Abstract
The non-coding part of our genome contains sequence motifs that can control gene transcription over distance. Here, we discuss functional genomics studies that uncover and characterize these sequences across the mammalian genome. The picture emerging is of a genome being a complex regulatory landscape. We explore the principles that underlie the wiring of regulatory DNA sequences and genes. We argue transcriptional control over distance can be understood when considering action in the context of the folded genome. Genome topology is expected to differ between individual cells, and this may cause variegated expression. High-resolution three-dimensional genome topology maps, ultimately of single cells, are required to understand the cis-regulatory networks that underlie cellular transcriptomes.
Keywords: enhancers, genome organization, genome topology, transcription
Introduction: gene regulation by remote DNA elements
Over 200 cell types exist within the human body, each being different in morphology and function, yet all containing the same genome. These differences are driven by cell-specific gene regulatory programs. Their proper execution not only relies on the protein-coding parts of genes, but also on that of non-coding sequences within and surrounding the genes. This was first realized when a deletion outside the β-globin gene was found to cause aberrant expression of the otherwise intact gene in a thalassaemia patient (Van der Ploeg et al, 1980). It is now well recognized that sequence alterations in the non-coding part of our genome, previously known as ‘junk DNA’, frequently drive the deregulation of critical genes to cause congenital and somatically acquired diseases (Kleinjan and van Heyningen, 2005). These alterations can locate at large distances from the relevant genes. One extreme example is where point mutations in the DNA located ∼1 Mb away from the Sonic Hedgehog (SHH) gene interfere with its regulation. This has been linked to preaxial polydactyly, a condition where patients suffer from limb malformations (Lettice et al, 2003). Chromosomal rearrangements may remove, mutate or separate distant regulatory sequences from genes, but may also lead to so-called ‘position effects’, where genes are placed adjacent to new regulatory sequences that drive their upregulation or downregulation. This is seen, for example, in B-cell lymphomas and T-cell leukaemias, when translocations juxtapose strong regulatory sequences of antigen receptor loci to proto-oncogenes. How frequent diseases are caused by genetic changes outside gene bodies is unknown, because most large-scale screening efforts only focus on exome integrity. Based on an examination of the database of genome-wide association studies (Hindorff et al, 2009), mutations in non-coding regions were estimated to contribute to a staggering 40% of disease cases (Visel et al, 2009b). No matter the exact frequency in disease, studies like these clearly underscore the importance of cis-linked regions in regulating gene expression and controlling normal development. Current research efforts in functional genomics, therefore, focus on uncovering the full regulatory potential of our genome.
Here, we will review studies that aim to identify and assign function to the regulatory DNA sequences of the mammalian genome and discuss how regulatory DNA elements and genes are wired to properly execute cell-specific transcription regulatory programs.
Identifying and classifying regulatory DNA sites in the genome
Regulatory DNA sequences in the genome can be identified through various methods. Perhaps, the most thorough assay is to screen for DNase I hypersensitive sites (HSs; Crawford et al, 2004; Dorschner et al, 2004). HSs typically are sites where the nucleosome fibre is locally disrupted, presumably through the action of DNA-binding proteins and associated factors. Any given human cell type appears to have ∼100 000 or more HSs (Boyle et al, 2008). This includes promoter sequences of active genes, but the majority is located away from transcriptional start sites in non-coding DNA.
To understand their function, we can test and categorize their activity. This is traditionally done using property defining assays, often in vitro, in which the isolated sequence element is placed in plasmids carrying a reporter gene. When transfected into cells, they may then activate or repress transcription, or neutralize transcriptional activation when placed between an activator and gene promoter. Accordingly, they are classified as an enhancer, silencer or insulator. A more laborious but often more revealing assay is to stably integrate an isolated site with a reporter gene in the genome, and measure transcriptional activity. By doing this in transgenic animals, one can also determine an element's tissue-specific activity and define other properties not appreciable in plasmid-based assays. Boundary elements, for example, are defined based on their property to protect against position effects when placed around an integrated transgene (Recillas-Targa et al, 2002), an ability not well appreciated using a plasmid-based assay. Similarly, locus control regions (LCRs), which often behave as enhancers in plasmid-based assays, have the additional capacity to confer tissue-specific, position-independent and copy number-dependent expression on linked reporter genes when stably integrated in transgenic animals (Grosveld et al, 1987). This defining property is not shared by classic enhancers and can only be appreciated when tested in transgenics.
While the categorization of DNA elements is useful, an activity picked up for a DNA motif in a reporter assay may not be relevant or detectable at its natural chromosomal context (Dillon and Sabbattini, 2000). This has been demonstrated for insulators, enhancers and LCRs (Epner et al, 1998; Bender et al, 2006; Splinter et al, 2006; Ahituv et al, 2007; Table I). Thus, DNA sequences may intrinsically harbour specific activities, and hence be classified accordingly, but the linear and, as we will argue below, three-dimensional chromosomal context often determines whether they exert this activity or not.
Table 1. Property defining activities measured for DNA sequences in reporter assays may not be detectable at their natural chromosomal context.
| Genomic site | Definition | Property defining reporter assay | Effect of genomic site deletion |
|---|---|---|---|
| β-Globin: HSs upstream of genes | LCR (Grosveld et al, 1987) | Confers position-independent, copy number-dependent transgene expression in mice | No heterochromatinization, basal non-activated gene expression (Epner et al, 1998)a |
| CTCF sites flanking β-globin locus | Insulators (Farrell et al, 2002) | Block enhancer activity and shield reporter genes in plasmid-based assays | No heterochromatinization, no transcription changes measurable (Bender et al, 2006; Splinter et al, 2006) |
| Ultra-conserved elements close to Dmrt3, Rcn1, Pola1 and Sox3 | Enhancers (Ahituv et al, 2007) | Tissue-specific reporter gene activation in transgenic mice | No transcription changes measurable (Ahituv et al, 2007) |
| aNote that the β-globin LCR is not necessary for maintenance of open chromatin in mice, but does appear necessary for this in humans (Forrester et al, 1990). | |||
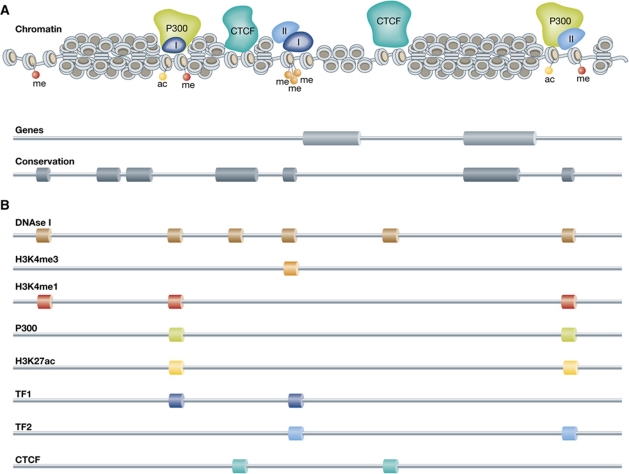
With 100 000 or more HSs per cell type, mammalian genomes are emerging as highly complex regulatory landscapes. Alternative strategies for the exposure and classification of regulatory sites confirm this idea. Genome-wide chromatin immunoprecipitation approaches (ChIP-chip and ChIP-seq) have been developed to identify specific classes of regulatory sites based on their unique chromatin signature. For example, enhancers were shown to have high H3K4me1 and promoters high H3K4me3 levels (Heintzman et al, 2007). Based on their H3K4me1 mark in two human cell types, ∼55 000 potential enhancer elements were uncovered (Heintzman et al, 2009). Nearly 80% of the enhancers were unique for the one or the other cell type, despite the two sharing 85% of their active genes. This underscores the idea that enhancers are tissue-specific elements acting on tissue-specific genes. At the same time, it raises the question whether all these sites are actively involved in gene regulation. A further subclassification of H3K4me1 enhancers is now made based on the shared presence of p300 protein or acetylated H3K27. Both signatures appear to separate the active from the poised pool of enhancers. In the case of p300, a co-activator protein that can acetylate H3K27, several thousand (instead of tens of thousands) active enhancers were identified in various primary mouse tissues (Visel et al, 2009a). Similar numbers of active enhancers were found based on the H3K27Ac profile (Creyghton et al, 2010; Rada-Iglesias et al, 2011). Collectively, these studies show that a given mammalian cell type contains thousands or more regulatory sites. With 200 different cell types, this confirms that our genome harbours a complex regulatory landscape (Figure 1).
