Abstract
The oncogenic cluster miR-17-92 encodes seven related microRNAs that regulate cell proliferation, apoptosis and development. Expression of miR-17-92 cluster is decreased upon cell differentiation. Here, we report a novel mechanism of the regulation of miR-17-92 cluster. Using transgenic PU.1−/− myeloid progenitors we show that upon macrophage differentiation, the transcription factor PU.1 induces the secondary determinant Egr2 which, in turn, directly represses miR-17-92 expression by recruiting histone demethylase Jarid1b leading to histone H3 lysine K4 demethylation within the CpG island at the miR-17-92 promoter. Conversely, Egr2 itself is targeted by miR-17-92, indicating existence of mutual regulatory relationship between miR-17-92 and Egr2. Furthermore, restoring EGR2 levels in primary acute myeloid leukaemia blasts expressing elevated levels of miR-17-92 and low levels of PU.1 and EGR2 leads to downregulation of miR-17-92 and restored expression of its targets p21CIP1 and BIM. We propose that upon macrophage differentiation PU.1 represses the miR-17-92 cluster promoter by an Egr-2/Jarid1b-mediated H3K4 demethylation mechanism whose deregulation may contribute to leukaemic states.
Keywords: AML, chromatin modification, differentiation, miR-17-92, PU.1
Introduction
Haematopoietic differentiation is a highly ordered multistep process comprising a gene circuitry that enables multipotential progenitors to generate various specialized and distinct blood cells. The successful generation of terminal blood cells is dependent upon coordinated regulation of gene expression by key regulators: transcription factors and microRNAs (Nerlov and Graf, 1998; Orkin and Zon, 2002; Rosenbauer and Tenen, 2007; Garzon and Croce, 2008). Despite their importance, the mutual interplay between these regulators is not completely understood. There exists homeostatic mechanisms, that if lost by aberrations in transcription factors (Okuda et al, 1996; Schmidt et al, 1998) or miRNA function (Volinia et al, 2010) can upset the balance of cell death, proliferation and differentiation resulting in a shift towards cancer. MicroRNAs are non-coding RNAs (19–23 nt) that interfere with the translation or RNA stability of up to hundreds of targets by complementary pairing to the target site(s) located within the 3′untranslated region of mRNAs. MicroRNAs are coded individually, or are clustered in polycistrons, and are transcribed by RNA Polymerase II as a single pri-miRNA transcript which is subsequently processed by RNAses Drosha and Dicer to generate individual mature miRNAs (Bartel, 2004).
The polycistron harbouring the miR-17-92 cluster (Oncomir-1) encodes six hairpin transcripts carrying six miRNAs (miR-17, miR-18a, miR-19a, miR-20a, miR-19b-1, and miR-92a) (Figure 1A), located on human chromosome 13 (mouse Ch14), within the third intron of the primary transcript C13orf25 (Ota et al, 2004). There are two additional paralogous clusters: miR-106a-363 and miR-106b-25, and also one related, an individually coded miR-92b. Collectively, these clusters encode 15 individual miRNAs each with similar or identical sequences to the miR-17-92 components that are highly conserved in vertebrates (Tanzer and Stadler, 2004). Previous work demonstrated crucial roles for miR-17-92 during cancer development. miR-17-92 is overexpressed in B-cell lymphomas (Ota et al, 2004; He et al, 2005; Tagawa and Seto, 2005), acute lymphoid and myeloid leukaemias (Dixon-McIver et al, 2008; Li et al, 2008), chronic myeloid leukaemia (Venturini et al, 2007), and solid tumours (Hayashita et al, 2005; Volinia et al, 2006; Petrocca et al, 2008a). In certain malignancies including human diffuse large B-cell lymphoma, overexpression of miR-17-92 results from amplification of the genomic region harbouring the miR-17-92 host gene (Ota et al, 2004); however, the mechanisms leading to the increased miR-17-92 expression in tumours remain poorly understood.
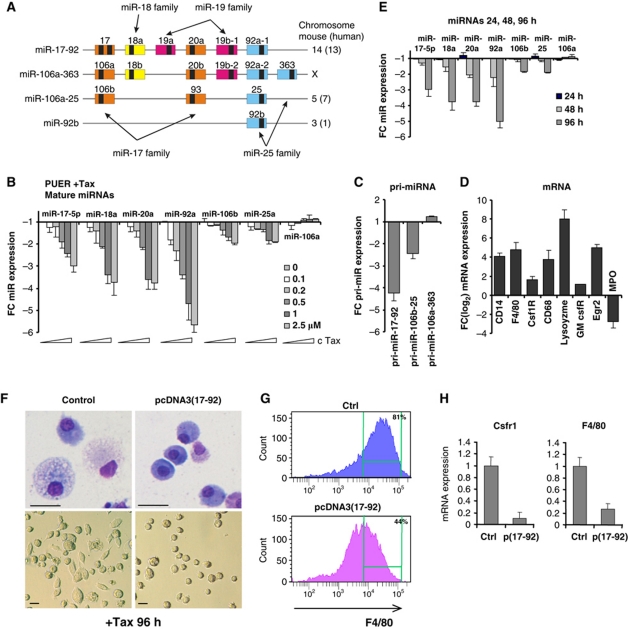
Figure 1.
The miR-17-92 and 106b-25 clusters are downregulated upon PU.1-mediated macrophage differentiation. (A) Schematic representation of the murine miR-17-92 cluster and its paralogues. Coloured boxes represent pre-miRNAs and black boxes mature miRNAs. Four families of miRs with the identical seed sequence are indicated by colour and arrows. (B) Expression of mature miRs of the miR-17-92 cluster evaluated by qPCR. PUER cells were stimulated by increasing concentration of Tamoxifen (X axis) for 96 h. (C, D) PUER cells were stimulated by 2.5 μM Tamoxifen for 96 h and expression of pri-miRNA transcripts (C) and macrophage-specific transcripts (D) was evaluated. (E) PUER cells were stimulated by 2.5 μM Tamoxifen for 24, 48 and 96 h and expression of mature miRNAs was analysed. Y axis (B–E) indicates fold change of normalized expression relative to untreated cells. Baseline represents expression value of untreated cells. Mean±s.d. (n=3). (F–H) Ectopic expression of miR-17-92 cluster inhibits PU.1-induced macrophage differentiation. PUER cells were co-transfected with expression vector encoding the miR-17-92 cluster (pCDNA3(17-92)) or empty vector (control) and subsequently stimulated by 2.5 μM Tamoxifen for 96 h. (F) Morphology of transfected PUER cells, evaluated by Wright-Giemsa staining (upper panels, magnification × 400, scale bar 10 μM) or phase contrast (lower panels, magnification × 100). (G) FACS analysis of F4/80 expression (X axis)/cell counts (Y axis). (H) mRNA expression of Csfr1 and F4/80 evaluated by qPCR. Mean±s.d. (n=3).
Recent studies identified among targets of miR-17-92 key members of anti-tumour guidance including cyclin-dependent kinase inhibitor CDKN1A (p21CIP1) (Ivanovska et al, 2008; Petrocca et al, 2008b), retinoblastoma-like 2 protein (p130, Rbl2) (Lu et al, 2007), transforming growth factor β1 (TGFβ1) (Petrocca et al, 2008a), the proapoptotic protein BIM (Bcl2 interacting mediator of cell death) (Koralov et al, 2008; Ventura et al, 2008; Xiao et al, 2008), and the tumour suppressor PTEN (Xiao et al, 2008). By blocking these factors, miR-17-92 promotes cell proliferation and survival. Within the haematopoietic compartment, miR-17-92 is highly expressed in stem cells and early progenitors while it is decreased upon the onset of myeloid and lymphoid differentiation (Fontana et al, 2007; Ventura et al, 2008; Xiao et al, 2008) and during in vitro differentiation of acute leukaemia blast cells (Kasashima et al, 2004; Schmeier et al, 2009). Conversely, sustained expression of miR-17-5p, -20a, and -106a has been shown to inhibit monocytic differentiation via targeting of myeloid transcription factor AML1 (acute myeloid leukaemia1, Runx1; Fontana et al, 2007).
Several lines of evidence document direct transcriptional regulation of the miR-17-92 cluster by specific transcription factors. For instance, MYC oncogene binds downstream of the CpG island of the miR-17-92 promoter and activates its expression in the human Burkitt's lymphoma cell line (O’Donnell et al, 2005). The miR-17-92 cluster is transactivated by the oncogene MYCN (Fontana et al, 2008) and several members of the E2F (Woods et al, 2007) transcription factors in human neuroblastoma and B-cell lymphoma by direct binding to the promoter region. The tumour suppressor protein p53 binds near the TATA box of the miR-17-92 promoter and represses its expression under hypoxia in human colorectal carcinoma cell line (Yan et al, 2009). MiR-17-92 expression is inhibited also by AML1 upon differentiation of cord blood myeloid progenitors (Fontana et al, 2007).
Macrophage differentiation is a complex and tightly regulated process initiated in early progenitors by PU.1 (Spi1), a key transcriptional factor essential for the myeloid and lymphoid development. PU.1-deficient mice lack macrophages, granulocytes, and B-lymphocytes and die during late fetal or early neonatal stage (Scott et al, 1994; McKercher et al, 1996; Back et al, 2004). Several microRNAs associated with macrophage development are transcriptional targets of PU.1, including miR-424, that stimulates monocytic development via targeting the transcription factor NFI-A (Rosa et al, 2007) and miR-146, that directs the differentiation of HSC into peritoneal macrophages (Ghani et al, 2011). Mice carrying a hypomorphic PU.1 allele reducing PU.1 expression to 20% develop acute myeloid leukaemia (AML), demonstrating a role of PU.1 in leukaemogenesis (Rosenbauer et al, 2004). AML is characterized by myeloid differentiation blockade and clonal leukaemic proliferation and may involve deregulation of transcription factors (Rosenbauer and Tenen, 2007) as well as microRNAs and epigenetic processes including deregulation of chromatin structure (Melnick and Licht, 2002).
