Abstract
Mitochondria are organelles with a complex architecture. They are bounded by an envelope consisting of the outer membrane and the inner boundary membrane (IBM). Narrow crista junctions (CJs) link the IBM to the cristae. OMs and IBMs are firmly connected by contact sites (CS). The molecular nature of the CS remained unknown. Using quantitative high-resolution mass spectrometry we identified a novel complex, the mitochondrial contact site (MICOS) complex, formed by a set of mitochondrial membrane proteins that is essential for the formation of CS. MICOS is preferentially located at the CJs. Upon loss of one of the MICOS subunits, CJs disappear completely or are impaired, showing that CJs require the presence of CS to form a superstructure that links the IBM to the cristae. Loss of MICOS subunits results in loss of respiratory competence and altered inheritance of mitochondrial DNA.
Keywords: contact site proteins, crista junction, MICOS, mitochondrial membrane proteome, molecular architecture of mitochondria
Introduction
The ability to determine the relationship between the molecular architecture of proteins and the functions of proteins has been key to progress in cell biology to a large extent. The relationship between the molecular architecture of cell organelles, the next higher level of cellular organization, and their function is much less understood. Organelles are composed of membranes and it is much more difficult to link the structure of membrane proteins and of lipids to a specific architecture. This is particularly challenging in the case of mitochondria because these organelles have a unique and quite complex membrane system, which is the basis for their numerous intricate functions (Scheffler, 2011). Mitochondria catalyse a plethora of metabolic reactions, in particular transducing energy by oxidative phosphorylation. Mitochondria replicate and inherit the mitochondrial genome and synthesize proteins and lipids. They are involved in apoptosis, cellular ageing and in a large number of diseases (Pellegrini and Scorrano, 2007; Wallace and Fan, 2009; Larsson, 2010). Mitochondria are dynamic organelles that have the ability to continuously divide and fuse (Griparic et al, 2004; Okamoto and Shaw, 2005; Hoppins and Nunnari, 2009). They move in the cell by association with the cytoskeleton (Boldogh and Pon, 2006; Chan et al, 2006; Westermann, 2010). They import proteins from the cytosol and lipids from the endoplasmic reticulum (ER).
The most prominent architectural elements of the mitochondria are their membranes (Frey and Mannella, 2000; Reichert and Neupert, 2002; Perkins et al, 2004; Mannella, 2006, 2008). Mitochondria are bounded by an envelope, which is comprised of the outer membrane (OM) and inner boundary membrane (IBM), two membranes of completely different composition and properties. The cristae protrude from the IBM into the inner space of the mitochondria, the matrix. The cristae and the IBM together make up the inner membrane (IM). The structure of the cristae, their arrangement in the matrix, their number and surface area are highly diverse in different types of cells, tissues and organisms (Fawcett, 1981). The connections between IBM and the cristae are the crista junctions (CJ). These are small, usually very short tubule or slot-like structures. The space between the OM and the IBM, the intermembrane space (IMS), is rather narrow in comparison to the space between cristae sheets, also called intracrista space. Furthermore, there are sites of firm interaction between OM and IBM that become apparent when isolated mitochondria are in a low-energy state or exposed to hypertonic medium. Under these conditions, the matrix condenses and the IBM retracts from the OM (Hackenbrock, 1968). These connections have been termed mitochondrial contact sites (CS), yet their molecular nature has remained elusive.
It has been discussed that these various membranes are shaped essentially by the lipid components (Renken et al, 2002). However, mutations have been described in which the overall shape of mitochondria as observed by light microscopy is altered (Shaw and Nunnari, 2002; Okamoto and Shaw, 2005). More recently, and important in the present context, mutations affecting mitochondrial ultrastructure as revealed by electron microscopy (EM) were reported (Dimmer et al, 2002; Meeusen et al, 2006; Tamai et al, 2008; Rabl et al, 2009). In most cases, the phenotype observed was loss of mitochondrial DNA and of respiratory competence. Thus, it seems reasonable to assume that there are groups of proteins that determine the various structural elements of mitochondria.
Here, we report on a search for proteins that determine CS by which the IBM and OM are attached to each other. We reasoned that it should be possible to isolate a fraction of the mitochondrial membranes that contains CS. To identify CS, we generated a novel marker by expressing in yeast cells a fusion protein that permanently spans both outer and IBM. We subfractionated mitochondria, separated vesicles and analysed the fractions by protein correlation profiling, a mass spectrometry-based organellar proteomics technique (Andersen et al, 2003; Foster et al, 2006; Dengjel et al, 2010). In this approach, we used an SILAC standard (Ong et al, 2002) to identify proteins with the same distribution as the marker. Resulting protein candidates were characterized in terms of their submitochondrial location, topology, importance for cell growth and mitochondrial respiratory competence. Six proteins were found that form a mitochondrial contact site (MICOS) complex. They are associated with the IM and interact with the OM by binding to the TOB/SAM complex and the Ugo1 protein. The MICOS complex is critically involved in the formation of CJs and cristae, as well as in several important functions of the mitochondria. We conclude that the MICOS complex represents the long searched molecular scaffold of mitochondrial contact sites.
Results
Identification of protein candidates of mitochondrial contact sites
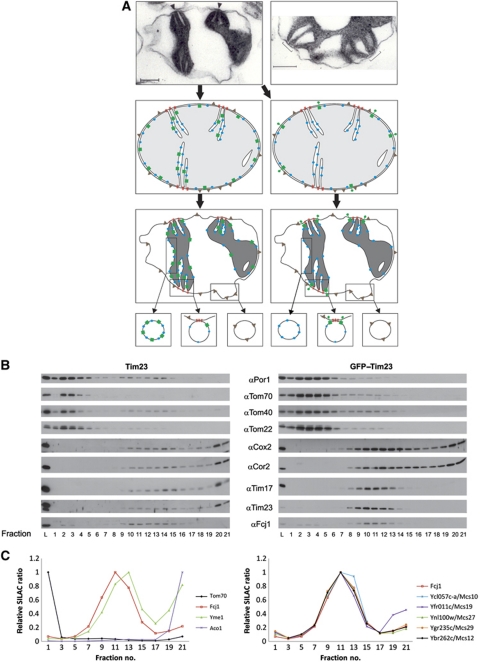
Examination of mitochondria by EM in their cellular environment yields images in which the OM and IBM are closely apposed to each other (‘orthodox state’). CS cannot be distinguished under these conditions. When mitochondria are isolated and subjected to hyperosmotic treatment, the IBM retracts, the matrix shrinks and the mitochondria assume a ‘condensed state’ (Hackenbrock, 1968). The IBM remains in close contact with the OM, however, only at few sites (Figure 1A, upper left panel). The gaps at these sites are filled with stain, indicating the presence of proteins (Figure 1A, upper right panel). Importantly, we observed that these contacts in most cases are present right next to where cristae merge with the IBM, at the CS. This is illustrated in a drawing in which an orthodox mitochondrion is reconstructed from a condensed mitochondrion (Figure 1A, middle panels). Taken together, these observations led us to hypothesize that CS forming proteins are present in these areas.