Figure 1.
The complex regulatory landscape of the genome. (A) DNA packaged into chromatin with regulatory proteins P300, transcription factors I and II, CTCF and histone modifications present. Underneath the chromatin fibre the position of genes and conserved elements, representing information that can be found in databases (e.g., UCSC, Ensembl). (B) The identification of accessible chromatin (DNAse I profile) and various chromatin marks (ChIP-seq) that are associated with regulatory elements yields a picture of a complex regulatory landscape.
The mammalian genome: a complex regulatory landscape
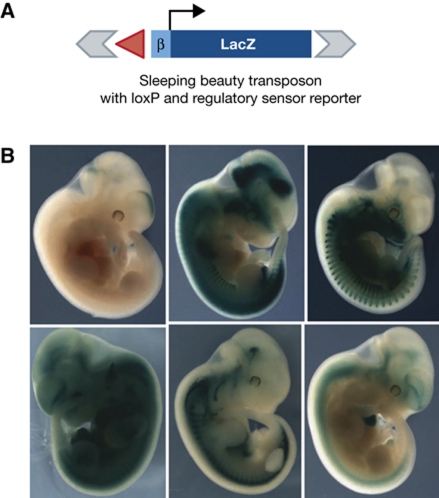
The term ‘regulatory landscape’ was first coined to describe the complex organization of regulatory sequences around the Hox loci (Spitz et al, 2003). A recent study provided further insight into the complexity of mammalian gene regulation (Ruf et al, 2011). In this study, hundreds of transgenic mice were generated, each carrying a transposable reporter cassette containing a LacZ reporter gene driven by a minimal promoter sequence (50 bp of the human β-globin gene) inserted at a random location in the genome. Each was then analysed for LacZ expression in E11.5 embryos, and categorized according to tissue specificity of expression. The minimal promoter by itself was insufficient to drive reporter gene expression, yet nearly 60% of the transgenics showed reporter activity, demonstrating that the majority of genomic locations harbour activating potential. Among the activated transgenics, only a very small percentage showed ubiquitous expression, while the great majority (>95%) demonstrated restricted, tissue-specific expression (Figure 2). Often, but not always, reporter expression followed that of the nearest endogenous gene. However, tissue-specific expression was also found for integration sites near genes with widespread expression patterns. This argues that also the expression of so-called housekeeping genes is modulated in a tissue-specific manner. The flanking transposon sites further enabled the investigators to locally hop around the reporter cassette and assess the regulatory impact at multiple locations within one chromosomal domain. Essentially, all possible outcomes were found. Sometimes, transposition over only a few kilobases resulted in clearly distinct expression patterns, whereas in other examples integration sites separated by hundreds of kilobases gave essentially the same expression profile (Ruf et al, 2011). This led the authors to propose that the genome harbours a ‘regulatory jungle’: chromosomal regions contain many regulatory sites that can activate gene expression over large distances and others that counteract this activity.
Figure 2.
Random integration of a reporter cassette reveals the regulatory potential of the mouse genome. (A) Schematic representation of the randomly integrated reporter cassette consisting of the LacZ gene driven by the β-globin minimal promoter. (B) Examples of LacZ expression patterns found in the different transgenic lines which, depending on the integration site, range from global to a highly restricted expression pattern. (Reproduced with permission from Ruf et al, 2011.)
How are all these linearly organized activities orchestrated such that genes are expressed at the right time and place? Genetic studies that manipulated the non-coding part of the genome to understand transcriptional control have delineated some primary rules of promoter–enhancer engagement that must be considered when addressing this question. So far, most of these studies focused on gene clusters such as the Hox and globin loci, aiming to understand how transcription of their individual genes is coordinated in time and space.
Rules of engagement in a complex regulatory landscape: (1) linear proximity
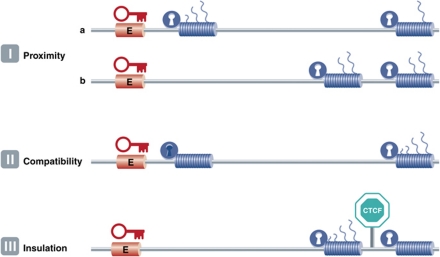
It is often assumed that enhancers act on the nearest genes in cis, and in many cases this is correct. Proximity on the linear DNA template, or genomic order, is a major determinant of selectivity: first-come, first-served (Figure 3, rule I). This was first demonstrated in plasmid-based competition assays, which showed that proximal genes have an advantage over distal genes to be activated by a shared enhancer (de Villiers et al, 1983; Wasylyk et al, 1983). The same principle was then shown to also apply to transcription regulation in the chromosomal context (Hanscombe et al, 1991; Dillon et al, 1997). Interestingly, order is no longer important when two competing genes are positioned close together at a large distance from a shared enhancer (Heuchel et al, 1989; Dillon et al, 1997). While linear proximity to a regulatory site is often a good predictor of target genes, many examples exist where enhancers ignore the nearest genes and specifically act on genes further away (de Laat and Grosveld, 2003). The previously mentioned limb bud-specific enhancer of the SHH gene is one such example, present in an intron of the Lmbr gene, but acting on the SHH gene 1 Mb away. Why do enhancers not always stick to the ‘first-come, first-served’ rule, but sometimes act instead on more distal genes?
Figure 3.
The three rules of engagement that dictate enhancer–promoter interactions. I: Proximity. (a) When multiple genes (cylinders) are compatible with (open lock) and relatively close to a shared enhancer (E), the most proximal gene is preferentially activated over the distal gene (represented by the number of transcripts originating from the gene). (b) This competitive advantage disappears when both genes are located far away from the shared enhancer. II: Compatibility. Enhancers ignore the ‘first-come, first-served’ rule when the proximal promoter is incompatible (closed lock) with the enhancer. Result: activation of the distal gene. III: Insulation. The presence of CTCF can block enhancer function across its binding site and prevent a compatible gene from being activated by the enhancer.
Rules of engagement in a complex regulatory landscape: (2) promoter specificity
Enhancer reporter assays in cell lines (in vitro) and mice (in vivo) usually analyse the capacity of test sequences to activate one and the same minimal or general promoter (Banerji et al, 1981; Visel et al, 2008; Ruf et al, 2011). The assumption is that enhancers and promoters show little specificity and will always act on each other. While this may be true for artificial constructs where the enhancer and minimal promoter are close together, it appears more complex in the context of the genome where regulatory sites do show target specificity. There are numerous examples of enhancers interacting with just a subset of equally nearby target promoters, and there are different possible explanations for this. Sometimes, an enhancer is thought to ignore a gene because its promoter is not accessible in the tissue where the enhancer is active. In the β-globin locus, for example, the LCR exclusively acts on the β-globin genes and totally ignores the nearby olfactory receptor genes, even when potentially interfering insulator sites are disrupted (Splinter et al, 2006). The LCR also ignores the nearby fetal globin genes to exclusively act on more distal adult β-globin genes at later stages of development. Inactivity, or promoter inaccessibility, may therefore be a reason for regulatory sites to skip nearby genes (Figure 3, rule II).