In this study, we investigated the regulation of miR-17-92 cluster upon PU.1-dependent differentiation of mouse myeloid progenitors. We describe the molecular mechanism required for macrophage differentiation-associated inhibition of miR-17-92 cluster that is mediated by the transcription factor Egr2 and the Jarid1b demethylase leading to histone H3 lysine K4 trimethyl (H3K4me3) demethylation of the miR-17-92 promoter. We further show that deregulation of this mechanism is also involved in leukaemogenesis.
Results
PU.1-dependent macrophage differentiation requires downregulation of miR-17-92
Fetal myeloid progenitors derived from PU.1−/− mutant mice can recapitulate macrophage development upon induction of the conditional transgene of PU.1 that is fused with the ligand binding domain of the Oestrogen Receptor (PUER), in response to Tamoxifen (Walsh et al, 2002; Supplementary Figure S1).
To identify miRNAs that are differentially regulated during PU.1-dependent macrophage differentiation, we compared the expression of miRNAs in uninduced and induced (2.5 μM Tamoxifen, 96 h) PUER cells using the TaqMan Low Density Array. The miRNAs encoded by miR-17-92 cluster as well as the paralogous miR-106b-25 clusters were identified to be among most abundant miRNAs in undifferentiated PUER cell, while they were among the most downregulated miRNAs upon PUER cell differentiation (Supplementary Tables 1 and 2). This observation was confirmed by qRT–PCR for the individual mature miRNAs and pri-miRNA transcripts in response to graded Tamoxifen concentrations. Four miRNAs representing the miR-17-92 cluster (miR-17-5p, miR-18a, miR-20a, and miR-92) were significantly downregulated in a dose-dependent manner (with a maximum of 3.5–6-fold by 2.5 μM Tamoxifen; Figure 1B) coincident with the induced transcript expression of macrophage-specific genes (Figure 1D). The pri-miR-17-92 transcript was downregulated to levels comparable to those of mature miRNAs (∼3–4-fold; Figure 1C), indicating that the miR-17-92 cluster is repressed at transcriptional level during macrophage differentiation. This finding is further supported by the similarities in the kinetics of downregulation of mature miRNAs (first detectable at 48 h, Figure 1E) and in the magnitude of their modulation (3.5–6-fold change). The levels of mature miRNAs and pri-miRNA of the paralogous miR-106b-25 cluster were also downregulated upon PU.1 activation, albeit to a lesser extent (∼2-fold), while the levels of the miR-106a-363 cluster were unchanged (Figure 1B and C, unrelated miRNA controls shown in Supplementary Figure S2).
To determine whether downregulation of miR-17-92 is a necessary requirement for macrophage differentiation, we overexpressed miR-17-92 (pCDNA3(17-92)) or control pCDNA3 vector in the PUER cells (Supplementary Figure S3A). Upon Tamoxifen induction, the ectopic miR-17-92 significantly inhibited PU.1-dependent macrophage differentiation, as demonstrated by altered cellular morphology (Figure 1F), reduced expression of the F4/80 cell surface protein (Figure 1G) and reduced expression of F4/80 and Csf1R (c-fms) transcripts (Figure 1H). We note that the differentiation block mediated by ectopic miR-17-92 is not caused by downregulation of the PUER protein (Supplementary Figure S3B).
Collectively, these data demonstrate that downregulation of miR-17-92 expression is a prerequisite for the PU.1-dependent macrophage development.
Downregulation of miR-17-92 is mediated by Egr2
As the expression of miR-17-92 becomes downregulated upon the induction of PU.1 activity, we first determined whether PU.1 could directly regulate this cluster. We failed to detect any PU.1 occupancy at the miR-17-92 region (±10 kb) by chromatin immunoprecipitation (ChIP, not shown), a fact supported by a recent genome-wide ChIP-Seq study (Heinz et al, 2010), and we have not detected PU.1-dependent regulation of the gene in a reporter assay (not shown). Therefore, we focused on the possibility of indirect regulation of miR-17-92. The genome-wide expression analysis of PUER cells stimulated with Tamoxifen indicated that among the early and strongest induced transcription factors was Early growth response 2 (Egr2, Krox20, Figure 2A). This was validated at the protein level (Figure 2A). Egr2 is a member of the family of closely related zinc finger transcriptional factors Egr1-4, implicated in the development and differentiation of the neural and the haematopoietic systems (Nguyen et al, 1993; Krishnaraju et al, 1995). Notably, Egr2 has been previously demonstrated to be required for macrophage (Laslo et al, 2006; Krysinska et al, 2007) and lymphocyte (Zhu et al, 2008; Li et al, 2011) development. The other members of the Egr family Egr3 and Egr4 are lowly expressed in PUER cells and their expression is not changed, while Egr1 is downregulated upon PUER cell differentiation (Supplementary Figure S4; Laslo et al, 2006).
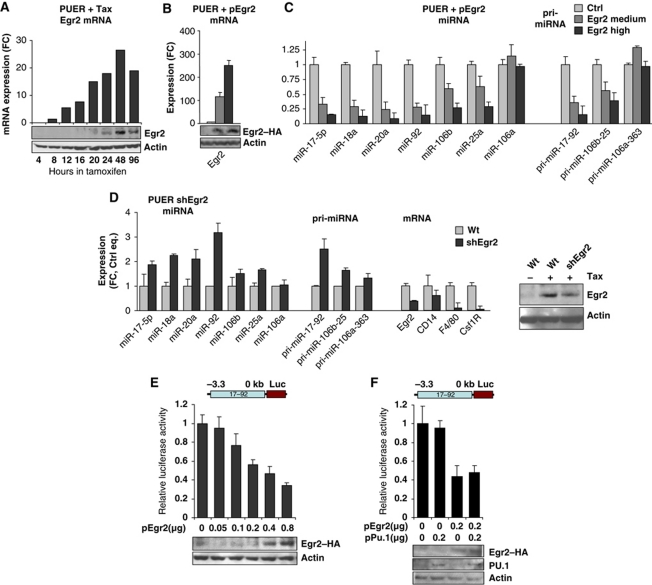
Figure 2.
Egr2 inhibits the miR-17-92 cluster. (A) Expression of Egr2 in PUER cells stimulated by 2.5 μM Tamoxifen for indicated time (hours), determined by gene expression array. Y axis represents normalized values of Egr2 mRNA relative to untreated cells. (B, C) Expression of mRNA, pri-miRNAs and mature miRNAs evaluated by qPCR. PUER cells were nucleofected with pEGR2 or control empty vector and pEGFP followed by sorting at 96 h for GFP-intermediate expression and GFP-highly positive cells. Mean±s.d. (n=3). (D) PUER WT and shEgr2 cells, stably expressing small hairpin inhibitory RNA against Egr2, were treated with 2.5 μM Tamoxifen for 96 h and expression of mRNA, miRNAs, pri-miRNAs was determined. (B–D) Expression values were normalized to Gapdh (mRNA) or Sno202 (miRs) and equalized to untreated cells. Mean±s.d.(n=3). (E, F) NIH 3T3 cells were transfected with pGL3(−3.2; 0) reporter vector and increasing amounts of pCB6-Egr2 (E) or pCB6-Egr2 and pXM-PU.1 (F), as indicated on X axis. The Y axis represents firefly luciferase activity, normalized to Renilla luciferase and relative to the empty vector. Mean±s.d. (n=3). The panels are accompanied by western blot analysis of Egr2 and Actin as a loading control (placed on the bottom).
To test if Egr2 mediates the repressive effects of PU.1 on the miR-17-92 cluster, we co-transfected the PUER cells with either an expression vector encoding Egr2 (pCB6-Egr2) or empty vector, both with pGFP. Both GFP medium- and GFP high-expressing cells were isolated by FACS sorting. Ectopic expression of Egr2 in unstimulated PUER progenitors (lacking endogenous Egr2 expression) strongly inhibited the basal expression of the miR-17-92 cluster (6–10-fold), both at mature miRNA and at cluster pri-miRNA levels. Interestingly, the level of miR-17-92 inversely correlated with the levels of ectopic Egr2 (Figure 2B and C). The paralogous miR-106b-25 cluster (unlike the miR-106a-363 cluster) was also downregulated by ectopic Egr2 albeit to a lesser extent (∼3-fold, Figure 2C).
To further test if Egr2 is dispensable for PU.1-induced miR-17-92 repression, we employed a PUER cell line that is stably transfected with a small hairpin RNA (shRNA) construct directed against Egr2 (shEgr2) (Laslo et al, 2006). The shEgr2 and parental PUER cells were stimulated with Tamoxifen and harvested at 96 h. The knockdown of Egr2 resulted in decreased repression of miR-17-92 cluster as manifested by increased levels of miR-17-92 mature miRNAs as well as of pri-miRNA concomitant with decreased levels of macrophage-specific transcripts (Figure 2D), indicating that Egr2 is indeed required for mR-17-92 repression. The association of Egr2 and miR-17-92 was further supported by experiment where Egr2 cDNA transfected into PUER cells partially rescued the block of differentiation imposed by overexpressed miR-17-92 (not shown).