Figure 1.
Identification of Mcs proteins. (A) Rationale for the analysis of distribution of membrane proteins in mitochondria. (Upper panels) Electron micrographs of sections of mitochondria from Neurospora crassa subjected to osmotic shrinking (‘condensed configuration’). Size bars, 200 nm. Arrowheads show regions of attachment between IM and OM (contact sites, CS); brackets indicate CS at higher magnification where two cristae invaginate in close neighbourhood. (Middle panels) ‘Orthodox configuration’ was reconstructed from the electron micrograph in the upper panel (see arrows). Left, mitochondria from wild type, expressing untagged Tim23; right, mitochondria from cells expressing GFP–Tim23. (Lower panels) Distribution of proteins indicated above after shrinking of mitochondria and theoretically generated vesicles. Symbols: red lines, putative proteins of CS; blue balls, IM proteins; green squares, Tim23; green dumbbells, GFP–Tim23; grey triangles, OM proteins. (B) Separation of vesicles of mitochondria from Tim23 and GFP–Tim23 expressing cells (left and right panels, respectively) by flotation equilibrium gradient centrifugation. Fractions were analysed by SDS–PAGE and immunodecorated with the indicated antibodies. L, load fraction. Odd numbered fractions of the gradients were analysed. (C) Analysis of mitochondrial fractions prepared as in (B) by quantitative mass spectrometry (SILAC). Distribution of marker proteins and of proteins qualifying as components of CS. (Left panel) Profiles of examples of OM proteins, IM proteins, matrix proteins and Fcj1. (Right panel) Profiles of six proteins that qualify as Mcs proteins. See also Supplementary Figure S1 for further controls.
On the basis of this reasoning, we devised an approach in which the proteins that are forming the CS could be identified. We designed a marker protein for these sites. Previous attempts to identify components of CS led to successful subfractionation of mitochondria but the absence of a marker prevented the identification of mitochondrial contact site (Mcs) proteins (Werner and Neupert, 1972; Pon et al, 1989). As a prerequisite, the marker must be permanently spanning OM and IBM. The design of this protein was based on our work on the structure and topology of yeast Tim23, an essential component of the protein conducting TIM23 translocase (Donzeau et al, 2000). We generated a fusion protein of GFP with Tim23, which locks Tim23 in the desired position, with the GFP domain being present on the mitochondrial surface (Harner et al, 2011). GFP–Tim23 expressed in cells lacking wild-type Tim23 has the intriguing property of complementing the function of the TIM23 translocase (Vogel et al, 2006). Figure 1A (middle panel) depicts the distribution of proteins of OM and IM, as well as of Tim23 in wild-type cells (left side, green squares) and of GFP–Tim23 in cells expressing this marker (right side, green dumbbells). The predicted different behaviour of these two proteins upon condensation of mitochondria will result in accumulation of GFP–Tim23 in CS, but not of Tim23. Therefore, vesicle fractions containing CS (Figure 1A, lower panels) will contain GFP–Tim23 as marker protein for the Mcs proteins.
Subfractionation of mitochondria and analysis of protein components by immunoblotting. To verify this concept, we subjected mitochondria to mild sonication and separation of fragments by density gradient flotation centrifugation. In a first set of experiments, we analysed the gradient fractions with antibodies against the designed marker and a number of OM and IM proteins. We compared mitochondria from wild-type cells and from cells expressing GFP–Tim23 instead of Tim23 (Figure 1B, left versus right panel). IM proteins were recovered at the bottom of the gradient and to a lower degree in the middle of the gradient of both wild-type and GFP–Tim23 expressing cells. Wild-type Tim23 was distributed in a similar manner as other IM proteins. In contrast, in mitochondria containing GFP–Tim23, this fusion protein was present exclusively in the middle of the gradient, being virtually absent in the bottom fractions. The submitochondrial location of intermediates of precursor proteins stalled upon import into isolated mitochondria was analysed previously by fractionation of the mitochondria and sucrose gradient centrifugation. The majority of the intermediates were recovered in fractions of intermediate density, in agreement with the localization of the TIM23 import machinery in these fractions shown here (Pon et al, 1989). OM proteins such as VDAC/porin and subunits of the TOM complex were present in the top fractions of the gradient and to a low degree in the fractions containing the GFP–Tim23 marker in both types of cells. Interestingly, Fcj1, a protein with an essential role in the formation of CJ (Rabl et al, 2009) showed a very similar distribution as GFP–Tim23. We concluded that we were able to resolve the CS containing vesicles and have identified a first component of this fraction.
Proteome of the CS fraction by quantitative mass spectrometry. In a further experiment, we used quantitative mass spectrometry (SILAC) to identify proteins specifically enriched in the CS fraction using protein correlation profiling via an internal SILAC standard. Cells were grown in media containing 12C and 14N (‘light’) or 13C and 15N (‘heavy’) lysine. Mitochondria were isolated, fragmented and subjected to gradient centrifugation as in Figure 1B. Gradient fractions 10–15 from the ‘heavy’ gradient were pooled and served as a metabolically labelled internal standard for SILAC-based protein quantitation. Individual fractions (the odd numbered ones of a total of 21) of the ‘light’ gradient were mixed with aliquots of this internal standard, digested with endoproteinase LysC to peptides and analysed by high-resolution mass spectrometry. Proteins were identified and quantified in each of the fractions based on their isotope ratios using the MaxQuant framework (Cox and Mann, 2008). Proteins of the various mitochondrial subcompartments showed characteristic profiles (Figure 1C; see also Supplementary Figure S1 and Supplementary Dataset File F1). IM proteins peaked in fraction 13 with a high isotope ratio in the bottom fraction. OM proteins, including subunits of the TOM complex, were prominent in the top fraction, and present to a minor extent in the middle of the gradient. Soluble matrix proteins were only present in the bottom fractions. Apparently, soluble proteins remained in IM vesicles or were released from the vesicles when experiencing the high osmotic pressure of the bottom fractions of the gradient. The procedure was highly reproducible and precise. The profiles of the same protein in two different mass spectrometry experiments were virtually identical. The same was true when comparing two different subunits of one complex (Supplementary Figure S1). The mitochondrial preparation in these experiments was intentionally not optimized for purity in order not to damage the architecture of mitochondria during isolation. Proteins of the ER originating either from contamination or from association of ER with the mitochondria showed a well-defined profile that can be easily discriminated from the profiles of mitochondrial proteins (Supplementary Figure S1).