Selective gene activation can also be explained by promoter competition, whereby the activation of one promoter precludes the activation of another equidistant promoter. This was shown clearly in Drosophila transgenic embryo assays, where different enhancers were demonstrated to prefer distinct classes of promoters, depending on the presence of certain core promoter elements (Ohtsuki et al, 1998; Butler and Kadonaga, 2001).
Promoter competition also occurs between mammalian genes. It manifests itself when the deletion of one or more genes impacts on the expression of remaining neighbouring genes, or when the deletion of a regulator causes downregulation of multiple genes. These phenomena have been described at the Hox (Spitz et al, 2003; Tschopp et al, 2009) and globin gene clusters (Forrester et al, 1990; Hanscombe et al, 1991; Epner et al, 1998; Sabatino et al, 1998; Lower et al, 2009). The implication of gene competition is two-fold: the first is that regulatory sites exist in the genome that can act on multiple endogenous genes. Evidence exist that the number of genes controlled by a given regulatory site probably depends on its chromosomal context. At its natural location, the β-globin LCR activates maximally two or three β-globin genes at any given developmental stage. However, when tested without globin genes at a defined new location in the genome it was found to activate 6–7 genes up to 150 kb away in cis (Noordermeer et al, 2008) and two genes in trans (see below) (Noordermeer et al, 2011). A second implication of gene competition is that enhancer–promoter interactions are, at least for some time, mutually exclusive. Convincing evidence for this was obtained in single-cell experiments that measured the ongoing transcriptional activity of LCR-competing β-globin genes in cells with traceable transcriptional history. The LCR was demonstrated to activate only a single gene at the time, but over time to dynamically flip-flop between competing globin genes (Wijgerde et al, 1995). This work provided insight into the mechanism behind competition between clustered genes for shared regulators.
In a genome described as a ‘regulatory jungle’ (Ruf et al, 2011), one might expect that regulators sometimes also ‘fortuitously’ activate more than their presumed target gene. Indeed, bystander activation has been observed in several cases. The B cell-specific human immunoglobulin-beta (Igβ) gene (or CD79b) is highly expressed, but presumably not functional, in pituitary due to its linear proximity to the LCR that is acting on the more distal growth hormone (hGH) gene in this tissue (Cajiao et al, 2004). A similar phenomenon was recently found at the human α-globin locus that is located in a gene-dense chromosomal region with many housekeeping genes. When a 500-kb region around the locus was analysed for gene expression levels, a gene called NME4, 300 kb apart, was found to be upregulated specifically in red blood cells where the α-globin genes are active. Subsequent analysis showed that the α-globin regulatory sequences were responsible for this, and that NME4 competes with the α-globin genes for these enhancers (Lower et al, 2009). Collectively, these data show that tissue-specific enhancers not always exclusively activate transcription of their real target genes but sometimes also that of unrelated genes nearby in cis. Based on arguments explained below, we predict this bystander activation will be seen even more often when transcriptome analysis is performed at the single-cell level rather than the cell population level.
Rules of engagement in a complex regulatory landscape: (3) insulators can block enhancer–promoter interactions
The original idea that led to the discovery of insulator elements was that the genome may be partitioned in physically separate chromatin domains that each has their own independent regulatory activities. If true, the assumption was that boundaries must exist that prevent regulatory cross talk between these domains. To test this hypothesis, two types of assays were developed. One investigated the ability of sequence elements flanking a reporter gene to overcome gene repression when integrated into heterochromatin (Kellum and Schedl, 1991). Another analysed whether an element can block enhancer activity when positioned in between the enhancer and a gene promoter (Kellum and Schedl, 1992). With the discovery of more and more regulatory sites in the mammalian genome that ignore nearby genes to act specifically on much more distal genes, the idea that regulatory activities are strictly separated along the linear chromosome template had to be adjusted (Dillon and Sabbattini, 2000; de Laat and Grosveld, 2003). Nevertheless, the assays were proven to be very useful to identify an intriguing new class of regulatory sites known as ‘insulators’. In mammals, one protein in particular is associated with insulator activity: CTCF (Chung et al, 1993; Bell et al, 1999). In in vitro reporter assays, CTCF bound to DNA often acts as an enhancer blocker (Figure 3, rule III). In vivo, CTCF binding sites are found next to genes that are active in an otherwise repressive chromatin surrounding, such as the human and mouse β-globin genes (Farrell et al, 2002) and near genes on the silenced X chromosome that escape X inactivation (Filippova et al, 2005). CTCF also acts as an allele-specific enhancer blocker to mediate imprinted gene expression at the H19-Igf2 locus (Bell and Felsenfeld, 2000; Hark et al, 2000). Interestingly, a single CTCF site can function as an insulator, but the introduction of a second CTCF site in between an enhancer and promoter often alleviates the enhancer blocking effect (Cai and Shen, 2001; Muravyova et al, 2001). ChIP-Seq experiments were carried out to obtain a genome-wide picture of CTCF binding sites. An estimated 20 000–87 000 CTCF binding sites exist in the human genome (Barski et al, 2007; Kunarso et al, 2010), meaning that on average a CTCF binding site is present every 35–155 kb. Most of these sites (50–90%) appear conserved between different cell types (Barski et al, 2007; Chen et al, 2008; Cuddapah et al, 2009; Kunarso et al, 2010). They are enriched at boundaries between repressive (H3K27me3-rich) and active (H3K27me3-poor) chromatin (Cuddapah et al, 2009), and at the borders between so-called lamin-associated domains (LADs) and non-LADs, being chromosomal regions that preferentially locate to the periphery or the interior of the cell nucleus (Guelen et al, 2008). Furthermore, genes separated by a CTCF site show a markedly reduced correlation in gene expression (Xie et al, 2007). Collectively, these studies demonstrate that, while not all CTCF sites will act as insulators, clearly their location, and that of possible other insulators, must be taken into account when considering the wiring of regulatory DNA networks in the genome. Interestingly, substantial overlap exists between the genomic binding sites of CTCF and cohesin (Parelho et al, 2008; Wendt et al, 2008), suggesting that cohesin, a protein complex that can hold two DNA helices together may assist CTCF in its function to separate regulatory activities.
In summary, genetic experiments have uncovered three rules of enhancer–promoter engagement: linear proximity matters, enhancer and promoter need to be compatible, and some sequences exist that can block enhancer activity. To understand the molecular mechanisms behind these rules, we need to know how regulatory sites exert activities over distance.
