (Figure 1A) reveals a distinct cluster for class II photolyases which does not only comprise all known orthologues from higher, multicellular eukaryotes like A. thaliana, O. sativa, D. melanogaster or P. tridactylus and some viruses, but also from several eubacteria and archaea. An analysis of the INTERPRO protein subfamily of class II photolyases (IPR008148) shows currently 117 eukaryotic, 91 eubacterial and 6 archaeal members including the euryarchaeal order of Methanosarcinales. Obviously, the occurrence of class II photolyases is much more restricted in prokaryotes than that of class I, because the class I-specific INTERPRO subfamily (IPR002081) comprises 1257 eubacterial and 35 archaeal orthologues. Moreover, among the class II photolyases, the subfamily members from plants and animals cluster together with the Methanosarcinal photolyases from M. mazei, M. acetivorans and M. barkeri into a common subbranch, where they exhibit pair-wise sequence identities of 45–48% (Figure 1C and D; Supplementary Figure S4B). Accordingly, one may postulate that at least some of the overall well-conserved class II photolyases were spread by horizontal gene transfer, for example, Methanosarcinales ↔ metazoans, and may hence be a result of late evolution. For comparison, the sequence identities between class II and microbial class I photolyases of Escherichia coli and A. nidulans (EcCPDI and AnCPDI) are 16% or less (Figure 1D; Supplementary Figure S4A). Noteworthy, for class II photolyases natural fusions with transcription factors, NAD-dependent reductases or chaperones can be identified in several genomes (Figure 1B). Structural and functional analysis of methanosarcinal photolyases may be hence rewarding as they are highly representative of related photolyases from plants, animals and viruses.
Figure 1.
Phylogenetic analysis of the class II photolyase subfamily. (A) The class II CPD photolyase subfamily forms a distinct cluster of light-dependent DNA-repair enzymes. The scale bar of the unrooted phylogenetic tree indicates amino-acid substitutions per site. (B) Several class II photolyases (UniProt identifiers depicted at left) show various kinds of domain fusions at the N- or C-terminal subdomains (blue and green). (C) Dendrogram of 98 non-redundant members of the class II photolyase subfamily. (D) Sequence identities of prominent members of class II and class I CPD photolyases. Class II CPD photolyases: Methanosarcina mazei (MmCPDII), Methanobacterium thermoautotrophicum (MtCPDII), Arabidopsis thaliana (AtCPDII), Oryza sativa (OsCPDII), Drosophila melanogaster (DmCPDII), Potorous tridactylus (PtCPDII), Chlorobium ferrooxidans (CfCPDII) and Halothermothrix orenii (HoCPDII); Class I CPD photolyases: Escherichia coli (EcCPDI) and Anacystis nidulans (AnCPDI). Phylogenetic analysis of class II photolyases is summarized in Supplementary data.
UV/Vis spectra, repair activity and DNA binding of M. mazei class II photolyase
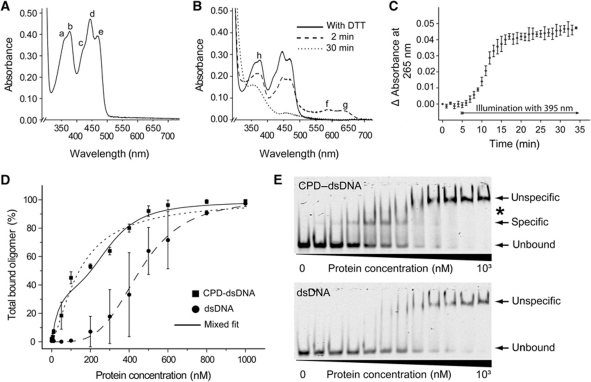
The Mm0852 gene from M. mazei, an orthologue of class II photolyases, was cloned and heterologously overexpressed in E. coli giving ∼50 mg protein per litre culture. After purification, the Mm0852 gene product was of a bright yellow colour indicating the presence of a flavin cofactor in the fully oxidized form. Its absorption spectrum exhibits peaks at 362, 377, 421, 444 and 469 nm (Figure 2A), which are characteristic for photolyase-bound FADox and differ to free FADox in solution as the latter one has peaks at 373 and 445 nm (Payne et al, 1990). The absence of any prominent absorption peak between 377 and 415 nm indicated that no antenna chromophore like MTHF is bound to the Mm0852 protein (now called MmCPDII).
Figure 2.
Spectroscopic characterization, repair activity and DNA binding. (A) UV/Vis spectrum of M. mazei photolyase (3.4 mg ml−1) shows fully oxidized FAD (a=362 nm; b=377 nm, c=421 nm; d=444 nm, e=469 nm). (B) UV/Vis spectra of photoreduction via illumination at 450 nm in the presence of reducing agent DTT. After addition of DTT (solid line), illumination leads to the neutral semiquinoid state of the photolyase (dashed line) with maxima at 590 nm (f) and 632 nm (g), respectively. The fully reduced state (dotted line) features a single maximum at 360 nm (h). (C) Complete repair of CPD lesions after onset of UV-A was achieved within 12 min. (D) Quantitative analysis of EMSA with M. mazei photolyase and CPD-dsDNA as well as undamaged DNA. The dashed curves correspond to non-linear fitting of the data to the simple Hill equation (Hill, 1910), whereas the solid line has been obtained by a mixed model for specific and non-specific DNA binding (see Supplementary data). (E) Scans of native EMSA-PAGE of IRDye700-labelled DNA probes incubated with M. mazei enzyme to determine dissociation constants. Observed photolyase•DNA complexes are marked with arrows. An additional distinct higher complex species is denoted with an asterisk.
Like other photolyases MmCPDII is capable to photoreduce its catalytic FAD to the active FADH− form. Illumination at 450 nm of MmCPDII in presence of the reducing agent dithiothreitol (DTT) led to facile build-up of the semiquinoid, neutral state of the flavin cofactor (FADH•) by showing absorption peaks at 590 and 632 nm. During further illumination, the formation of fully reduced cofactor (FADH−) was accompanied by loss of absorption except at 360 nm (Figures 2B and 6C). Analogous intra-protein photoreductions of FAD have been described not only for various class I photolyases (Sancar, 2003) and A. thaliana cryptochrome 3 (Song et al, 2006), but also for plant class II photolyases (Okafuji et al, 2010).
Catalytic activity of the recombinant MmCPDII as CPD photolyase was demonstrated by a modified repair assay using UV-damaged oligo(dT)18 (Jorns et al, 1985) and monitoring increased absorption at 265 nm due to repair of the CPD lesion upon illumination at 395 nm (Figure 2C). The MmCPDII enzyme showed unequivocal DNA-repair activity upon UV-A illumination as known from other photolyases. However, its DNA-binding characteristics differs significantly from class I photolyases. Electrophoretic mobility shift assays (EMSAs) using the experimental set-up established for A. thaliana cryptochrome 3 (Pokorny et al, 2008) were used to estimate the dissociation constants of MmCPDII to CPD-damaged and undamaged duplex DNA (Figure 2D and E). Undamaged 50mer DNA duplexes formed higher molecular mass complexes with MmCPDII at enzyme concentrations exceeding 200 nM. In contrast, CPD lesion comprising DNA duplexes showed a smaller and defined complex species already at 10 nM and above, which at higher concentrations is consumed by the large, non-specific DNA–enzyme complexes. Quantitation of the non-specific duplex DNA binding shows a dissociation constant of KD,NS=455±7 nM and a cooperativity that is consistent with the binding of four enzyme molecules per duplex DNA (n=3.9±0.2). Given the length of the oligonucleotides and the apparent lack of ladder-like complex intermediates this means that MmCPDII coaggregates along non-damaged duplex DNA and covers here about 12 base pairs or a stretch of 42 Å length. Non-specific binding by MmCPDII is much more prominent than in class I photolyases, where only high micromolar dissociation constants have been determined (EcCPDI: KD,NS=∼100 μM). Specific binding of UV-damaged duplex DNA is clearly stronger; a simple Hill analysis already gives a dissociation constant of KD,S=141±15 nM (n=1.4±0.2). Because the binding curve gives evidence between 100 and 400 nM of competing non-specific binding, a mixed model with specific 1:1 binding and cooperative non-specific binding appears to be more appropriate. This model gives specific and non-specific dissociation constants of KD,S=44±12 nM and KD,NS=305±27 nM (nD,NS=3.7±0.9), respectively. Compared with class I enzymes like EcCPDI (KD,S=∼1 nM; Sancar, 2003) specific DNA binding is by two orders of magnitude lower and the specificity ratios KD,NS/KD,S, ∼104 for EcCPDI, are reduced to a factor of 10 for MmCPDII. However, this is no shortcoming of class II photolyases, because the enzyme concentrations available in vivo can be expected to be too limiting to allow competing, cooperative non-specific binding to duplex DNA.
