Abstract
MicroRNAs (miRNAs) are ∼22 nt non-coding RNAs that typically bind to the 3′ UTR of target mRNAs in the cytoplasm, resulting in mRNA destabilization and translational repression. Here, we report that miRNAs can also regulate gene expression by targeting non-coding antisense transcripts in human cells. Specifically, we show that miR-671 directs cleavage of a circular antisense transcript of the Cerebellar Degeneration-Related protein 1 (CDR1) locus in an Ago2-slicer-dependent manner. The resulting downregulation of circular antisense has a concomitant decrease in CDR1 mRNA levels, independently of heterochromatin formation. This study provides the first evidence for non-coding antisense transcripts as functional miRNA targets, and a novel regulatory mechanism involving a positive correlation between mRNA and antisense circular RNA levels.
Keywords: CDR1 , circular RNA, miR-671, natural antisense transcript, nuclear microRNA
Introduction
MicroRNAs (miRNA) are known to regulate gene expression by binding to the 3′ UTR of target mRNAs in the cytoplasm, resulting in mRNA destabilization and translational repression (Humphreys et al, 2005; Pillai et al, 2005; Petersen et al, 2006; Chendrimada et al, 2007). It is becoming evident that miRNAs are at some level involved in most, if not all, cellular pathways, and deregulated miRNA expression patterns are a hallmark of diseases such as cancer (Guil and Esteller, 2009). In some cases, miRNAs have also been demonstrated to direct Ago2-dependent cleavage of mRNAs with near perfect complementary miRNA target sites in an RNA interference (RNAi)-like manner (Hutvagner and Zamore, 2002; Yekta et al, 2004), and certain miRNAs may function by targeting sites in the 5′ UTR and ORF of mRNAs (Forman et al, 2008; Orom et al, 2008; Tay et al, 2008). In addition, functional RISC activity has been detected in the nucleus of human cells (Langlois et al, 2005; Robb et al, 2005), and a subset of miRNAs are predominantly nuclear (Hwang et al, 2007; Liao et al, 2010), suggesting that miRNAs may have a variety of biological functions distinct from canonical 3′ UTR target mRNA repression.
In fission yeast and plants, small interfering RNAs (siRNAs) targeted to promoter regions induce RNAi-dependent transcriptional gene silencing (TGS) by directing local heterochromatin formation (Verdel et al, 2009). It has been shown that promoter-targeting siRNAs can induce TGS in cultured human cell lines as well (Morris et al, 2004), and recently an endogenous mammalian TGS mechanism involving miRNAs has been proposed (Gonzalez et al, 2008; Kim et al, 2008). In human cells, siRNA-directed TGS is usually accompanied by H3K27 and/or H3K9 methylation and occasionally with DNA hypermethylation (Svoboda et al, 2004; Suzuki et al, 2005; Ting et al, 2005; Weinberg et al, 2006; Hawkins et al, 2009). It is also associated with increased localization of Argonaute proteins to chromosomal DNA, consistent with a possible role for the miRNA machinery, but the underlying mechanisms are poorly understood (Janowski et al, 2006; Kim et al, 2006; Gonzalez et al, 2008). Non-coding transcription around the targeted promoter seems to be required, and the siRNA probably directs the effect via RNA–RNA interactions in the nucleus (Han et al, 2007; Schwartz et al, 2008; Napoli et al, 2009).
High-throughput technologies have in recent years led to the uncovering of a plethora of gene proximal sense and antisense non-coding transcription. Natural antisense transcripts (NATs) refer to gene overlapping antisense transcription, which is detected in ∼70% of protein coding genes in mouse (Katayama et al, 2005), suggesting an important widespread role in regulation. NATs have mostly been associated with gene silencing, but recent reports of activation demonstrate that the relationship between sense and antisense levels is more complex (Katayama et al, 2005; Morris et al, 2008). Several mechanisms for NAT gene regulation have been proposed, including: (i) inhibition of translation by blocking mRNA association with ribosomes (Ebralidze et al, 2008), (ii) stabilization of mRNA by duplex formation (Faghihi et al, 2008), (iii) perturbation of splicing leading to intron inclusion (Beltran et al, 2008), (iv) epigenetic silencing by deposition of silent state chromatin marks similar to TGS (Yap et al, 2010), and (v) a collision model where antisense transcription interferes with polymerase activity on the sense strand (Crampton et al, 2006). Moreover, NAT expression has been implicated in cancer, as p15 expression is known to be epigenetically regulated by a long antisense ncRNA (Yu et al, 2008). Thus, NATs seem to be an important regulator in mammalian systems.
Here, we aimed to investigate whether miRNAs can modulate gene expression by targeting NATs. A bioinformatic scan for possible miRNA targets in promoter proximal non-coding transcripts identified Cerebellar Degeneration-Related protein 1 (CDR1) as a promising candidate target locus of miR-671. We show that miR-671 predominantly localizes to the nucleus and directs Ago2-mediated cleavage of an antisense transcript of the CDR1 gene, leading to a concomitant decrease in steady-state CDR1 mRNA levels. Characterization of the CDR1 NAT revealed an unexpected non-linear alternative splicing (NAS) event, producing a circular exonic RNA that appears to stabilize the CDR1 mRNA. This study for the first time demonstrates that non-coding antisense transcripts can act as direct miRNA targets, and also suggests a novel gene regulatory mechanism involving a positive correlation between mRNA and circular antisense RNA levels.
Results
miR-671 negatively regulates CDR1
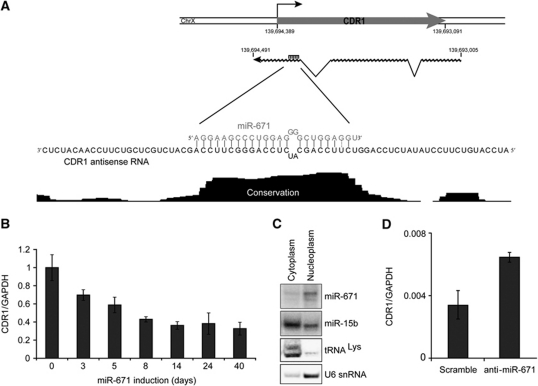
In order to identify miRNAs with a possible role in TGS, we scanned promoter proximal regions for highly complementary and phylogenetically conserved miRNA target sites. A putative target in the intronless gene CDR1 (also known as CDR34; Dropcho et al, 1987; Furneaux et al, 1989) was identified for miR-671 (Figure 1A). The target site is highly complementary to mature miR-671, is situated in the antisense direction proximal to the CDR1 transcription start site, and is associated with a peak of conservation exactly spanning the predicted miR-671 target sequence (Figure 1A).