Chromatin looping and spatial interactions between regulatory sites
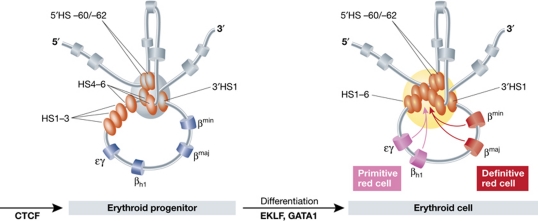
The idea that DNA topology can play an important role in long-range gene activation was originally based on observations on bacterial and phage repressor proteins, like the Gal, AraC and λ repressor proteins. They were found to only function when homo-multimerized and bound to separated operator sequences and this was shown by electron microscopy to result in looping out of the intervening DNA fibre (Ptashne, 1986). The first direct evidence in eukaryotes for spatial interactions between regulatory sites and their target genes was provided by two independent studies on the mouse β-globin locus. One study involved the use of RNA-TRAP to demonstrate a chromatin loop between an active β-globin gene and HS2 of the LCR (Carter et al, 2002). The other study applied 3C technology and showed that not only HS2 but also other β-globin regulatory sites participate in the interactions with the genes (Tolhuis et al, 2002). The spatial chromatin entity they formed was called an Active Chromatin Hub (ACH; Figure 4). 3C (chromosome conformation capture) technology turned out to be the method of choice for unraveling the three-dimensional regulatory circuitries at gene loci. The technique relies on crosslinking and ligation of spatially proximal DNA sequences, which are then identified and quantified with PCR strategies (Dekker et al, 2002). In subsequent studies on the β-globin locus, it was demonstrated that during development the genes dynamically switch their physical interactions with the LCR in relation to their change in gene expression (Palstra et al, 2003) and that transcription factors mediate these long-range DNA interactions (Drissen et al, 2004; Vakoc et al, 2005). 3C also provided evidence for chromatin loops between distal enhancers and target genes at other loci, including the interleukin (Spilianakis and Flavell, 2004), α-globin (Vernimmen et al, 2007) and immunoglobulin loci (Liu and Garrard, 2005), and showed that chromatin loops can also be involved in gene repression (Horike et al, 2005; Comet et al, 2006, 2011; Tiwari et al, 2008; Bantignies et al, 2011). In a recent study, both microscopy and 3C data showed that the distal limb bud enhancer of SHH loops to the gene (Amano et al, 2009).
Figure 4.
A schematic representation of the three-dimensional structure of the mouse β-globin locus during differentiation. In erythroid progenitors, a ‘Chromatin Hub’ (CH) is formed by the clustering of the CTCF sites that surround the β-globin locus. Upon differentiation, the remaining hypersensitive sites of the LCR and the globin gene that is activated participate in this clustering to form an ‘Active Chromatin Hub’ (ACH). Hypersensitive sites are represented by the ovals and genes by the cylinders. In grey, the olfactory receptor genes that surround the locus are depicted. A developmental switch in gene usage occurs between primitive and definitive red cells.
Collectively, these and other studies firmly establish that regulatory sites act on genes via chromatin looping. This knowledge argues that we must know the topological constraints of chromatin in order to understand how transcription is controlled in the regulatory jungle of the genome. Question therefore is how do separated genomic sites find each other?
Rules of engagement in three dimensions: proximity matters
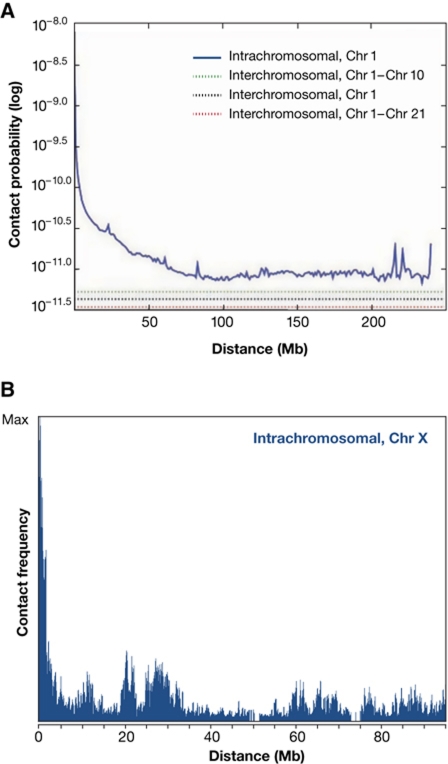
When we ignore all biological activity exerted by DNA-binding proteins and associated factors, a chromatin-packed chromosome fibre is expected to fold and behave essentially as a polymer with a given flexibility. This implies that random collisions will take place between pairs of sites present on a chromosome, with contact probabilities that exponentially decrease with increasing site separation on the linear template (Rippe et al, 1995). This is exactly what is measured in vivo by Hi-C technology (Lieberman-Aiden et al, 2009; Duan et al, 2010; Tanizawa et al, 2010; Figure 5A). Hi-C is a high-throughput genomic variant of 3C that analyses interactions between all genomic sites. A smooth, continuous and exponential decline in contact probability is seen with increased site separation when Hi-C measurements for all pairs of sites are plotted in relation to their genomic distance. Beyond certain distances the curves plateau indicating that contact probabilities then no longer depend on the amount of intervening DNA. For human chromosomes, this seems to happen only beyond 90 Mb of DNA or more. Of relevance, the data also show that two given sites on one chromosome, no matter their linear site separation, are much more likely to contact each other than to contact a given site on a different chromosome (Lieberman-Aiden et al, 2009).
Figure 5.
The relationship between contact frequencies (spatial distance) and the position of sequences on the DNA template. (A) Hi-C experiments showing the average contact probabilities between all pairs of sequences (within and between chromosomes). The average profile shows that with an increasing linear distance the interaction frequencies drop exponentially. (Reproduced with permission from Lieberman-Aiden et al, 2009.) (B) 4C experiment that measures interaction frequencies for an individual locus (‘viewpoint’) versus all other genomic sequences. The same global anti-correlation between distance and contact frequency is seen, but on top specific interactions (peaks) are observed at sequences that are preferentially contacted by the viewpoint.
Thus, Hi-C data confirm what was predicted from polymer physics: any given site in the genome has the ability to contact neighbouring sequences, the chance of which decreases exponentially with increasing site separation on the linear template. This very basic concept of chromosome flexibility can be assumed to underlie the first rule of engagement: linear proximity matters. Without activities interfering with local chromatin flexibility, any regulatory site is much more likely to contact a gene nearby than a gene far away on the chromosome. The genetic observation that linear order no longer matters when two similar genes are close to each other but far apart from a shared enhancer (Dillon et al, 1997) is also in line with a polymer-like behaviour of chromatin: both will have a roughly equal chance of colliding into the enhancer.
Rules of engagement in three dimensions: affinity matters
While polymer physics dictates, and Hi-C data shows, that collisions between linearly proximal sequences will be frequent, they will be aspecific and equally transient for all pairs of sites unless two sites have increased affinity. This can be mediated, for example, via the proteins associated with them. Upon collision of two such sites, a stabilization of the contact will occur, which will be measurable as a chromatin loop. This is what is seen by 4C technology (Figure 5B). 4C technology is a different high-throughput version of 3C technology that produces high-resolution DNA contact maps for individual genomic sites. A 4C profile typically shows the same anti-correlation between contact probability and genomic distance as seen by Hi-C, but the curves are no longer smooth: peaks and valleys of contacts are imposed on it (Simonis et al, 2006, 2007). A peak in a 4C contact profile identifies a preferred interaction site and reveals a chromatin loop between this site and the selected target sequence (‘viewpoint’). Since 3C-based methods provide average topology impressions of populations of cells, such a loop must have been present in a relevant proportion of the cells the moment they were fixed. In several instances, it was demonstrated that the stability of such chromatin loops indeed relies on the associated proteins (Drissen et al, 2004; Vakoc et al, 2005; Splinter et al, 2006).