Overall structure of M. mazei class II photolyase
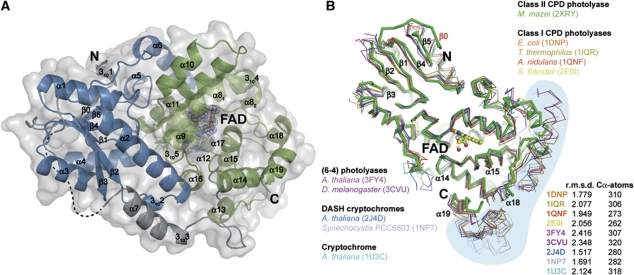
The crystal structure of the M. mazei class II photolyase at 1.5 Å resolution reveals the overall fold of the photolyase-cryptochrome family. Like class I (Park et al, 1995; Tamada et al, 1997; Komori et al, 2001; Fujihashi et al, 2007) and (6-4) photolyases (Maul et al, 2008; Hitomi et al, 2009) as well as cryptochromes (Brudler et al, 2003; Brautigam et al, 2004; Huang et al, 2006; Klar et al, 2007), the structure of MmCPDII is organized in an N-terminal α/β subdomain and a C-terminal all-helical subdomain (Figure 3A). The C-terminal FAD-binding subdomain (P232-Y461, α8-α19, 3104, 3105) contains the catalytic cofactor FAD in the U-shaped conformation commonly observed for photolyases and cryptochromes. The N-terminal subdomain (M3-S185) corresponds with its secondary structure elements α1-α6, β0-β5, 3101, 3102 to the Rossmann fold and lacks any kind of bound antenna chromophore.
Figure 3.
Overall structure of the M. mazei class II photolyase and structural comparison of members of the photolyase-cryptochrome family. (A) The N-terminal DNA-photolyase domain is shown in blue. C-terminal FAD-binding domain (green) contains the catalytic cofactor FAD (yellow) with SigmaA-weighted 2Fobs−Fcalc electron density contoured at 1σ. Domains are connected by a linker (grey) containing an α-helix (α7) with a 310-helical extension (3103). Not defined part of the linker by electron density is illustrated with a dashed line. Colouring of domain limits corresponds to Pfam database (Finn et al, 2010). (B) The M. mazei class II photolyase reveals a common overall fold to superimposed structures of the photolyase-cryptochrome family.
Despite low pair-wise sequence identities to class I photolyases (Figure 1D), a comparison of MmCPDII with structures of the photolyase-cryptochrome family shows a highly preserved fold with overall r.m.s.d. of <2.5 Å for the superimposed Cα traces (Figure 3B). However, three features discriminate MmCPDII from other known members of the photolyase/cryptochrome family: the Rossmann-like fold harbours a sixth β-strand (β0) at the N-terminus; the catalytic subdomain is C-terminally truncated by ∼50 residues including the long C-terminal helix observed in class I photolyases (Figure 3B); and finally, the linker region connecting both subdomains (V186-E231) is significantly longer than in class I photolyases (EcCPDI: 25 aa, TtCPDI: 20 aa, AnCPD: 36 aa). Parts of the linker are apparently flexible as indicated by a lack of electron density for E189-V197 (Figure 3A). Such a lack of structural definition was also observed for the linker region of the class II photolyase from M. barkeri (E187-L226) whose structure was used for phasing of MmCPDII by molecular replacement (see Materials and methods and Supplementary Table SI). Overall, recombinant MmCPDII as used for crystallization and structure solution proved to be functional in photoreduction of the catalytic FAD cofactor, binding and repair of CPD lesions (Figure 2).
Structure of the M. mazei photolyase•CPD–DNA complex
To elucidate the recognition mode of CPD lesions within class II photolyases a stoichiometric complex between MmCPDII and a 14-meric DNA duplex with a synthetic CPD lesion in its central position has been crystallized under safe-light conditions (Mailliet et al, 2009). The MmCPDII•CPD–DNA structure at 2.2 Å resolution (Supplementary Figure S1A) reveals two complexes per asymmetric unit, where the dsDNA adopts a typical B-type conformation, but is severely kinked at the lesion site. The duplex at the 5′-side of the CPD lesion can be hence referred as 5′-arm, the other as 3′-arm. Both complexes are associated with each other by quasi-continuous arrangement of their bound DNA duplexes along the 3′-arms (Supplementary Figure S1B). Well-defined electron density for the DNA strand comprising the synthetic CPD lesion as well as for the counter strand is observed in both complexes with only a few nucleotides missing at the free termini of the 5′-arms (complex A: A1-T2; complex B: A1-C3 of the CPD strand and complex B: T14 of the counter strand, respectively). A comparison with the non-bound MmCPDII structure shows that complexation with dsDNA causes no major structural changes within the enzyme, as the r.m.s.d. between the uncomplexed photolyase and both MmCPDII•CPD–DNA complexes are rather low with 0.43 Å for complex A (396 Cα atoms) and 0.41 Å for complex B (352 Cα atoms), respectively. Similarly, only local structural changes within the enzyme upon CPD-DNA binding have previously been reported for the A. nidulans class I photolyase (Mees et al, 2004). Although both MmCPDII•CPD-DNA complexes are structurally similar, the 3′-arms of the duplex DNA remarkably exhibit deviating orientations of about 11° thus indicating plasticity in the recognition of dsDNA along the enzyme's surface (Supplementary Figure S1C). Nevertheless, as most of the protein–DNA interactions are preserved in both complexes, all following structural analyses used complex A unless otherwise stated.
The CPD lesion of the kinked dsDNA is flipped out of the duplex into the active site of MmCPDII and bound there in a similar fashion as reported before for the complexes of the class I photolyase from A. nidulans (Mees et al, 2004) and the DASH cryptochrome from A. thaliana (Pokorny et al, 2008). However, the synthetic CPD lesion within the active site of MmCPDII is still intact with unbroken C5-C5 and C6-C6 bonds as indicated by unambiguous electron density in both complexes (Figure 4B) and differs in this regard from the CPD-DNA complexes reported for AnCPDI and Atcry3, where the CPD lesions have been repaired in situ by exposure to X-rays. The prevalence of an intact CPD lesion within the active site of MmCPDII is not caused by a general lack of photochemical reactivity of MmCPDII crystals. MmCPDII crystals readily undergo photoreduction at room temperature but not at cryogenic temperatures (Supplementary Figure S2E). Furthermore, MmCPDII crystals suffer from X-ray-induced reduction of the FAD chromophore (Supplementary Figure S2F), a behaviour that has been reported before for AnCPDI (Kort et al, 2004).
