Abstract
Brain wiring depends on cells making highly localized and selective connections through surface protein–protein interactions, including those between NetrinGs and NetrinG ligands (NGLs). The NetrinGs are members of the structurally uncharacterized netrin family. We present a comprehensive crystallographic analysis comprising NetrinG1–NGL1 and NetrinG2–NGL2 complexes, unliganded NetrinG2 and NGL3. Cognate NetrinG–NGL interactions depend on three specificity-conferring NetrinG loops, clasped tightly by matching NGL surfaces. We engineered these NGL surfaces to implant custom-made affinities for NetrinG1 and NetrinG2. In a cellular patterning assay, we demonstrate that NetrinG-binding selectivity can direct the sorting of a mixed population of NGLs into discrete cell surface subdomains. These results provide a molecular model for selectivity-based patterning in a neuronal recognition system, dysregulation of which is associated with severe neuropsychological disorders.
Keywords: brain patterning, crystal structure, protein engineering, schizophrenia, synaptic adhesion
Introduction
The intricate choreography of cell populations required for embryonic morphogenesis and adult tissue homeostasis is directed by several distinct families of cell guidance cues. One major family, the netrins, interacts with diverse single pass cell surface receptors to mediate cell repulsion, attraction or adhesion in tissues, which include the nervous system, vasculature, lung, pancreas, mammary gland and muscle (Lai Wing Sun et al, 2011). The neuronal NetrinG1 and NetrinG2 are vertebrate specific and by binding their cognate NetrinG ligand (NGL) partners (Woo et al, 2009b), they promote thalamocortical axon outgrowth (Lin et al, 2003), induce and maintain excitatory synapse formation (Kim et al, 2006) and contribute to subdendritic segmentation in the hippocampus and cortex (Nishimura-Akiyoshi et al, 2007). Abnormal expression of NetrinGs is associated with behavioural phenotypes in mice (Zhang et al, 2008) and with schizophrenia, bipolar disease, temporal lobe epilepsy and Rett syndrome in humans (Aoki-Suzuki et al, 2005; Borg et al, 2005; Eastwood and Harrison, 2008; Ohtsuki et al, 2008; Pan et al, 2010). The third member of the NGL family, NGL3, does not bind netrins, instead regulating excitatory synapse formation through interaction with the type IIa receptor protein tyrosine phosphatases (Woo et al, 2009a; Kwon et al, 2010).
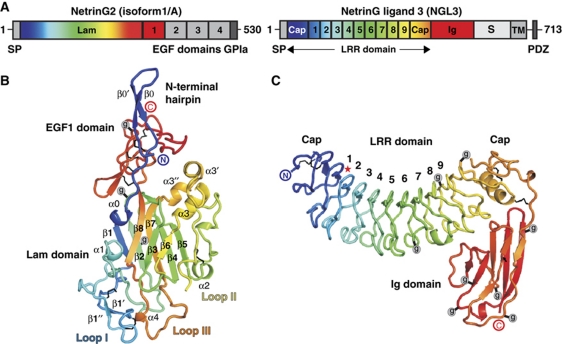
Sequence analysis predicts that the NetrinGs contain an extracellular N-terminal laminin-like domain (Lam) and, depending on the splice isoform, up to four epidermal-growth-factor-like (EGF) domains, and a C-terminal glycosylphosphatidylinisotol (GPI) anchor (Figure 1A) (Nakashiba et al, 2000, 2002; Yin et al, 2002). The N-terminal Lam domain is characteristic of the classic, secreted, netrin cell guidance factors and of the extracellular matrix laminin proteins (Supplementary Figures S1 and S2). NGLs are type I transmembrane (TM) proteins composed of nine leucine-rich repeats (lrr) forming the lrr-rich domain (LRR), an immunoglobulin-like domain (Ig), a highly glycosylated region (S), a TM helix and a cytoplasmic domain containing a PDZ-binding motif (Figure 1A). In the absence of any structural information for the NetrinGs, the NGLs or their recognition complexes, the determinants of binding selectivity and mode of interaction required for function have remained uncharacterized.
Figure 1.
Crystal structures of unliganded NetrinG2Lam–EGF1 and NGL3LRR–Ig. (A) Domain organization of human NetrinG2 (isoform 1/A) and NGL3. Domain nomenclature as in main text. SP, signal peptide; GPIa, GPI anchor; Cap, N- and C-terminal Cap regions. (B) Ribbon diagram of NetrinG2Lam–EGF1 (residues Y19–A345). Jelly-roll β-strands are labelled β1–β8. Three predicted N-linked glycosylation sites are marked with g enclosed in a grey circle. Black lines denote disulphide bridges. N- and C-termini are indicated (encircled N and C). (C) Ribbon diagram of NGL3LRR–Ig (residues S58–N454). lrrs are numbered 1–9. Q96, essential for binding of NGL3 to LAR (Kwon et al, 2010) is marked with a red star. Other labelling and colouring as for (B).
Results
Crystallographic analysis of NetrinG and NGL proteins
We present crystal structures of a NetrinG1Lam–EGF1+NGL1LRR–Ig complex, NetrinG2Lam–EGF1, a NetrinG2Lam–EGF1+NGL2LRR–Ig complex and NGL3LRR–Ig determined at resolutions of 3.3, 2.2, 2.6 and 3.1 Å, respectively (Table I).
Table 1. Data collection and refinement statistics.