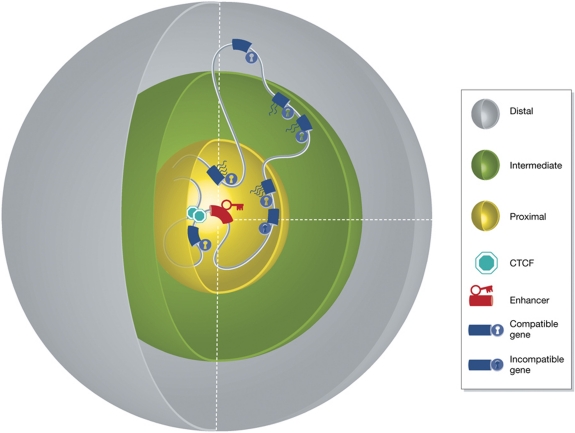
One can speculate that the formation and stabilization of a given chromatin loop has two important consequences: one is that for the duration of the interaction the two sites involved may not be available for direct interactions with other genomic sites. The second is that this structure imposes constraints on the flexibility of the intervening and surrounding genomic sites. The latter is expected to be relevant for insulator function, as will be discussed below. The first would clearly be relevant for promoter competition: as long as an enhancer is stably engaged in an interaction with one gene, it cannot interact with another gene (Wijgerde et al, 1995). Thus, the genetically deduced rule of engagement stating that enhancer–promoter compatibility is essential may well be dependent on the affinities between enhancer- and promoter-bound proteins and their ability to stably form a chromatin loop. The outcome of promoter competition, that is, the number of genes activated and their eventual transcript levels, will therefore depend on their order on the linear template, their relative distance to and their affinity for the enhancer (Figure 6).
Figure 6.
Spatial distance, enhancer–promoter compatibility and sites of insulation are the main determinants of enhancer-mediated gene expression. This figure represents a three-dimensional model of chromatin ‘in action’, explaining all three rules of engagement by considering regulatory action in three dimensions. Genes proximal to the enhancer in 3D space (in the yellow sphere) have a high probability to be contacted, but activation depends on compatibility and whether or not a gene is isolated from the enhancer (CTCF sites). Compatible genes further away in 3D (in the green sphere) have a decreased probability to be contacted and activated, while compatible genes that too far (grey sphere) will not be activated.
Rules of engagement in three dimensions: insulators and DNA flexibility
If chromatin looping between enhancers and promoters is critical for gene activation, would insulators exert their blocking activity also by acting on DNA topology? An increasing body of evidence suggests they do. A role for insulators in organizing genome topology was first appreciated from microscopy observation in Drosophila, showing that insulator sequences separated in the genome come together in the nuclear space in so-called insulator bodies (Gerasimova et al, 2000). Direct looping between neighbouring insulator sites was first demonstrated by 3C technology for CTCF sites at the H19-Igf2 locus and the β-globin locus (Kurukuti et al, 2006; Splinter et al, 2006). Binding of CTCF to an internal site of the imprinted H19-Igf2 locus was shown to create a loop encompassing the Igf2 gene that presumably isolated the gene from enhancers outside the loop (Kurukuti et al, 2006). At the β-globin locus, flanking CTCF sites were demonstrated to form a large chromatin loop containing both enhancers and genes that was further folded later during differentiation into smaller loops to bring stage-specific genes in closer proximity to their enhancers (Splinter et al, 2006). Insulators not only block the activating effects of enhancers, but they can also interfere with the spreading of repression mediated by polycomb group proteins (Sigrist and Pirrotta, 1997; Mallin et al, 1998). It was noticed that chromatin coating by polycomb proteins will stop at an insulator site, but may continue into more downstream regions beyond a second insulator site (Comet et al, 2006). The abrupt block at the first site suggests that insulators act as a physical barrier. The continued spreading beyond the second site suggests these downstream regions are brought into spatial proximity of the upstream source of polycomb protein, which indeed was shown to happen as a consequence of loop formation between the two intervening insulator sites (Comet et al, 2011). These and other detailed transgenic studies (e.g., Maksimenko et al, 2008), together with genomic evidence that CTCF sites often locate to borders of spatially separated chromatin domains (Guelen et al, 2008), have established that insulator sequences strongly influence chromosome topology. They induce chromatin loops that will affect the flexibility of neighbouring and intervening chromatin sites. The impact hereof on gene regulation can be positive, negative or neutral, depending on the regulatory sites they spatially bring together or separate. Further evidence for this was recently provided in a study that performed large-scale mapping of chromatin loops between CTCF sites (Handoko et al, 2011). We expect that within chromosomal segments at any given time enhancer–promoter combination will compete with interspersed CTCF sites for chromatin loop formation, the outcome of which again will depend on linear proximity and affinity between interacting sites.
DNA interactions and gene expression in single cells
For genes that are crucial for cellular identity correct expression must be controllable in all relevant cells. Transcription occurs in bursts, as was shown in live cell imaging studies (Chubb et al, 2006). Chromatin looping is thought to bring additional DNA-binding sites for relevant transcription factors in close proximity to gene promoters; this will increase the local concentration of these factors and consequently, transcription efficiency will increase (Droge and Muller-Hill, 2001; de Laat and Grosveld, 2003). Whether enhancers increase the frequency or amplitude of transcription bursts is not yet known, but it is relevant to ask what the maximum amount of intervening DNA is that can be bridged by an enhancer to contact and activate a gene in every single cell. This question is pertinent also because overall DNA topology is thought to be relatively stable during interphase, both at the level of the genome (Chubb et al, 2002; Gerlich et al, 2003) and at that of individual chromosomes (Muller et al, 2010).
The previously mentioned study that analysed ectopically integrated LCRs in the mouse genome aimed to get insight into the capacity of regulatory sites to reach and activate genes over ultra-long distances in individual cells. By 4C technology, it was shown that the LCR had little impact on overall genome topology, as the many long-range genomic contacts made by the integration site were similar with and without the integrated LCR (Noordermeer et al, 2011). Transcriptome analysis then demonstrated that none of the contacted genes benefited from the presence of the new LCR, with the exception of two endogenous mouse β-globin genes located on another chromosome. They were upregulated not in every single cell but exclusively in so-called ‘jackpot’ cells that showed the interchromosomal interaction. The work uncovered a number of principles behind long-range gene activation. First, it showed that the ability of regulatory sites to search the nuclear interior for preferred target genes is severely limited by their chromosomal context. This is likely to be true for all genomic positions, although the degree of constraint may vary between sites. Second, the data showed that promoter–enhancer compatibility is essential to transform spurious contacts between enhancers and promoters into productive interactions that drive gene activation; of all contacted genes, only two natural target genes of the LCR increased their transcription. Third, the data demonstrated that relatively stable cell-specific genome conformations can induce variegated gene expression. Cells that have their genome folded such that it brings together a regulatory site with a responsive gene can have different transcript levels of this gene than otherwise identical cells (Noordermeer et al, 2011). The phenomenon was referred to as ‘Spatial Effect Variegation’ (SEV). SEV may happen in trans, as shown in this artificial situation, but may also work in cis. Possibly, previous work already provided one example for SEV in cis. The interaction over 1 Mb between the limb bud enhancer and SHH was shown to occur in the expressing limb bud cells in a manner not dependent on the enhancer per se: its deletion abrogated expression without changing the DNA configuration (Amano et al, 2009).
We envision that indeed beyond a certain chromosomal distance regulatory sites will not be able to independently find a target gene, but for contact instead depend on the topological constraints imposed by the remainder sequences of the chromosome. The overall genome topology may or may not bring these sites in relative proximity in single cells. Depending on the nature of the interacting regions, transcription may be activated or repressed in these cells, causing variegated expression across the cell population. It is tempting to speculate that such SEV, when acting on key developmental regulators, may provide cells with a mechanism to make autonomous cell fate decisions, without the need for external signalling.