Importantly, in contrast to the profile of IM proteins, Fcj1 showed a peak of isotope ratio in fraction 11 and a rather low ratio in the bottom fractions. We examined the ∼350 mitochondrial membrane proteins, which were detected in the SILAC analysis for having a profile like Fcj1 and thereby GFP–Tim23. This screen yielded a list of five proteins whose gradient profiles are shown in Figure 1C, right panel. These proteins are present in the Saccharomyces cerevisiae genome database (http://www.yeastgenome.org), where they are listed as mitochondrial proteins of unknown function. Three of them are present in a collection of proteins, resulting from a screen for altered inheritance of mitochondrial DNA (AIM) (Hess et al, 2009), which comprises a total of 46 entries.
We conclude that we have identified a set of six proteins, which, according to their co-fractionation with the marker protein, qualify as components of the IBM/CS. From now on, we address these proteins as Mcs proteins and by their calculated molecular mass in kDa. These proteins are Mcs29 (Ygr235c), Mcs27 (Ynl100w, AIM37), Mcs19 (Yfr011c, AIM13), Mcs12 (Ybr262c, AIM5), Mcs10 (Ycl057c-a) and Fcj1 (Ykr016w, AIM28).
The MICOS complex
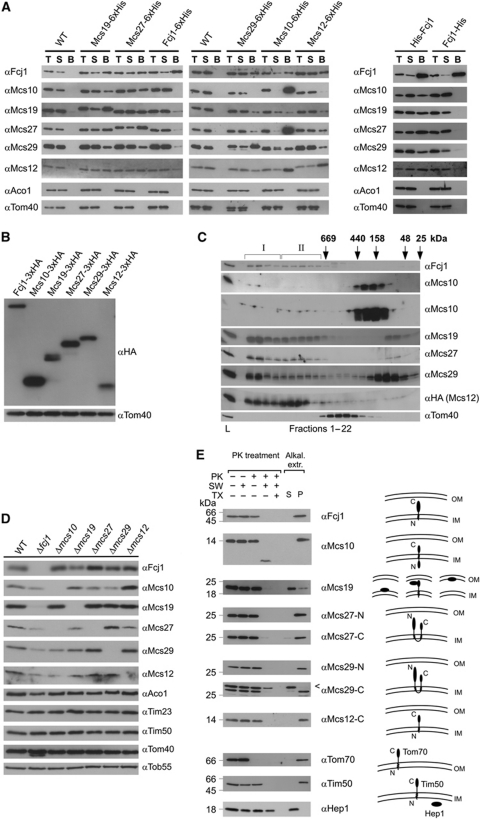
Co-isolation of Mcs proteins. To investigate whether the proteins identified interact with each other, we performed both co-isolation and molecular sizing experiments. A summary of the interactions observed upon co-isolation is presented in Figure 2A. All six proteins showed interactions between each other but not with any of the other mitochondrial proteins tested. Interestingly, Fcj1 could be co-isolated together with all five C-terminally His-tagged Mcs proteins. However, pull down of C-terminally tagged Fcj1 led to co-isolation of only trace amounts of Mcs proteins (Figure 2A, left panel). This was surprising, in particular since C-terminally tagged Fcj1 was able to rescue the deletion of Fcj1. Therefore, we suspected that the presence of the tag leads to a weakened interaction of Fcj1 with other Mcs proteins. We then expressed N- or C-terminally tagged Fcj1 versions from a plasmid in an Fcj1 deletion strain. Indeed, all the Mcs proteins were co-isolated only when the tag was present at the N-terminus (Figure 2A, right panel). This observation indicates the importance of the conserved C-terminal domain of Fcj1 for interaction with the other complex components. Altogether, these results demonstrate the existence of a complex network of physical interactions between the various Mcs proteins. Whether the intensities observed in this assay are a reflection of the strength of binding or are influenced by the experimental conditions is not clear.
Figure 2.
The MICOS complex. (A) Co-isolation of Mcs proteins. His-tagged versions of the Mcs proteins were expressed under control of their own promoters (left panel) or from the pYX242 plasmid (right panel) in the respective deletion strain. Mitochondria were isolated, solubilized with digitonin and incubated with Ni-NTA beads. Total (T, 5% of total), supernatant (S, 5% of total) and bound material (B, 100%) were analysed by SDS–PAGE and immunodecoration with the indicated antibodies. Mitochondria from wild-type cells served as control. (B) Relative abundance of Mcs proteins. HA-tagged versions of the Mcs proteins were expressed under their own promoters. Equal amounts of mitochondria were analysed by SDS–PAGE and immunoblotting with antibodies against the HA-tag (αHA) and against Tom40 (αTom40), the loading control. (C) Molecular sizing of Mcs proteins from wild-type mitochondria. Mitochondria were lysed with digitonin and lysates subjected to gel filtration on a Superose 6 column. The fractions were analysed by SDS–PAGE and immunoblotting using the indicated antibodies; in case of Msc12, a strain was used which expressed HA-tagged Msc12 and immunoblotting was with αHA antibody. Msc10 blots were exposed for two different time periods. The TOM complex (Tom40) was decorated as a control. I and II, MICOS complex I and II. Positions of marker proteins for calibration are indicated with arrows. L, load (10% of material applied to column). See also Supplementary Figure S2A. (D) Steady-state levels of Mcs proteins in cells in which one of the MCS genes was deleted. Mitochondria were analysed by SDS–PAGE and immunoblotting. See also Supplementary Figure S2B. (E) Membrane integration and orientation of Mcs proteins. (Left) Mitochondria from wild-type cells were left untreated or treated with proteinase K (PK) either directly, after subjecting them to osmotic swelling (SW) or after lysis with Triton X-100 (TX); bars indicate the apparent molecular masses of the full-length proteins. (Right) Mitochondria were exposed to alkaline extraction at pH 12. Soluble (S) and membrane integrated (P, pellet) material were separated by centrifugation. Aliquots were subjected to SDS–PAGE and immunodecoration with antibodies against the indicated proteins. Arrowhead, unspecific cross-reaction.