| NetrinG2Lam–EGF (native) | NetrinG2Lam–EGF (selenomethionine) | NGL3LRR | NGL3LRR–Ig | NetrinG1Lam–EGF+ NGL1LRR Ig | NetrinG2Lam–EGF+ NGL2LRR–Ig | |
|---|---|---|---|---|---|---|
| PDB accession code | 3ZYG | — | 3ZYN | 3ZYO | 3ZYJ | 3ZYI |
| Data collection | ||||||
| Space group | C2 | P1 | C2 | H32 | F222 | I222 |
| Cell dimensions | ||||||
| a, b, c (Å) | 111.5, 64.9, 120.1 | 64.4, 64.5, 120.9 | 157.1, 57.3, 100.4 | 131.3, 131.3, 174.9 | 184.4, 204.4, 288.1 | 75.4, 153.4, 158.7 |
| α, β, γ (deg) | 90, 103.1, 90 | 82.5, 75.1, 60.4 | 90, 117.8, 90 | 90, 90, 120 | 90, 90, 90 | 90, 90, 90 |
| Resolution (Å)a | 29.1–2.2 (2.4–2.2) | 30–2.4 (2.5–2.4) | 53–3.2 (3.28–3.2) | 38.6–3.1 (3.27–3.1) | 29.4–3.25 (3.4–3.25) | 28.9–2.6 (2.7–2.6) |
| Rmeas (%)a | 17.3 (61.1) | 23.1 (109) | 20.6 (79.7) | 13.6 (93.3) | 20.6 (76.6) | 9.2 (94.2) |
| I/σIa | 6.4 (2.35) | 7.0 (1.8) | 8.2 (2.2) | 11.2 (2.6) | 8.8 (1.5) | 16.6 (2.1) |
| Completeness (%)a | 98.9 (96.6) | 95.5 (86.6) | 99.6 (99.8) | 99.9 (100) | 91.3 (73.3) | 98.7 (98.2) |
| Redundancya | 3.6 (3.4) | 5.3 (4.6) | 3.6 (3.6) | 7.3 (7.4) | 5.7 (2.7) | 6.2 (6.3) |
| Refinement | ||||||
| Resolution (Å)a | 29.2–2.2 (2.26–2.2) | — | 53–3.2 (3.28–3.2) | 38.6–3.1 (3.18–3.1) | 29.4–3.25 (3.33–3.25) | 28.9–2.6 (2.7–2.6) |
| No. reflections | 42 207 (2863) | — | 12 344 (809) | 10 223 (705) | 39 039 (2266) | 28 340 (2924) |
| Rwork/Rfree | 0.246/0.291 | — | 0.217/0.268 | 0.229/0.269 | 0.257/0.268 | 0.246/0.287 |
| No. atoms | 5383 | — | 5027 | 3115 | 11 398 | 5450 |
| Protein | 5117 | — | 4999 | 3100 | 11 262 | 5411 |
| Ligand/ion | 58 | — | 28 | 15 | 136 | 39 |
| Water | 208 | — | 0 | 0 | 0 | 0 |
| Average B-factors | ||||||
| Protein | 25.7 | — | 42.2 | 103 | 102 | 58.7 |
| Ligand/ion | 27.2 | — | 54.8 | 122.7 | 105 | 80.2 |
| Water | 21.4 | — | — | — | — | — |
| R.m.s.d. | ||||||
| Bond lengths (Å) | 0.01 | — | 0.006 | 0.005 | 0.007 | 0.008 |
| Bond angles (deg) | 1.11 | — | 0.986 | 0.857 | 0.93 | 1.00 |
| aValues in parentheses are for the highest-resolution shell. | ||||||
The core of the 269-residue NetrinG2 Lam domain comprises a β-sandwich with a ‘jelly-roll’ topology (Richardson, 1981) decorated by elaborate insertions including a helical region (Supplementary Figure S3; Figure 1B). The extended N-terminus forms a β-hairpin, which partially abuts the 58-residue C-terminal EGF1 domain. Extensive disulphide bonding in the Lam and EGF1 domains stabilize the intricate arrangement. The structure is most similar to that recently reported for the Lam domain of mouse LamininA5 (Hussain et al, 2011) (root mean square deviation, r.m.s.d., of 2.05 Å for 220 Cα atoms; Supplementary Figure S3).
The 391-residue NGL3LRR–Ig structure reveals an arrangement in which a C2-type Ig domain folds back onto the convex face of a horseshoe-shaped LRR domain (Figure 1C). In its domain composition, NGL3 resembles the neuronal Nogo-66 receptor-binding glycoprotein Lingo-1, although the Lingo-1 LRR domain is larger by 3 lrrs (Mosyak et al, 2006) (Supplementary Figure S4). In common with all LRR domain proteins, the NGL3 lrrs are sandwiched by N-terminal and C-terminal cap structures.
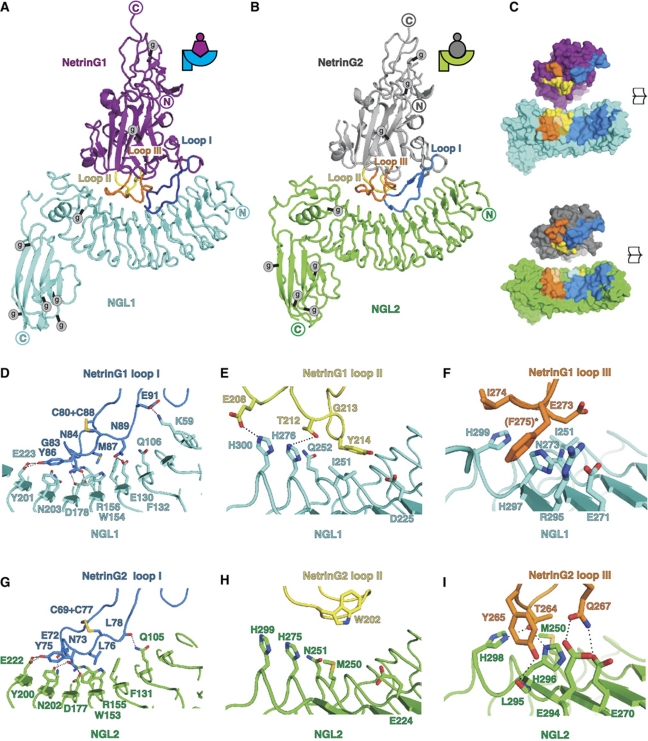
Our complex structures show that the concave faces of the NGL1 and NGL2 LRR domains bind to their cognate NetrinGs through a complementarily shaped binding site on the Lam domain (Figure 2A and B), reminiscent of the hand-clasp mode described for other LRR-ligand binding interactions (Fan and Hendrickson, 2005; Bella et al, 2008). The total buried surface area (Krissinel and Henrick, 2007) in the complexes is ∼2410 Å2 for NetrinG1Lam–EGF1+NGL1LRR–Ig and ∼2080 Å2 for NetrinG2Lam–EGF1+NGL2LRR–Ig. Multiangle light scattering (MALS) measurements support a 1:1 stochiometry for both complexes (Supplementary Figure S5). In the crystal structures, the NetrinG and NGL C-termini point in opposite directions, an arrangement suitable for the trans interactions required for a functional role mediating binding events between neurons. The NGL-binding site on NetrinGs is essentially composed of the three long loops (I, II and III) located between strands β1–β2, β5–β6 and β7–β8, respectively (Figure 2A and B). In both complexes, loop I provides extensive interactions, primarily to conserved NGL residues. Loop II interacts via two non-conserved hydrogen bonds and hydrophobic interactions in the NetrinG1–NGL1 structure. In the NetrinG2–NGL2 complex, however, it provides little direct contact. Conversely, while NetrinG1 loop III provides only a few hydrophobic contacts to NGL1, NetrinG2 loop III binds to NGL2 with five hydrogen bonds plus additional hydrophobic interactions (Figure 2C–I). The conserved core of interactions contributed by loop I suggest an ancestral NetrinG–NGL interaction diversified with addition of homologue-specific interactions via loops II and III. Structural superposition of NetrinG2Lam–EGF1 in its unliganded and NGL2-bound forms reveals an induced-fit binding mechanism affecting loop I (Cα r.m.s.d. 4.1 Å2, contrasting with the overall r.m.s.d. of 1.6 Å2) and loop II, which is disordered in the unbound form. Overlay of the two NGL-bound structures of NetrinG1 and NetrinG2 shows that conformational differences are, again, concentrated in the NGL-binding loops, consistent with a role as determinants of binding specificity (for loops I+II+III, the Cα r.m.s.d. is 3.3 Å2, whereas the overall Cα r.m.s.d. is 1.8 Å2). In contrast to the NetrinGs, the structures of NGL1LRR–Ig (complexed), NGL2LRR–Ig (complexed) and NGL3LRR–Ig (unliganded) are similar, suggesting that the structure of NGLLRR–Igs does not undergo major rearrangement upon ligand binding. There are, however, some subtle differences in the relative orientations of the Ig and LRR domains between the three NGL family members (Supplementary Figure S4). In combination, these data define a generic binding architecture for the NetrinG–NGL system, which comprises the docking of three flexible NetrinG loops onto a relatively rigid concave surface on NGL.