Figure 1.
miR-671 represses CDR1 expression. (A) Schematic representation of the CDR1 locus. CDR1 mRNA (chrX: 139,693,091–139,694,389, hg18) and the antisense RNA (chrX: 139,693,005–139,694,491, hg18) derived from EST accessions (for additional detail, see Supplementary Figure S3). The enlargement shows the miR-671 antisense target region along with vertebrate conservation from UCSC Genome Browser (17-way conservation). (B) CDR1 expression in HEK293-eGFP-671 cells at indicated time points from 0 to 40 days of tetracycline induction. Expression was determined by qRT–PCR on random hexamer primed total RNA using the primer set CDR1(4)FW/CDR1(4)RE (normalized to 18S, relative to uninduced levels). (C) Northern blot showing subcellular localization of miR-671 in nuclear and cytoplasmic fractions of HEK293 cells transiently transfected with pJEBB-671. miR-15b is included as a localization control for an endogenously expressed miRNA and U6 snRNA and tRNALys are nuclear- and cytoplasm-specific RNAs, respectively. (D) CDR1 expression in HEK293 cells transfected with anti-mir-671 or scrambled control determined by qPCR (as in B).
To investigate whether CDR1 levels were affected by miR-671 expression, we established stable tetracycline inducible miR-671 and negative control miR-769 expression HEK293 cell lines (HEK293-eGFP-671 and HEK293-eGFP-769, respectively) using the FRT/TO system (Supplementary Figure S1). CDR1 expression levels were subsequently evaluated in time course induction of HEK293-eGFP-671 and HEK293-eGFP-769 cells. As seen in Figure 1B, CDR1 levels dropped to 65% after 3 days and stabilized at <40% throughout long-term miR-671 induction. Gene expression microarray data confirmed these results, with CDR1 mRNA levels dropping to 47 and 36% after 3 and 40 days, respectively (Supplementary Figure S2A and B), whereas negative control miR-769 induction had no or little effect on CDR1 expression (Supplementary Figure S2B and C).
An miRNA gene regulatory effect mediated via a non-coding antisense transcript would presumably be a nuclear event, similar to previous reports of TGS. Northern blot analysis of transiently transfected miR-671 showed that miR-671 indeed localizes predominantly to the nucleus (Figure 1C), in agreement with a recent analysis of subcellular miRNA abundance (Liao et al, 2010). In this study, miR-671 was found to be among the most nuclear enriched miRNAs, with a nuclear to cytoplasmic ratio of 6. This suggests that the miR-671 directed downregulation of CDR1 is likely to be mediated by a nuclear mechanism.
We observed that HEK293 cells express miR-671 endogenously (Supplementary Figure S1B), consistent with a publicly available high-throughput sequencing data set of HEK293 small RNAs (∼250 miR-671-5p reads per million, GSM416733; Mayr and Bartel, 2009). Thus, removing the miRNA by an anti-mir-671, we propose, should activate CDR1. As expected, CDR1 levels increased upon anti-mir-671 transfection compared with a scramble control (Figure 1D), thereby demonstrating an endogenous involvement of miR-671 in CDR1 regulation in HEK293 cells.
Non-linear splicing and circularization of the antisense transcript
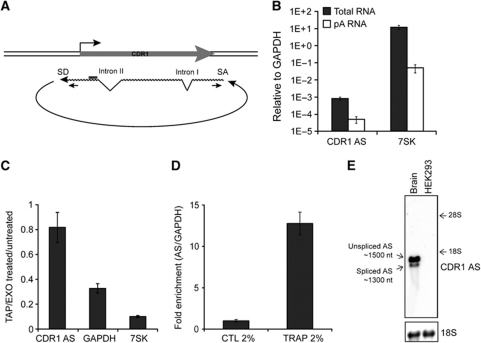
We went on to characterize the CDR1 antisense transcript in more detail. Searching the UCSC genome browser EST database for the existence of NATs overlapping the miR-671 target region, several accessions in and around the CDR1 locus were identified (Supplementary Figure S3). Interestingly, at least 23 ESTs appear to derive from an unusual NAS mechanism (Gingeras, 2009), with the splice acceptor (SA) 1485 bp upstream of the splice donor (SD) (Figure 2A; Supplementary Figure S3A and B). These splice site sequences are conserved and resemble consensus mRNA splicing motifs (Supplementary Figure S3A), suggesting that the NAS event is mediated by the canonical spliceosomal apparatus giving rise to 5′–3′ exon–exon linkage, similar to the circular SRY transcript in mice, which is essential for male sex determination (Capel et al, 1993). Moreover, the CDR1 antisense RNA harbours at least two canonical co-linear introns with consensus splice sites (Figure 2A; Supplementary Figure S3C and D).
Figure 2.
Characterization of the circular CDR1 antisense RNA. (A) Schematic illustration of the circular antisense RNA including the NAS splice donor (SD) and splice acceptor (SA) sites and the position of the NAS-specific primer set. (B) qRT–PCR for total or poly(A)-enriched RNA from HEK293 using primers specific for the NAS RNA. 7SK is included as a poly(A) tail-deficient control RNA. (C) NAS-specific qRT–PCR for RNA treated with TAP (Tobacco Acid Pyrophosphatase) and EXO (Terminator 5′ Phosphate-Dependent Exonuclease) normalized to untreated RNA. GAPDH mRNA and 7SK RNA are included as controls for TAP and EXO efficiencies. (D) Total HEK293 RNA was mixed with solidifying agarose and subjected to electrophoresis. In this system, circular RNA species will be physically trapped, whereas linear RNAs remain free to migrate. A no-electrophoresis control was included to determine gel-extraction efficiency, and −fold enrichment in CDR1 AS was quantified by NAS-specific qRT–PCR relative to total INPUT RNA and normalized to GAPDH. (E) Agarose northern blot with 2 μg RNA from HEK293 cells or human brain showing distinct AS migration using a CDR1 NAS-specific probe (top panel). Unspliced and spliced AS represent splicing of canonical intron II (see A). The approximate RNA sizes were deduced based on ribosomal RNA migration. 18S (bottom panel) serves as loading control.