To test whether the inhibitory effect of Egr2 on miR-17-92 levels is mediated through the miR-17-92 promoter, NIH3T3 fibroblasts were co-transfected with increasing amounts of the Egr2 plasmid together with a luciferase-expressing vector (pGL3) containing the upstream regulatory region (−3.3 to 0 kb, relative to the first nucleotide of pre-miR-17-5p) of the miR-17-92 cluster, designated pGL3(−3.3; −0). Increasing concentrations of Egr2 led to gradual repression of the reporter activity (Figure 2E), demonstrating repressive activities of Egr2 upon the miR-17-92 promoter. To further test if PU.1 enhances the Egr2-mediated repression of miR-17-92, the reporter pGL3(−3.3; 0) was co-transfected into the NIH3T3 cells with the expression vectors encoding Egr2 and PU.1 (Figure 2F). While Egr2 alone repressed the reporter activity, co-transfection of PU.1 with Egr2 did not further intensify the repression, indicating that PU.1 is dispensable for the Egr2-mediated direct repression of miR-17-92.
Altogether, these data indicate that during macrophage differentiation Egr2 protein is activated by PU.1, but regardless of PU.1, Egr2 alone can transcriptionally inhibit the miR-17-92 cluster.
Egr2 binding to the miR-17-92 cluster promoter results in H3K4 demethylation
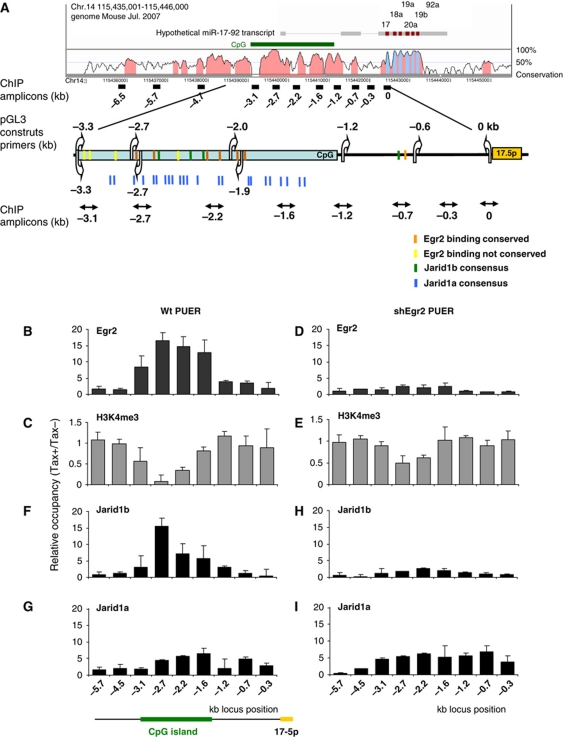
There is a long CpG island (CG content ∼80%) within the region spanning −3.3 to −1.2 kb (relative to the first nucleotide of the pre-miR-17-5p sequence), representing a putative recognition element for the transcriptional regulation of the miR-17-92 cluster (Figure 3A). A sequence motif analysis of the upstream region up to −10 kb revealed multiple putative Egr2 binding sites. Importantly, six of them are conserved in human, mouse, and rat and are located within the CpG island (between −2.8 and −1.8 kb).
Figure 3.
Egr2 binds the miR-17-92 promoter and recruits Jarid1b to demethylate H3K4me3. (A) Vista plot (http://genome.lbl.gov/vista/) and scheme of putative promoter region of miR-17-92 cluster. Vertical arrows indicate positions of primers utilized for amplification of DNA fragments of miR-17-92 upstream region used in reporter assays, horizontal arrows indicate ChIP qPCR amplicons. Colour boxes represent binding and consensus sites of Egr2, Jarid1a and Jarid1b. (B–I) ChIP analysis of miR-17-92 promoter region. PUER cells (B–E) or shEgr2 PUER cells (D–I) were differentiated by 2.5 μM Tamoxifen for 96 h and chromatin was precipitated with indicated antibodies. Y axis represents occupancy of Tamoxifen treated relative to Tamoxifen untreated cells. H3K4me3 values were further equalized to Histone H3. X axis represents the position of ChIP amplicons relative to start of miR-17-5p. Mean±s.d. (n=3).
To test if Egr2 occupies this putative regulatory region of miR-17-92 cluster, ChIP was performed on crosslinked chromatin from Tamoxifen stimulated (for 96 h) and parental PUER cells using anti-Egr2 antibody. Quantitative PCR at nine amplicons spanning 6.5 kb of the miR-17-92 regulatory region revealed specific occupancy of Egr2 (∼20-fold) within the −3.3 to −1.6 kb region that overlaps with the CpG island (Figure 3B).
Nab2 is a known corepressor of Egr2 (Svaren et al, 1996) and a potential candidate for Egr2-mediated repression of miR-17-92. To determine if Nab2 is required for the repression of miR-17-92, we used a PUER cell line variant that stably expresses an shRNA against Nab2 (shNab2 PUER) (Laslo et al, 2006). Surprisingly, upon macrophage differentiation of shNab2 cells, the expression of miR-17-92 was repressed to comparable levels as in control cells (Supplementary Figure S5A). Furthermore, as Nab2 is often associated with the NUrD repressive complex (Srinivasan et al, 2006) which is sensitive to histone deacetylase inhibitor Trichostatin (TSA), we treated PUER cells with Trichostatin and failed to observe any dysregulation in the expression of miR-17-92 upon PUER cell differentiation (Supplementary Figure S5B). These observations demonstrate that while Egr2 clearly represses miR-17-92, it does so in an Nab2-independent manner.
To test whether Egr2 occupancy affects the chromatin modification of the miR-17-92 gene, we determined the levels of histone 3 lysine 9 acetylation (H3K9Ac) and histone 3 lysine 4 trimethylation (H3K4Me3), both established marks of transcriptionally active chromatin. ChIP assay demonstrates that the H3K4Me3 (Figure 3C) but not H3K9Ac (not shown) pattern is significantly reduced near the −2.7 kb locus during macrophage differentiation. The observed significant decrease of H3K4 methylation occurs at the region of miR-17-92 CpG island occupied by Egr2. To address whether Egr2 indeed mediates the H3K4 demethylation of the miR-17-92 locus, we measured the level of H3K4 trimethylation within the Tamoxifen-treated shEgr2 cells. ChIP analysis demonstrated that the Egr2 knockdown resulted not only in the expected loss of Egr2 occupancy at the miR-17-92 CpG but also led to a significant increase of H3K4 methylation compared with the parental PUER cells (Figure 3D and E).
Collectively, these experiments demonstrate that Egr2 directly associates with the CpG island upstream of the miR-17-92 gene and mediates histone H3K4 demethylation of this region.
Egr2 recruits Jardi1b demethylase onto miR-17-92 cluster
Demethylation of histones H3K4 at gene promoters is an active process of transcriptional repression and may represent a mechanism by which Egr2 regulates the miR-17-92 cluster. However, as Egr2 does not possess any demethylation activity, we asked whether it recruits a demethylase(s) in order to repress miR-17-92 locus. The Jumonji C domain-containing proteins of Jarid1a-c family are capable of efficient and specific H3K4 demethylation in vivo (Secombe and Eisenman, 2007). Genome-wide expression analysis identified that two Jarid1 demethylases, Jarid1a (Rbp2, Kdm5a) and Jarid1b (Plu1, Kdm5b) are upregulated at transcript levels during early macrophage differentiation of the PUER cells (Supplementary Figure S6). Interestingly, the CpG island of miR-17-92 contains multiple Jarid1a (GGGCGG) and Jarid1b (GCACA/C) consensus sequences (Scibetta et al, 2007; Lopez-Bigas et al, 2008; see schematic in Figure 3A). ChIP analysis revealed specific occupancy of both Jarid1a and Jarid1b to the miR-17-92 promoter region upon Tamoxifen-induced differentiation of PUER cells. Jarid1b occupancy (∼15-fold compared with untreated controls) was detected at the −3.1- to −2.2-kb region, culminating at −2.7 kb position of the CpG island (Figure 3F), matching thus the positional occupancy patterns of Egr2 and of decreased H3K4 methylation, while Jarid1a occupancy exhibited a more diffuse profile (Figure 3G).
To test if Egr2 is required for the recruitment of Jarid1a and Jarid1b to the miR-17-92 promoter, we determined their occupancy in the Tamoxifen-induced shEgr2 cells. Knockdown of Egr2 correlated with significant (∼6-fold) depletion of Jarid1b (but not of Jarid1a, see Figure 3H and I) as well as with an increase in H3K4 trimethylation state of miR-17-92 CpG island (−3.1 to −2.2 kb) (Figure 3E). These data demonstrated Jarid1b recruitment to the miR-17-92 promoter region that depends on Egr2 recruitment within the same region and leads to subsequent H3K4me3 demethylation.