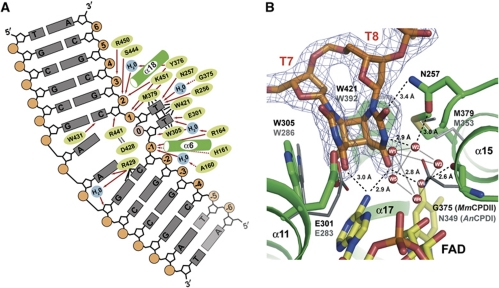
Figure 4.
Detailed view of the CPD lesion within the binding pocket of the M. mazei class II photolyase. (A) The schematic diagram illustrates that the class II photolyase is almost exclusively interacting with the CPD-comprising DNA strand. Dashed arrows indicate interactions with the protein backbone, whereas solid arrows characterize interactions with side chains. Black lines represent other mentioned interactions. Faded nucleotides are not defined by electron density. (B) Comparison of the active site of the MmCPDII•CPD–DNA complex (green) with AnCPDI (grey) bound to analogue CPD lesion (not shown). The class II enzyme features a cluster of water molecules (W1-W6) at the 3′-thymine replacing the corresponding asparagine residue (MmCPDII: G375, AnCPDI: N349). Electron density (SigmaA-weighted 2Fobs-Fcalc) is contoured at 1σ.
Whereas the structures of AnCPDI and Atcry3 with in situ repaired CPD lesions show almost coplanar thymine bases with tilt angles of ∼13° (Mees et al, 2004; Pokorny et al, 2008), the cyclobutane ring of the CPD lesion within MmCPDII closely resembles with a tilt of 34° intact CPD lesions known from other structures. For example, in the crystal structure of the synthetic CPD lesion itself (Butenandt et al, 1998) the thymine bases adopt an interplanar angle of 57° (Supplementary Figure S3). Furthermore, in a set of four independent structures of the human polymerase η complexed to CPD–DNA (Biertümpfel et al, 2010), the interplanar angles of the CPD lesion vary position-dependent between 34° and 56° (Supplementary Figure S3). Apparently, the conformational plasticity of the CPD lesion is not causing a loss of the CB+ pucker of the cyclobutane ring upon binding to the active site of MmCPDII and only the class I and DASH-like photolyases are capable to select a CPD conformation that is already prone to breakage of the C5-C5 and C6-C6 bonds by X-ray-driven repair. In MmCPDII, the steric distortion induced by the active site onto the CPD lesion might be hence more relaxed due to the differences within the binding pocket of the thymine dimer including a unique water cluster (see below), although an influence of the crystallization matrix cannot be excluded.
The C4 carbonyl groups of the 5′- and 3′-thymines of the CPD lesion in the MmCPDII structure form hydrogen bonds with the N6-amino group of the adenine from the catalytic FAD. This adenine is at least a crucial part of the docking site for the CPD lesion, if not even of the electron transfer pathway between the lesion and the isoalloxazine moiety. Theoretical calculations on electron transfer (Prytkova et al, 2007; Acocella et al, 2010) infer that the adenine drives the transfer to the CPD lesion from the 8-methyl group of FADH− rather indirectly by acting as an electrostatic ‘bouncer’ or as a structural organizer of the bound lesion. Accordingly, the hydrogen bonds between the thymines and the N6-amino group are maintained in the X-ray repaired states of the AnCPDI and Atcry3 CPD–DNA complexes, which correspond to structural snapshots after the first two nanoseconds of DNA repair.
Despite the division into class I, class II and cryDASH-subfamilies for CPD photolyases the active sites share a high degree of structural homology as shown by a comparison between the CPD binding mode in MmCPDII with that of thymine dimers in AnCPDI and Atcry3 complexes (Mees et al, 2004; Pokorny et al, 2008). Overall, the CPD lesion is surrounded by hydrophobic side chains (Figure 4A and B). In MmCPDII, the conserved tryptophans W305 and W421 form the L-shaped walling of the active site that clamps the CPD lesion together with the side chain of the conserved M379 (Figure 4A and B). Upon repair the 5′-thymine base is expected to remain in place upon breakage of the C5-C5 and C6-C6 bonds by maintaining the π-stacking interactions with the indole moiety of W305 (AnCPDI: W285), whereas the 3′-thymine dissociates by ca. 1 Å away towards the thioether group of M379.
The currently accepted mechanism for the repair of CPD lesions by photolyases implies a transient radical form of the CPD lesion that is formed upon the light-driven electron transfer from the excited catalytic cofactor FADH−*. Prior to decomposition into single bases the CPD radical anion is predicted to be stabilized by proton transfer from a neutral glutamic acid residue at the bottom of the active site (Mees et al, 2004; Masson et al, 2009). In MmCPDII, this residue is found to be E301 (Figure 4A and B) as it forms hydrogen bonds with the 5′-thymine of the lesion like the corresponding residue in other photolyase families. Mutational studies of the homologous glutamate (E384A) in the S. cerevisiae class I photolyase showed a 50% reduced binding affinity to CPD-DNA and a significant decrease in quantum yield of repair by 60% (Vande Berg and Sancar, 1998).
A different situation is given for the interaction between the 3′-thymine base and the enzyme. In other CPD photolyases, the side chain of a conserved asparagine (AnCPDI: N349, Atcry3: N391) forms hydrogen bonds with the N3 amide and the C4 carbonyl and is here postulated to support catalysis by stabilizing the anionic thymine radical after bond breakage. No such interaction exists in class II photolyases because this position is replaced by a glycine residue (MmCPDII: G375). Instead, a cluster of six water molecules occupies the space between the CPD lesion and the active site's walling (Figure 4B). One water molecule (W6) replaces the side chain of the missing asparagine by taking over the bridging interaction between the C4-carbonyl group of the 3′-thymine and the C2′-hydroxyl of the FAD's ribityl group. The other five water molecules are sandwiched as a square-pyramidal cluster between the 3′-thymine and the diphosphate group of FAD. This water cluster is already organized in the active site prior to DNA binding, because these water molecules are observed in the high-resolution structure of the uncomplexed MmCPDII as well. The recognition of the 3′-thymine by a structurally flexible water cluster may allow the CPD lesion to adopt a wider range for the tilt angles of its base planes. Interestingly, G375 is conserved in all metazoan photolyases including plants and animals (30/30), whereas half of the prokaryotic class II photolyases (29/59) harbour an asparagine like class I or even a serine residue (3/59) (Supplementary Figure S4A).
A conserved asparagine residue (N257) interacts directly via a hydrogen bond with the C2 carbonyl group of the 3′-thymine and indirectly via the water cluster (W1-W5) with its N3-atom (Figure 4B). Currently, it is not clear whether this interaction is a result of the intact CPD lesion or a slightly altered binding mode, as the corresponding residue in the structures of CPD photolyases complexed to repaired CPD lesions (AnCPDI: N233, Atcry3: N277) was not found to participate directly in damage recognition. Another residue close to the surface of the CPD binding site, R256 (AnCPDI: R232, Atcry3: R276), is adjacent to the 3′-thymine and may allow electrostatic stabilization of a transiently formed radical anion (Figure 4A). Interestingly, QM/MM calculations on the splitting mechanism of the CPD lesion within AnCPDI indicated a strong interaction between the C2-carbonyl of the 3′-thymine and the latter residues (Masson et al, 2009). In class II enzymes, the stabilizing interactions may not only involve these two amino acids, but also the active site's water cluster next to it. The latter may even contribute to transient protonation of the radical anion species at the 3′-thymine base. Such a proton transfer from a water cluster is not without precedence in light-triggered proteins. In the proton-pumping bacteriorhodopsins, a cluster of water molecules mediates the terminal release of a proton during the photocycle (Essen et al, 1998; Garczarek et al, 2005).