The predicted NAS event was assessed in HEK293 cells by RT–PCR with three different NAS-specific primer sets (Supplementary Figure S3C) and subsequent sequencing of the resulting amplicons confirmed the non-linear nature of the CDR1 antisense transcript and the optional removal of intron II by canonical linear splicing (‘Unspliced’ and ‘Spliced’; Supplementary Figure S3C and D).
Next, 3′ RACE analysis failed to reveal any polyadenylation of the CDR1 antisense RNA. This observation was confirmed by decreased antisense transcript levels in poly(A)-enriched RNA relative to GAPDH, indicating that a poly(A)-tail is absent in the antisense species (Figure 2B). This posed the question of whether NAS occurs in trans, forming antisense concatamers, or whether it may be a cis-acting event resulting in a circular NAT.
In order to distinguish the linear or circular structure of the antisense transcript, total HEK293 RNA was subjected to treatment with Tobacco Acid Pyrophosphatase (TAP), which removes the 5′ cap, and subsequently with Terminator 5′-phosphate-dependent exonuclease (EXO), thereby assaying for the presence of a free 5′ end. In contrast to linear and capped control RNAs (7SK RNA and GAPDH mRNA) that were degraded, the CDR1 antisense transcript proved to be resistant to the TAP/EXO treatment (Figure 2C). Circularization was further confirmed by TRAP electrophoresis (Figure 2D); a system in which a solution containing RNA and melted agarose is allowed to solidify in the well of a preformed gel, thereby physically trapping circular species upon electrophoresis (Schindler et al, 1982). Indeed, the CDR1 antisense was strongly enriched in the TRAP fraction relative to input RNA, further confirming its circular nature (Figure 2D). Moreover, as in RT–PCR, northern blotting RNA from human brain, where CDR1 NAT is highly expressed (cf. Supplementary Figure S9), clearly revealed two distinct antisense species (Figure 2E), which reflects optional inclusion of intron II in the circular RNA. The distinct migration of AS species according to predicted size of circular RNA (1485 and 1301 nt, respectively) on agarose northern blotting (Figure 2E), similar to the circular SRY RNA (Capel et al, 1993), emphasizes the existence and prevalence of circular CDR1 NAT RNA in vivo.
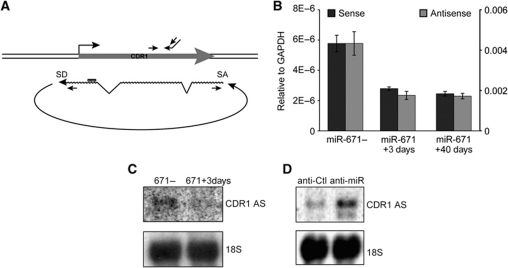
Strand-specific quantification of CDR1 mRNA and circular AS
To directly assess the effect of miR-671 on both antisense RNA and CDR1 mRNA, we designed a strand-specific qRT–PCR approach (Figure 3A). Interestingly, the antisense levels were several orders of magnitude higher than CDR1 mRNA levels, indicating that the non-coding antisense transcript is the predominant RNA species of the CDR1 locus (Figure 3B). However, relative downregulation of the CDR1 sense and antisense transcripts upon miR-671 induction was similar (Figure 3B). This suggests that removal of the antisense transcript is directly correlated with reduced mRNA levels. Furthermore, the upregulatory and downregulatory effects on CDR1 NAT associated with miR-671 overexpression and inhibition, respectively, were verified by northern blotting (Figure 3C and D).
Figure 3.
Strand-specific quantification of CDR1 mRNA and the antisense RNA. (A) Schematic illustration of the CDR1 locus with primers used for strand-specific qRT–PCR of CDR1 mRNA (above) and antisense RNA (below), respectively. (B) Expression of the CDR1 mRNA (black) and NAS antisense RNA (grey) in uninduced, 3 and 40 days induced HEK293-eGFP-671 cells determined by strand-specific qRT–PCR. Left and right axes denote relative levels of CDR1 mRNA and CDR1 AS, respectively. (C, D) Northern blot showing CDR1 AS levels after 3 days miR-671 induction (C) or upon 100 nM anti-miR-671-5p treatment (D) compared with uninduced and anti-Control treatment, respectively. 18S serves as loading control.
Given the speculation that miRNAs may be involved in TGS, we next investigated whether the mode of CDR1 repression was epigenetic. Surprisingly, no clear accumulation of the repressive H3K27me3 polycomb mark or depletion of active H3K9 acetylation (H3K9ac) was observed by ChIP-qPCR upon miR-671 induction (Supplementary Figure S4A–C). The CDR1 locus also remained largely unmethylated and, although CDR1 was activated overall, further depletion of DNA methylation by 5-aza-dC failed to inhibit miR-671 directed CDR1 repression (Supplementary Figure S4D and E). Consistently, CDR1 mRNA and antisense levels increased when HEK293-671 cells were treated with anti-miR-671-5p following 3 days tetracycline induction (Supplementary Figure S5A and B), suggesting that the CDR1 regulation is a transient phenomenon rather than dependent on a stable, epigenetic mechanism. As such, the primary mechanism of miR-671-directed CDR1 silencing appears to be distinct from TGS.
Positive correlation between CDR1 antisense and mRNA levels
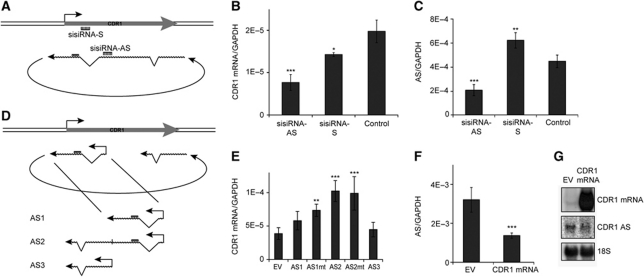
To confirm the dependency of the CDR1 mRNA on the NAT, and to further support that the antisense is the direct target of miR-671, small internally segmented interfering RNAs (sisiRNA) (Bramsen et al, 2007) was employed to target strictly either the antisense transcript (sisiRNA-AS) or the sense mRNA (sisiRNA-S) (Figure 4A). Strand-specific qRT–PCR showed a significant decrease of CDR1 mRNA using either sisiRNA-AS or sisiRNA-S compared with a mock siRNA (Control) (Figure 4B). In contrast, only the antisense-specific sisiRNA (sisiRNA-AS) repressed the level of NAT (Figure 4C), and a significant increase in AS levels was in fact observed as a consequence of mRNA knockdown (Figure 4C). Concordant with the effects observed upon miR-671 expression, this further demonstrates that the CDR1 mRNA is dependent on antisense RNA levels, and that miR-671 mediated antisense removal accounts for the reduction in steady-state CDR1 mRNA levels.