Future prospects
Functional genomics strives to assign function to all the relevant sites of the genome. Over the last decade, large-scale efforts have produced chromosome maps with the linear distribution of nucleosomes, transcription factor binding sites and chromatin modifications. Collectively, these studies demonstrated that what was previously known as junk DNA in fact appears a regulatory jungle. In order to understand the laws of the jungle, linear information must now be converted into spatial relationships. For this, highly detailed 3D topology maps need to be generated for all regulatory sites individually. They are expected to reveal the function of regulatory sites in gene deserts, to uncover the cis-regulatory networks of individual genes and to distinguish the functionally active from inactive regulatory sites. Detailed DNA interaction maps should also uncover the 3D chromosome scaffolds created by insulator sequences, and how the one regulatory site is hampered and the other facilitated in its action by the 3D configuration. Finally, this eventually needs to be done at the single-cell level, as we predict an unknown portion of the genome will only reveal its regulatory potential at the level of the individual cell.
Acknowledgments
We would like to thank Patrick Wijchers and Elzo de Wit for useful comments. This work was financially supported by grants from the Dutch Scientific Organization (NWO) (91204082 and 935170621), an European Research Council Starting Grant (209700, ‘4C’) and by InteGeR FP7 Marie Curie ITN (contract number PITN-GA-2007-214902).
Footnotes
The authors declare that they have no conflict of interest.
References
- Ahituv N, Zhu Y, Visel A, Holt A, Afzal V, Pennacchio LA, Rubin EM (2007) Deletion of ultraconserved elements yields viable mice. PLoS Biol 5: e234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amano T, Sagai T, Tanabe H, Mizushina Y, Nakazawa H, Shiroishi T (2009) Chromosomal dynamics at the Shh locus: limb bud-specific differential regulation of competence and active transcription. Dev Cell 16: 47–57 [DOI] [PubMed] [Google Scholar]
- Banerji J, Rusconi S, Schaffner W (1981) Expression of a beta-globin gene is enhanced by remote SV40 DNA sequences. Cell 27 (2 Part 1): 299–308 [DOI] [PubMed] [Google Scholar]
- Bantignies F, Roure V, Comet I, Leblanc B, Schuettengruber B, Bonnet J, Tixier V, Mas A, Cavalli G (2011) Polycomb-dependent regulatory contacts between distant Hox loci in Drosophila. Cell 144: 214–226 [DOI] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K (2007) High-resolution profiling of histone methylations in the human genome. Cell 129: 823–837 [DOI] [PubMed] [Google Scholar]
- Bell AC, Felsenfeld G (2000) Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature 405: 482–485 [DOI] [PubMed] [Google Scholar]
- Bell AC, West AG, Felsenfeld G (1999) The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell 98: 387–396 [DOI] [PubMed] [Google Scholar]
- Bender MA, Byron R, Ragoczy T, Telling A, Bulger M, Groudine M (2006) Flanking HS-62.5 and 3′ HS1, and regions upstream of the LCR, are not required for beta-globin transcription. Blood 108: 1395–1401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle AP, Davis S, Shulha HP, Meltzer P, Margulies EH, Weng Z, Furey TS, Crawford GE (2008) High-resolution mapping and characterization of open chromatin across the genome. Cell 132: 311–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler JE, Kadonaga JT (2001) Enhancer-promoter specificity mediated by DPE or TATA core promoter motifs. Genes Dev 15: 2515–2519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai HN, Shen P (2001) Effects of cis arrangement of chromatin insulators on enhancer-blocking activity. Science 291: 493–495 [DOI] [PubMed] [Google Scholar]
- Cajiao I, Zhang A, Yoo EJ, Cooke NE, Liebhaber SA (2004) Bystander gene activation by a locus control region. EMBO J 23: 3854–3863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter D, Chakalova L, Osborne CS, Dai YF, Fraser P (2002) Long-range chromatin regulatory interactions in vivo. Nat Genet 32: 623–626 [DOI] [PubMed] [Google Scholar]
- Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB, Wong E, Orlov YL, Zhang W, Jiang J, Loh YH, Yeo HC, Yeo ZX, Narang V, Govindarajan KR, Leong B, Shahab A, Ruan Y, Bourque G, Sung WK et al. (2008) Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell 133: 1106–1117 [DOI] [PubMed] [Google Scholar]
- Chubb JR, Boyle S, Perry P, Bickmore WA (2002) Chromatin motion is constrained by association with nuclear compartments in human cells. Curr Biol 12: 439–445 [DOI] [PubMed] [Google Scholar]
- Chubb JR, Trcek T, Shenoy SM, Singer RH (2006) Transcriptional pulsing of a developmental gene. Curr Biol 16: 1018–1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung JH, Whiteley M, Felsenfeld G (1993) A 5′ element of the chicken beta-globin domain serves as an insulator in human erythroid cells and protects against position effect in Drosophila. Cell 74: 505–514 [DOI] [PubMed] [Google Scholar]
- Comet I, Savitskaya E, Schuettengruber B, Negre N, Lavrov S, Parshikov A, Juge F, Gracheva E, Georgiev P, Cavalli G (2006) PRE-mediated bypass of two Su(Hw) insulators targets PcG proteins to a downstream promoter. Dev Cell 11: 117–124 [DOI] [PubMed] [Google Scholar]
- Comet I, Schuettengruber B, Sexton T, Cavalli G (2011) A chromatin insulator driving three-dimensional Polycomb response element (PRE) contacts and Polycomb association with the chromatin fiber. Proc Natl Acad Sci USA 108: 2294–2299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford GE, Holt IE, Mullikin JC, Tai D, Blakesley R, Bouffard G, Young A, Masiello C, Green ED, Wolfsberg TG, Collins FS (2004) Identifying gene regulatory elements by genome-wide recovery of DNase hypersensitive sites. Proc Natl Acad Sci USA 101: 992–997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, Boyer LA, Young RA, Jaenisch R (2010) Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci USA 107: 21931–21936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuddapah S, Jothi R, Schones DE, Roh TY, Cui K, Zhao K (2009) Global analysis of the insulator binding protein CTCF in chromatin barrier regions reveals demarcation of active and repressive domains. Genome Res 19: 24–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Laat W, Grosveld F (2003) Spatial organization of gene expression: the active chromatin hub. Chromosome Res 11: 447–459 [DOI] [PubMed] [Google Scholar]
- de Villiers J, Olson L, Banerji J, Schaffner W (1983) Analysis of the transcriptional enhancer effect. Cold Spring Harb Symp Quant Biol 47 (Part 2): 911–919 [DOI] [PubMed] [Google Scholar]
- Dekker J, Rippe K, Dekker M, Kleckner N (2002) Capturing chromosome conformation. Science 295: 1306–1311 [DOI] [PubMed] [Google Scholar]
- Dillon N, Sabbattini P (2000) Functional gene expression domains: defining the functional unit of eukaryotic gene regulation. Bioessays 22: 657–665 [DOI] [PubMed] [Google Scholar]
- Dillon N, Trimborn T, Strouboulis J, Fraser P, Grosveld F (1997) The effect of distance on long-range chromatin interactions. Mol Cell 1: 131–139 [DOI] [PubMed] [Google Scholar]
- Dorschner MO, Hawrylycz M, Humbert R, Wallace JC, Shafer A, Kawamoto J, Mack J, Hall R, Goldy J, Sabo PJ, Kohli A, Li Q, McArthur M, Stamatoyannopoulos JA (2004) High-throughput localization of functional elements by quantitative chromatin profiling. Nat Methods 1: 219–225 [DOI] [PubMed] [Google Scholar]