Relative abundance of Mcs proteins in mitochondria. We checked the relative abundance of the Mcs proteins by tagging them with a C-terminal HA-tag and expressing them under their endogenous promoters from the chromosome. Isolated mitochondria were analysed by SDS–PAGE (polyacrylamide gel electrophoresis) and immunoblotting with an antibody against the HA-tag. The experimental approach used obviously provides only a rough estimate of the abundance due to the limitations of the method. Yet, it appears that Fcj1, Mcs12, Mcs19 and Mcs29 are of roughly equal abundance, whereas Mcs27 and, in particular, Mcs10 are present at higher levels (Figure 2B).
Complexes formed by the Mcs proteins. In order to analyse complexes formed by the Mcs proteins, we subjected mitochondria to lysis with digitonin followed by gel filtration (Figure 2C). All of the Mcs proteins were present in two large complexes of ∼1.5 and 0.7 MDa apparent molecular mass, which we termed MICOS I and MICOS II, respectively. Some of the Mcs proteins, however, were not completely or only partly recovered in MICOS I and II. In particular, very little of Mcs10 was found with the large complexes, but rather in fractions of about 200 kDa. These species probably contain oligomers of Mcs10 since untagged Mcs10 could be co-isolated with tagged Mcs10 when expressed together (not shown). Mcs29 was recovered only to a lower extent in MICOS I and II; however, more was present in the low molecular mass range. Finally, Mcs12 was present to a higher extent in MICOS II than in MICOS I. The TOM complex that served as a control eluted with an apparent mass of 400–500 kDa as to be expected (Kunkele et al, 1998). To test whether the absence of the majority of Mcs10 from the large complexes was due to a minor contribution to MICOS I and II or whether Mcs10 easily dissociates from the other Mcs proteins upon solubilization of mitochondria, we performed Blue native (BN)-PAGE. Immunoblotting with antibodies against Fcj1, Mcs27 or Mcs29 revealed two high molecular mass complexes of roughly 1.5 and 0.7 MDa. The sizes of these two complexes matched very well with those obtained upon gel filtration. The antibody against Mcs10 did not recognize its antigen upon BN-PAGE; therefore, we analysed mitochondria harbouring his-tagged Mcs10. Immunoblotting using antibody against the His-tag (penta-His; Qiagen) revealed again two high molecular complexes with similar sizes as observed upon immunodecoration with the other Mcs proteins (Supplementary Figure S2A). This indicates that Mcs10 has the tendency to dissociate from the complex after detergent solubilization of mitochondria and gel filtration.
Steady-state levels of Mcs proteins in deletion strains. We further asked as to whether deletion of one of the Mcs proteins would affect the steady-state levels of the other Mcs proteins (Figure 2D). Indeed, severe reduction of the levels of proteins of the MICOS complex occurred (with the exception of deletion of Mcs12), but not of other proteins of the different mitochondrial subcompartments. Deletion of Fcj1, in particular, resulted in a strong reduction of the levels of all other subunits of the MICOS complex. Interestingly, not only downregulation was observed; deletion of Mcs29 caused upregulation of Mcs27. These results suggest that the expression of Mcs proteins is subject to a regulatory network.
The MICOS complex fell apart when one of the Mcs proteins was missing as observed by filtration analysis (Supplementary Figure S2B). In most cases, MICOS I and II were either completely absent or strongly reduced and the level of lower mass complexes increased (Δfcj1, Δmcs10, Δmcs19, Δmcs29, Δmcs12). Upon deletion of Mcs27, a shift from MICOS I to MICOS II was observed. We conclude that the six different Mcs proteins cooperate in an intricate manner to generate the MICOS complex.
Topology of Mcs proteins. In order to determine the topology of the Mcs proteins, we performed protease accessibility and alkaline extraction assays followed by immunoblotting (Figure 2E). This list includes the previously established topology of Fcj1 (Rabl et al, 2009). Mcs10 is integrated into the IM with a predicted transmembrane segment located about 50 residues from the N-terminus. A topology in which the N-terminal part is located in the matrix and the C-terminal part in the IMS is supported by the loss of C-terminal His or HA tags upon protease treatment of mitoplasts (not shown). However, in view of the absence of a classical targeting signal other topologies cannot be excluded. Mcs19 is present in the IMS; it is apparently not an integral membrane protein, as it could be extracted from membranes at alkaline pH, a notion supported by the absence of a predicted membrane anchor. Since it is not lost during hypo-osmotic swelling, most of Mcs19 is likely bound to the IM. The relatively high isotope ratio at the top of the gradient (see Figure 1C) raises the possibility that part of the protein is associated with the OM. On the other hand, the comparatively high ratio at the bottom of the gradient may suggest that part of it is present in cristae membranes. Mcs27 and Mcs29 are also integral proteins of the IM. They have two predicted transmembrane spanning helices; the hydrophilic parts are present in the IMS. Mcs12 is anchored to the IM with its N-terminus exposing a hydrophilic domain of about 70 residues into the IMS. In summary, the six proteins found to participate in CS formation are inserted into or associated with the IM.