Figure 4.
Effect of CDR1 antisense-specific knockdown and overexpression. (A) Schematic illustration showing the target sites for strand-specific sisiRNAs. (B, C) HEK293 cells were transfected with 20 nM strand-specific sisiRNAs targeting the CDR1 antisense RNA (sisiRNA-AS) or mRNA (sisiRNA-S), or a mock control siRNA (Control). After 48 h, RNA levels were evaluated by qRT–PCR using mRNA-specific (B) or NAS-specific (C) primers, respectively. (D) Schematic illustration showing antisense vector design. (E) HEK293 cells were transfected with a vector expressing an antisense transcript harbouring the miR-671 target site (AS1), a vector expressing NAS-mimicking antisense RNA (AS2), a vector expressing a non-target site part of AS (AS3), or an empty vector (EV). The ASmt vectors carry a perfect miR-769 target site instead of the putative miR-671 target. RNA was harvested after 48 h, and the CDR1 mRNA level was quantified by strand-specific qRT–PCR. (F, G) NAS-specific qRT–PCR (F) and northern blot (G) with RNA extracted from cells transfected with a CDR1 mRNA expression vector or an empty vector (EV). Upper, middle, and lower panels show CDR1 mRNA, CDR1 AS, and 18S levels, respectively (G) (*P<0.05; **P<0.01; ***P<0.001).
To strengthen the effect of depleting either the AS or mRNA species, we overexpressed CDR1 mRNA or NAT mimics. Indeed, ectopic expression of a linear transcript mimicking the NAS product encompassing the circularized antisense region (AS2; Figure 4D) increased CDR1 mRNA levels compared with a non-NAS truncated construct (AS1 and AS3) or an empty vector (EV) control (Figure 4E). This confirms that CDR1 mRNA steady-state levels are stabilized by antisense expression, and indicates that non-linear adjoining of the exonic sequences by NAS is an important functional feature of the CDR1 antisense transcript. The effect was similar when expressing AS2 with a target-site mutation (AS2mt; Figure 4E), rejecting an miR-671 sponge effect as the responsible cause. In contrast to the antisense-directed increase in CDR1 mRNA, transiently overexpressing the mRNA resulted in decreased NAT levels based on qRT–PCR and northern blotting (Figure 4F and G, respectively), also in accordance with the observed increase after mRNA depletion (Figure 4C). This suggests a sense–antisense-based feedback mechanism where the antisense RNA stimulates or stabilizes the sense mRNA with subsequent negative impact on antisense levels. Targeting specifically the predominant unspliced isoforms of the circular RNA or the NAS-derived exon–exon junction using sisiRNA against intron II (sisi-Unspliced) or against the NAS junction (sisi-NAS), respectively, resulted in potent knockdown of antisense and CDR1 mRNA comparable to sisiRNA-AS (Supplementary Figure S6A and B). This suggests that canonical splicing of intron II in CDR1 NAT is not important for its function and verifies the involvement of circular AS in mRNA regulation. Moreover, to eliminate the possibility that the miR-671-3p arm was responsible for the effect, we tested an miR-671-3p sisiRNA mimic. Here, no detectable changes in CDR1 levels were observed (Supplementary Figure S6A and B), which in agreement with anti-miR-671-5p treatment confirms a dispensatory role of the miR-671-3p arm.
Ago2-mediated cleavage of the CDR1 antisense transcript
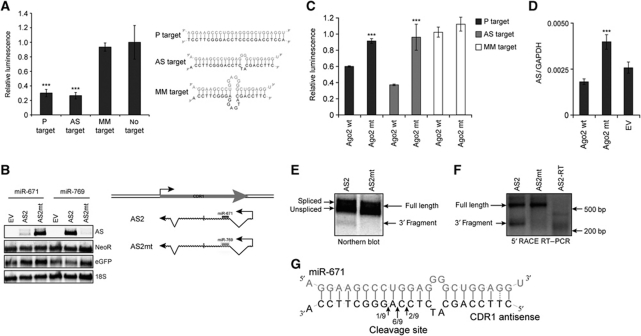
The concept of circular RNAs poses the interesting question of how these species are regulated in vivo. Normal RNA turnover typically involves 5′ and 3′ exonucleolytic activity at unprotected ends (Schmid and Jensen, 2008), but this mechanism would probably be insufficient for circular RNA decay. In luciferase reporter assays, comparable levels of luminescence reduction were observed for the predicted endogenous miR-671 target site (AS target) and a mutated fully complementary miR-671 target (P target), whereas no difference in the effect was observed between a mismatched target (MM target) and the no-target vector (Figure 5A). This demonstrates that the putative antisense target site has similar potency to a perfect complementary sequence, which is indicative of an siRNA-like effect.
Figure 5.
Ago2-mediated cleavage of the CDR1 antisense transcript. (A) Firefly luciferase reporters (pISO) with a perfect miR-671 target (P target), the endogenous CDR1 antisense target (AS target), a mismatched target (MM target) or no target were co-transfected with a Renilla luciferase expression vector (pcDNA3-RL) and miR-671 (pJEBB-671) or miR-769 (pJEBB-769). Relative luminescence represents the Firefly/Renilla ratio for pJEBB-671 relative to pJEBB-769, normalized to the no target control. (B) Northern blot with RNA from HEK293 cells co-transfected with empty vector (EV), antisense wild-type (AS2) or antisense with an miR-769 target-site (AS2mt) and pJEBB-671 or pJEBB-769. Transient AS expression was determined with an AS-specific probe (upper panel). The NeoR probe reflects transfection efficiency of the AS expression vector (second panel), the eGFP probe shows ectopic gene expression from the transfected miRNA vector (third panel) and 18S serves as loading control (lower panel). (C) Set-up as in (A) but with transient co-transfection of Ago2-wild-type (Ago2-wt) or Ago2-slicer mutant (Ago2-D669A) expression vectors (Supplementary Figure S7A). (D) Antisense-specific qRT–PCR on RNA from HEK293 cells transiently transfected with miR-671 along with EV, Ago2 wild-type (Ago2-wt), or Ago2-slicer mutant (Ago2-D669A) expression vectors (Supplementary Figure S7B). (E) Northern blot on RNA from HEK293 cells transfected with either AS2 or AS2mt, after siRNA-mediated Xrn1 knockdown, showing AS2 full-length and 3′ cleavage fragment. (F) 5′ RACE on RNA from (E) showing 5′ ends of 3′ cleavage fragment and full-length AS2. (G) Clonal sequencing of cleavage fragments obtained from 5′ RACE (***P<0.001).