- Drissen R, Palstra RJ, Gillemans N, Splinter E, Grosveld F, Philipsen S, de Laat W (2004) The active spatial organization of the beta-globin locus requires the transcription factor EKLF. Genes Dev 18: 2485–2490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Droge P, Muller-Hill B (2001) High local protein concentrations at promoters: strategies in prokaryotic and eukaryotic cells. Bioessays 23: 179–183 [DOI] [PubMed] [Google Scholar]
- Duan Z, Andronescu M, Schutz K, McIlwain S, Kim YJ, Lee C, Shendure J, Fields S, Blau CA, Noble WS (2010) A three-dimensional model of the yeast genome. Nature 465: 363–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epner E, Reik A, Cimbora D, Telling A, Bender MA, Fiering S, Enver T, Martin DI, Kennedy M, Keller G, Groudine M (1998) The beta-globin LCR is not necessary for an open chromatin structure or developmentally regulated transcription of the native mouse beta-globin locus. Mol Cell 2: 447–455 [DOI] [PubMed] [Google Scholar]
- Farrell CM, West AG, Felsenfeld G (2002) Conserved CTCF insulator elements flank the mouse and human beta-globin loci. Mol Cell Biol 22: 3820–3831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippova GN, Cheng MK, Moore JM, Truong JP, Hu YJ, Nguyen DK, Tsuchiya KD, Disteche CM (2005) Boundaries between chromosomal domains of X inactivation and escape bind CTCF and lack CpG methylation during early development. Dev Cell 8: 31–42 [DOI] [PubMed] [Google Scholar]
- Forrester WC, Epner E, Driscoll MC, Enver T, Brice M, Papayannopoulou T, Groudine M (1990) A deletion of the human beta-globin locus activation region causes a major alteration in chromatin structure and replication across the entire beta-globin locus. Genes Dev 4: 1637–1649 [DOI] [PubMed] [Google Scholar]
- Gerasimova TI, Byrd K, Corces VG (2000) A chromatin insulator determines the nuclear localization of DNA. Mol Cell 6: 1025–1035 [DOI] [PubMed] [Google Scholar]
- Gerlich D, Beaudouin J, Kalbfuss B, Daigle N, Eils R, Ellenberg J (2003) Global chromosome positions are transmitted through mitosis in mammalian cells. Cell 112: 751–764 [DOI] [PubMed] [Google Scholar]
- Grosveld F, van Assendelft GB, Greaves DR, Kollias G (1987) Position-independent, high-level expression of the human beta-globin gene in transgenic mice. Cell 51: 975–985 [DOI] [PubMed] [Google Scholar]
- Guelen L, Pagie L, Brasset E, Meuleman W, Faza MB, Talhout W, Eussen BH, de Klein A, Wessels L, de Laat W, van Steensel B (2008) Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature 453: 948–951 [DOI] [PubMed] [Google Scholar]
- Handoko L, Xu H, Li G, Ngan CY, Chew E, Schnapp M, Lee CW, Ye C, Ping JL, Mulawadi F, Wong E, Sheng J, Zhang Y, Poh T, Chan CS, Kunarso G, Shahab A, Bourque G, Cacheux-Rataboul V, Sung WK et al. (2011) CTCF-mediated functional chromatin interactome in pluripotent cells. Nat Genet 43: 630–638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanscombe O, Whyatt D, Fraser P, Yannoutsos N, Greaves D, Dillon N, Grosveld F (1991) Importance of globin gene order for correct developmental expression. Genes Dev 5: 1387–1394 [DOI] [PubMed] [Google Scholar]
- Hark AT, Schoenherr CJ, Katz DJ, Ingram RS, Levorse JM, Tilghman SM (2000) CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature 405: 486–489 [DOI] [PubMed] [Google Scholar]
- Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW, Ching KA, Antosiewicz-Bourget JE, Liu H, Zhang X, Green RD, Lobanenkov VV, Stewart R, Thomson JA, Crawford GE, Kellis M et al. (2009) Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature 459: 108–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, Wang W, Weng Z, Green RD, Crawford GE, Ren B (2007) Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet 39: 311–318 [DOI] [PubMed] [Google Scholar]
- Heuchel R, Matthias P, Schaffner W (1989) Two closely spaced promoters are equally activated by a remote enhancer: evidence against a scanning model for enhancer action. Nucleic Acids Res 17: 8931–8947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hindorff LA, Junkins HA, Mehta JP, Manolio TA (2009) A catalog of published genome-wide association studies. Available at http://www.genome.gov/gwastudies
- Horike S, Cai S, Miyano M, Cheng JF, Kohwi-Shigematsu T (2005) Loss of silent-chromatin looping and impaired imprinting of DLX5 in Rett syndrome. Nat Genet 37: 31–40 [DOI] [PubMed] [Google Scholar]
- Kellum R, Schedl P (1991) A position-effect assay for boundaries of higher order chromosomal domains. Cell 64: 941–950 [DOI] [PubMed] [Google Scholar]
- Kellum R, Schedl P (1992) A group of scs elements function as domain boundaries in an enhancer-blocking assay. Mol Cell Biol 12: 2424–2431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinjan DA, van Heyningen V (2005) Long-range control of gene expression: emerging mechanisms and disruption in disease. Am J Hum Genet 76: 8–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunarso G, Chia NY, Jeyakani J, Hwang C, Lu X, Chan YS, Ng HH, Bourque G (2010) Transposable elements have rewired the core regulatory network of human embryonic stem cells. Nat Genet 42: 631–634 [DOI] [PubMed] [Google Scholar]
- Kurukuti S, Tiwari VK, Tavoosidana G, Pugacheva E, Murrell A, Zhao Z, Lobanenkov V, Reik W, Ohlsson R (2006) CTCF binding at the H19 imprinting control region mediates maternally inherited higher-order chromatin conformation to restrict enhancer access to Igf2. Proc Natl Acad Sci USA 103: 10684–10689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lettice LA, Heaney SJ, Purdie LA, Li L, de Beer P, Oostra BA, Goode D, Elgar G, Hill RE, de Graaff E (2003) A long-range Shh enhancer regulates expression in the developing limb and fin and is associated with preaxial polydactyly. Hum Mol Genet 12: 1725–1735 [DOI] [PubMed] [Google Scholar]
- Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, Sandstrom R, Bernstein B, Bender MA, Groudine M, Gnirke A, Stamatoyannopoulos J, Mirny LA, Lander ES, Dekker J (2009) Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326: 289–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Garrard WT (2005) Long-range interactions between three transcriptional enhancers, active Vkappa gene promoters, and a 3′ boundary sequence spanning 46 kilobases. Mol Cell Biol 25: 3220–3231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lower KM, Hughes JR, De Gobbi M, Henderson S, Viprakasit V, Fisher C, Goriely A, Ayyub H, Sloane-Stanley J, Vernimmen D, Langford C, Garrick D, Gibbons RJ, Higgs DR (2009) Adventitious changes in long-range gene expression caused by polymorphic structural variation and promoter competition. Proc Natl Acad Sci USA 106: 21771–21776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maksimenko O, Golovnin A, Georgiev P (2008) Enhancer-promoter communication is regulated by insulator pairing in a Drosophila model bigenic locus. Mol Cell Biol 28: 5469–5477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallin DR, Myung JS, Patton JS, Geyer PK (1998) Polycomb group repression is blocked by the Drosophila suppressor of Hairy-wing [su(Hw)] insulator. Genetics 148: 331–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller I, Boyle S, Singer RH, Bickmore WA, Chubb JR (2010) Stable morphology, but dynamic internal reorganisation, of interphase human chromosomes in living cells. PLoS One 5: e11560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muravyova E, Golovnin A, Gracheva E, Parshikov A, Belenkaya T, Pirrotta V, Georgiev P (2001) Loss of insulator activity by paired Su(Hw) chromatin insulators. Science 291: 495–498 [DOI] [PubMed] [Google Scholar]