Egr2 and Jarid1b target the narrowed down region of miR-17-92 CpG Island
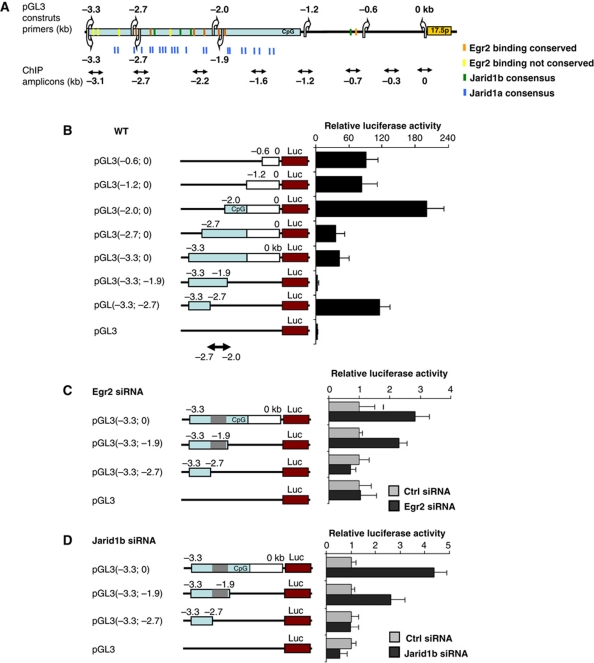
To functionally delineate the region of miR-17-92 promoter targeted by Egr2-Jarid1b, various deletion fragments from −3.3 to 0 kb relative to the start of pre-miR-17-5p sequence were subcloned into the promoterless Luciferase pGL3 vector (see Figures 3A, 4A and B). Equimolar ratios of the miR-17-92 pGL3 vectors were transfected into PUER cells and the luciferase activity was determined upon 96 h of Tamoxifen stimulation. The constructs that contained extending miR-17-92 promoter regions: (−0.6 to 0), (−1.2 to 0) and (−2.0 to 0) kb stimulated luciferase activity and reached a maximum of 200-fold for the pGL3(−2.0; 0) construct as compared with empty vector (Figure 4B). Notably, upon further prolongation of the construct pGL3(−2.0; 0) by 0.7 and 1.3 kb of the distal portions of the CpG island, creating pGL3(−2.7; 0) and pGL3(−3.3; −0) constructs, we observed a significant repression of reporter activity (∼6-fold). This finding strongly suggests that the region within the −2.7 to −2.0 kb of miR-17-92 gene conveys the repressive effects.
Figure 4.
Egr2 and Jarid1b regulate miR-17-92 promoter. (A) Schematic representation of the miR-17-92 upstream region. Arrows and boxes are same as Figure 3A. (B) PUER cells were transfected with pGL3 constructs (breakpoints are shown in parentheses in kb relative to the first nucleotide of 17-5p pre-miR), and differentiated by 2.5 μM Tamoxifen for 96 h. (C, D) PUER cells were co-transfected with indicated pGL3 constructs and siRNA inhibiting Egr2 (C) or Jarid1b (D) and treated with 2.5 μM Tamoxifen for 96 h. The Y axis represents firefly luciferase activity of indicated reporter vectors, normalized to Rennila luciferase and relative to the activity of empty vector (A) or to the activity of Ctrl siRNA-treated cells (B, C). CpG island is visualized by blue colour the repressive region (−2.7 to −2.0kb) by grey colour. Mean±s.d. (n=3).
To further test this observation, we cloned major portion of the miR-17-92 CpG island into the pGL3 vector creating pGL3(−3.3; −1.9) construct. Upon transfection into PUER cells, this construct failed to activate the luciferase activity compared with empty vector. Strikingly, a deletion of the newly identified repressive region (−2.7 to −2.0) creating pGL3(−3.3, −2.7) construct led to a dramatic increase (∼120-fold) in the luciferase activity (Figure 4B). These observations demonstrate that the repressive activity associated with macrophage differentiation is located at the interval (−2.7 to −2.0) while the (−3.3 to −2.7)-kb region conveys the stimulatory activity. We noted that the repressive region harbours several conserved Egr2 binding sites and multiple Jarid1a and Jarid1b consensus sequences, often mutually overlapping. Furthermore, this region is occupied by Egr2 and Jarid1 and exhibits H3K4 demethylation (Figure 3). However, due to a very high CG content (80–90%) of the CpG island in the close proximity of Egr2 binding sites, we have not succeeded to discern the individual Egr2 motifs in order to generate their individual mutants by PCR-based mutagenesis.
To further validate whether the (−2.7 to −2.0) region is a target region for Egr2-mediated repression, the PUER cells were transfected with these reporters: pGL3(−3.3; 0) containing the repressive (−2.7 to −2.0) and two activation (−3.3 to −2.7), (−2.0 to 0) regions, pGL3(−3.3; −1.9) containing repressive (−2.7 to −2.0) and activation (−3.3 to −2.7) regions, and finally pGL3(−3.3; −2.7) lacking repressive region and containing a single activation region (see schematic in Figure 4A and C). The experiment was done in the presence of siRNA inhibiting Egr2 (or control siRNA) and 2.5 μM Tamoxifen for 96 h. Inhibition of the Egr2 levels (by ∼75%) caused an increase in the luciferase activity of the reporter constructs containing the minimal repressive region: pGL3 (−3.3 to 0) and pGL3 (−3.3 to −1.9), while it did not affect the activity of the construct (−3.3 to −2.7) which lacks the minimal repressive region (Figure 4C).
To determine if Jarid1 demethylases are required for the repression of the (−2.7 to −2.0) region, the PUER cells were co-transfected with pGL3(−3.3; 0), pGL3(−3.3; −1.9) and pGL3(−3.3; −2.7) reporters described in the previous paragraph along with siRNA inhibiting either Jarid1a or Jarid1b or the control siRNA. The cells were subsequently treated by 2.5 μM Tamoxifen. The knockdown of Jarid1b (Figure 4D), but not that of Jarid1a (not shown), led to an increase in luciferase activity of the constructs containing minimal repressive region while it did not affect the activity of the construct which lacks the minimal repressive region. This demonstrates the requirement of Jarid1b for the repression of the miR-17-92 cluster conveyed through the (−2.7 to −2.0) region.
Collectively, the data presented above show that the repression of miR-17-92 is mediated via the (−2.7; −2.0) region within the upstream CpG island and that this region is directly regulated by Egr2 and Jarid1b which both are required for the repression of miR-17-92 during macrophage differentiation.
Egr2 is both a repressor and a target of miR-17-92
To identify gene targets of miR-17-92, we employed a bioinformatic target prediction using TargetScan (http://www.Targetscan.org). Interestingly, among the predicted targets was Egr2 itself which led us to postulate a double-negative feedback loop between these two factors. Eight of fifteen miRNAs of the miR-17-92 cluster and its paralogues are predicted to target the Egr2 3′UTR in two separate binding sites (Figure 5A). MiR-17-5p, -20a, -20b, -93, -106a and miR-106b are predicted to bind to the Egr2 3′UTR in the position 419–425 bp, each displaying an 8-mer seed sequence (the maximum compatibility) while miR-92a, -92b, -25 and miR-363 are predicted to bind to the 3′UTR position 612–618 bp and display a 7-mer seed sequence. Both predicted binding sites in the Egr2 3′UTR are conserved among mammals and surrounded by stretches of adenines additionally suggesting that they are indeed functional miR target sequences (Grimson et al, 2007). Similar results were obtained by using PicTar and MirBase databases.
Figure 5.
Egr2 is a target of the miR-17-92 cluster. (A) Schematic representation of pGL3(EGR2 3′UTR) construct and pairing of miRs of miR-17-92, 106b-25 and 106a-363 clusters to the WT (top) and mutated Egr2 3′UTR (bottom). Boxes represent predicted binding sites of indicated miRs. (B) Unstimulated PUER progenitors, expressing high level of miR-17-92 were transfected with pGL3-Egr2 3′UTR wild-type (wt) or mutated (mut) vector and co-transfected with miR-17-5p or with miR-17-5p and miR20a microRNA inhibitors or scrambled control (indicated bellow). The ratio of normalized luciferase activity (at 48 h) of wt versus mut pGL3-Egr2 3′UTR vector is shown. Mean±s.d. (n=3). (C) 3T3 cells were transfected with the pCDNA(17-92) or empty vector and cultured 12 h in absence of serum. The levels of Egr2 protein at 1.5 h following the serum stimulation were evaluated by western blotting. (D) Model of mutual regulation of Egr2 and miR-17-92 upon macrophage differentiation.
To test whether Egr2 is post-transcriptionally inhibited by miR-17-92, a 900-bp fragment of murine Egr2 3′UTR containing two wild-type binding sites of miR-17-92 or their mutants were subcloned into the pGL3-Promoter vector, downstream of the firefly luciferase gene (Figure 5A). This created a wild-type (pGL3-Egr2 3′UTRwt) and mutant (pGL3-Egr2 UTRmut) reporter vectors. These vectors were transfected into PUER cells and luciferase activity was measured after 96 h of Tamoxifen stimulation. Transfection of wild-type (pGL3-Egr2 3′UTRwt) reporter vector resulted in ∼60% decrease of luciferase activity, due to the inhibitory activity of endogenous miR-17-92, as compared with the mutant vector. Furthermore, ectopic expression of miR-17-92 resulted in a further decrease of luciferase activity. Conversely, co-transfection of miRNA inhibitors of either miR-17-5p or miR-17-5p and miR-20a, but not of the scrambled control, abrogated the inhibitory effects of the miR-17-92 and resulted in partial rescue of the luciferase activity (Figure 5B). Similar results were reproduced in HeLa cells (data not shown). 3T3 cells temporarily express Egr2 upon serum activation of serum-starved cells (Svaren et al, 1996). To further validate that Egr2 is translationally repressed by miR-17-92, 3T3 cells were transfected with the pCDNA(17-92) or empty vector and cultured 12 h in absence of serum followed by 1.5 h serum stimulation. Ectopic expression of miR-17-92 significantly inhibited the levels of Egr2 protein (Figure 5C).
The data in this paragraph demonstrate that Egr2 mRNA is targeted by miR-17-92. As Egr2 is also a repressor of the miR-17-92 expression, Egr2 and the miR-17-92 create double-negative feedback loop that may be involved in macrophage differentiation characterized by a bistable state (Tsang et al, 2007; Martinez et al, 2008), where Egr2 represses miR-17-92 cluster in differentiating cells and in turn, miR-17-92 cluster negatively regulates Egr2 in proliferating progenitor cells (Figure 5C).