Alternative binding modes of UV-damaged duplex DNA by photolyases
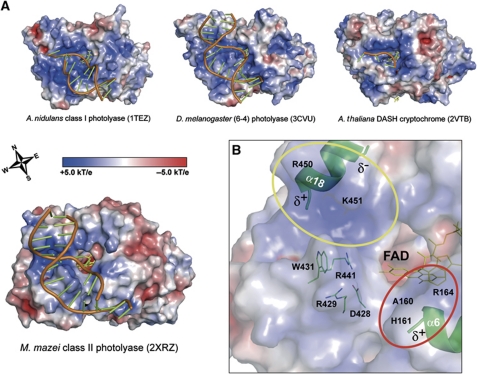
The interface of MmCPDII with dsDNA is with 1242 Å2 and 1354 Å2 for the complexes A and B, respectively, larger than for AnCPDI (1123 Å2) or the (6-4) photolyase from D. melanogaster (1182 Å2) and also differs in terms of the electrostatic surface properties (Figure 5A). Focusing on the CPD binding pocket, the positively charged surface area of MmCPDII that is capable of DNA interactions is spatially more restricted, but sufficient to provide stabilizing electrostatic interactions with the deoxyribophosphate backbone along the phosphates P−1, P+1, P+2 and, to a lesser extent, P+3. In MmCPDII, the canyon-like DNA-binding surface is built up along its ‘Western’ and ‘Southern’ rim (Figure 5A) by the basic side chains of R429, R441, R450 and K451 from the loop between helices α17 and α18 and from the short helix α18. This elongated and basic ridge forms numerous interactions with the DNA backbone along the major groove (yellow circle in Figure 5B). Interestingly, the ‘Western’ rim of class I photolyases as AnCPDI (Mees et al, 2004) or (6-4) photolyases (Maul et al, 2008) is built up by a distinct structural element, a long C-terminal helix preserved only in class I and (6-4) photolyases, but missing in MmCPDII and in cry3 from A. thaliana (Pokorny et al, 2008; Supplementary Figure S1D). The lack of a pronounced Western rim may explain, why the latter two enzymes are more prone to non-specific duplex DNA binding than class I and (6-4) photolyases.
Figure 5.
Surface features of the photolyase-cryptochrome family bound to DNA lesions. Electrostatic potentials were calculated with APBS (Baker et al, 2001) using a concentration of 0.1 M for monovalent ions and a solvent radius of 1.4 Å. (A) Electrostatic surface properties of photolyase subclasses and a DASH cryptochrome. The wind rose indicates the orientation of the canyon-like DNA-binding surface. (B) Electrostatic surface properties of the MmCPDII•CPD–DNA complex illustrate the areas (coloured circles) of the photolyase responsible for interactions with the sugar-phosphate backbone of the CPD-DNA.
Further interactions with the deoxyribophosphate backbone of the CPD lesion comprising DNA strand are given at position P−1 by the basic side chain of R164 and the amide groups of A160 and H161, respectively (red circle in Figure 5B). Additional electrostatic stabilizations of the recognized phosphates occur in both areas via the dipole moments of helices α6 and α18, respectively (Mees et al, 2004).
Compared with uncomplexed double-stranded CPD–DNA, the overall deformation of bound duplex DNA (Supplementary Figure S1C) is hallmarked by an increased kink from 27–30° (Pearlman et al, 1985; Husain et al, 1988; Park et al, 2002) to ∼50° as result of the binding event. Although the overall kinking is almost the same as in the AnCPDI•CPD–DNA complex due to the double flip of the thymine dimer into the active site, an alternative view reveals an additional dislocation by 45° of the 3′-arm of the CPD–DNA in the MmCPDII•CPD–DNA complex. As the 3′-arm makes most of the specific interactions between its P+1, P+2, P+3 phosphates and the enzyme surface, there are no major changes for the backbone torsions of the DNA apart from a different pucker of the deoxyribose of C9. Nevertheless, the altered binding mode of the 3′-arm causes interactions, which are specific for class II but not for class I photolyases.
Binding of duplex CPD-DNA goes along with the double flip of the thymine dimer into the active site and partial unwinding of duplex DNA, thus creating free space with a section of ∼120 Å2. The π-stacking of the complementary adenines among each other remains intact, but the contact to adjacent bases is partly disordered. In the AnCPDI•CPD–DNA complex, the hole caused by unwinding and double flip is partly occupied by the loop connecting α17 and α18 (Mees et al, 2004). In MmCPDII, interactions between the side chain of R429 and the N3 atom of the purine ring of the counter base A8 as well as intimate van-der-Waals packing with the neighbouring base of G9 suggest a stabilizing role of this residue for the flip of the 5′-thymine into the active site. In complex A, W431 is rotated approximately by 90° as compared with the uncomplexed state of MmCPDII and the indole nitrogen forms there a hydrogen bond with the C2 carbonyl of a neighbouring cytidine. The position of the rigid salt bridge D428/R441 is altered in such a way that R441 gets into distance for stabilizing π-stacking interactions with W431. This combination closely fits into the hole between the CPD lesion and the counter bases and might hence act like a bolt in unwinding the duplex DNA and flipping the thymine dimer into the catalytic position, respectively (Supplementary Figure S1E).
FAD-binding site and electron transfer pathway in class II photolyases
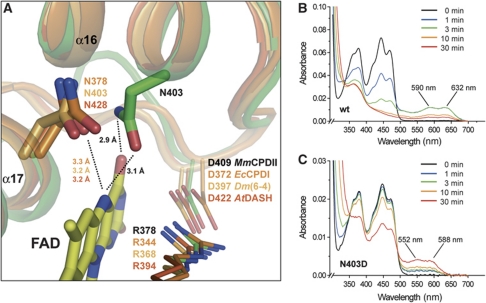
The binding site of the catalytic FAD in MmCPDII structurally resembles other members of the photolyase-cryptochrome family. Half of the residues lining the FAD site (8/16) are hence conserved like the salt bridge R378-D409 on top of the isoalloxazine ring (Figure 6A). However, there are changes along the FAD-binding site of class II photolyases which affect their mechanism for photoreduction. The first change relates to the stabilizing interaction between the N5 atom of the isoalloxazine ring and an asparagine side chain nearby. In the structure of MmCPDII the highly conserved, but class II-specific asparagine N403 on helix α16 projects its side chain to form a putative H-bond between its carboxamide nitrogen and the O4 oxygen of the FAD cofactor (Figure 6A). Furthermore, the carboxamide's oxygen is suitably positioned to stabilize protonation of the N5 nitrogen of FAD during photoreduction. Other photolyases including the DASH cryptochromes utilize instead a topologically distinct asparagine (e.g., EcCPDI: N378) that is part of helix α17 (Figure 6A) for stabilizing the neutral FADH• radical during photoreduction and DNA repair (Xu et al, 2008). In plant cryptochromes, this asparagine is replaced by an aspartate causing photocycle arrest in the signaling-competent FADH• radical state. In class II photolyases, the corresponding position is occupied by a conserved glycine (MmCPDII: G415; Supplementary Figure S4). Interestingly, for charge compensation during photoreduction to FADH− the side chain of N403 is in H-bonding distance to the surface-exposed D404, whereas in class I enzymes this role is taken up by a glutamate one turn of the α16-helix upward (EcCPDI: E363, AnCPDI: E371, TtCPDI: E332).
Figure 6.
FAD-binding site of a class II photolyase. Structural comparison of M. mazei class II photolyase (green) with members of photolyases and DASH cryptochromes (shades of orange) shows (A) a different location of the conserved asparagine residue, which is crucial for stabilizing the neutral radical state of the catalytic cofactor. UV/Vis spectra of photoreduction via illumination at 450 nm in the presence of the reducing agent DTT of MmCPDII wild-type (B) and the N403D mutant (C).