Figure 2.
Similar surfaces with distinct binding properties mediate NetrinG–NGL interactions. (A) Ribbon diagram of NetrinG1Lam–EGF1 (purple) in complex with NGL1LRR–Ig (cyan). Main NGL1-binding loops are coloured separately. Predicted N-linked glycosylation sites are marked with g enclosed in a grey circle. (B) Ribbon diagram of NetrinG2Lam–EGF1 (grey) in complex with NGL2LRR–Ig (green). Predicted N-linked glycosylation sites and NGL2-binding loops are indicated. (C) Open surface views showing NetrinG1–NGL1 and NetrinG2–NGL2 interfaces. NGL1 (cyan) and NGL2 (green) residues in the binding interface are coloured according to the NetrinG loop they contact: blue (loop I), yellow (loop II), orange (loop III), mustard (loop I and loop III), light orange (loop II and loop III), light blue (loop I and loop II). (D–I) Views of selected residues in the NetrinG1–NGL1 and NetrinG2–NGL2 interfaces. Hydrogen bonds are shown as dotted lines. Colours as for (A, B). The side chain of NetrinG1 F275, labelled with an asterisk, is depicted for completeness although poorly defined in the electron density (F). The backbone carbonyl oxygen of NetrinG2 L295, but not the side chain, is shown in stick representation (I).
Wild-type NetrinG–NGL binding affinities
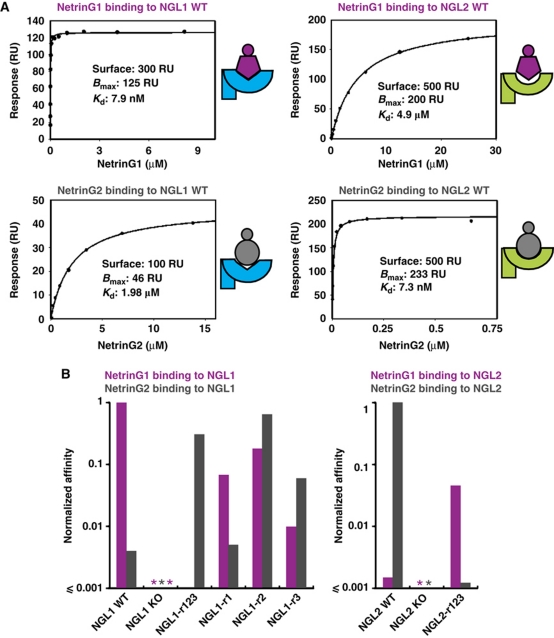
NetrinG1/NGL1 and NetrinG2/NGL2 have, in general, been observed to segregate to non-overlapping regions in the brain (Nakashiba et al, 2002; Yin et al, 2002; Kim et al, 2006; Nishimura-Akiyoshi et al, 2007). We refer to these cognate pairs as ‘intra-class’ binding partners and to interactions between members of different pairs as ‘cross-class’ binding. Using surface plasmon resonance (SPR) equilibrium experiments, we measured intra-class affinities of 7.9 nM (NetrinG1–NGL1) and 7.3 nM (NetrinG2–NGL2), values lying within the range reported previously (Lin et al, 2003; Nishimura-Akiyoshi et al, 2007; Zhang et al, 2008). We found that the introduction of an N-linked glycosylation site in the NetrinG-binding site on NGL1 (A205T) and NGL2 (G204T) abolished NetrinG–NGL binding, in agreement with the binding interface revealed by the crystal structures (Figure 3). Interestingly, we were able to measure previously undetected cross-class binding affinities, albeit in the low micromolar range: 4.9 μM (NetrinG1–NGL2) and 2.0 μM (NetrinG2–NGL1) (Figure 4; Supplementary Figure S6). It remains to be established whether these low affinity interactions play any functional role in vivo. Consistent with previous reports (Kim et al, 2006), we find that the affinity between NGL3 and either NetrinG is very weak (Kd>100 μM).
Figure 3.
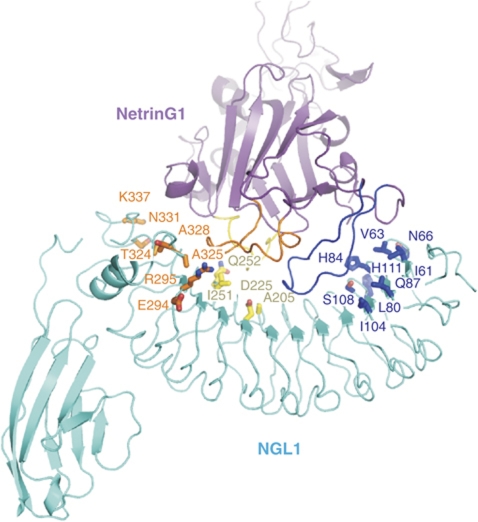
Non-conserved residues in the concave LRR surfaces of NGL1 and NGL2 were swapped to create proteins with modified binding specificities. Differences between NGL1 and NGL2 are concentrated at the N-terminal and C-terminal regions of the binding surface, with only few variations found in the central region. In the NGL1-r1 mutant, non-conserved residues in the vicinity of the NetrinG loop I binding site were swapped (shown in blue, stick representation). These include NGL1 residues I61, V63, N66, L80, H84, Q87, I104, S108 and H111, equivalent to NGL2 residues V60, T62, G65, Y79, M83, N86, V103, G107 and S110. In NGL1-r2, residues around the loop II binding site were swapped (shown in yellow). These include NGL1 residues A205, D225, I251 and Q252, equivalent to NGL2 residues G204, E224, M250 and N251. In NGL1-r3, residues including those in the loop III binding site were swapped (NGL1 residues E294, R295, T324, A325, A328, N331 and K337, equivalent to NGL2 residues V293, E294, S323, T324, G327, H330 and R336, shown in orange). In NGL1-r123, all non-conserved residues in the binding interface were swapped. Vice versa, in NGL2-r123, the equivalent residues were swapped to match those found in NGL1. The NetrinG-binding specificities of these mutants were analysed using SPR (Figure 4; Supplementary Figure S8).
Figure 4.
Manipulation of NetrinG–NGL recognition by structure-guided grafting of specificity. (A) SPR experiments for the interaction of WT NetrinG1Lam–EGF1/4 and NetrinG2Lam–EGF1 with WT NGL1/2LRR–Ig reveals intra-class binding Kds in the low nanomolar range and cross-class binding Kds in the low micromolar range. (B) The affinities depicted were normalized using the Kd value for WT intra-class binding. Binding to NGL constructs by NetrinG1 is shown in purple, binding by NetrinG2 is shown in grey. Introduction of an N-linked glycosylation site in the interface results in the loss of NetrinG binding (NGL1 KO and NGL2 KO), as indicated with an asterisk. NetrinG1Lam–EGF1/4 contains an extra EGF domain compared with NetrinG2Lam–EGF1. Addition of this domain does not affect the affinity of NetrinG1 to NGL1LRR–Ig or NGL2LRR–Ig (unpublished observation).