The mutual regulation between Egr2 and miR-17-92 cluster is observable in AML
AML is characterized by a block of myeloid differentiation that involves monocytic maturation. Previous reports have shown increased expression of miR-17-92 in patients with myeloid malignancies including AML and preleukaemia (myelodysplastic syndromes, MDSs) (Li et al, 2008; Pons et al, 2009). These observations prompted us to investigate a possible contribution of the repressive mechanism imposed by Egr2 on the miR-17-92 locus to the pathology of AML. We first determined the expression of the miR-17-92 by TaqMan qRT–PCR in peripheral blood mononuclear cells from AML patients (N=27) (the clinical data are shown in Supplementary Table 3), and from six healthy controls. Based on the expression levels of three representative members of miR-17-92 cluster, miR-17-5p, 20a and 92, the patients were subdivided into two groups with either normal (NORM) or elevated (HIGH) levels of miR-17-92. The group HIGH contained samples where at least two of the three miRNAs tested were elevated >1.5-fold compared with the average value of the control samples. In all, 14 (52%) out of 27 AML patients overexpressed miR-17-5p, miR-20a and miR-92 (P<0.001 compared with control and P<0.0001 compared with NORM group; Figure 6A; Supplementary Figure S7A). A majority of AML patients with increased expression of miR-17-92 cluster were undifferentiated AMLs: 6 of them were classified as M1 (42%) (myeloblastic leukaemia without maturation) and 3 (21%) as M0 (AML with minimal myeloid differentiation) according to FAB classification, indicating that miR-17-92 cluster may be involved in early differentiation blockade during pathogenesis of AML. Increased levels of miR-17-92 were seen in most AML samples associated with the downregulation of differentiation-associated mRNA: CSF1R, CD14 (Supplementary Figure S5 and data not shown). Next, we evaluated expression of the proapoptotic protein BIM (Bcl2 interacting mediator of cell death) and the tumour suppressor p21 (Cdkn1A), both established targets of miR-17-92 (Fontana et al, 2008; Ventura et al, 2008; Xiao et al, 2008; Petrocca et al, 2008b). The group with elevated levels of miR-17-92 expressed significantly lower levels of the BIM (P<0.001) and p21 mRNAs (P<0.01) displaying negative correlation (Spearman) between miR-17-92 and both BIM (r=−0.7216, P<0.0001) and p21 (r=−0.4481, P=0.0191) expression, suggesting that BIM and p21 are targets of miR17-92 in AML and their deregulation may be involved in pathogenesis of AML (Figure 6B).
Figure 6.
The miR-17-92 cluster is upregulated in a subset of AML patients. Expression of miR-17-5p, 20a, 92, PU.1, EGR2 (A), BIM and p21 (B) was analysed in 27 AML patients PBMCs by qPCR. Depending on the expression levels of miR-17-5p, 20a and 92a, the patients were subdivided into two groups with either elevated (HIGH) or normal (NORM) levels of miR-17-92 (X axis). Values are normalized to RNU44 or GAPDH and are relative to average expression value of healthy controls (CTRL) (n=6). Mean±s.e.m. values are shown. (C) AML patient cells were nucleofected by 5 μg of pEGR2 or empty vector (control) and 1 μg pEGFP, sorted by FACS and analysed after 96 h by qPCR. Values are normalized to RNU44 or GAPDH. Mean±s.e.m. (n=2). *P<0.05, **P<0.01, *** P<0.001.
In this study, we have identified a mechanism of negative regulation of miR-17-92 cluster by PU.1-mediated induction of Egr2 and we asked if this mechanism is deregulated in AML and whether elevated levels of miR-17-92 in AML patient cells are associated with altered PU.1 and Egr2 levels. Interestingly, the group of AML patients with elevated miR-17-92 levels had significantly lower expression of PU.1 (P<0.01) and EGR2 (P<0.025, correlation of miR-17-92/Egr2 expression r=−0.402, P=0.018) compared with healthy controls (Figure 6A). Conversely patients’ cells with elevated EGR2 expression had decreased levels of the miR-17-92 cluster (Figure 6A; Supplementary Figure S5), indicating that the overexpression of miR-17-92 can result from the lack of inhibition by repressive mechanism imposed by Egr2. To test whether Egr2 is indeed capable of inhibiting miR-17-92 cluster in AML, the primary AML cells (characterized by elevated miR-17-92 and decreased Egr2 expression) were transfected with the expression vector encoding Egr2. Significant downregulation of miR-17-5p, miR-20a, miR-92 representing miR-17-92 cluster was achieved concurrently with the upregulation of mRNA levels of validated miR-17-92 targets BIM and p21, originally found downregulated in these AML patients (Figure 6C). Ectopic Egr2 expression in primary AML blasts also stimulated the expression of CSF1R and the myeloid surface marker CD11B (Supplementary Figure S7B). In addition, to demonstrate that Egr2 could inhibit miR-17-92 and induce leukaemic cells along the myeloid lineage, we have used the human AML cell line HL60 (M2 according to FAB classification) that expresses high levels of miR-17-92 and low levels of Egr2 (Supplementary Figure S7A). Upon overexpression of Egr2, we observed a downregulation of pri-miR-17-92 along with concomitant stimulation of p21, BIM and CD11B and other myeloid genes (CSF1R, CSF3R), but not CD14 (Supplementary Figure S8). The HL60 cells transfected by Egr2 displayed altered cellular morphology that is characteristic of a differentiating cell characterized by increased cytoplasmic/nuclear ratio and vacuolysed cytoplasm. We also observed mild stimulation of apoptosis supporting possibility of suppression of leukaemic growth by Egr2.
Altogether, these data indicate that Egr2 repressive mechanism is deregulated in subset of AML patients overexpressing miR-17-92 and that ectopic expression of Egr2 in AML primary cells inhibits miR-17-92 leading to derepression of its targets. The PU.1-EGR2-miR-17-92 pathway may thus serve as both normal differentiation route and a checkpoint for leukaemogenesis.
Discussion
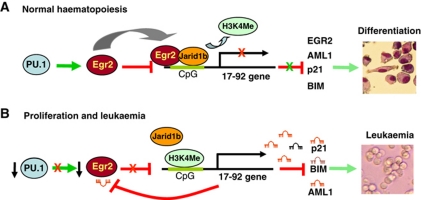
In the present study, we describe a molecular mechanism underlying transcriptional silencing of miR-17-92 cluster during PU.1-dependent differentiation of myeloid progenitors. The following model (Figure 7A) is proposed: during macrophage differentiation PU.1 stimulates expression of secondary determinant Egr2. Subsequently, Egr2 binds to the CpG island upstream of miR-17-92 cluster and recruits Histone 3 Lysine 4 (H3K4) demethylase Jarid1b to demethylate H3K4Me3 at a discrete segment of the CpG island leading to a state that is not permissive for miR-17-92 transcription. This results in the repression of miR-17-92 expression and a release of miR-17-92 targets (including p21, Bim and Aml1) from a post-transcriptional blockade that enables them to facilitate macrophage differentiation.
Figure 7.
Model of regulation of miR-17-92 cluster. (A) Upon macrophage differentiation, PU.1 stimulates expression of transcription factor Egr2, which in turn recruits demethylase Jarid1b to demethylate H3K4 of miR-17-92 cluster promoter, leading to downregulation of miR-17-92 expression, thus releasing a transcriptional block of its targets that include important differentiation factors and cell-cycle inhibitors resulting in macrophage differentiation. (B) In proliferating progenitor cells or in AML, low levels of PU.1 are not capable of stimulating Egr2. Low levels of Egr2 are not able to repress expression of miR-17-92 cluster. Elevated levels of miR-17-92 stimulate proliferation and survival of the cells and silence factors required for myeloid differentiation or cell-cycle arrest and apoptosis.
Several lines of evidence document that miR-17-92 is expressed in proliferating and undifferentiated cells (Marson et al, 2008; Ventura et al, 2008; Xiao et al, 2008). Here, we show that miR-17-92 cluster is downregulated in myeloid progenitors undergoing macrophage differentiation initiated by PU.1. The downregulation step appears important as the ectopic expression of miR-17-92 prevents macrophage differentiation. This is consistent with previously published data demonstrating that downregulation of miR-17-5p, -20a and -106a is required for monocyte differentiation of cord blood haematopoietic progenitors (Fontana et al, 2007). Furthermore, we observed in our model a downregulation of all members of miR-17-92 cluster including miR-18a, miR-19a and miR-92 but not miR-106a. The miR-17-92 downregulation is also observed upon phorbol myristate (PMA and TPA)-induced differentiation of promyelocytic leukaemia HL60 cells (Kasashima et al, 2004), and also during normal lymphoid development (Ventura et al, 2008; Xiao et al, 2008), indicating the importance of miR-17-92 downregulation for haematopoietic differentiation.
During differentiation of myeloid progenitors along the macrophage cell fate, Egr2 cooperates with Jarid1b to demethylate H3K4me3 at upstream regulatory region of miR-17-92 cluster leading to its repression. It is well documented that H3K4 trimethylation facilitates RNAII polymerase recruitment and transcription initiation, while H3K4 demethylation leads to a transcriptional silencing (Bernstein et al, 2002, 2005; Sims et al, 2007). Jarid1b occupancy at miR-17-92 cluster was previously detected in mouse ESC (Dey et al, 2008). The potential role of Jarid1 demethylases in haematopoiesis and leukaemogenesis was documented by identification of a fusion gene containing H3K4Me3 binding domain of Jarid1a and Nucleoporin 98 (NUP98) in human AML (van Zutven et al, 2006; Reader et al, 2007), supported by genetic mouse model (Wang et al, 2009).
Jarid1 demethylases bind preferentially specific sequences in GC-rich DNA regions (Scibetta et al, 2007; Lopez-Bigas et al, 2008) and specifically recognize histone H3K4 trimethyl (Wang et al, 2009), thus one possible model is that Jarid1 directly binds and demethylate regions enriched by its consensus motives near methylated H3K4. However, as we did not observe chromatin occupancy of Jarid1b at miR-17-92 cluster promoter in absence of Egr2, we propose that Jarid1b is targeted to the miR-17-92 promoter (and possibly other genes) via additional factors such as Egr2 and affinity of Jarid1b to its consensus sequence and to H3K4me3 have an additive role.