To demonstrate the crucial role of asparagine N403 during photoreduction, two different mutagenesis approaches were performed. The replacement of asparagine N403 for either a non-polar alanine or a hydrophobic leucine causes complete loss of the catalytic FAD during purification by size exclusion chromatography (unpublished observations). The liability of FAD binding in N403A and N403L mutants emphasizes the stabilizing role of asparagine N403 in class II photolyases. In contrast, the N403D mutant has an FAD incorporation of at least 70% as judged from 280/450 nm absorption ratios of the purified enzyme. The replacement of N403 by an aspartate provokes a photocycle arrest in the FADH• state of the N403D mutant upon illumination (Figure 6C). The ready formation of the FADH• state, but lack of further reduction to FADH− is reminiscent of the photochemistry of plant cryptochromes. The absorption peaks of the FADH• species are blue shifted from 590/632 nm of the wild type to 550/588 nm of the N403D mutant, the blue-shifted absorption of the FADH• species is also a hallmark of plant cryptochromes. Overall, this implies that independent of its origin the placement of an anionic charge close to the N5 nitrogen of the catalytic FAD is sufficient for photocycle arrest and blue shift of the FADH• state.
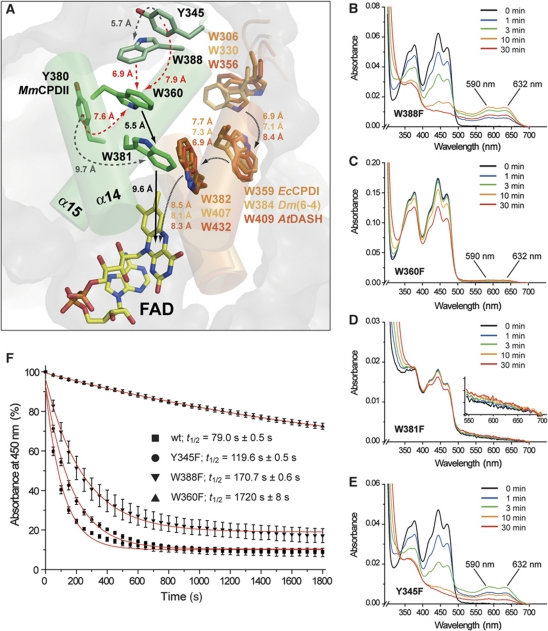
The second major structural change affects photoreduction as well, especially the pathway along which electrons are transferred to oxidized FAD species upon light excitation. In members of the photolyase-cryptochrome family other than class II (Park et al, 1995; Klar et al, 2007; Muller and Carell, 2009), this pathway extends from a tryptophan near the isoalloxazine ring on helix α17 via a residue on α16 to a surface-exposed tryptophan with intervening distances of 7–8 Å. In the M. mazei photolyase as well as other class II enzymes this linear pathway is missing. According to sequence and structure conservation an analogous tryptophan triad (W388 → W360 → W381 → FAD) can be postulated leading from the protein surface to the catalytic cofactor using class II-specific tryptophans located on helices α14 and α15 (Figure 7A). This new type of a tryptophan triad has been postulated before for the O. sativa class II photolyase (W406 → W378 → W399) (Okafuji et al, 2010). Apart from a slightly larger distance between the proximal tryptophan W381 and the isoalloxazine ring (9.6 versus 8.3 Å; Figure 7A), the intervening distances between the tryptophans are smaller or similar as in class I photolyases, for example, W381 and W360 are only 5.5 Å apart and the terminal tryptophan W388 is within 6.9 Å of W360. To prove the function of this electron transfer pathway W388, W360 and W381 as well as the tyrosines Y345 and Y380 next to it were mutated to phenylalanine. All mutants showed proper incorporation of the catalytic FAD comparable to wild type except the W381F mutant which had an incorporation of ∼30% as judged by its 280/450 nm ratio. Nevertheless, all UV/Vis spectra are similar to the wild type (Figure 7B–E). As expected ablation of the proximal tryptophan, W381, in the W381F mutant causes complete loss of photoreduction activity indicating that this innermost tryptophan is crucial for catalytic activity. Mutation of the medial tryptophan, W360, gives a 22-fold decrease of photoreduction activity relative to wild type (Figure 7F). Particularly, no major build-up of the semiquinoid FADH• species can be observed for W360F (Figure 7C), indicating that a transiently formed FADH•/W381+• radical pair cannot be efficiently reduced by W360F. In contrast, the surface-exposed variant W388F still has a residual activity of 46% and can be thus not regarded as an essential part of a tryptophan triad.
Figure 7.
Electron transfer pathway along the tryptophan dyad of the M. mazei class II photolyase. (A) The conserved tryptophans W381, W360 and W388 are located on different structural elements in MmCPDII than in other members of the photolyase-cryptochrome family. Tyrosine residues that are possibly involved as branches in the electron transfer pathway and exposed along the protein surface are widely conserved in the class II photolyase subfamily (Supplementary Figure S4B). (B–E) UV/Vis spectra of MmCPDII pathway mutants in the presence of the reducing agent DTT. Photoreduction was performed as described. The inlay of Figure 6D illustrates that the innermost mutant W381F is locked in the oxidized state, remaining changes are caused by aggregation. (F) During photoreduction under identical conditions, absorbance at 450 nm was recorded and fitted by a first-order exponential decay to determine half times of wild type and mutants. The Y380F mutant has wild-type activity (t1/2=74.3±0.6 s, data not shown). W388 is not essential for photoreduction due to its 46% residual activity.
The residual activities of 4.6 and 46% for the W360F and W388F mutants, respectively, indicate that class II photolyases do not depend on a linear tryptophan triad like their counterparts in the photolyase/cryptochrome family, but just on a tryptophan dyad. As its outmost residue, W360, is still buried below the protein surface, class II photolyases have either to bypass or use a branched electron transfer pathway to reduce the oxidized W360+• radical. Bypassing could give rise to the 4.6% residual activity of the W360F mutant, for example, by using Y380 as an alternative electron donor for the transiently formed FADH•/W381+• radical pair species. W360 is suitable as a branch point for the electron transfer pathway, because this residue is close to at least three aromatic residues for accepting electrons: W388, Y345 and Y380. Together with the likewise surface-exposed residue Y442 all of these residues are highly conserved in class II photolyases (Supplementary Figure S4). A redundant role for these residues is underlined by the observation that the Y345F and Y380F mutants have only slightly affected rates for photoreduction (Figure 7F). Overall, the conserved surface-exposed tyrosines may be hence involved in collecting electrons for transfer to the catalytic cofactor by acting as multiple, redundant termination points of this pathway.
Conclusions
The first structure of a class II photolyase from the archaeon M. mazei reveals unique structural features in this phylogenetically divergent photolyase clade. Amino acids crucial for photoreduction and DNA-repair activity are either functionally conserved but positioned on different secondary structure elements or replaced by analogous elements like the water cluster involved in 3′-thymine recognition. These differences or a late arrival of class II photolyases during evolution may explain why no cryptochrome-like photoreceptors have been yet discovered which are derived from class II photolyases.
The availability of an unrepaired crystalline MmCPDII•CPD–DNA complex provides access to detailed studies about structural changes of amino acids involved in DNA repair and the back-flipping of repaired DNA, respectively. Finally, the loss of a bulky C-terminal element commonly used by microbial class I photolyases as constitutive element of the binding groove for dsDNA may imply that the C-terminally truncated class II photolyases of eukaryotes have improved access to UV-damaged DNA outside the nucleosome-packaged regions (Thoma, 2005). Given the proven potential of class II photolyases to improve the UV resistance of plants, viruses and animals (Schul et al, 2002; Jans et al, 2005; Kaiser et al, 2009), the structure-based engineering of these photolyases, for example, for the binding of different antenna chromophores, or the design of class-specific photolyase inhibitors is now a feasible route for future research.