Engineering of protein binding affinities
To dissect the precise determinants of NGL specificity, we introduced point mutations in the NetrinG-binding surface of NGL1, which transplanted the corresponding NGL2 surface (Figure 3; Supplementary Figure S7). A swap of nine non-conserved residues in the loop I binding region (NGL1-r1 mutant), does not result in a major increase of affinity to NetrinG2 (<2-fold), although binding to NetrinG1 is decreased >10-fold (Figure 4; Supplementary Figure S8). Thus, as we hypothesised based on the structural results, the interactions provided by loop I are not the main determinants for binding specificity. In contrast, when we transplanted the NGL2 surfaces corresponding to the binding regions for loop II (NGL1-r2 mutant, switching four residues) or loop III (NGL1-r3 mutant, switching seven residues), we observed a reversal of the wild-type (WT) binding preference to favour cross-class binding (Figure 4; Supplementary Figure S8). This is consistent with the interactions mediated by regions 2 and 3 providing the major components of binding specificity. For all three individual NGL1-r1, -r2 and -r3 mutants, the binding selectivity is decreased compared with WT NGL1, consistent with these mutant binding surfaces comprising a patchwork of NGL1 and NGL2 elements. A combined transplant of all three NGL2 regions into NGL1 (NGL1-r123 mutant), or conversely, of NGL1 regions into NGL2 (NGL2-r123 mutant), results in a complete switch in NetrinG specificity (Figure 4; Supplementary Figure S8).
As expected based on the crystal structures of the complexes, we were able to impair binding in SPR assays by introducing an N-linked glycosylation site in the NGL-binding site of NetrinG1 or NetrinG2 loop I (Supplementary Figure S8). We then attempted to manipulate specificities in the NetrinG1 and NetrinG2 proteins by swapping one, two or all three main NGL-binding loops. This panel of mutants displays a plethora of NGL-binding properties, summarized and discussed in Supplementary Figure S8. Thus, while the rigid scaffold of the NGL LRR domain can be subdivided into a mosaic of essentially discrete NetrinG-binding regions, the flexible loops of the NetrinGs cannot be treated as stand-alone structural elements.
NetrinG-binding controls cell surface NGL patterns
Analyses of NetrinG-deficient mice have demonstrated that NetrinG1 and NetrinG2, expressed on different sets of axonal projections, are essential for localizing NGL1 and NGL2, co-expressed in the receiving dendrite, to distinct dendritic subdomains in hippocampal or cortical neurons (Nishimura-Akiyoshi et al, 2007). We developed an assay to test the ability of NetrinGs to induce and maintain such compartmentalization of NGLs at the surface of HEK293T cells. Cells over-expressing either GPI-anchored NetrinG1 or NetrinG2 were plated alongside cells co-expressing full-length (TM) NGL1 and NGL2. We found that, indeed, contact with NetrinG1- or NetrinG2-expressing cells led to the exclusive targeting of NGL1 or NGL2 to the cell–cell interface, respectively (Figure 5A and B; Supplementary Movie). We then exploited our findings on the determinants of NetrinG-binding specificity to switch the NetrinG-binding properties of NGL1 in live cells. We co-expressed full-length NGL1-r123 with NGL2, and found that both proteins were targeted to cell–cell interfaces with NetrinG2-, but not NetrinG1-, presenting cells (Figure 5C and D). Thus, the low affinity interactions measured for cross-class binding are not enough to build up intra-cellular NGL patterns efficiently. Further, the specificity determinants revealed by combined structural and biophysical analyses are sufficient (even when transplanted into the cross-class NGL) to control molecular sorting in a cellular context through trans recognition events. Thus, the binding affinity and selectivity conferred on the NetrinG–NGL interaction by the specificity determinants we have identified provide the properties necessary to generate and maintain the reported compartmentalization of NGL1 and NGL2 to distinct dendritic subdomains (Figure 6), as proposed for a ‘lock-in’ type model (Nishimura-Akiyoshi et al, 2007).
Figure 5.
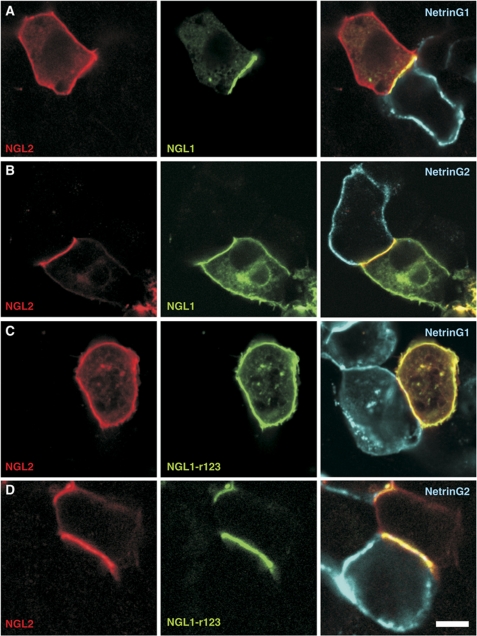
NetrinG-binding properties determine subcellular NGL localization. HEK293T cells co-expressing full-length TM NGL1-mVenus (green) and NGL2-mCherry (red) were grown alongside cells expressing GPI-anchored NetrinGs (blue). (A) Predominantly NGL1, and not NGL2, localizes to interfaces with NetrinG1-expressing cells. (B) NGL2 localizes to interfaces with NetrinG2-expressing cells. (C) NGL2 and NGL1-r123 (full-length) have less propensity to localize at cell–cell contacts with NetrinG1-presenting cells. (D) Both NGL2 and NGL1-r123 localize at interfaces with NetrinG2-presenting cells. The scale bar corresponds to 10 μm (A), 9.3 μm (B), 8.6 μm (C), 6 μm (D).
Figure 6.
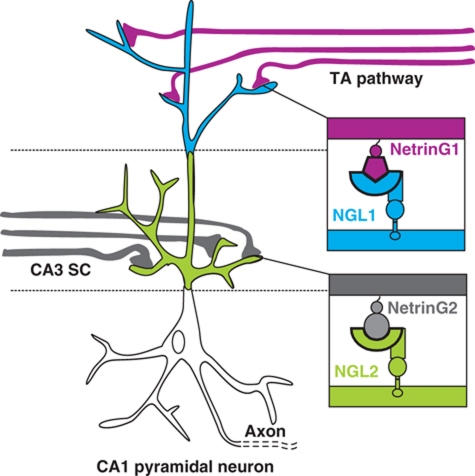
Schematic illustrating the structurally encoded NetrinG–NGL binding selectivity driving subcellular NGL localization in hippocampal CA1 pyramidal cells. CA1 dendrites receive inputs from apposing layers enriched in either NetrinG1-expressing axons (from the temporoammonic pathway, TA) or NetrinG2-expressing axons (Schaffer collaterals, SC, coming from the CA3 hippocampal region). These inputs localize NGL1 and NGL2 to distinct subdendritic regions via intra-class NGL–NetrinG interaction (Nishimura-Akiyoshi et al, 2007; Woo et al, 2009b).