Accordingly, as determined by northern blot analysis, ectopically expressed AS2 was nearly completely removed by miR-671, but not affected by miR-769 (Figure 5B). Vice versa, substituting the AS2 miR-671 target site with a sequence perfectly complementary to miR-769, led to transcript removal upon miR-769 expression. Thus, the presence of the putative miR-671 target site can account for the observed decrease in CDR1 antisense levels. Therefore, the presence of a perfect or near perfect miRNA target site is sufficient for transcript removal, further indicating that the CDR1 NAS antisense is cleaved and degraded in a miRNA-dependent manner (Figure 5B).
To confirm target cleavage, we investigated whether miR-671-mediated repression of the CDR1 antisense depends on Ago2-slicer activity. Two Ago2-slicer mutants (Ago2-D669A and Ago2-D597A, otherwise functional; Liu et al, 2004), were co-expressed to trans-dominate endogenous Ago2 slicing. This completely blocked the knockdown of the perfect, as well as the antisense target luciferase reporters (Figure 5C; Supplementary Figure S7A), thus confirming slicer-mediated miR-671-dependent cleavage of the luciferase reporters. Concordantly, ectopic expression of the Ago2-slicer mutants rescued the repression of endogenous CDR1 antisense (Figure 5D; Supplementary Figure S7B), suggesting that slicer activity is not only needed in reporter knockdown but also in turnover of the endogenous AS. Hence, we conclude that miR-671 directs Ago2-slicer-dependent cleavage and removal of the CDR1 NAS antisense transcript, which leads to a subsequent destabilization of the CDR1 mRNA.
Finally, 5′ RACE analysis was employed to pinpoint the actual cleavage position in the CDR1 antisense transcript. Initial attempts to detect endogenous cleavage products were unsuccessful, probably due to the unstable nature of cleaved, unprotected RNA. To overcome this problem, the 5′ exoribonuclease XRN1 was transiently knocked down in order to stabilize the cleavage fragments (Supplementary Figure S8). Wild-type and mutant linear NAS antisense mimics (AS2 and AS2mt) were subsequently transiently co-expressed with miR-671 (Figure 5E). Under these conditions, the 3′ cleavage fragment was readily observed by northern blot for the wild-type target site (Figure 5E). Cloning and sequencing of this 5′ RACE product revealed that the antisense transcript is predominantly cleaved between nucleotides 10 and 11 counting from the miRNA 5′ end, which is consistent with typical siRNA-directed target cleavage (Figure 5F and G; Martinez et al, 2002). This proves that sequence-specific miR-671-mediated cleavage regulates homeostasis of the circular CDR1 antisense.
The circular CDR1 NAT is highly expressed in brain
Consistent with previous report of CDR1 being brain specific (Dropcho et al, 1987), expression profiling of the CDR1 NAT across 20 different human tissues revealed high expression in brain and spinal cord tissues (Supplementary Figure S9A, lanes 1, 12, and 19). In fact, quantitative analysis of circular AS in brain tissue revealed a surprisingly high level of expression (Supplementary Figure S9C), indicating not only that it is biologically functional, but also that the circularization itself must be very efficient (Figure 2E). CDR1 mRNA was detectable in human brain tissue as well, but below northern detection limits in HEK293 cells (Supplementary Figure S9B).
A similar circular NAT species was observed by northern blotting total RNA extracted from mouse cerebrum and cerebellum (Supplementary Figure S9D). As in human brain tissue, we found 200-fold higher mouse NAT levels in cerebellum compared with TATA-box Binding Protein (TBP; Supplementary Figure S9E). Hence, the CDR1 NAT likely belongs to one of the highest expressed polymerase II transcript at all in cerebellum. This observation in combination with high conservation of the CDR1 antisense splice motifs and miRNA target site in mammals strongly suggest an evolutionary selection pressure for preserving this unusual regulatory mechanism.
Discussion
There is no general model for antisense regulation of gene expression; however, certain recurring phenomena are emerging. An intrinsic feature of antisense RNA is the obvious ability to form a duplex with the sense mRNA. Although this could be the case for several NATs, duplex formation itself does not necessarily give rise to any particular mRNA phenotype. Rather, NATs probably shield functional mRNA sequences from exerting their function, as reported for ZEB2 (Beltran et al, 2008), protect intrinsically unstable mRNAs, or change mRNA destiny due to the presence of specific antisense motifs (Matsui et al, 2008).
To better understand the relationship between sense and antisense transcripts, the effects of removing or overexpressing one or the other strand must be investigated, but this has only been done in detail for limited sense–antisense pairs (Katayama et al, 2005; Faghihi et al, 2008). Here, we attempted to ectopically express the CDR1 NAT, but due to difficulties in recreating the circularization event from plasmid borne DNA, we used an engineered linear species mimicking the NAS sequence. This led to stabilization of the CDR1 mRNA, consistent with the observations that both miR-671 expression and NAT turnover are associated with reduced CDR1 mRNA levels. Conversely, we also confirmed a dependency of the sense mRNA on antisense levels by sisiRNA-mediated strand-specific knockdown. However, the mechanistic link between miR-671 cleavage of the circular NAT and repression of the sense mRNA is unclear. One hypothesis could be that the abundance of CDR1 NAS antisense may titrate an miRNA from acting on the CDR1 mRNA, that is a sponge model (Poliseno et al, 2010). However, a search for single 7mer putative target seeds sequences, shared between CDR1 3′UTR and the antisense, was negative (based on miRBase ver. 17), suggesting that CDR1 NAS antisense is not acting as a decoy. As the mechanism is epigenetically independent and the CDR1 mRNA can be increased by ectopic AS expression, we argue that the effect is more likely to be post-transcriptional and that the NAT transcript confers CDR1 mRNA stability possibly via direct base pairing as observed previously (Faghihi et al, 2008).