Submitochondrial location of the Mcs proteins
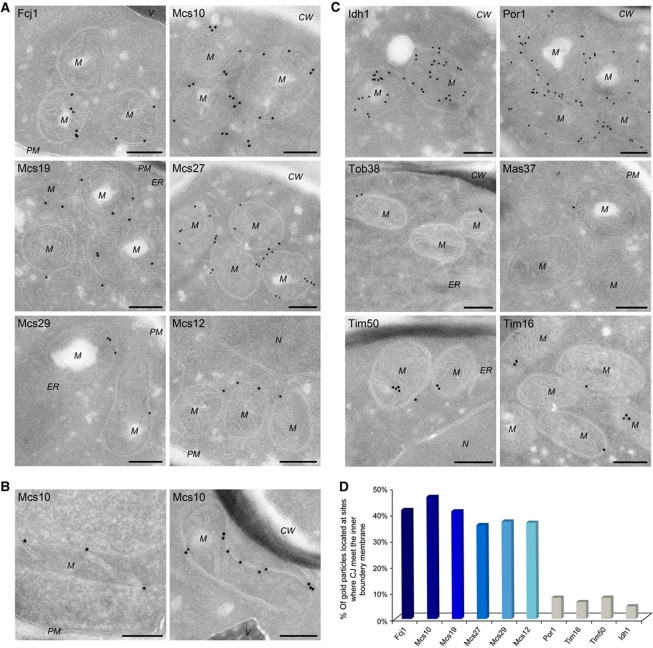
To determine the location of the various Mcs proteins at the submitochondrial level, immunolabelling with gold particles of cryosections was performed. It should be noted that with this procedure, gold particles can be present at a distance of up to 20 nm from the epitopes. Cells expressing the HA-tagged Mcs proteins were grown on lactate or glycerol (Figure 3A; see also Supplementary Figure S3A). Fcj1 was predominantly found at the mitochondrial envelope, very often in close proximity to CJ, in agreement with our previous work. Most intriguingly, the Mcs10 protein had a very similar distribution. It was also associated with the mitochondrial envelope and in most cases present at CJ. The observed immune reactions are specific as background labelling in wild-type cells was negligible (not shown). To analyse the distribution of Mcs10 in more detail, cells were also grown on glycerol where fewer cristae are present. Under these conditions, a close association of Mcs10 with CJ was conspicuous (Figure 3B). A third protein with a similar distribution is Mcs19, which, however, was also found at cristae. The latter location appears to be consistent with its higher levels in gradient fractions that correspond to crista membranes (cf Figure 1C, right panel, and see below Figure 4A). Mcs27, Mcs29 and Mcs12 also showed a preferential location at the envelope with very few gold particles found in the interior of mitochondria. Notably, the immunoreactivity of the cryosections was in agreement with the estimated levels of the Mcs proteins (Figure 2B).
Figure 3.
Localization of Mcs proteins by immuno-EM. Cells expressing C-terminally HA-tagged versions of Mcs proteins, were processed for immuno-EM, cryosections were labelled with anti-HA antibodies and protein A bound gold particles, with the exception of Por1, for which specific antibodies were used. (A) Distribution of Mcs proteins in cells grown on lactate. (B) Mcs10 localization in cells grown on glycerol. (C) Distribution of proteins of various mitochondrial subcompartments in cells grown on lactate. Matrix (Idh1); OM, (Por1, Tob38 and Mas37/Tob37); inner boundary and crista membrane (Tim50 and Tim16). CW, cell wall; M, mitochondrion; N, nucleus; PM, plasma membrane; V, vacuole. Size bars, 200 nm. Additional examples in Supplementary Figure S3. (D) Quantitative analysis of the distribution of Mcs and control proteins at sites where the CJ meet the IBM. For each labelling, the percentage of gold particles present at the CJ was determined.
Figure 4.
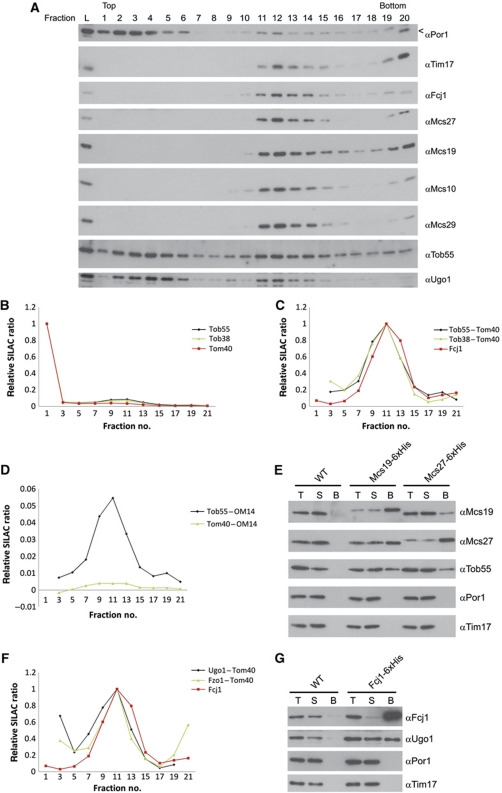
Interaction of Mcs proteins with components of the OM. (A) Wild-type mitochondria were fractionated and subjected to density flotation centrifugation as in Figure 1B. Fractions were subjected to SDS–PAGE and immunodecoration with antibodies against Por1, Tim17, Mcs proteins, Tob55 and Ugo1. (B) Analysis of the distribution on flotation gradients of Tob55 and Tob38 compared with Tom40 by mass spectrometry. (C) Differential profiles of Tob55 and Tob38 after subtraction of the profile of Tom40. (D) Subtraction of the profiles of two OM proteins, Tom40 and OM14, from each other. (E) Co-isolation of Tob55 with Mcs19 and Mcs27. For details see Figure 2A. (F) Density gradient profiles of Ugo1 and Fzo1 processed as in (C). (G) Co-isolation of Ugo1 with Fcj1. Analysis as in (E), with the exception that Triton X-100 was used for solubilization. Arrow head, unspecific cross reaction.
The distribution of a number of other mitochondrial proteins was studied for comparison (Figure 3C). Isocitrate dehydrogenase (Idh1), an abundant matrix protein was distributed all over the internal space of the mitochondria. VDAC/porin (Por1) was only found on or close to the OM. Likewise, Tob38/Sam35 and Mas37/Tob37/Sam37, components of the TOB complex, were present at the OM. Tim50 and Tim16, components of the TIM23 protein import machinery, in contrast, were found at the envelope membranes and to a lower degree in the interior of the mitochondria, in agreement with a previous study (Vogel et al, 2006) (see also Supplementary Figure S3B). Quantitative analysis of the immunolabelling of the Mcs proteins showed that some 35–45% of the gold particles were present at CJ; in contrast, only 5–8% of those marking control proteins were found at CJ (Figure 3D; Supplementary Table SI). Since CJs are small structures, they are not always visible in case of grazing sections; therefore, the number of Mcs proteins at CJs may be underestimated. On the other hand, a fraction of Mcs proteins may also be present outside CJ.
In summary, immuno-EM revealed that the MICOS complex is indeed located predominantly or entirely at the mitochondrial envelope strongly supporting the results of analysis of mitochondrial subfractions. Most interestingly, the MICOS complex is predominantly located at CJs.