Discussion
Multiple mechanisms are required to direct brain development, controlling patterning and neuronal circuit formation at the laminar, cellular, subcellular and synaptic levels (Sanes and Yamagata, 2009; Williams et al, 2010; Tessier-Lavigne and Kolodkin, 2011). NetrinG–NGL interactions have been implicated in functions at each of these levels (Lin et al, 2003; Kim et al, 2006; Nishimura-Akiyoshi et al, 2007), culminating in trans-synaptic adhesion and synaptogenesis (Kim et al, 2006). Our results reveal the hand-clasp architecture and define the specificity-conferring elements of the NetrinG–NGL adhesive interaction. The selectivity of adhesive interactions has been shown to drive sorting at the cellular level, for example, cadherin-mediated motor neuron segregation (Patel et al, 2006) and Dscam isoform-coded dendritic self-avoidance (Meijers et al, 2007; Sawaya et al, 2008). However, the molecular systems that drive subcellular compartmentalization events are as yet poorly resolved. Molecules known to perform this role include members of the Ig superfamily (Ango et al, 2004) and semaphorins plus their cognate plexin receptors (Suto et al, 2007). In such cases, pre-established patterns of molecules on receiving dendrites determine the subcellular localization of synapses with in-coming axons (Ango et al, 2004; Suto et al, 2007). In contrast, the NetrinG–NGL system exemplifies how subcellular molecular patterns, that is, NGL1-rich versus NGL2-rich regions on dendrites, can be induced by pre-synaptic components, the layer-specific NetrinG cues. Our results on NetrinG–NGL provide a paradigm whereby the discrimination generated by adhesive binding affinity drives the sorting of a mixed population of molecules into discrete cell surface subdomains.
Materials and methods
Cloning and vectors
We cloned hexahistidine-tagged constructs of human NetrinG1Lam–EGF1/4 (GenBank: BC030220.1, residues M1-H418, corresponding to domains Lam+EGF1+EGF4, thus equalling isoform 1 minus the GPI-anchor region), human NetrinG2Lam–EGF1 (GenBank: BC013770.1, residues M1-A345, corresponding to domains Lam+EGF1), human NGL1LRR–Ig (GenBank: BC041374.2, residues A44-T444, corresponding to domains LRR+Ig), human NGL2LRR–Ig (GenBank: NP_071426.1, residues M1-A444, corresponding to domains LRR+Ig), mouse NGL3LRR and NGL3LRR–Ig (GenBank: BC060263, residues M1-A365, corresponding to the LRR domain and residues M1-V455, corresponding to domains LRR+Ig) into the multiple cloning site of pHLsec or pHL-Avitag3 (Aricescu et al, 2006) using standard PCR methods and the restriction sites EcoRI and KpnI (NetrinG1, NetrinG2, NGL2, NGL3) or AgeI and KpnI (NGL1). The NetrinG1 construct used for crystallization harbours two threonine to alanine mutations (T350A+T353A) to avoid predicted O-linked glycosylation.
We generated 23 mutant and hybrid NGL and NetrinG proteins, including the following:
Knockout constructs (KO) harbouring N-linked glycosylation sites in the NetrinG–NGL binding interfaces designed to disrupt binding (NetrinG1 P85A+Y86T, NetrinG2 P74A+Y75T, NGL1 A205T, NGL2 G204T).
NetrinG1Lam–EGF1/4 and NetrinG2Lam–EGF1 proteins where one or more NGL-binding loops are swapped. To swap loop I, we exchanged residues 81–90 (AMGNPYMCNN) in NetrinG1 and residues 70–79 (SHENPYLCSN) in NetrinG2. For loop II, we swapped residues 212–220 (TGYTTNSKI) in NetrinG1 and residues 201–209 (RWAGSKKEKH) in NetrinG2. For loop III, we exchanged NetrinG1 residues 271–279 (VGEIFVDEL) and NetrinG2 residues 261–269 (LGGTYVQRE).
NGLLRR–Ig constructs, in which one or more surface regions within the LRR concave face were exchanged between NGL1 and NGL2. The residues manipulated are depicted in Figure 3.
Production of NetrinG and NGL proteins
We expressed NetrinG, NGL1 and NGL2 ectodomain constructs transiently in GnTI-deficient HEK293S cells (Aricescu et al, 2006; Chang et al, 2007), then dialysed the cell medium against phosphate-buffered saline (PBS), loaded it onto a Ni-containing affinity column (His-Trap, GE Healthcare), washed with 20 mM Tris pH 7.5, 300 mM NaCl, 40 mM imidazole and eluted the protein with 20 mM Tris pH 7.5, 300 mM NaCl, 500 mM imidazole. For crystallization, we incubated proteins overnight at room temperature with 1:20 recombinant endoglycosidase F1 (Grueninger-Leitch et al, 1996; Chang et al, 2007). To remove the endoglycosidase, we diluted the protein 1:10 in 20 mM Tris pH 7.5, 300 mM NaCl, re-loaded it on a Ni-containing column, washed and re-eluted. The eluted protein was concentrated and loaded on a Superdex 200 column (GE Healthcare) previously equilibrated with 200 mM NaCl and 20 mM Tris pH 7.5. We expressed NGL3 constructs similarly, but after two-fold dilution in PBS pH 7.5, we loaded cell medium directly onto a Ni affinity column. We washed the column with PBS pH 7.5 containing 30 mM imidazole and eluted the protein by increasing the imidazole concentration to 500 mM. After 1:20 endoglycosidase F1 treatment for 3 h at 37°C, we exchanged the buffer to 50 mM HEPES, 50 mM NaCl pH 7.5, loaded the protein onto a heparin affinity column (HiTrap heparin HP, GE Healthcare) and eluted it with a sodium chloride gradient. We further purified NGL3 proteins by size-exclusion chromatography (Superdex 75, GE Healthcare) in 10 mM HEPES, 150 mM NaCl pH 7.5.
We produced complexes of NetrinG and NGL by mixing cognate binding partners in a 1:1 molar ratio prior to injection on a Superdex 200 column. We pooled fractions from the main protein peak, supplemented the samples with 100 mM non-detergent sulfobetaine 256 (Hampton) and concentrated for crystallization. Final protein concentrations were 5.3 mg/ml (NetrinG2Lam–EGF1), 4.1 mg/ml (NetrinG1Lam–EGF1/4+NGL1LRR–Ig), 7.5 mg/ml (NetrinG2Lam–EGF1+NGL2LRR–Ig), 1.5 mg/ml (NGL3LRR) and 4 mg/ml (NGL3LRR–Ig). We expressed selenomethionine-labelled NetrinG2Lam–EGF1 according to previously published protocols (Aricescu et al, 2006) and carried out purification and crystallization as for the native protein.
Protein crystallization
Crystals grew in 100 nl+100 nl sitting nanodrops by the vapour diffusion method at 20°C (Walter et al, 2005). The crystallization conditions were 20% MME550, 0.1 M NaCl, 0.1 M bicine pH 9 (NetrinG2Lam–EGF1), 2% PEG 400, 2 M ammonium sulphate, 0.1 M HEPES pH 7.5+additive=PEG400 (NetrinG1Lam–EGF1/4+NGL1LRR–Ig), 50% v/v 2-methyl-2,4-pentanediol, 0.2 M ammonium di-hydrogen phosphate, 0.1 M Tris pH 8.5 (NetrinG2Lam–EGF1+NGL2LRR–Ig), 0.8 M succinic acid, pH 7.0 (NGL3LRR) and 0.1 M NaH2PO4, 0.1 M KH2PO4, 2 M NaCl, 0.1 M MES pH 6.5 (NGL3LRR–Ig). We collected X-ray diffraction images at the Diamond Light Source beamlines I02, I03 and I04 and processed the data using XDS (Kabsch, 1993), HKL2000 (Otwinowski and Minor, 1997), xia2 (Winter, 2010) and programs from the CCP4 suite (Collaborative Computational Project 4, 1994).