H3K4 demethylation within the miR-17-92 promoter region occurs near −2.7 kb. The murine −2.7 kb region is fully complementary to human sequence that contains a putative transcriptional start site (TSS) of pri-miR-17-92 (Woods et al, 2007), therefore supporting our hypothesis that murine pri-miR-17-92 TSS is located at −2.7 kb region, where H3K4 methylation change occurs (Supplementary Figure S9). This is consistent with genome-wide studies showing enrichment of H3K4 trimethylation in close proximity (∼1 kb) to TSS of transcriptionally active genes (Bernstein et al, 2005; Barski et al, 2007) and by recent strategies identifying microRNA promoter regions according to H3K4 methylation (Marson et al, 2008).
The presence of epigenetic events other than H3K4 demethylation is suggested by existence of functional interlink between the loss of H3K4Me3 and DNA methylation of CpG islands (Ooi et al, 2007). According to our data, the repression of miR-17-92 occurs concurrently with the Egr2 recruitment and also relatively rapidly (as early as within 48 h). Thus, the DNA methylation may not be involved in the initial phase of miR-17-92 repression.
In contrast to Egr2, Egr1 is expressed in undifferentiated PUER cell and is strongly downregulated upon PUER cell differentiation. This evidence suggests that Egr1 in a context of PU.1-dependent myeloid differentiation does not substitute for Egr2 in repressing miR-17-92. We, however, cannot exclude that in other cellular context Egr1 can inhibit miR-17-92 transcription.
MiR-17-92 and its paralogues, the miR-106a-363 and miR-106b-25 clusters, encode microRNAs with identical or similar sequences that can target common mRNAs. Knockout studies provide evidence for the partial functional redundancy of the miR-17-92 cluster and its paralogues (Ventura et al, 2008). Although we focused particularly on the transcription regulation of the miR-17-92 cluster, our expression data suggest that both miR-17-92 and miR-106b-25 clusters share common yet varying features of regulation (distinct from the miR-106a-363 cluster) (Figures 1B, 2C and D). This is supported by high homology of the promoter regions of these clusters and by earlier observation, showing that the miR-17-92 and miR-106b-25 clusters display similar expression patterns (Ventura et al, 2008; Xiao et al, 2008). The overlap in regulation is also supported by our data in a subset of AML patients that displayed elevated levels of miR-17-92 cluster and concomitantly elevated levels of miR-106b-25 cluster (data not shown).
Recently, several transcription factors such as Myc, Mycn, E2f1-3, p53 and Aml1 were postulated to transcriptionally regulate miR-17-92 cluster (O’Donnell et al, 2005; Fontana et al, 2007, 2008; Sylvestre et al, 2007; Yan et al, 2009). Based on the expression pattern and the data from reporter assays, it is likely that the repression of miR-17-92 cluster in differentiating PUER cells is mediated by active Egr2 repression rather than by decreased activation by Myc, Mycn, E2f1-3 or increased repression by p53 or Aml1 (see Supplementary data and Supplementary Figure S10).
Altered expression of microRNAs including miR-17-92 is often associated with leukaemogenesis including AML and MDS (Li et al, 2008; Pons et al, 2009). We show that the miR-17-92 cluster is downregulated during macrophage differentiation while ectopic expression of miR-17-92 blocks this differentiation indicating that in leukaemogenesis, overexpression of miR-17-92 may contribute to the leukaemic blockade. We report here that miR-17-92 cluster is overexpressed in significant fraction of AML patients. The same AML patients had simultaneously downregulated expression of miR-17-92 repressors PU.1 and Egr2. This indicates that PU.1-EGR2-miR-17-92 pathway important for macrophage differentiation may be dysregulated in this AML patients and the overexpresion of miR-17-92 could be potentially caused by the ineffective inhibitory mechanisms imposed by EGR2. This notion is supported by the finding that ectopic expression of EGR2 inhibits miR-17-92 levels in AML blast cells and HL60 AML cell line and by our unpublished observation, that PU.1 hypomorphic mouse (Rosenbauer et al, 2004), developing AML has upregulated miR-17-92 cluster. Based on our herein presented data validating Egr2 as a direct target of miR-17-92, we hypothesize that miR-17-92-overexpressing AML blasts are blocked from differentiation by oncogenic mechanisms among them elimination of remaining levels of EGR2 represents a likely candidate.
In AML patients characterized by elevated miR-17-92 levels, the following regulatory states are considered (Figure 7B). First, low levels of PU.1 are not sufficient to activate EGR2. Low levels of EGR2 cannot repress miR-17-92 cluster by Jarid1b-mediated H3K4 demethylation. Elevated miR-17-92 cluster in turn reduces remaining EGR2 levels. Second, mechanisms leading to upregulation of miR-17-92 may also involve other upstream factors (such as MYC or MYCN) or miR-17-92 cluster gene amplification, or various combinations of the abovementioned mechanisms. Elevated levels of miR-17-92 downregulate EGR2 and prevent repression of miR-17-92. In both regulatory states, elevated levels of miR-17-92 cluster in AML may stimulate cell proliferation and survival capacity by a post-transcriptional blockade of the key factors required for myeloid differentiation, apoptosis and cell-cycle arrest.
This study characterizes a novel macrophage differentiation-associated gene circuitry comprising the transcription factors PU.1 and Egr2 and the miR-17-92 microRNA cluster. This pathway is required for normal macrophage differentiation and is deregulated in AML. As such, the mutual regulation between transcription factors and microRNAs regulate the bistable state between progenitor maintenance and myeloid development, where repression of inhibitory miRNA by histone demethylation of its promoter is associated with macrophage differentiation. Manipulation of this described pathway may thus serve as molecular target of differentiation therapy of human leukaemias.
Materials and methods
Cell culture
PUER (Walsh et al, 2002), shEgr2, shNab2 (Laslo et al, 2006) and HL60 cells were cultured as described. Macrophage differentiation was induced by 2.5 μM Tamoxifen (Sigma) for 96 h if not stated different. In untreated samples, the vehicle without Tamoxifen was added. For HDAC inhibition, 30 nM Trichostatin was used. Primary cells were obtained upon informed consent. Peripheral blood mononuclear cells were isolated by Ficol gradient.
DNA constructs plasmids
A 1.1-kb BamHI/XhoI genomic fragment encompassing the miR-17-92 cluster was cloned into the pCDNA3 vectors to create pGL3(17-92).
The miR-17-92 promoter fragments were PCR amplified from genomic DNA, digested with BglII and HindIII and cloned into pGL3 basic vector (Promega).
An 872 nt long XbaI fragment of the Egr2 3′UTR, including two miR-17-92 binding sites, was cloned into the pGL3 promo vector (Promega) to generate pGL3-Egr2 3′UTRwt. This vector was mutated by inverse PCR using primers containing the two mutated miR-17-92 binding sites. The primer sequences and details are shown in Supplementary data.
Transient transfections and reporter assays
PUER, HL60 and primary cells were transfected by AMAXA (LONZA) using Mouse stem cell kit with 2 μg of pCDNA3(17-92) or 4 μg of pCB6-Egr2 and 1 μg plasmid pGFP. In siRNA experiments, the cells were transfected with a pool of four siRNAs targeting EGR2, Jarid1a and Jarid1b (Smart Pool siRNA, Dharmacon) at 800 nM concentration.
For reporter experiments, PUER cells were transfected by 1 μg pGl3(17-92) promoter or Egr2 3′UTR reporter vector, 0.33 μg of pRL-TK control vector, alternatively together with 300 nM miR hairpin inhibitors (Dharmacon). NIH3T3 cells were transfected by jetPEI (Polyplus Transfection) in 24-well plate by 0.1 μg pGl3(17-92) promoter or Egr2 3′UTR reporter vector, 10 ng of pRL-TK control vector and indicated amounts of pCB6-Egr2 and pCMV-PU.1 vectors and analysed by Dual Luciferase Assay (Promega). Firefly activity was normalized to Renilla luciferase activity.
RNA isolation and qPCR
Total RNA was extracted by TRIzol reagent (Invitrogen) with enhanced precipitation (20 ng/ml of linear polyacrylamide). Expression of mature miRNAs was determined using TaqMan qRT–PCR (Applied Biosystems), data are calculated using 2−dCt equation. MicroRNA levels were normalized to Sno202 (mouse) or RNU44 (human). For pri-miR and mRNA, Gapdh was used as a load control (for more details, see Supplementary data).
Microarray profiling and data analysis
mRNA expression profiling in PUER cells was performed on 100 ng of total RNA using the Affymetrix 3′-IVT Express kit and GeneChip MG-430A 2.0. microRNAs were isolated by mirVana miRNA Isolation Kit (Ambion) and analysed on TaqMan MicroRNA Array (Rodent A and B v2.0, Applied Biosystems).
Entire array data set will be published in public database GEO (http://www.ncbi.nlm.nih.gov/geo/) upon acceptance of the manuscript.
Chromatin immunoprecipitation
ChIP was performed on 2 × 107 cells as described (Stopka et al, 2005). Antibodies used were PU.1 (sc-352, Santa Cruz Biotech), Egr-2 (Covance, PRB-236P), Jarid1a (ab26049), Jarid1b (ab50958), H3K9Ac (Upstate, 07-353), and H3K4me3 (ab8580) H3 (ab1791) (Abcam). The value of enrichment is shown as ratio of Tamoxifen+ and Tamoxifen− treated cells. Determinations of H3K9Ac and H3K4me3 were equalized to the histone H3.