The high prevalence of non-linear splicing suggests that the circular structure is an essential feature of the antisense. This probably renders the NAT resistant towards normal exonucleolytic RNA degradation, which explains the requirement for endonucleolytic Ago2-dependent cleavage for CDR1 NAT turnover. The only well-known previous example of a functional circular RNA species in mammals is the SRY ncRNA, which is required for male sex determination during mouse development, but little is known about the actual molecular mechanisms involved (Capel et al, 1993). Interestingly, a circular RNA arising from the ANRIL antisense transcript of the INK4a-ARF-INK4b locus was recently identified in human cells (Burd et al, 2010). As for CDR1, this circular antisense RNA was also positively correlated with the corresponding mRNA, but no function or mode of regulation was proposed for this species. Rather, the circularization seemed to arise from re-splicing of a lariat structure formed by canonical alternative splicing of ANRIL. It is possible that the CDR1 antisense is circularized in a similar manner, but there is no existing indication that the CDR1 NAS antisense is positioned in an intron of a larger transcript, which argues against this type of mechanism in the non-linear splicing of the CDR1 antisense.
In summary, we report NATs as a novel bona fide miRNA target class by demonstrating that the predominantly nuclear miR-671 targets and cleaves an antisense transcript of the CDR1 locus, leading to a concomitant decrease in steady-state CDR1 mRNA levels. As such, we also provide evidence for a novel regulatory mechanism, in which miRNA-dependent regulation of antisense transcripts is directly coupled to a decrease in mRNA expression. Moreover, the CDR1 NAT predominantly engages in a cis-acting NAS event, producing a highly abundant, circularized species and it likely serves as a component in sense mRNA stabilization. To date, no naturally prevalent and functional NAS or circular RNA has been reported in human cells. Additionally, the NAS-specific splice sites and the miR-671 target site are conserved between mouse and human, strongly suggesting that the circularization event and miR-671-mediated CDR1 repression are components of a novel, biologically relevant, and evolutionarily conserved regulatory mechanism.
Materials and methods
Gene expression analyses
Total RNA was extracted from cells in culture using TRIzol® reagent (Invitrogen) according to the standard procedures. cDNA for qRT–PCR expression analyses was synthesized from total RNA by SuperScriptIII® (Invitrogen) according to the supplied protocol using random hexamer primer or a gene-specific primer cocktail for strand-specific RT (CDR1 ssRT, GAPDH RT RE, and 7SK RT RE). qPCR was performed either with Platinum® SYBR® Green qPCR Supermix UDG (Invitrogen) on a MxPro3000 cycler (Stratagene, Palo Alto, CA) or with SYBR® Green PCR Master Mix (Applied Biosystems) on a LightCycler® 480 (Roche) according to the standard procedures. mRNA levels were normalized to GAPDH (SYBR) or 18S rRNA (TaqMan® assay, Applied Biosystems) (2−ΔCt). CDR1 ssRT FW/CDR1 ssRT RE primers were used for strand-specific CDR1 mRNA quantification whereas NAS-specific antisense levels were measured by CDR1(8)FW/CDR1(10)RE. qRT–PCR on RNA from mouse tissue was performed using mmuNAS FW/mmuNAS RE to amplify antisense RNA and TBP FW/TBP RE for normalization.
Subcellular fractionation and northern blot
Subcellular fractionation was performed as described by Hwang et al. In all, 13 μg RNA from the nucleoplasmic fraction, 20 μg from the cytoplasmic fraction, or 30 μg whole cell RNA was loaded onto a 12% denaturing PAGE and transferred onto an Amersham hybond™-N+ membrane (GE Healthcare). The membrane was hybridized with 32P-labelled DNA oligos (listed in Supplementary Table 1) in church buffer (0.5 M NaPO4, 7% SDS, 1 mM EDTA, 1% BSA, pH 7.5) at 37°C and washed in SSC buffer (2 × SSC, 0.1% SDS) at room temperature. The membranes were exposed on phosphorimager screens and analysed using Bio-Rad Quantity One® software (Bio-Rad, Hercules, CA). Agarose northern blots were performed with 0.5–10 μg RNA separated in 1.2% agarose. Subsequent hybridization and wash were carried out at 50°C, and otherwise conducted as described above. Human total RNA master panel II (Clontech, Mountain View, CA) was used for tissue atlas northern blots.
TAP and EXO treatment
HEK293 total RNA (5 μg) was incubated with or without 10 U TAP (Epicentre Biotechnologies, Madison, WI) according to the manufacturer's protocol. RNA was subsequently incubated with 10 U Terminator™ 5′-phosphate-dependent Exonuclease (EXO) (Epicentre Biotechnologies) according to the supplied protocol, purified and used directly in qRT–PCR.
Circular RNA gel trap
In all, 10 μg total RNA was incubated with 2% low-melting agarose at 65°C for 5 min. The liquid agarose–RNA mixture was loaded directly onto a solidified 2% agarose gel and subjected to electrophoresis (5 V/cm for 3 h). As a control, the agarose–RNA mixture was allowed to solidify without subsequent electrophoresis. Trapped and control RNA were recovered by standard phenol–chloroform extraction and ethanol precipitation. Half the output was used in RNA quantification as described above. CDR1 antisense levels were assessed by qRT–PCR, normalized to GAPDH and quantified as –fold enrichment relative to total input RNA.
sisiRNA transfection
sisiRNAs specifically targeting the CDR1 mRNA or the AS transcript were designed as described in Bramsen et al (2007) (Supplementary Table 1, LNA-modified nucleotides underlined, RiboTask, Odense, Denmark). For sisiRNA annealing, the SS 5′, SS 3′, and AS oligos were mixed in 1 × Dharmacon annealing buffer (Dharmacon, Lafayette, CO) and incubated at 95°C for 1 min followed by 1 h at 37°C. sisiRNA concentration was quantified using the Quant-iT™ RiboGreen® RNA Assay Kit (Invitrogen) on FLUOstar luminometer (BMG Labtech). siBCR/ABL-1 (Howard et al, 2006) was used as a negative control. HEK293 cells were transfected with 20 nM sisiRNA and Lipofectamine™ 2000 (Invitrogen) according to the manufacturer's protocol. Growth medium was added 4 h after transfection and total RNA was harvested at 48 h and quantified as described above.
Supplementary Material
Acknowledgments
We thank Mr Claus Bus and Mrs Rita Rosendahl for technical assistance and Dr Anne F Nielsen for critical review of the manuscript. We additionally thank Greg Hannon Lab for providing us with Ago2-wt and -slicer mutant expression vectors and Jens Lykke-Andersen for providing us with anti-Xrn1 and anti-Upf1 antibodies. This work was supported by the SIROCCO EU consortium, the Danish Council for Independent Research∣Natural Sciences, and the Australian National Health and Medical Research Council (NH&MRC) and National Breast Cancer Foundation (NBCF). TBH and EDW were supported by the Carlsberg foundation.