Materials and methods
Cloning, overexpression and purification of MmCPDII
The Mm0852 gene encoding the class II photolyase of M. mazei Go1 (UniProt entry Q8PYK9_METMA) was amplified from genomic DNA with primers 5′-CTAGGCCATATGAATCCGAAACGTATCAGG-3′ and 5′-CCCCTCGAGCTACAAAGCTGAATATTTTTC-3′ (Invitrogen) introducing recognition sites for NdeI and XhoI (underlined). The PCR product was inserted between NdeI and XhoI sites of the vector pET-28a (Novagen) yielding an N-terminally His6-tagged fusion protein. Sequencing showed a point mutation within the Mm0852 sequence leading to the amino-acid replacement M377T as compared with the annotated sequence. This residue on helix α15 was later found to be non-relevant for DNA or cofactor binding. E. coli BL21-Gold (DE3) (Stratagene) cells were transformed with the pET-28a-MmCPDII plasmid. Cells were grown at 25°C for 24 h in terrific broth medium and gene expression was controlled by autoinduction. After cell harvesting, pellets were resuspended in buffer I (50 mM NaH2PO4, 300 mM NaCl, pH 8.0), frozen in liquid nitrogen and stored at −80°C. Cell disruption was performed in the presence of lysozyme, EDTA and PMSF with an emulsifier (Avestin). After centrifugation, the supernatant was applied onto an Ni-NTA column (Qiagen) and the recombinant photolyase was eluted at a concentration of 125 mM imidazole. After control on a 12% SDS-PAGE, yellowish fractions were combined before concentrating the protein with an Amicon Ultra concentrator (Millipore, 30 kDa cutoff). As a final polishing step, size exclusion chromatography with a Superdex200 column and buffer II (10 mM Tris–HCl, 100 mM NaCl, pH 8.0) was used and yielded >95% pure protein. M. mazei photolyase was concentrated with an Amicon Ultra concentrator (Millipore) and stored at 4°C.
The generation and production of mutant MmCPDII enzymes as well as binding assays and spectral and kinetic characterization have been described in Supplementary data.
Crystallization and structure determination of M. mazei photolyase
Initial crystallization attempts were performed with a Cartesian robot system using commercially available screens (Qiagen) in 96-well format. Microcrystals were obtained at 4°C and a protein concentration of 7.7 mg ml−1 within 24 h in a condition containing 0.5 M lithium sulphate and 7.5% (w/v) PEG 8000. Optimization was done using the hanging drop vapour diffusion method in 24-well format. Yellowish crystals for data collection were obtained at 4°C and a protein concentration of 7 mg ml−1 after 3–5 days in a condition containing 0.5 M lithium sulphate and 7.5% PEG 6000 (Supplementary Figure S2A and B). Crystals were soaked in the latter solution containing 35% glycerol as cryoprotectant and flash frozen in liquid nitrogen. X-ray data were collected from a single crystal at 100 K at the beamline ID29, European Synchrotron Radiation Facility (ESRF), Grenoble, France, with an ADSC Q315r detector. M. mazei photolyase was crystallized in space group P43212 with unit cell parameters a=b=69.77 Å, c=243.08 Å and α=β=γ=90°. Data reduction was carried out using XDS and XSCALE (Kabsch, 1993). The structure was solved by molecular replacement using PHASER (McCoy et al, 2007) of the CCP4 package (CCP4, 1994) and structural data of the Methanosarcina barkeri class II photolyase (Supplementary Table SI). The PHASER solution with data up to 2.5 Å yielded one molecule per asymmetric unit with z scores of 16.9 for the rotation and 38.5 for the translation function. Semiautomatic tracing and model rebuilding were done by ARP/wARP (Langer et al, 2008). Further refinement using COOT (Emsley et al, 2010) and REFMAC5 (Murshudov et al, 1997) at 1.5 Å resolution led to R-factors of R=18.5% and Rfree=21.0%.
Crystallization and structure determination of the MmCPDII•CPD–DNA complex
A 14-meric ssDNA oligonucleotide comprising a CPD lesion (5′-ATCGGCT<>TCGCGCA-3′) with a synthetic formacetal-linked thymine dimer (Mees et al, 2004) was provided by Andreas Glas, Ludwig-Maximilians University, Munich. The CPD-DNA oligonucleotide was annealed with a complementary single-stranded oligonucleotide (Metabion) by cooling down the 1:1 mixture from 95°C to room temperature. All further steps were done under safe red light to avoid light-induced DNA repair. For initial crystallization attempts, M. mazei photolyase with a final protein concentration of 6.2 mg ml−1 was incubated with a 1.25-fold molar excess of hybridized dsDNA oligonucleotide in the dark at 4°C for at least 12 h. Several commercially available screens (Qiagen) were performed with a Cartesian robot system in 96-well format and kept in darkness at 4°C. First microcrystals were obtained after 2–3 days in a condition containing 0.2 M ammonium sulphate, 0.1 M sodium acetate pH 4.6 and 10% PEG 4000. Optimization was done using the hanging drop vapour diffusion technique in 24-well format. Cocrystals for data collection grew in the dark within 24 h at 18°C in a condition containing 0.2 M ammonium sulphate, 0.1 M sodium acetate pH 4.6, 10% PEG 4000 and 10% glycerol (Supplementary Figure S2C and D). Cryosoaking of cocrystals was done in crystallization buffer supplemented with 35% glycerol as cryoprotectant. Cocrystals were flash frozen in liquid nitrogen. Diffraction data were collected from a single crystal at 100 K at the beamline ID14-1, European Synchrotron Radiation Facility (ESRF), Grenoble, France, with an ADSC Q210 detector. The CPD-DNA•photolyase complex crystallized in space group P212121 with unit cell parameters a=71.7 Å, b=116.2 Å, c=168.3 Å and α=β=γ=90°. After data reduction using XDS and XSCALE (Kabsch, 1993), molecular replacement was performed by PHASER (McCoy et al, 2007) using structural data of the uncomplexed M. mazei photolyase with data up to 2.5 Å. Two molecules per asymmetric unit were found with z scores of 21.2 for rotation and 36.0 for translation function for the first molecule and z scores of 17.4 and 66.4 for the second molecule, respectively. Refinement at 2.2 Å using COOT (Emsley et al, 2010) and REFMAC5 (Murshudov et al, 1997) led to a final model with R-factors of R=17.0% and Rfree=20.8% for the complex.
The data collection and refinement statistics are summarized in Table I. Protein interfaces and intermolecular interactions were calculated using the Protein Interfaces, Surfaces and Assemblies Service at the European Bioinformatics Institute (http://www.ebi.ac.uk/msd-srv/prot_int/pistart.html) (Krissinel and Henrick, 2007). Secondary structures of MmCPDII were assigned by the program STRIDE (Heinig and Frishman, 2004); PYMOL0.99 (DeLano, 2002) was used for graphical analysis and visualization.
Table 1. Crystallographic statistics of MmCPDII and CPD-DNA•photolyase complex.
| Data collection and processing | MmCPDII | MmCPDII•CPD–DNA |
|---|---|---|
| X-ray source | ID29 | ID14-1 |
| ESRF, Grenoble, France | ||
| Detector | ADSC Q315r | ADSC Q210 |
| Wavelength (Å) | 0.88567 | 0.93400 |
| Space group | P43212 | P212121 |
| Cell dimensions (a, b, c Å) | 69.77, 69.77, 243.08 | 71.70, 116.20, 168.30 |
| Resolution (Å) | 19.94-1.50 (1.58–1.50) | 29.48-2.20 (2.32–2.20) |
| Total reflections | 515668 | 354320 |
| Multiplicity | 5.4 (5.4) | 4.9 (4.9) |
| Unique reflections | 95566 | 72130 |
| Rmerge (%) | 8.5 (39.3) | 7.9 (68.7) |
| Completeness (%) | 98.4 (98.8) | 99.9 (100.0) |
| I/σ(I) | 11.8 (4.4) | 17.5 (2.6) |
| Mosaicity (deg) | 0.197 | 0.099 |
| Wilson B-factor (Å2) | 12.6 | 35.6 |
| Refinement statistics | ||
| Resolution (Å) | 15.0-1.50 | 29.5-2.20 |
| Rfactor, Rfree (%) | 18.5, 21.0 | 17.0, 20.8 |
| Reflections (working, test set) | 94 089, 1416 | 70 962, 1094 |
| Completeness for range (%) | 98.3 | 99.9 |
| R.m.s.d. from ideal | ||
| Bond lengths (Å) | 0.012 | 0.012 |
| Bond angles (deg) | 1.378 | 1.363 |
| Total number of atoms | 4270 | 8978 |
| Mean B value (Å2) | 11.4 | 29.7 |
Data deposition
The coordinates and structure factors have been deposited at the Protein Data Bank (PDB), Research Collaboratory for Structural Bioinformatics (RCSB), with accession codes 2XRY for the MmCPDII structure and 2XRZ for the MmCPDII•CPD-DNA complex, respectively.