Interaction of Mcs proteins with OM proteins
In a search for components of the mitochondrial OM that might be interaction partners of the Mcs proteins, we checked the gradient fractions of Figure 1 for OM proteins that overlap with the characteristic profile of Mcs proteins. Subunits of the TOM complex and OM45 were present in the middle of the gradient to a very low degree. They probably represent the pieces of OM that adhere to the IBM in the CS fractions. There were conspicuous exceptions. The levels of components of the TOB/SAM complex that mediates the insertion of β-barrel proteins into the mitochondrial OM (Kozjak et al, 2003; Paschen et al, 2003) were significantly higher in the fractions containing the Mcs proteins as seen both with immunoblotting and mass spectrometry (Figure 4A and B). A more detailed evaluation was performed by subtracting the profile of proteins such as the TOM complex subunit Tom40. Thereby, a clear coincidence of the profiles of TOB/SAM components with that of the Mcs proteins was observed (Figure 4C). The precision of the SILAC measurements allows this operation; subtraction of two OM proteins from each other yielded a reproducible zero line (Figure 4D). We conclude that a fraction of the TOB/SAM complex and Mcs proteins belong to a common structure.
To analyse whether TOB/SAM components can be co-isolated with Mcs proteins, mitochondria were solubilized with the mild detergent digitonin and interaction with Mcs proteins was studied (Figure 4E). Indeed, Tob55 was observed to bind to both Mcs27 and Mcs19. In a separate study, the interaction of Tob55 with Fcj1 was analysed. Fcj1-His interacted with Tob55, however, in a rather inefficient way. A mutational study of Fcj1 revealed a role of the C-terminus of Fcj in the interaction with Tob55 (Körner, Reichert et al, unpublished data). No interaction was seen with components of the TOM complex and other OM proteins such as OM45 (not shown). We conclude that a fraction of the TOB/SAM complexes in the OM is present in CS and interacts with Mcs proteins that are associated with the IM.
Two further proteins of the OM were found that shared the gradient profile of the TOB/SAM proteins, Ugo1 and Fzo1 (Figure 4A and F). When the SILAC profiles of typical OM proteins such as Tom40 or OM45 were subtracted from the SILAC profiles of Ugo1 and Fzo1, a peak in the characteristic Mcs protein fraction 11 remained, indicating that a fraction of these latter proteins were present in CS. Interaction of Ugo1 with Fcj1 was observed by co-isolation (Figure 4G). The OM protein Ugo1 is required for mitochondrial fusion and is associated with the fusion protein Fzo1 (Sesaki and Jensen, 2001, 2004; Wong et al, 2003). Thus, a fraction of Ugo1 seems to play a role in tethering Fcj1 to the OM and thereby to localize Fzo1 to CS.
To corroborate these results, we analysed vesicles by sucrose gradient centrifugation that were generated from mitochondria of a Δfcj1 strain (Supplementary Figure S4A). Strikingly, Mcs proteins and Ugo1 showed a different distribution in these cells compared with wild type (Figure 4A). Mcs10 and Mcs29 were shifted from fractions of intermediate density to fractions of high density so that their profiles were very similar to that of Tim17, an IM protein. The subpopulation of Ugo1 that was present in vesicles of intermediate density from wild-type mitochondria was almost completely absent in the gradient fractions of vesicles of Δfcj1 mitochondria. These results confirm the role of Fcj1 in bringing together proteins of the IM with proteins of the OM, and also substantiate our concept of the enrichment of Mcs proteins in gradient fractions of intermediate density.
We also studied the ultrastructure of mitochondria in Δugo1 cells (Supplementary Figure S4B). These cells did not grow on non-fermentable carbon source and therefore were grown on glucose and compared with wild-type and Δmcs10 cells grown on glucose. Their mitochondria were grossly altered. In particular, the number of CJs was extremely low; crista-like structures were almost completely absent. Apparently, Ugo1 has a role in the fusion of mitochondria, but is also critically involved in determining the architecture of mitochondria as shown here.
Functional characterization of Mcs proteins
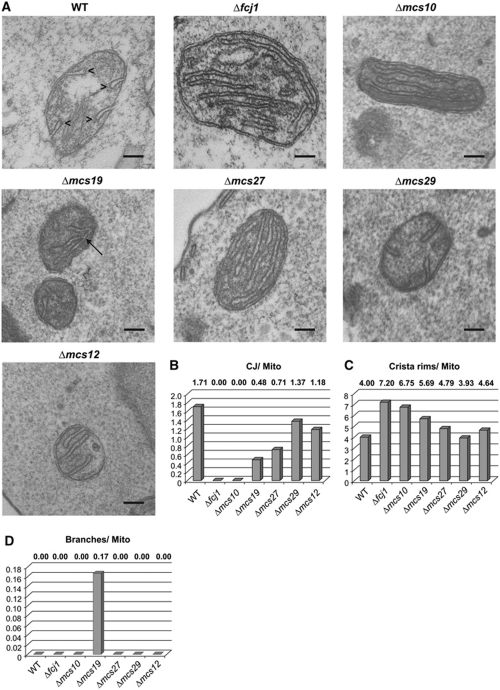
Role of Mcs proteins for the architecture of mitochondria. Figure 5 shows representative EM micrographs of cells in which each of the MCS genes was deleted, together with a quantitative evaluation of morphological parameters (see also Supplementary Figure S5 and Supplementary Table SII). The deletion of Fcj1 led to virtually complete loss of CJ, to an increase of cristae stacks and to an increased number of crista rims/endings/apexes as previously reported (Rabl et al, 2009). Likewise, deletion of Mcs10 caused virtually complete loss of CJ, accumulation of crista stacks and an increase of crista rims. The deletion of Mcs19 led to a massive loss of CJ and cristae showed bizarre shapes with frequent kinks. Notable are the abundant branches of the cristae. In Δmcs27 mitochondria, CJ were reduced by about 60%, there were crista stacks but much fewer than in Δfcj1 mitochondria. Deletion of Mcs29 resulted in a slight reduction of the number of CJ. Yet in contrast to Δfcj1 mitochondria, stacks were sometimes observed to be connected to the IBM by CJ. Upon deletion of Mcs12, fewer CJ were seen than in wild type, but similar to Mcs27 some of the crista stacks were connected to the IBM.
Figure 5.
Morphological roles of Mcs proteins. (A) Electron micrographs of mitochondria of wild-type cells and of cells lacking the various Mcs proteins. Cells were fixed with glutaraldehyde and sections contrasted with OsO4. Size bar, 100 nm. (B–D) Quantitative evaluation of images. Average numbers of (B) CJs, (C) crista rims and (D) crista branches per mitochondrial profile. Additional examples and quantifications in Supplementary Figures S4B and S5 and Supplementary Table SII. Arrowheads, crista rims; arrow, crista branch.
In conclusion, these data show that the proteins found in the search for components of CS are characterized by a complete or partial deficiency in CJ and altered crista morphology, such as increased number of crista rims or crista branching.