Author contributions:TBH and EDW conceived, designed and performed the experiments, analysed the data, and wrote the manuscripts. JBB and SBV aided in manuscript preparation and performed additional experiments. ALS performed and analysed microarray data. SJC and JK supervised the project.
Footnotes
The authors declare that they have no conflict of interest.
References
- Beltran M, Puig I, Pena C, Garcia JM, Alvarez AB, Pena R, Bonilla F, de Herreros AG (2008) A natural antisense transcript regulates Zeb2/Sip1 gene expression during Snail1-induced epithelial-mesenchymal transition. Genes Dev 22: 756–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bramsen JB, Laursen MB, Damgaard CK, Lena SW, Babu BR, Wengel J, Kjems J (2007) Improved silencing properties using small internally segmented interfering RNAs. Nucleic Acids Res 35: 5886–5897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burd CE, Jeck WR, Liu Y, Sanoff HK, Wang Z, Sharpless NE (2010) Expression of linear and novel circular forms of an INK4/ARF-associated non-coding RNA correlates with atherosclerosis risk. PLoS Genet 6: e1001233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capel B, Swain A, Nicolis S, Hacker A, Walter M, Koopman P, Goodfellow P, Lovell-Badge R (1993) Circular transcripts of the testis-determining gene Sry in adult mouse testis. Cell 73: 1019–1030 [DOI] [PubMed] [Google Scholar]
- Chendrimada TP, Finn KJ, Ji X, Baillat D, Gregory RI, Liebhaber SA, Pasquinelli AE, Shiekhattar R (2007) MicroRNA silencing through RISC recruitment of eIF6. Nature 447: 823–828 [DOI] [PubMed] [Google Scholar]
- Crampton N, Bonass WA, Kirkham J, Rivetti C, Thomson NH (2006) Collision events between RNA polymerases in convergent transcription studied by atomic force microscopy. Nucleic Acids Res 34: 5416–5425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dropcho EJ, Chen YT, Posner JB, Old LJ (1987) Cloning of a brain protein identified by autoantibodies from a patient with paraneoplastic cerebellar degeneration. Proc Natl Acad Sci USA 84: 4552–4556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebralidze AK, Guibal FC, Steidl U, Zhang P, Lee S, Bartholdy B, Jorda MA, Petkova V, Rosenbauer F, Huang G, Dayaram T, Klupp J, O'Brien KB, Will B, Hoogenkamp M, Borden KL, Bonifer C, Tenen DG (2008) PU.1 expression is modulated by the balance of functional sense and antisense RNAs regulated by a shared cis-regulatory element. Genes Dev 22: 2085–2092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faghihi MA, Modarresi F, Khalil AM, Wood DE, Sahagan BG, Morgan TE, Finch CE, St Laurent G III, Kenny PJ, Wahlestedt C (2008) Expression of a noncoding RNA is elevated in Alzheimer's disease and drives rapid feed-forward regulation of beta-secretase. Nat Med 14: 723–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forman JJ, Legesse-Miller A, Coller HA (2008) A search for conserved sequences in coding regions reveals that the let-7 microRNA targets Dicer within its coding sequence. Proc Natl Acad Sci USA 105: 14879–14884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furneaux HM, Dropcho EJ, Barbut D, Chen YT, Rosenblum MK, Old LJ, Posner JB (1989) Characterization of a cDNA encoding a 34-kDa Purkinje neuron protein recognized by sera from patients with paraneoplastic cerebellar degeneration. Proc Natl Acad Sci USA 86: 2873–2877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingeras TR (2009) Implications of chimaeric non-co-linear transcripts. Nature 461: 206–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez S, Pisano DG, Serrano M (2008) Mechanistic principles of chromatin remodeling guided by siRNAs and miRNAs. Cell Cycle 7: 2601–2608 [DOI] [PubMed] [Google Scholar]
- Guil S, Esteller M (2009) DNA methylomes, histone codes and miRNAs: tying it all together. Int J Biochem Cell Biol 41: 87–95 [DOI] [PubMed] [Google Scholar]
- Han J, Kim D, Morris KV (2007) Promoter-associated RNA is required for RNA-directed transcriptional gene silencing in human cells. Proc Natl Acad Sci USA 104: 12422–12427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins PG, Santoso S, Adams C, Anest V, Morris KV (2009) Promoter targeted small RNAs induce long-term transcriptional gene silencing in human cells. Nucleic Acids Res 37: 2984–2995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard KA, Rahbek UL, Liu X, Damgaard CK, Glud SZ, Andersen MO, Hovgaard MB, Schmitz A, Nyengaard JR, Besenbacher F, Kjems J (2006) RNA interference in vitro and in vivo using a novel chitosan/siRNA nanoparticle system. Mol Ther 14: 476–484 [DOI] [PubMed] [Google Scholar]
- Humphreys DT, Westman BJ, Martin DI, Preiss T (2005) MicroRNAs control translation initiation by inhibiting eukaryotic initiation factor 4E/cap and poly(A) tail function. Proc Natl Acad Sci USA 102: 16961–16966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutvagner G, Zamore PD (2002) A microRNA in a multiple-turnover RNAi enzyme complex. Science 297: 2056–2060 [DOI] [PubMed] [Google Scholar]
- Hwang HW, Wentzel EA, Mendell JT (2007) A hexanucleotide element directs microRNA nuclear import. Science 315: 97–100 [DOI] [PubMed] [Google Scholar]
- Janowski BA, Huffman KE, Schwartz JC, Ram R, Nordsell R, Shames DS, Minna JD, Corey DR (2006) Involvement of AGO1 and AGO2 in mammalian transcriptional silencing. Nat Struct Mol Biol 13: 787–792 [DOI] [PubMed] [Google Scholar]
- Katayama S, Tomaru Y, Kasukawa T, Waki K, Nakanishi M, Nakamura M, Nishida H, Yap CC, Suzuki M, Kawai J, Suzuki H, Carninci P, Hayashizaki Y, Wells C, Frith M, Ravasi T, Pang KC, Hallinan J, Mattick J, Hume DA et al. (2005) Antisense transcription in the mammalian transcriptome. Science 309: 1564–1566 [DOI] [PubMed] [Google Scholar]