Supplementary Material
Acknowledgments
We are grateful to Volker Müller for genomic DNA of Methanosarcinales; Andreas Glas for samples of the CPD–DNA; Petra Gnau for technical support during crystallization; Jo Mailliet and Ying Zhang for molecular biological work on methanosarcinal photolyases; staff of the beamlines ID14-1 and ID29 at the European Synchrotron Radiation Facility (ESRF), Grenoble, France, for support with data collection. Special thanks are due to David von Stetten and Antoine Royant for their excellent support with in crystallo UV/Vis spectroscopy. This work was supported by a grant of the Volkswagen-Stiftung to L-OE and TC, and the DFG grant BA 985/10-3 to AB.
Author contributions: SK, RP, AB and L-OE designed research; SK, YG and RP performed research; TC contributed new reagents/analytic tools; SK, YG, RP, AB and L-OE analysed data; SK, AB and L-OE wrote the paper.
Footnotes
The authors declare that they have no conflict of interest.
References
- Acocella A, Jones GA, Zerbetto F (2010) What is adenine doing in photolyase? J Phys Chem B 114: 4101–4106 [DOI] [PubMed] [Google Scholar]
- Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA (2001) Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci USA 98: 10037–10041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biertümpfel C, Zhao Y, Kondo Y, Ramón-Maiques S, Gregory M, Lee JY, Masutani C, Lehmann AR, Hanaoka F, Yang W (2010) Nature 465: 1044–1048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brautigam CA, Smith BS, Ma Z, Palnitkar M, Tomchick DR, Machius M, Deisenhofer J (2004) Structure of the photolyase-like domain of cryptochrome 1 from Arabidopsis thaliana. Proc Natl Acad Sci USA 101: 12142–12147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brudler R, Hitomi K, Daiyasu H, Toh H, Kucho K, Ishiura M, Kanehisa M, Roberts VA, Todo T, Tainer JA, Getzoff ED (2003) Identification of a new cryptochrome class. Structure, function, and evolution. Mol Cell 11: 59–67 [DOI] [PubMed] [Google Scholar]
- Butenandt J, Eker APM, Carell T (1998) Synthesis, crystal structure, and enzymatic evaluation of a DNA-photolesion isostere. Chem Eur J 4: 642–654 [Google Scholar]
- CCP4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr 50: 760–763 [DOI] [PubMed] [Google Scholar]
- DeLano WL (2002) The PyMOL Molecular Graphics System. DeLano Scientific: San Carlos, CA, USA http://www.pymol.org
- Emsley P, Lohkamp B, Scott WG, Cowtan K (2010) Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66: 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essen LO (2006) Photolyases and cryptochromes: common mechanisms of DNA repair and light-driven signaling? Curr Opin Struct Biol 16: 51–59 [DOI] [PubMed] [Google Scholar]
- Essen LO, Siegert R, Lehmann WD, Oesterhelt D (1998) Lipid patches in membrane protein oligomers: crystal structure of the bacteriorhodopsin-lipid complex. Proc Natl Acad Sci USA 95: 11673–11678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falciatore A, Bowler C (2005) The evolution and function of blue and red light photoreceptors. Curr Top Dev Biol 68: 317–350 [DOI] [PubMed] [Google Scholar]
- Finn RD, Mistry J, Tate J, Coggill P, Heger A, Pollington JE, Gavin OL, Gunasekaran P, Ceric G, Forslund K, Holm L, Sonnhammer EL, Eddy SR, Bateman A (2010) The Pfam protein families database. Nucleic Acids Res 38 (Database issue): D211–D222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujihashi M, Numoto N, Kobayashi Y, Mizushima A, Tsujimura M, Nakamura A, Kawarabayasi Y, Miki K (2007) Crystal structure of archaeal photolyase from Sulfolobus tokodaii with two FAD molecules: implication of a novel light-harvesting cofactor. J Mol Biol 365: 903–910 [DOI] [PubMed] [Google Scholar]
- Garczarek F, Brown LS, Lanyi JK, Gerwert K (2005) Proton binding within a membrane protein by a protonated water cluster. Proc Natl Acad Sci USA 102: 3633–3638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heelis PF, Kim ST, Okamura T, Sancar A (1993) The photo repair of pyrimidine dimers by DNA photolyase and model systems. J Photochem Photobiol B 17: 219–228 [DOI] [PubMed] [Google Scholar]
- Heinig M, Frishman D (2004) STRIDE: a web server for secondary structure assignment from known atomic coordinates of proteins. Nucleic Acids Res 32(Web Server issue): W500–W502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrischk AK, Fruhwirth SW, Moldt J, Pokorny R, Metz S, Kaiser G, Jager A, Batschauer A, Klug G (2009) A cryptochrome-like protein is involved in the regulation of photosynthesis genes in Rhodobacter sphaeroides. Mol Microbiol 74: 990–1003 [DOI] [PubMed] [Google Scholar]
- Hill AV (1910) The possible effects of the aggregation of the molecules of hæmoglobin on its dissociation curves. J Physiol 40 (Suppl): i–vii [Google Scholar]
- Hitomi K, DiTacchio L, Arvai AS, Yamamoto J, Kim ST, Todo T, Tainer JA, Iwai S, Panda S, Getzoff ED (2009) Functional motifs in the (6-4) photolyase crystal structure make a comparative framework for DNA repair photolyases and clock cryptochromes. Proc Natl Acad Sci USA 106: 6962–6967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang YH, Baxter R, Smith BS, Partch CL, Colbert CL, Deisenhofer J (2006) Crystal structure of cryptochrome 3 from Arabidopsis thaliana and its implications for photolyase activity. Proc Natl Acad Sci USA 103: 17701–17706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husain I, Griffith J, Sancar A (1988) Thymine dimers bend DNA. Proc Natl Acad Sci USA 85: 2558–2562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwamatsu Y, Aoki C, Takahashi M, Teranishi M, Ding Y, Sun C, Kumagai T, Hidema J (2008) UVB sensitivity and cyclobutane pyrimidine dimer (CPD) photolyase genotypes in cultivated and wild rice species. Photochem Photobiol Sci 7: 311–320 [DOI] [PubMed] [Google Scholar]
- Jans J, Schul W, Sert YG, Rijksen Y, Rebel H, Eker APM, Nakajima S, van Steeg H, de Gruijl FR, Yasui A, Hoeijmakers JHJ, van der Horst GTJ (2005) Powerful skin cancer protection by a CPD-photolyase transgene. Curr Biol 15: 105–115 [DOI] [PubMed] [Google Scholar]
- Jorns MS, Sancar GB, Sancar A (1985) Identification of oligothymidylates as new simple substrates for Escherichia coli DNA photolyase and their use in a rapid spectrophotometric enzyme assay. Biochemistry 24: 1856–1861 [DOI] [PubMed] [Google Scholar]
- Kabsch W (1993) Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J Appl Crystallogr 26: 795–800 [Google Scholar]
- Kaiser G, Kleiner O, Beisswenger C, Batschauer A (2009) Increased DNA repair in Arabidopsis plants overexpressing CPD photolyase. Planta 230: 505–515 [DOI] [PubMed] [Google Scholar]