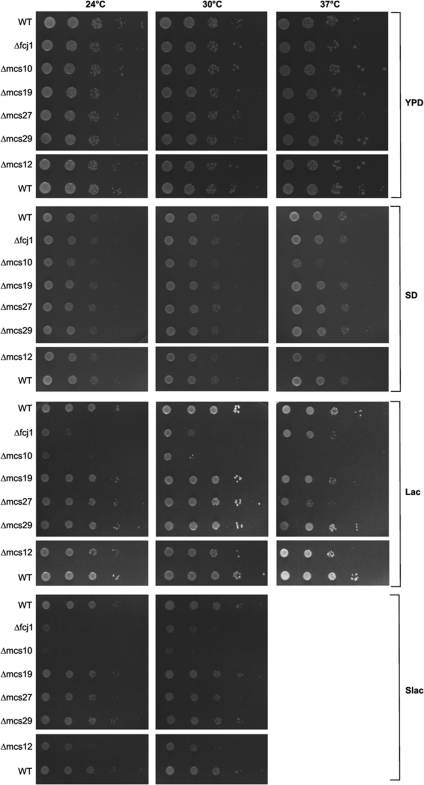
Growth behaviour of cells. Each one of the strains depleted of the various Mcs proteins grew like wild type on rich glucose-containing (YPD) medium (Figure 6). On fermentable carbon sources, some of the deletion mutants showed severe (Δfcj1, Δmcs10) or mild (Δmcs27 and Δmcs12) growth defects, partly depending on growth temperature. Thus, deletion of the MCS genes leads to loss or reduction of the capacity of mitochondria for oxidative phosphorylation, demonstrating that the architecture of mitochondria is key to basic metabolic processes of the mitochondria.
Figure 6.
Growth characteristics of cells lacking Mcs proteins. Cells were spotted in 10-fold dilution steps on agar plates containing one of the following media: glucose and yeast extract (YPD); glucose-containing synthetic medium (SD); lactate-containing medium (Lac); lactate-containing synthetic medium (Slac). Plates were incubated at the indicated temperatures. None of the strains grew on Slac at 37°C.
Discussion
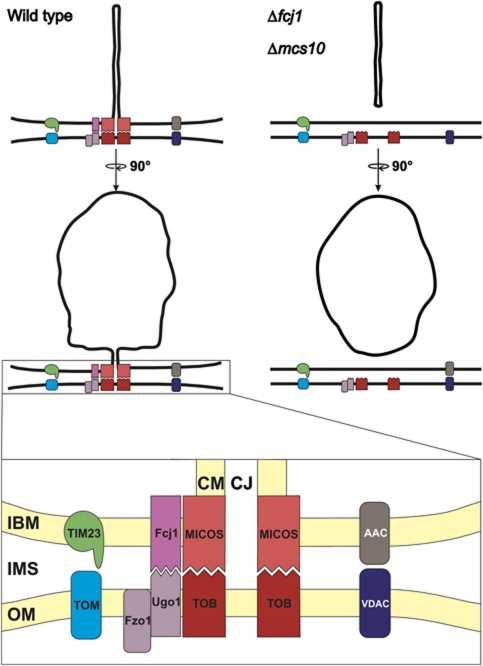
In this work, we describe a novel complex with an important role in determining the architecture and function of mitochondria (Figure 7). This large multisubunit complex, which we term MICOS complex, is anchored to the IBM and extends across the IMS to reach specific OM proteins, the TOB/SAM and the Ugo1–Fzo1 complexes. Interaction of the Ugo1–Fzo1 complex with the IM has been reported previously (Fritz et al, 2001); however, the interaction partner in the IM remained obscure so far. Why the TOB/SAM complex serves as an anchor for Mcs proteins is an intriguing question. Perhaps equivalent interactions were present in the gram-negative ancestors of the mitochondria, the elusive Bayer's junctions (Bayer, 1991), and maintained during evolution of mitochondria for similar or new purposes.
Figure 7.
Working model of the role of the MICOS complex in mitochondrial architecture. (Upper panels) Mitochondrion of wild-type cells (left) and of cells lacking Fcj1 or Mcs10 (right) in two orientations. (Lower panel) Blow-up of CS with interaction of MICOS-TOB and of Fcj1–Ugo1–Fzo1. Dynamic interaction of TOM and TIM protein import complexes, and of AAC (ADP/ATP carrier) and VDAC/porin bridging the IMS, supported by CS. For further explanation and abbreviations see Discussion.
MICOS is preferentially located at sites where the cristae originate from the IBM. Two of the six Mcs proteins identified, Mcs10 and Fcj1, are essential for forming CJ, the others affect the presence of CJ to different degrees. Among these, Mcs19 is not only important for the presence of CJ, but also for the formation of intact cristae, as its depletion leads to appearance of branches in cristae. It remains to be determined whether Ugo1 and Fzo1 are present in the MICOS complex like the other Mcs proteins. Ugo1 was found also in association with Mgm1, a dynamin-like GTPase (Wong et al, 2003; Sesaki and Jensen, 2004). It is also possible that they form a separate complex with different composition and function.
Our findings shed new light on the architectural organization of mitochondria and at the same time raise a number of intriguing questions. They strongly support our initial hypothesis on the existence of firm contacts between OM and IBM at sites where the CJs merge with the IBM. The MICOS complex appears to be important for mitochondrial architecture, dynamics and function in several respects. First, it is necessary for the formation of CJs. Second, it is critical for the capacity of mitochondria for oxidative phosphorylation and inheritance of mitochondrial DNA, as highlighted by the presence of four of the MCS genes in the AIM collection (Hess et al, 2009). Third, changes in the bouyant density of membrane vesicles from mitochondria depleted of Fcj1 would suggest that the transfer of membrane lipids to and between mitochondrial membranes is dependent on the MICOS complex (unpublished results).
The definition of the CS as important specific architectural elements leads us to some speculations regarding so far unexplained processes in mitochondria. We speculate that the MICOS complex may control the lateral diffusion of newly imported IM proteins into the cristae after their insertion into the IBM. As a consequence, the MICOS complex may influence the composition and structure of both the IBM and the crista membrane. Furthermore, by shaping cristae junctions, the MICOS complex might be involved in the release of components of the intracrista space, an important process in apoptosis in higher eukaryotes. Such a role of CS and several others have been previously proposed (Frey and Mannella, 2000; Mannella, 2008).