Statistical analysis
Data are presented as mean; error bars indicate the standard deviation (s.d.) or the standard error of mean (s.e.m.). The data sets were compared using Mann–Whitney and Kruskal–Wallis two-tailed tests and Spearman correlation test.
Supplementary Material
Acknowledgments
We thank Drs R Dahl (University of New Mexico), J Svaren (University of Wisconsin), EV Benevolenskaya (University of Illinois at Chicago) and J Taylor-Papadimitriou (King's College London School of Medicine, UK) for kindly providing us with reagents. We thank number of clinicians including Dr M Trneny for useful discussion. This work was supported by grants from Czech Ministries of Health (NS10310-3/2009), Education (NPVII 2B06077, MSM 0021620806, LC 06044, SVV-2011-262507), Czech Republic (301/06/1093) and Industry/Trade (FR-TI2/509). We thank the Genome Technology Center at NYU Langone Medical Center, New York (grant NIH/NCI P30 CA016087-30), for expert assistance with the microarray gene expression profiling.
Author contributions: VP, TS wrote the manuscript. JZ, PL, EN and AJ revised the manuscript. VP, TS, JZ, PL designed experiments. VP, KV, JK, JR, FS performed the experiments and analysed data.
Footnotes
The authors declare that they have no conflict of interest.
References
- Back J, Dierich A, Bronn C, Kastner P, Chan S (2004) PU.1 determines the self-renewal capacity of erythroid progenitor cells. Blood 103: 3615–3623 [DOI] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K (2007) High-resolution profiling of histone methylations in the human genome. Cell 129: 823–837 [DOI] [PubMed] [Google Scholar]
- Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116: 281–297 [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Humphrey E.L, Erlich R.L, Schneider R, Bouman P, Liu J.S, Kouzarides T, Schreiber S.L. (2002) Methylation of histone H3 Lys 4 in coding regions of active genes. Proc Natl Acad Sci USA 99: 8695–8700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Kamal M, Lindblad-Toh K, Bekiranov S, Bailey DK, Huebert DJ, McMahon S, Karlsson EK, Kulbokas EJ 3rd, Gingeras TR, Schreiber SL, Lander ES (2005) Genomic maps and comparative analysis of histone modifications in human and mouse. Cell 120: 169–181 [DOI] [PubMed] [Google Scholar]
- Dey BK, Stalker L, Schnerch A, Bhatia M, Taylor-Papidimitriou J, Wynder C (2008) The histone demethylase KDM5b/JARID1b plays a role in cell fate decisions by blocking terminal differentiation. Mol Cell Biol 28: 5312–5327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon-McIver A, East P, Mein CA, Cazier JB, Molloy G, Chaplin T, Andrew Lister T, Young BD, Debernardi S (2008) Distinctive patterns of microRNA expression associated with karyotype in acute myeloid leukaemia. PLoS One 3: e2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontana L, Fiori ME, Albini S, Cifaldi L, Giovinazzi S, Forloni M, Boldrini R, Donfrancesco A, Federici V, Giacomini P, Peschle C, Fruci D (2008) Antagomir-17-5p abolishes the growth of therapy-resistant neuroblastoma through p21 and BIM. PLoS One 3: e2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontana L, Pelosi E, Greco P, Racanicchi S, Testa U, Liuzzi F, Croce CM, Brunetti E, Grignani F, Peschle C (2007) MicroRNAs 17-5p-20a-106a control monocytopoiesis through AML1 targeting and M-CSF receptor upregulation. Nat Cell Biol 9: 775–787 [DOI] [PubMed] [Google Scholar]
- Garzon R, Croce CM (2008) MicroRNAs in normal and malignant hematopoiesis. Curr Opin Hematol 15: 352–358 [DOI] [PubMed] [Google Scholar]
- Ghani S, Riemke P, Schonheit J, Lenze D, Stumm J, Hoogenkamp M, Lagendijk A, Heinz S, Bonifer C, Bakkers J, Abdelilah-Seyfried S, Hummel M, Rosenbauer F (2011) Macrophage development from hematopoietic stem cells requires PU.1 coordinated microRNA expression. Blood 118: 2275–2284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP (2007) MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell 27: 91–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashita Y, Osada H, Tatematsu Y, Yamada H, Yanagisawa K, Tomida S, Yatabe Y, Kawahara K, Sekido Y, Takahashi T (2005) A polycistronic microRNA cluster, miR-17-92, is overexpressed in human lung cancers and enhances cell proliferation. Cancer Res 65: 9628–9632 [DOI] [PubMed] [Google Scholar]
- He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, Powers S, Cordon-Cardo C, Lowe SW, Hannon GJ, Hammond SM (2005) A microRNA polycistron as a potential human oncogene. Nature 435: 828–833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK (2010) Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 38: 576–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanovska I, Ball AS, Diaz RL, Magnus JF, Kibukawa M, Schelter JM, Kobayashi SV, Lim L, Burchard J, Jackson AL, Linsley PS, Cleary MA (2008) MicroRNAs in the miR-106b family regulate p21/CDKN1A and promote cell cycle progression. Mol Cell Biol 28: 2167–2174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasashima K, Nakamura Y, Kozu T (2004) Altered expression profiles of microRNAs during TPA-induced differentiation of HL-60 cells. Biochem Biophys Res Commun 322: 403–410 [DOI] [PubMed] [Google Scholar]
- Koralov SB, Muljo SA, Galler GR, Krek A, Chakraborty T, Kanellopoulou C, Jensen K, Cobb BS, Merkenschlager M, Rajewsky N, Rajewsky K (2008) Dicer ablation affects antibody diversity and cell survival in the B lymphocyte lineage. [see comment]. Cell 132: 860–874 [DOI] [PubMed] [Google Scholar]
- Krishnaraju K, Nguyen HQ, Liebermann DA, Hoffman B (1995) The zinc finger transcription factor Egr-1 potentiates macrophage differentiation of hematopoietic cells. Mol Cell Biol 15: 5499–5507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krysinska H, Hoogenkamp M, Ingram R, Wilson N, Tagoh H, Laslo P, Singh H, Bonifer C (2007) A two-step, PU.1-dependent mechanism for developmentally regulated chromatin remodeling and transcription of the c-fms gene. Mol Cell Biol 27: 878–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laslo P, Spooner CJ, Warmflash A, Lancki DW, Lee HJ, Sciammas R, Gantner BN, Dinner AR, Singh H (2006) Multilineage transcriptional priming and determination of alternate hematopoietic cell fates. Cell 126: 755–766 [DOI] [PubMed] [Google Scholar]
- Li S, Symonds AL, Zhu B, Liu M, Raymond MV, Miao T, Wang P (2011) Early growth response gene-2 (Egr-2) regulates the development of B and T cells. PLoS One 6: e18498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Lu J, Sun M, Mi S, Zhang H, Luo RT, Chen P, Wang Y, Yan M, Qian Z, Neilly MB, Jin J, Zhang Y, Bohlander SK, Zhang DE, Larson RA, Le Beau MM, Thirman MJ, Golub TR, Rowley JD et al. (2008) Distinct microRNA expression profiles in acute myeloid leukemia with common translocations. Proc Natl Acad Sci USA 105: 15535–15540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Bigas N, Kisiel TA, Dewaal DC, Holmes KB, Volkert TL, Gupta S, Love J, Murray HL, Young RA, Benevolenskaya EV (2008) Genome-wide analysis of the H3K4 histone demethylase RBP2 reveals a transcriptional program controlling differentiation. Mol Cell 31: 520–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Thomson JM, Wong HY, Hammond SM, Hogan BL (2007) Transgenic over-expression of the microRNA miR-17-92 cluster promotes proliferation and inhibits differentiation of lung epithelial progenitor cells. Dev Biol 310: 442–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marson A, Levine SS, Cole MF, Frampton GM, Brambrink T, Johnstone S, Guenther MG, Johnston WK, Wernig M, Newman J, Calabrese JM, Dennis LM, Volkert TL, Gupta S, Love J, Hannett N, Sharp PA, Bartel DP, Jaenisch R, Young RA (2008) Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell 134: 521–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez NJ, Ow MC, Barrasa MI, Hammell M, Sequerra R, Doucette-Stamm L, Roth FP, Ambros VR, Walhout AJ (2008) A C. elegans genome-scale microRNA network contains composite feedback motifs with high flux capacity. Genes Dev 22: 2535–2549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKercher SR, Torbett BE, Anderson KL, Henkel GW, Vestal DJ, Baribault H, Klemsz M, Feeney AJ, Wu GE, Paige CJ, Maki RA (1996) Targeted disruption of the PU.1 gene results in multiple hematopoietic abnormalities. EMBO J 15: 5647–5658 [PMC free article] [PubMed] [Google Scholar]
- Melnick A, Licht JD (2002) Histone deacetylases as therapeutic targets in hematologic malignancies. Curr Opin Hematol 9: 322–332 [DOI] [PubMed] [Google Scholar]
- Nerlov C, Graf T (1998) PU.1 induces myeloid lineage commitment in multipotent hematopoietic progenitors. Genes Dev 12: 2403–2412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen HQ, Hoffman-Liebermann B, Liebermann DA (1993) The zinc finger transcription factor Egr-1 is essential for and restricts differentiation along the macrophage lineage. Cell 72: 197–209 [DOI] [PubMed] [Google Scholar]