Structure solution and model refinement
We used a fragment containing nine lrr repeats of the Lingo-1 crystal structure (PDB accession number 2ID5), with side chains trimmed to Cβ atoms using Chainsaw (Collaborative Computational Project 4, 1994), as the initial search model for molecular replacement with the 3.2 Å NGL3LRR data set in PHASER (McCoy et al, 2007). The resultant model consists of two copies in the asymmetric unit. We used Phenix Autobuild (Terwilliger et al, 2008) and manual adjustments in Coot (Emsley and Cowtan, 2004) to rebuild the core of the LRR domain and used the resultant model together with an NMR structure of the Ig domain of human NGL3 (PDB accession number 2DL9), for molecular replacement with the 3.1 Å NGL3LRR–Ig data set in PHASER (McCoy et al, 2007). Using Phenix Autobuild (Terwilliger et al, 2008) and parallel refinement of both NGL3LRR and NGL3LRR–Ig structures with Phenix (Adams et al, 2010), Buster (Blanc et al, 2004) and Refmac (Murshudov et al, 1997) resulted in final models containing residues 57–366 and 55–366 for molecules A and B of NGL3LRR and 58–454 (with the exception of two disordered loops, 68–69 and 335–338) of NGL3LRR–Ig.
We derived initial phases for the NetrinG2Lam–EGF1/NGL2LRR–Ig complex data (space group I222) from molecular replacement using the model of NGL3LRR–Ig and that of human HSPB11 (PDB accession code 1TVG) (McCoy et al, 2007; Ramelot et al, 2009), which we predicted, using Fugue (Shi et al, 2001), to be structurally related to the NetrinG2 Lam domain. The resulting electron density map allowed manual building (Emsley and Cowtan, 2004) and refinement (Blanc et al, 2004) of an initial NetrinG2Lam–EGF1 model, which we used as molecular replacement model with the selenomethionine derivative data for unliganded NetrinG2Lam–EGF1 in Phaser (McCoy et al, 2007). This selenomethionine data set was analysed using space group P1 and displayed translational non-crystallographic symmetry (Patterson off-origin peak=64% of origin). The molecular replacement solution was used to determine selenium positions with PHASER (McCoy et al, 2007) and the resulting maps and selenium substructure were then used to manually improve (Emsley and Cowtan, 2004) the NetrinG2Lam–EGF1 model. We took the resultant model for molecular replacement with the high-resolution native data set of NetrinG2Lam–EGF1 (indexed according to the space group C2, with Patterson off-origin peak=32% of origin), fully built (Emsley and Cowtan, 2004) and refined (Blanc et al, 2004) it, and then fed it back into the NetrinG2Lam–EGF1/NGL2LRR–Ig complex structure for refinement (Blanc et al, 2004) of the complex. The resulting model served to phase the NetrinG1/NGL1LRR–Ig data (space group F222) by molecular replacement (McCoy et al, 2007). We fully refined all models using autoBuster (Blanc et al, 2004). In contrast to the NetrinG2Lam–EGF1 construct, the NetrinG1Lam–EGF1/4 construct comprises two EGF domains. In the crystal, this second EGF domain points towards a solvent channel. Although there is space to accommodate the second EGF domain in the lattice, there was no interpretable electron density, suggesting that it is flexibly connected to the rest of the protein and adopts multiple conformations in the crystal. We therefore refer to the crystal structure resulting from the NetrinG1Lam–EGF1/4 construct as NetrinG1Lam–EGF1. Interestingly, constructs lacking this second EGF domain failed to crystallize in our hands, further supporting the presence of full-length NetrinG1Lam–EGF1/4 in the crystal. An N-linked sugar that was not cleaved during the deglycosylation process contributes to crystal packing and consequently has well-ordered electron density for one of the two NetrinG1Lam–EGF1 copies in the asymmetric unit. We used Molprobity (Davis et al, 2007) to assess the quality of all protein models. Diffraction data and refinement details are summarized in Table I. We used Coot to superpose molecules for the calculation of root mean square deviation (r.m.s.d.) between Cα atoms. To calculate r.m.s.d. values for NetrinG loops I–III, we defined NetrinG1 residues 74–82 and NetrinG2 residues 63–82 as loop I, NetrinG1 residues 209–219 and NetrinG2 residues 198–209 as loop II and NetrinG1 residues 270–280 and NetrinG2 residues 260–270 as loop III. The shape correlation statistic (Sc), a parameter that describes the shape complementarity of two protein surfaces (Lawrence and Colman, 1993), is 0.62 for the NetrinG1Lam–EGF1+NGL1LRR–Ig complex and 0.69 for NetrinG2Lam–EGF1+NGL2LRR–Ig, consistent with tight interactions.
Multiangle light scattering
We purified protein samples by size-exclusion chromatography and concentrated to ∼0.5–1 mg/ml. Separation for MALS was achieved using an analytical Superdex S200 10/30 column (GE Healthcare) and the eluate was passed through online static light scattering (DAWN HELEOS II, Wyatt Technology, Santa Barbara, CA), differential refractive index (Optilab rEX, Wyatt Technology) and Agilent 1200 UV (Agilent Technologies) detectors. We analysed data using the ASTRA software package (Wyatt Technology).
SPR experiments
Mutant and WT proteins were tested in SPR equilibrium experiments where the native membrane topology was mimicked by coupling of proteins to a streptavidin-coated chip via a C-terminal-linked biotin label. We used a Biacore T100 machine (GE Healthcare) at 25°C in a running buffer comprising 10 mM HEPES, pH 7.5, 150 mM NaCl, 0.005% (v/v) polysorbate 20. All proteins underwent size-exclusion chromatography in running buffer before SPR experiments. To mimic the native membrane insertion topology, we biotinylated proteins enzymatically at an engineered C-terminal tag (Aricescu et al, 2006) and attached them via the biotin label to streptavidin that was covalently coupled to the Biacore chip surface (O’Callaghan et al, 1999). The signal from experimental flow cells was corrected by subtraction of a blank and reference signal from a mock-coupled flow cell in Scrubber2 (BioLogic). In all experiments analysed, the experimental trace returned to baseline after a regeneration step with 2 M MgCl2. Kd and maximum analyte binding (Bmax) values were obtained by non-linear curve fitting of a 1:1 Langmuir interaction model (bound=Bmax/(Kd+C), where C is the analyte concentration calculated as monomer). The NetrinG1 construct used for crystallization harbours two threonine to alanine mutations (T350A+T353A) to avoid predicted O-linked glycosylation. As expected from their distance to the NGL-binding site, we found these mutations do not affect the affinity to NGL1 in SPR experiments. Compared with NetrinG2, the NetrinG1 construct we used in crystallization and SPR experiments contains an additional EGF domain. As expected from the crystal structures, we found that removal of EGF4 does not affect the NetrinG1–NGL1 binding affinity (unpublished observation).