- Noordermeer D, Branco MR, Splinter E, Klous P, van Ijcken W, Swagemakers S, Koutsourakis M, van der Spek P, Pombo A, de Laat W (2008) Transcription and chromatin organization of a housekeeping gene cluster containing an integrated beta-globin locus control region. PLoS Genet 4: e1000016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noordermeer D, de Wit E, Klous P, van de Werken H, Simonis M, Lopez-Jones M, Eussen B, de Klein A, Singer RH, de Laat W (2011) Variegated gene expression caused by cell-specific long-range DNA interactions. Nat Cell Biol 13: 944–951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsuki S, Levine M, Cai HN (1998) Different core promoters possess distinct regulatory activities in the Drosophila embryo. Genes Dev 12: 547–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palstra RJ, Tolhuis B, Splinter E, Nijmeijer R, Grosveld F, de Laat W (2003) The beta-globin nuclear compartment in development and erythroid differentiation. Nat Genet 35: 190–194 [DOI] [PubMed] [Google Scholar]
- Parelho V, Hadjur S, Spivakov M, Leleu M, Sauer S, Gregson HC, Jarmuz A, Canzonetta C, Webster Z, Nesterova T, Cobb BS, Yokomori K, Dillon N, Aragon L, Fisher AG, Merkenschlager M (2008) Cohesins functionally associate with CTCF on mammalian chromosome arms. Cell 132: 422–433 [DOI] [PubMed] [Google Scholar]
- Ptashne M (1986) Gene regulation by proteins acting nearby and at a distance. Nature 322: 697–701 [DOI] [PubMed] [Google Scholar]
- Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J (2011) A unique chromatin signature uncovers early developmental enhancers in humans. Nature 470: 279–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Recillas-Targa F, Pikaart MJ, Burgess-Beusse B, Bell AC, Litt MD, West AG, Gaszner M, Felsenfeld G (2002) Position-effect protection and enhancer blocking by the chicken beta-globin insulator are separable activities. Proc Natl Acad Sci USA 99: 6883–6888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rippe K, von Hippel PH, Langowski J (1995) Action at a distance: DNA-looping and initiation of transcription. Trends Biochem Sci 20: 500–506 [DOI] [PubMed] [Google Scholar]
- Ruf S, Symmons O, Uslu VV, Dolle D, Hot C, Ettwiller L, Spitz F (2011) Large-scale analysis of the regulatory architecture of the mouse genome with a transposon-associated sensor. Nat Genet 43: 379–386 [DOI] [PubMed] [Google Scholar]
- Sabatino DE, Cline AP, Gallagher PG, Garrett LJ, Stamatoyannopoulos G, Forget BG, Bodine DM (1998) Substitution of the human beta-spectrin promoter for the human agamma-globin promoter prevents silencing of a linked human beta-globin gene in transgenic mice. Mol Cell Biol 18: 6634–6640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigrist CJ, Pirrotta V (1997) Chromatin insulator elements block the silencing of a target gene by the Drosophila polycomb response element (PRE) but allow trans interactions between PREs on different chromosomes. Genetics 147: 209–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonis M, Klous P, Splinter E, Moshkin Y, Willemsen R, de Wit E, van Steensel B, de Laat W (2006) Nuclear organization of active and inactive chromatin domains uncovered by chromosome conformation capture-on-chip (4C). Nat Genet 38: 1348–1354 [DOI] [PubMed] [Google Scholar]
- Simonis M, Kooren J, de Laat W (2007) An evaluation of 3C-based methods to capture DNA interactions. Nat Methods 4: 895–901 [DOI] [PubMed] [Google Scholar]
- Spilianakis CG, Flavell RA (2004) Long-range intrachromosomal interactions in the T helper type 2 cytokine locus. Nat Immunol 5: 1017–1027 [DOI] [PubMed] [Google Scholar]
- Spitz F, Gonzalez F, Duboule D (2003) A global control region defines a chromosomal regulatory landscape containing the HoxD cluster. Cell 113: 405–417 [DOI] [PubMed] [Google Scholar]
- Splinter E, Heath H, Kooren J, Palstra RJ, Klous P, Grosveld F, Galjart N, de Laat W (2006) CTCF mediates long-range chromatin looping and local histone modification in the beta-globin locus. Genes Dev 20: 2349–2354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanizawa H, Iwasaki O, Tanaka A, Capizzi JR, Wickramasinghe P, Lee M, Fu Z, Noma K (2010) Mapping of long-range associations throughout the fission yeast genome reveals global genome organization linked to transcriptional regulation. Nucleic Acids Res 38: 8164–8177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari VK, McGarvey KM, Licchesi JD, Ohm JE, Herman JG, Schubeler D, Baylin SB (2008) PcG proteins, DNA methylation, and gene repression by chromatin looping. PLoS Biol 6: 2911–2927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolhuis B, Palstra RJ, Splinter E, Grosveld F, de Laat W (2002) Looping and interaction between hypersensitive sites in the active beta-globin locus. Mol Cell 10: 1453–1465 [DOI] [PubMed] [Google Scholar]
- Tschopp P, Tarchini B, Spitz F, Zakany J, Duboule D (2009) Uncoupling time and space in the collinear regulation of Hox genes. PLoS Genet 5: e1000398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vakoc CR, Letting DL, Gheldof N, Sawado T, Bender MA, Groudine M, Weiss MJ, Dekker J, Blobel GA (2005) Proximity among distant regulatory elements at the beta-globin locus requires GATA-1 and FOG-1. Mol Cell 17: 453–462 [DOI] [PubMed] [Google Scholar]
- Van der Ploeg LH, Konings A, Oort M, Roos D, Bernini L, Flavell RA (1980) gamma-beta-Thalassaemia studies showing that deletion of the gamma- and delta-genes influences beta-globin gene expression in man. Nature 283: 637–642 [DOI] [PubMed] [Google Scholar]
- Vernimmen D, De Gobbi M, Sloane-Stanley JA, Wood WG, Higgs DR (2007) Long-range chromosomal interactions regulate the timing of the transition between poised and active gene expression. EMBO J 26: 2041–2051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A, Blow MJ, Li Z, Zhang T, Akiyama JA, Holt A, Plajzer-Frick I, Shoukry M, Wright C, Chen F, Afzal V, Ren B, Rubin EM, Pennacchio LA (2009a) ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature 457: 854–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A, Prabhakar S, Akiyama JA, Shoukry M, Lewis KD, Holt A, Plajzer-Frick I, Afzal V, Rubin EM, Pennacchio LA (2008) Ultraconservation identifies a small subset of extremely constrained developmental enhancers. Nat Genet 40: 158–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A, Rubin EM, Pennacchio LA (2009b) Genomic views of distant-acting enhancers. Nature 461: 199–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasylyk B, Wasylyk C, Augereau P, Chambon P (1983) The SV40 72 bp repeat preferentially potentiates transcription starting from proximal natural or substitute promoter elements. Cell 32: 503–514 [DOI] [PubMed] [Google Scholar]
- Wendt KS, Yoshida K, Itoh T, Bando M, Koch B, Schirghuber E, Tsutsumi S, Nagae G, Ishihara K, Mishiro T, Yahata K, Imamoto F, Aburatani H, Nakao M, Imamoto N, Maeshima K, Shirahige K, Peters JM (2008) Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature 451: 796–801 [DOI] [PubMed] [Google Scholar]
- Wijgerde M, Grosveld F, Fraser P (1995) Transcription complex stability and chromatin dynamics in vivo. Nature 377: 209–213 [DOI] [PubMed] [Google Scholar]
- Xie X, Mikkelsen TS, Gnirke A, Lindblad-Toh K, Kellis M, Lander ES (2007) Systematic discovery of regulatory motifs in conserved regions of the human genome, including thousands of CTCF insulator sites. Proc Natl Acad Sci USA 104: 7145–7150 [DOI] [PMC free article] [PubMed] [Google Scholar]