- Kim DH, Saetrom P, Snove O Jr, Rossi JJ (2008) MicroRNA-directed transcriptional gene silencing in mammalian cells. Proc Natl Acad Sci USA 105: 16230–16235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DH, Villeneuve LM, Morris KV, Rossi JJ (2006) Argonaute-1 directs siRNA-mediated transcriptional gene silencing in human cells. Nat Struct Mol Biol 13: 793–797 [DOI] [PubMed] [Google Scholar]
- Langlois MA, Boniface C, Wang G, Alluin J, Salvaterra PM, Puymirat J, Rossi JJ, Lee NS (2005) Cytoplasmic and nuclear retained DMPK mRNAs are targets for RNA interference in myotonic dystrophy cells. J Biol Chem 280: 16949–16954 [DOI] [PubMed] [Google Scholar]
- Liao JY, Ma LM, Guo YH, Zhang YC, Zhou H, Shao P, Chen YQ, Qu LH (2010) Deep sequencing of human nuclear and cytoplasmic small RNAs reveals an unexpectedly complex subcellular distribution of miRNAs and tRNA 3′ trailers. PLoS One 5: e10563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song JJ, Hammond SM, Joshua-Tor L, Hannon GJ (2004) Argonaute2 is the catalytic engine of mammalian RNAi. Science 305: 1437–1441 [DOI] [PubMed] [Google Scholar]
- Martinez J, Patkaniowska A, Urlaub H, Luhrmann R, Tuschl T (2002) Single-stranded antisense siRNAs guide target RNA cleavage in RNAi. Cell 110: 563–574 [DOI] [PubMed] [Google Scholar]
- Matsui K, Nishizawa M, Ozaki T, Kimura T, Hashimoto I, Yamada M, Kaibori M, Kamiyama Y, Ito S, Okumura T (2008) Natural antisense transcript stabilizes inducible nitric oxide synthase messenger RNA in rat hepatocytes. Hepatology 47: 686–697 [DOI] [PubMed] [Google Scholar]
- Mayr C, Bartel DP (2009) Widespread shortening of 3′UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell 138: 673–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris KV, Chan SW, Jacobsen SE, Looney DJ (2004) Small interfering RNA-induced transcriptional gene silencing in human cells. Science 305: 1289–1292 [DOI] [PubMed] [Google Scholar]
- Morris KV, Santoso S, Turner AM, Pastori C, Hawkins PG (2008) Bidirectional transcription directs both transcriptional gene activation and suppression in human cells. PLoS Genet 4: e1000258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napoli S, Pastori C, Magistri M, Carbone GM, Catapano CV (2009) Promoter-specific transcriptional interference and c-myc gene silencing by siRNAs in human cells. EMBO J 28: 1708–1719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orom UA, Nielsen FC, Lund AH (2008) MicroRNA-10a binds the 5′UTR of ribosomal protein mRNAs and enhances their translation. Mol Cell 30: 460–471 [DOI] [PubMed] [Google Scholar]
- Petersen CP, Bordeleau ME, Pelletier J, Sharp PA (2006) Short RNAs repress translation after initiation in mammalian cells. Mol Cell 21: 533–542 [DOI] [PubMed] [Google Scholar]
- Pillai RS, Bhattacharyya SN, Artus CG, Zoller T, Cougot N, Basyuk E, Bertrand E, Filipowicz W (2005) Inhibition of translational initiation by Let-7 MicroRNA in human cells. Science 309: 1573–1576 [DOI] [PubMed] [Google Scholar]
- Poliseno L, Salmena L, Zhang J, Carver B, Haveman WJ, Pandolfi PP (2010) A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature 465: 1033–1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robb GB, Brown KM, Khurana J, Rana TM (2005) Specific and potent RNAi in the nucleus of human cells. Nat Struct Mol Biol 12: 133–137 [DOI] [PubMed] [Google Scholar]
- Schindler CW, Krolewski JJ, Rush MG (1982) Selective trapping of circular double-stranded DNA molecules in solidifying agarose. Plasmid 7: 263–270 [DOI] [PubMed] [Google Scholar]
- Schmid M, Jensen TH (2008) The exosome: a multipurpose RNA-decay machine. Trends Biochem Sci 33: 501–510 [DOI] [PubMed] [Google Scholar]
- Schwartz JC, Younger ST, Nguyen NB, Hardy DB, Monia BP, Corey DR, Janowski BA (2008) Antisense transcripts are targets for activating small RNAs. Nat Struct Mol Biol 15: 842–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K, Shijuuku T, Fukamachi T, Zaunders J, Guillemin G, Cooper D, Kelleher A (2005) Prolonged transcriptional silencing and CpG methylation induced by siRNAs targeted to the HIV-1 promoter region. J RNAi Gene Silencing 1: 66–78 [PMC free article] [PubMed] [Google Scholar]
- Svoboda P, Stein P, Filipowicz W, Schultz RM (2004) Lack of homologous sequence-specific DNA methylation in response to stable dsRNA expression in mouse oocytes. Nucleic Acids Res 32: 3601–3606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tay Y, Zhang J, Thomson AM, Lim B, Rigoutsos I (2008) MicroRNAs to Nanog, Oct4 and Sox2 coding regions modulate embryonic stem cell differentiation. Nature 455: 1124–1128 [DOI] [PubMed] [Google Scholar]
- Ting AH, Schuebel KE, Herman JG, Baylin SB (2005) Short double-stranded RNA induces transcriptional gene silencing in human cancer cells in the absence of DNA methylation. Nat Genet 37: 906–910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdel A, Vavasseur A, Le Gorrec M, Touat-Todeschini L (2009) Common themes in siRNA-mediated epigenetic silencing pathways. Int J Dev Biol 53: 245–257 [DOI] [PubMed] [Google Scholar]
- Weinberg MS, Villeneuve LM, Ehsani A, Amarzguioui M, Aagaard L, Chen ZX, Riggs AD, Rossi JJ, Morris KV (2006) The antisense strand of small interfering RNAs directs histone methylation and transcriptional gene silencing in human cells. RNA 12: 256–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap KL, Li S, Munoz-Cabello AM, Raguz S, Zeng L, Mujtaba S, Gil J, Walsh MJ, Zhou MM (2010) Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Mol Cell 38: 662–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yekta S, Shih IH, Bartel DP (2004) MicroRNA-directed cleavage of HOXB8 mRNA. Science 304: 594–6 [DOI] [PubMed] [Google Scholar]
- Yu W, Gius D, Onyango P, Muldoon-Jacobs K, Karp J, Feinberg AP, Cui H (2008) Epigenetic silencing of tumour suppressor gene p15 by its antisense RNA. Nature 451: 202–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.