- Kanai S, Kikuno R, Toh H, Ryo H, Todo T (1997) Molecular evolution of the photolyase-blue-light photoreceptor family. J Mol Evol 45: 535–548 [DOI] [PubMed] [Google Scholar]
- Klar T, Pokorny R, Moldt J, Batschauer A, Essen LO (2007) Cryptochrome 3 from Arabidopsis thaliana: structural and functional analysis of its complex with a folate light antenna. J Mol Biol 366: 954–964 [DOI] [PubMed] [Google Scholar]
- Kleiner O, Butenandt L, Carell T, Batschauer A (1999) Class II DNA photolyase from Arabidopsis thaliana contains FAD as a cofactor. Eur J Biochem 264: 161–167 [DOI] [PubMed] [Google Scholar]
- Komori H, Masui R, Kuramitsu S, Yokoyama S, Shibata T, Inoue Y, Miki K (2001) Crystal structure of thermostable DNA photolyase: pyrimidine-dimer recognition mechanism. Proc Natl Acad Sci USA 98: 13560–13565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kort R, Komori H, Adachi S, Miki K, Eker A (2004) DNA apophotolyase from Anacystis nidulans: 1.8 A structure, 8-HDF reconstitution and X-ray-induced FAD reduction. Acta Crystallogr D Biol Crystallogr 60: 1205–1213 [DOI] [PubMed] [Google Scholar]
- Krissinel E, Henrick K (2007) Inference of macromolecular assemblies from crystalline state. J Mol Biol 372: 774–797 [DOI] [PubMed] [Google Scholar]
- Langer G, Cohen SX, Lamzin VS, Perrakis A (2008) Automated macromolecular model building for X-ray crystallography using ARP/wARP version 7. Nat Protoc 3: 1171–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C, Todo T (2005) The cryptochromes. Genome Biol 6: 220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas-Lledo JI, Lynch M (2009) Evolution of mutation rates: phylogenomic analysis of the photolyase/cryptochrome family. Mol Biol Evol 26: 1143–1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mailliet J, Psakis G, Schroeder C, Kaltofen S, Durrwang U, Hughes J, Essen LO (2009) Dwelling in the dark: procedures for the crystallography of phytochromes and other photochromic proteins. Acta Crystallogr D Biol Crystallogr 65: 1232–1235 [DOI] [PubMed] [Google Scholar]
- Masson F, Laino T, Rothlisberger U, Hutter J (2009) A QM/MM investigation of thymine dimer radical anion splitting catalyzed by DNA photolyase. Chemphyschem 10: 400–410 [DOI] [PubMed] [Google Scholar]
- Maul MJ, Barends TR, Glas AF, Cryle MJ, Domratcheva T, Schneider S, Schlichting I, Carell T (2008) Crystal structure and mechanism of a DNA (6-4) photolyase. Angew Chem Int Ed Engl 47: 10076–10080 [DOI] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ (2007) Phaser crystallographic software. J Appl Crystallogr 40: 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mees A, Klar T, Gnau P, Hennecke U, Eker AP, Carell T, Essen LO (2004) Crystal structure of a photolyase bound to a CPD-like DNA lesion after in situ repair. Science 306: 1789–1793 [DOI] [PubMed] [Google Scholar]
- Muller M, Carell T (2009) Structural biology of DNA photolyases and cryptochromes. Curr Opin Struct Biol 19: 277–285 [DOI] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr 53: 240–255 [DOI] [PubMed] [Google Scholar]
- Okafuji A, Biskup T, Hitomi K, Getzoff ED, Kaiser G, Batschauer A, Bacher A, Hidema J, Teranishi M, Yamamoto K, Schleicher E, Weber S (2010) Light-induced activation of class II cyclobutane pyrimidine dimer photolyases. DNA Repair 9: 495–505 [DOI] [PubMed] [Google Scholar]
- Ozturk N, Kao YT, Selby CP, Kavakli IH, Partch CL, Zhong D, Sancar A (2008) Purification and characterization of a type III photolyase from Caulobacter crescentus. Biochemistry 47: 10255–10261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H, Zhang K, Ren Y, Nadji S, Sinha N, Taylor JS, Kang C (2002) Crystal structure of a DNA decamer containing a cis-syn thymine dimer. Proc Natl Acad Sci USA 99: 15965–15970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park HW, Kim ST, Sancar A, Deisenhofer J (1995) Crystal structure of DNA photolyase from Escherichia coli. Science 268: 1866–1872 [DOI] [PubMed] [Google Scholar]
- Payne G, Wills M, Walsh C, Sancar A (1990) Reconstitution of Escherichia coli photolyase with flavins and flavin analogues. Biochemistry 29: 5706–5711 [DOI] [PubMed] [Google Scholar]
- Pearlman DA, Holbrook SR, Pirkle DH, Kim SH (1985) Molecular models for DNA damaged by photoreaction. Science 227: 1304–1308 [DOI] [PubMed] [Google Scholar]
- Pokorny R, Klar T, Hennecke U, Carell T, Batschauer A, Essen LO (2008) Recognition and repair of UV lesions in loop structures of duplex DNA by DASH-type cryptochrome. Proc Natl Acad Sci USA 105: 21023–21027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prytkova TR, Beratan DN, Skourtis SS (2007) Photoselected electron transfer pathways in DNA photolyase. Proc Natl Acad Sci USA 104: 802–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancar A (2003) Structure and function of DNA photolyase and cryptochrome blue-light photoreceptors. Chem Rev 103: 2203–2237 [DOI] [PubMed] [Google Scholar]
- Schul W, Jans J, Rijksen YM, Klemann KH, Eker AP, de Wit J, Nikaido O, Nakajima S, Yasui A, Hoeijmakers JH, van der Horst GT (2002) Enhanced repair of cyclobutane pyrimidine dimers and improved UV resistance in photolyase transgenic mice. EMBO J 21: 4719–4729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selby CP, Sancar A (2006) A cryptochrome/photolyase class of enzymes with single-stranded DNA-specific photolyase activity. Proc Natl Acad Sci USA 103: 17696–17700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song SH, Dick B, Penzkofer A, Pokorny R, Batschauer A, Essen LO (2006) Absorption and fluorescence spectroscopic characterization of cryptochrome 3 from Arabidopsis thaliana. J Photochem Photobiol B 85: 1–16 [DOI] [PubMed] [Google Scholar]
- Tamada T, Kitadokoro K, Higuchi Y, Inaka K, Yasui A, de Ruiter PE, Eker AP, Miki K (1997) Crystal structure of DNA photolyase from Anacystis nidulans. Nat Struct Biol 4: 887–891 [DOI] [PubMed] [Google Scholar]
- Thoma F (2005) Repair of UV lesions in nucleosomes – intrinsic properties and remodeling. DNA Repair 4: 855–869 [DOI] [PubMed] [Google Scholar]
- Vande Berg BJ, Sancar GB (1998) Evidence for dinucleotide flipping by DNA photolyase. J Biol Chem 273: 20276–20284 [DOI] [PubMed] [Google Scholar]
- Xu L, Mu W, Ding Y, Luo Z, Han Q, Bi F, Wang Y, Song Q (2008) Active site of Escherichia coli DNA photolyase: Asn378 is crucial both for stabilizing the neutral flavin radical cofactor and for DNA repair. Biochemistry 47: 8736–8743 [DOI] [PubMed] [Google Scholar]
- Yasui A, Eker AP, Yasuhira S, Yajima H, Kobayashi T, Takao M, Oikawa A (1994) A new class of DNA photolyases present in various organisms including aplacental mammals. EMBO J 13: 6143–6151 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.