- O’Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT (2005) c-Myc-regulated microRNAs modulate E2F1 expression. [see comment]. Nature 435: 839–843 [DOI] [PubMed] [Google Scholar]
- Okuda T, van Deursen J, Hiebert SW, Grosveld G, Downing JR (1996) AML1, the target of multiple chromosomal translocations in human leukemia, is essential for normal fetal liver hematopoiesis. Cell 84: 321–330 [DOI] [PubMed] [Google Scholar]
- Ooi SK, Qiu C, Bernstein E, Li K, Jia D, Yang Z, Erdjument-Bromage H, Tempst P, Lin SP, Allis CD, Cheng X, Bestor TH (2007) DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature 448: 714–717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orkin SH, Zon LI (2002) Hematopoiesis and stem cells: plasticity versus developmental heterogeneity. Nat Immunol 3: 323–328 [DOI] [PubMed] [Google Scholar]
- Ota A, Tagawa H, Karnan S, Tsuzuki S, Karpas A, Kira S, Yoshida Y, Seto M (2004) Identification and characterization of a novel gene, C13orf25, as a target for 13q31-q32 amplification in malignant lymphoma. Cancer Res 64: 3087–3095 [DOI] [PubMed] [Google Scholar]
- Petrocca F, Vecchione A, Croce CM (2008a) Emerging role of miR-106b-25/miR-17-92 clusters in the control of transforming growth factor beta signaling. Cancer Res 68: 8191–8194 [DOI] [PubMed] [Google Scholar]
- Petrocca F, Visone R, Onelli MR, Shah MH, Nicoloso MS, de Martino I, Iliopoulos D, Pilozzi E, Liu CG, Negrini M, Cavazzini L, Volinia S, Alder H, Ruco LP, Baldassarre G, Croce CM, Vecchione A (2008b) E2F1-regulated microRNAs impair TGFbeta-dependent cell-cycle arrest and apoptosis in gastric cancer. Cancer Cell 13: 272–286 [DOI] [PubMed] [Google Scholar]
- Pons A, Nomdedeu B, Navarro A, Gaya A, Gel B, Diaz T, Valera S, Rozman M, Belkaid M, Montserrat E, Monzo M (2009) Hematopoiesis-related microRNA expression in myelodysplastic syndromes. Leuk Lymphoma 50: 1854–1859 [DOI] [PubMed] [Google Scholar]
- Reader JC, Meekins JS, Gojo I, Ning Y (2007) A novel NUP98-PHF23 fusion resulting from a cryptic translocation t(11;17) (p15;p13) in acute myeloid leukemia. Leukemia 21: 842–844 [DOI] [PubMed] [Google Scholar]
- Rosa A, Ballarino M, Sorrentino A, Sthandier O, De Angelis FG, Marchioni M, Masella B, Guarini A, Fatica A, Peschle C, Bozzoni I (2007) The interplay between the master transcription factor PU.1 and miR-424 regulates human monocyte/macrophage differentiation. Proc Natl Acad Sci USA 104: 19849–19854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbauer F, Tenen DG (2007) Transcription factors in myeloid development: balancing differentiation with transformation. Nat Rev Immunol 7: 105–117 [DOI] [PubMed] [Google Scholar]
- Rosenbauer F, Wagner K, Kutok JL, Iwasaki H, Le Beau MM, Okuno Y, Akashi K, Fiering S, Tenen DG (2004) Acute myeloid leukemia induced by graded reduction of a lineage-specific transcription factor, PU.1. Nat Genet 36: 624–630 [DOI] [PubMed] [Google Scholar]
- Schmeier S, MacPherson CR, Essack M, Kaur M, Schaefer U, Suzuki H, Hayashizaki Y, Bajic VB (2009) Deciphering the transcriptional circuitry of microRNA genes expressed during human monocytic differentiation. BMC Genomics 10: 595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt M, Nagel S, Proba J, Thiede C, Ritter M, Waring JF, Rosenbauer F, Huhn D, Wittig B, Horak I, Neubauer A (1998) Lack of interferon consensus sequence binding protein (ICSBP) transcripts in human myeloid leukemias. Blood 91: 22–29 [PubMed] [Google Scholar]
- Scibetta AG, Santangelo S, Coleman J, Hall D, Chaplin T, Copier J, Catchpole S, Burchell J, Taylor-Papadimitriou J (2007) Functional analysis of the transcription repressor PLU-1/JARID1B. Mol Cell Biol 27: 7220–7235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott EW, Simon MC, Anastasi J, Singh H (1994) Requirement of transcription factor PU.1 in the development of multiple hematopoietic lineages. Science 265: 1573–1577 [DOI] [PubMed] [Google Scholar]
- Secombe J, Eisenman RN (2007) The function and regulation of the JARID1 family of histone H3 lysine 4 demethylases: the Myc connection. Cell Cycle 6: 1324–1328 [DOI] [PubMed] [Google Scholar]
- Sims RJ 3rd, Millhouse S, Chen CF, Lewis BA, Erdjument-Bromage H, Tempst P, Manley JL, Reinberg D (2007) Recognition of trimethylated histone H3 lysine 4 facilitates the recruitment of transcription postinitiation factors and pre-mRNA splicing. Mol Cell 28: 665–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stopka T, Amanatullah DF, Papetti M, Skoultchi AI (2005) PU.1 inhibits the erythroid program by binding to GATA-1 on DNA and creating a repressive chromatin structure. Embo J 24: 3712–3723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan R, Mager GM, Ward RM, Mayer J, Svaren J (2006) NAB2 represses transcription by interacting with the CHD4 subunit of the nucleosome remodeling and deacetylase (NuRD) complex. J Biol Chem 281: 15129–15137 [DOI] [PubMed] [Google Scholar]
- Svaren J, Sevetson BR, Apel ED, Zimonjic DB, Popescu NC, Milbrandt J (1996) NAB2, a corepressor of NGFI-A (Egr-1) and Krox20, is induced by proliferative and differentiative stimuli. Mol Cell Biol 16: 3545–3553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sylvestre Y, De Guire V, Querido E, Mukhopadhyay UK, Bourdeau V, Major F, Ferbeyre G, Chartrand P (2007) An E2F/miR-20a autoregulatory feedback loop. J Biol Chem 282: 2135–2143 [DOI] [PubMed] [Google Scholar]
- Tagawa H, Seto M (2005) A microRNA cluster as a target of genomic amplification in malignant lymphoma. Leukemia 19: 2013–2016 [DOI] [PubMed] [Google Scholar]
- Tanzer A, Stadler PF (2004) Molecular evolution of a microRNA cluster. J Mol Biol 339: 327–335 [DOI] [PubMed] [Google Scholar]
- Tsang J, Zhu J, van Oudenaarden A (2007) MicroRNA-mediated feedback and feedforward loops are recurrent network motifs in mammals. Mol Cell 26: 753–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Zutven LJ, Onen E, Velthuizen SC, van Drunen E, von Bergh AR, van den Heuvel-Eibrink MM, Veronese A, Mecucci C, Negrini M, de Greef GE, Beverloo HB (2006) Identification of NUP98 abnormalities in acute leukemia: JARID1A (12p13) as a new partner gene. Genes Chromosomes Cancer 45: 437–446 [DOI] [PubMed] [Google Scholar]
- Ventura A, Young AG, Winslow MM, Lintault L, Meissner A, Erkeland SJ, Newman J, Bronson RT, Crowley D, Stone JR, Jaenisch R, Sharp PA, Jacks T (2008) Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell 132: 875–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venturini L, Battmer K, Castoldi M, Schultheis B, Hochhaus A, Muckenthaler MU, Ganser A, Eder M, Scherr M (2007) Expression of the miR-17-92 polycistron in chronic myeloid leukemia (CML) CD34+ cells. Blood 109: 4399–4405 [DOI] [PubMed] [Google Scholar]
- Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, Prueitt RL, Yanaihara N, Lanza G, Scarpa A, Vecchione A, Negrini M, Harris CC, Croce CM (2006) A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA 103: 2257–2261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volinia S, Galasso M, Costinean S, Tagliavini L, Gamberoni G, Drusco A, Marchesini J, Mascellani N, Sana ME, Abu Jarour R, Desponts C, Teitell M, Baffa R, Aqeilan R, Iorio MV, Taccioli C, Garzon R, Di Leva G, Fabbri M, Catozzi M et al. (2010) Reprogramming of miRNA networks in cancer and leukemia. Genome Res 20: 589–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh JC, DeKoter RP, Lee HJ, Smith ED, Lancki DW, Gurish MF, Friend DS, Stevens RL, Anastasi J, Singh H (2002) Cooperative and antagonistic interplay between PU.1 and GATA-2 in the specification of myeloid cell fates. Immunity 17: 665–676 [DOI] [PubMed] [Google Scholar]
- Wang GG, Song J, Wang Z, Dormann HL, Casadio F, Li H, Luo JL, Patel DJ, Allis CD (2009) Haematopoietic malignancies caused by dysregulation of a chromatin-binding PHD finger. Nature 459: 847–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods K, Thomson JM, Hammond SM (2007) Direct regulation of an oncogenic micro-RNA cluster by E2F transcription factors. J Biol Chem 282: 2130–2134 [DOI] [PubMed] [Google Scholar]
- Xiao C, Srinivasan L, Calado DP, Patterson HC, Zhang B, Wang J, Henderson JM, Kutok JL, Rajewsky K (2008) Lymphoproliferative disease and autoimmunity in mice with increased miR-17-92 expression in lymphocytes. Nat Immunol 9: 405–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan HL, Xue G, Mei Q, Wang YZ, Ding FX, Liu MF, Lu MH, Tang Y, Yu HY, Sun SH (2009) Repression of the miR-17-92 cluster by p53 has an important function in hypoxia-induced apoptosis. EMBO J 28: 2719–2732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu B, Symonds AL, Martin JE, Kioussis D, Wraith DC, Li S, Wang P (2008) Early growth response gene 2 (Egr-2) controls the self-tolerance of T cells and prevents the development of lupuslike autoimmune disease. J Exp Med 205: 2295–2307 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.