Cell surface patterning assay
Full-length human NGL1 (WT or harbouring the NGL1-r123 mutations) and NGL2 were fused to C-terminal mVenus-His or mCherry-His tags by subcloning into the appropriate pHLsec vector backbone. Full-length NetrinG1 isoform 1 (including the GPI anchor) or NetrinG2Lam–EGF1 fused to residues C502–R530 (equivalent to the C-terminal NetrinG2 GPI-anchor sequence) were subcloned into pHLsec without further tags. We grew HEK293T cells in Dulbecco's modified Eagle's Medium (DMEM high glucose; Sigma) supplemented with L-glutamine, non-essential amino acids (Gibco) and 10% (v/v) fetal calf serum (FCS; Sigma) and transfected plasmid DNA (full-length NGL1+NGL2, NetrinG1 isoform 1 or NetrinG2 GPI-fusion) using lipofectamine 2000 following the recommended protocol (Invitrogen). Final concentrations were 2 μg plasmid DNA per ml, 5 μl lipofectamine per ml and 1.5% (v/v) FCS. After 4–5 h, we replaced the transfection mix with original growth medium, re-suspended the cells and plated these on poly-L-lysine-coated glass bottom culture plates (MatTek). To observe trans interactions of different constructs, we mixed separately transfected cells at this stage. After 4% paraformaldehyde fixation for 10 min, to visualize NetrinG-expressing cells, the samples were stained with PBS containing 10 μg/ml His-tagged NGL1LRR–Ig or NGL2LRR–Ig, 5 μg/ml mouse anti-Penta-His (Qiagen) and 6 μg/ml Alexa647-conjugated anti-mouse (Invitrogen). We took multicoloured fluorescent images (1024 × 1024 pixel, 4 scans per frame, sequential scanning) with an LSM510 inverted confocal microscope system and Plan-NEOFLUAR × 40/1.3 oil DIC immersion lens (Carl Zeiss Ltd., Welwyn Garden City, UK), excitation 488 nm and emission 500–550 nm by-pass filter, excitation 543 nm and emission 565–615 nm by-pass filter or excitation 633 nm and emission 650 nm long-pass filter, optical slice 0.1 μm.
Supplementary Material
Acknowledgments
We thank Y Zhao and W Lu for protein expression; B Janssen and T Malinauskas for assistance with SPR experiments; M Jones and TS Walter for technical support; G Sutton for aiding multiangle light scattering experiments; and B Janssen, C Siebold and DI Stuart for critical reading of the manuscript. We are grateful to the staff of the Diamond Light Source for assistance with diffraction data collection. This research was funded by Cancer Research UK (CR-UK) and the UK Medical Research Council. ES is funded by an IEF Marie Curie fellowship, CHC was the recipient of a Wellcome Trust DPhil studentship, ARA is a UK Medical Research Council Career Development Award Fellow and EYJ is a Cancer Research UK Principal Research Fellow.
Author contributions: ES and CHC conducted crystallographic studies, SPR experiments and multiangle light scattering analyses. ES and PVP performed cellular assays and imaging. KH performed crystal mounting and aided X-ray data collection. All authors contributed to data analysis, discussion and preparation of the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66: 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ango F, di Cristo G, Higashiyama H, Bennett V, Wu P, Huang ZJ (2004) Ankyrin-based subcellular gradient of neurofascin, an immunoglobulin family protein, directs GABAergic innervation at purkinje axon initial segment. Cell 119: 257–272 [DOI] [PubMed] [Google Scholar]
- Aoki-Suzuki M, Yamada K, Meerabux J, Iwayama-Shigeno Y, Ohba H, Iwamoto K, Takao H, Toyota T, Suto Y, Nakatani N, Dean B, Nishimura S, Seki K, Kato T, Itohara S, Nishikawa T, Yoshikawa T (2005) A family-based association study and gene expression analyses of netrin-G1 and -G2 genes in schizophrenia. Biol Psychiatry 57: 382–393 [DOI] [PubMed] [Google Scholar]
- Aricescu AR, Lu W, Jones EY (2006) A time- and cost-efficient system for high-level protein production in mammalian cells. Acta Crystallogr D Biol Crystallogr 62: 1243–1250 [DOI] [PubMed] [Google Scholar]
- Bella J, Hindle KL, McEwan PA, Lovell SC (2008) The leucine-rich repeat structure. Cell Mol Life Sci 65: 2307–2333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc E, Roversi P, Vonrhein C, Flensburg C, Lea SM, Bricogne G (2004) Refinement of severely incomplete structures with maximum likelihood in BUSTER-TNT. Acta Crystallogr D Biol Crystallogr 60: 2210–2221 [DOI] [PubMed] [Google Scholar]
- Borg I, Freude K, Kubart S, Hoffmann K, Menzel C, Laccone F, Firth H, Ferguson-Smith MA, Tommerup N, Ropers HH, Sargan D, Kalscheuer VM (2005) Disruption of Netrin G1 by a balanced chromosome translocation in a girl with Rett syndrome. Eur J Hum Genet 13: 921–927 [DOI] [PubMed] [Google Scholar]
- Chang VT, Crispin M, Aricescu AR, Harvey DJ, Nettleship JE, Fennelly JA, Yu C, Boles KS, Evans EJ, Stuart DI, Dwek RA, Jones EY, Owens RJ, Davis SJ (2007) Glycoprotein structural genomics: solving the glycosylation problem. Structure 15: 267–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collaborative Computational Project 4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr 50: 760–763 [DOI] [PubMed] [Google Scholar]
- Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, Murray LW, Arendall WB III, Snoeyink J, Richardson JS, Richardson DC (2007) MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res 35: W375–W383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eastwood SL, Harrison PJ (2008) Decreased mRNA expression of netrin-G1 and netrin-G2 in the temporal lobe in schizophrenia and bipolar disorder. Neuropsychopharmacology 33: 933–945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60: 2126–2132 [DOI] [PubMed] [Google Scholar]
- Fan QR, Hendrickson WA (2005) Structure of human follicle-stimulating hormone in complex with its receptor. Nature 433: 269–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grueninger-Leitch F, D’Arcy A, D’Arcy B, Chene C (1996) Deglycosylation of proteins for crystallization using recombinant fusion protein glycosidases. Protein Sci 5: 2617–2622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain SA, Carafoli F, Hohenester E (2011) Determinants of laminin polymerization revealed by the structure of the alpha5 chain amino-terminal region. EMBO Rep 11: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W (1993) Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J Appl Crystallogr 26: 795–800 [Google Scholar]
- Kim S, Burette A, Chung HS, Kwon SK, Woo J, Lee HW, Kim K, Kim H, Weinberg RJ, Kim E (2006) NGL family PSD-95-interacting adhesion molecules regulate excitatory synapse formation. Nat Neurosci 9: 1294–1301 [DOI] [PubMed] [Google Scholar]
- Krissinel E, Henrick K (2007) Inference of macromolecular assemblies from crystalline state. J Mol Biol 372: 774–797 [DOI] [PubMed] [Google Scholar]
- Kwon SK, Woo J, Kim SY, Kim H, Kim E (2010) Trans-synaptic adhesions between netrin-G ligand-3 (NGL-3) and receptor tyrosine phosphatases LAR, protein-tyrosine phosphatase delta (PTPdelta), and PTPsigma via specific domains regulate excitatory synapse formation. J Biol Chem 285: 13966–13978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai Wing Sun K, Correia JP, Kennedy TE (2011) Netrins: versatile extracellular cues with diverse functions. Development 138: 2153–2169 [DOI] [PubMed] [Google Scholar]
- Lawrence MC, Colman PM (1993) Shape complementarity at protein/protein interfaces. J Mol Biol 234: 946–950 [DOI] [PubMed] [Google Scholar]
- Lin JC, Ho WH, Gurney A, Rosenthal A (2003) The netrin-G1 ligand NGL-1 promotes the outgrowth of thalamocortical axons. Nat Neurosci 6: 1270–1276 [DOI] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ (2007) Phaser crystallographic software. J Appl Crystallogr 40: 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijers R, Puettmann-Holgado R, Skiniotis G, Liu JH, Walz T, Wang JH, Schmucker D (2007) Structural basis of Dscam isoform specificity. Nature 449: 487–491 [DOI] [PubMed] [Google Scholar]
- Mosyak L, Wood A, Dwyer B, Buddha M, Johnson M, Aulabaugh A, Zhong X, Presman E, Benard S, Kelleher K, Wilhelm J, Stahl ML, Kriz R, Gao Y, Cao Z, Ling HP, Pangalos MN, Walsh FS, Somers WS (2006) The structure of the Lingo-1 ectodomain, a module implicated in central nervous system repair inhibition. J Biol Chem 281: 36378–36390 [DOI] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr 53: 240–255 [DOI] [PubMed] [Google Scholar]
- Nakashiba T, Ikeda T, Nishimura S, Tashiro K, Honjo T, Culotti JG, Itohara S (2000) Netrin-G1: a novel glycosyl phosphatidylinositol-linked mammalian netrin that is functionally divergent from classical netrins. J Neurosci 20: 6540–6550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashiba T, Nishimura S, Ikeda T, Itohara S (2002) Complementary expression and neurite outgrowth activity of netrin-G subfamily members. Mech Dev 111: 47–60 [DOI] [PubMed] [Google Scholar]
- Nishimura-Akiyoshi S, Niimi K, Nakashiba T, Itohara S (2007) Axonal netrin-Gs transneuronally determine lamina-specific subdendritic segments. Proc Natl Acad Sci USA 104: 14801–14806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Callaghan CA, Byford MF, Wyer JR, Willcox BE, Jakobsen BK, McMichael AJ, Bell JI (1999) BirA enzyme: production and application in the study of membrane receptor-ligand interactions by site-specific biotinylation. Anal Biochem 266: 9–15 [DOI] [PubMed] [Google Scholar]
- Ohtsuki T, Horiuchi Y, Koga M, Ishiguro H, Inada T, Iwata N, Ozaki N, Ujike H, Watanabe Y, Someya T, Arinami T (2008) Association of polymorphisms in the haplotype block spanning the alternatively spliced exons of the NTNG1 gene at 1p13.3 with schizophrenia in Japanese populations. Neurosci Lett 435: 194–197 [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol 276: 307–326 [DOI] [PubMed] [Google Scholar]
- Pan Y, Liu G, Fang M, Shen L, Wang L, Han Y, Shen D, Wang X (2010) Abnormal expression of netrin-G2 in temporal lobe epilepsy neurons in humans and a rat model. Exp Neurol 224: 340–346 [DOI] [PubMed] [Google Scholar]
- Patel SD, Ciatto C, Chen CP, Bahna F, Rajebhosale M, Arkus N, Schieren I, Jessell TM, Honig B, Price SR, Shapiro L (2006) Type II cadherin ectodomain structures: implications for classical cadherin specificity. Cell 124: 1255–1268 [DOI] [PubMed] [Google Scholar]
- Ramelot TA, Raman S, Kuzin AP, Xiao R, Ma LC, Acton TB, Hunt JF, Montelione GT, Baker D, Kennedy MA (2009) Improving NMR protein structure quality by Rosetta refinement: a molecular replacement study. Proteins 75: 147–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson JS (1981) The anatomy and taxonomy of protein structure. Adv Protein Chem 34: 167–339 [DOI] [PubMed] [Google Scholar]
- Sanes JR, Yamagata M (2009) Many paths to synaptic specificity. Annu Rev Cell Dev Biol 25: 161–195 [DOI] [PubMed] [Google Scholar]
- Sawaya MR, Wojtowicz WM, Andre I, Qian B, Wu W, Baker D, Eisenberg D, Zipursky SL (2008) A double S shape provides the structural basis for the extraordinary binding specificity of Dscam isoforms. Cell 134: 1007–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Blundell TL, Mizuguchi K (2001) FUGUE: sequence-structure homology recognition using environment-specific substitution tables and structure-dependent gap penalties. J Mol Biol 310: 243–257 [DOI] [PubMed] [Google Scholar]
- Suto F, Tsuboi M, Kamiya H, Mizuno H, Kiyama Y, Komai S, Shimizu M, Sanbo M, Yagi T, Hiromi Y, Chedotal A, Mitchell KJ, Manabe T, Fujisawa H (2007) Interactions between plexin-A2, plexin-A4, and semaphorin 6A control lamina-restricted projection of hippocampal mossy fibers. Neuron 53: 535–547 [DOI] [PubMed] [Google Scholar]
- Terwilliger TC, Grosse-Kunstleve RW, Afonine PV, Moriarty NW, Zwart PH, Hung LW, Read RJ, Adams PD (2008) Iterative model building, structure refinement and density modification with the PHENIX AutoBuild wizard. Acta Crystallogr D Biol Crystallogr 64: 61–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tessier-Lavigne M, Kolodkin AL (2011) Neuronal Guidance: The Biology of Brain Wiring. New York: Cold Spring Harbor Laboratory Press [Google Scholar]
- Walter TS, Diprose JM, Mayo CJ, Siebold C, Pickford MG, Carter L, Sutton GC, Berrow NS, Brown J, Berry IM, Stewart-Jones GB, Grimes JM, Stammers DK, Esnouf RM, Jones EY, Owens RJ, Stuart DI, Harlos K (2005) A procedure for setting up high-throughput nanolitre crystallization experiments. Crystallization workflow for initial screening, automated storage, imaging and optimization. Acta Crystallogr D Biol Crystallogr 61: 651–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams ME, de Wit J, Ghosh A (2010) Molecular mechanisms of synaptic specificity in developing neural circuits. Neuron 68: 9–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter G (2010) xia2: an expert system for macromolecular crystallography data reduction. J Appl Cryst 43: 186–190 [Google Scholar]
- Woo J, Kwon SK, Choi S, Kim S, Lee JR, Dunah AW, Sheng M, Kim E (2009a) Trans-synaptic adhesion between NGL-3 and LAR regulates the formation of excitatory synapses. Nat Neurosci 12: 428–437 [DOI] [PubMed] [Google Scholar]
- Woo J, Kwon SK, Kim E (2009b) The NGL family of leucine-rich repeat-containing synaptic adhesion molecules. Mol Cell Neurosci 42: 1–10 [DOI] [PubMed] [Google Scholar]
- Yin Y, Miner JH, Sanes JR (2002) Laminets: laminin- and netrin-related genes expressed in distinct neuronal subsets. Mol Cell Neurosci 19: 344–358 [DOI] [PubMed] [Google Scholar]
- Zhang W, Rajan I, Savelieva KV, Wang CY, Vogel P, Kelly M, Xu N, Hasson B, Jarman W, Lanthorn TH (2008) Netrin-G2 and netrin-G2 ligand are both required for normal auditory responsiveness. Genes Brain Behav 7: 385–392 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.