Our results also bring up the exciting possibility that the connection of cristae with the IBM is not static but dynamic. It has been suggested that the width of CJ may limit the diffusion of proteins like cytochrome c from the intracrista space into the space between IMS (Scorrano et al, 2002). We speculate that CJ can undergo a time dependent opening and closing, governed by fusion and fission at the CS. The MICOS complex could be a scaffold for maintenance of defined sites of fusion and participate in controlling the equilibrium of fusion and fission. Such a dynamic structure of the mitochondria would have far reaching implications for the efficiency of oxidative phosphorylation. Cristae in a closed state would be able to maintain the proton gradient generated by the respiratory chain to a much higher extent than cristae, which are continuously open to the IMS and therefore to the cytosolic compartment. The question of whether the proton gradient usually measured in mitochondria is sufficient for optimal ATP production is an issue in mitochondrial bioenergetics. Several possible mechanisms have been proposed that might prevent proton equilibration with the bulk phase (Mulkidjanian et al, 2005; Strauss et al, 2008).
Protein translocases of OM and IBM, the TOM and TIM23 complexes, were shown to interact closely, yet transiently. The MICOS complex may not be essential for this process, but CS could increase the efficiency of matching TOM with the TIM and TOB complexes (Figure 7). Likewise, export of ATP from the mitochondria by a joint action of the ADP/ATP translocase, VDAC/porin and hexokinase (Brdiczka et al, 2006) may not need the presence of CS. On the other hand, the MICOS complex might increase the efficiency of their interactions by providing a scaffold for matching of the protein complexes residing in IBM and OM.
Is the complex which we describe here a common structure in all eukaryotes? Mcs10 is a highly conserved protein (Supplementary Figure S6A). Its sequence is remarkable in several aspects. It is a small protein with a single predicted transmembrane segment, containing a GX3G motive, followed by a GXGXGXG motif in which the X residues are hydrophobic. The C-terminal motif is particularly interesting. Since Mcs10 apparently forms homo-oligomers both elements might be involved in the self-association. Fcj1 was found previously to have sequence similarity to mammalian mitofilin and homologues are present in metazoa. Downregulation of mitofilin led to mitochondria with altered cristae and absence of CJ. Mitofilin was therefore recognized as a protein controlling crista morphology (Odgren et al, 1996; John et al, 2005; Rabl et al, 2009). Mcs29 and Mcs27 proteins might be evolutionarily related (Supplementary Figure S6B). Both contain two predicted transmembrane segments at similar positions. However, we found no obvious homologues in higher eukaryotes. Mcs19 also does not show related sequences in higher organisms, even among fungi conservation is very limited. However, the very C-terminal part shows somewhat higher similarity. Interestingly, a CX10C motif in the latter part is conserved among fungal species. It could possibly serve as a motif for the disulphide relay system for import into the IMS (Hell, 2008). Mcs12 also belongs to the group of Mcs proteins with low-sequence similarity even among fungi.
Interestingly, two recent reports describe proteins that appear to play a role in the ultrastructure of mitochondria of higher eukaryotes. ChChd3 in mammalian cells is located on the IM facing the IMS and is involved in maintaining cristae integrity and mitochondrial function (Darshi et al, 2011). None of the Mcs proteins can be recognized as a homologue; however, both Mcs19 and ChChd3 have a myristoylation motif and pairs of cysteine residues close to the C-terminus. A homologue of this protein in Caenorhabditis elegans, CHCH-3, was suggested to have a role as a chaperone. Furthermore, the protein MOMA-1 in C. elegans was proposed to display low-sequence similarity to Mcs27 and Mcs29 (Head et al, 2011). Although we could not detect significant sequence similarity (Supplementary Figure S6B), the overall structures of Mcs27, Mcs29 and MOMA-1 are similar. MOMA-1 was found mainly in the OM (Head et al, 2011), in contrast to Mcs27 and Mcs29, which are integrated into the IM of yeast mitochondria. It will be important to clarify as to whether the submitochondrial location of MOMA-1 is different from that of Mcs27 and Mcs29 or the sequence similarity is too low to predict homology.
Irrespective of possible homologies, there are similarities in the observed interactions, knockdown phenotypes and possible functions between ChChd3 and MOMA-1 and the respective yeast proteins. The mammalian ChChd3 interacts with mitofilin and Sam50/Tob55; morphological aberrations such as clustering of mitochondria around the nucleus, fragmented and tubular cristae and reduced opening diameter of CJ were observed (Xie et al, 2007; Darshi et al, 2011). Mutations in MOMA-1 and in the mitofilin homologue in C. elegans, IMMT-1, led to altered crista morphology. Furthermore, the phenotypes of knockdown studies suggested an interaction of MOMA-1, the ChChd3 homologue, CHCH-3, and IMMT-1.
Thus, in view of these similarities, our discovery and biochemical and functional characterization of the MICOS complex are relevant not only for lower eukaryotes but apparently for the whole eukaryotic world. We believe that it opens the door for a profound and detailed analysis of the molecular basis of mitochondrial architecture, also in higher eukaryotic cells.
Materials and methods
Yeast strains and cell growth
For analysis of submitochondrial fractions, strains W303 {leu2-3,112 trp1-1 can1-100 ura3-1 ade2-1 his3-11,15} and W303Δtim23 harbouring pRS315GFP–Tim23 under the endogenous Tim23 promoter were used (Vogel et al, 2006). For the SILAC analysis of submitochondrial fractions, YPH499 {ura3-52, lys2-801amber, ade2-101ocre, trp1-Δ63, his3-Δ200, leu2-Δ1} was used because of its lysine auxotrophy. Chromosomal manipulations (knockouts, C-terminal 6 × His- and 3 × HA-tagging) were performed in the YPH499 background (Longtine et al, 1998; Knop et al, 1999). For the generation of deletion strains, the coding regions of FCJ1, YCL057C-A/MCS10, YFR011C/MCS19, YNL100W/MCS27, YGR235C/MCS29 and YBR262C/MCS12 were replaced by a HIS3 cassette using the pFA6a-His3MX6 plasmid as the PCR template. The 6 × His-tags were introduced using the pYM9 plasmid as the PCR template. The 3 × HA-tags were introduced using the pYM2 vector as the PCR template. All strains expressing tagged versions were tested for growth and all were found to grow like wild type. Wild-type morphology of mitochondria in cells expressing the HA-fusion proteins was also confirmed by EM (see Figure 3). Strains were grown on 2% lactate medium (Lac) (containing 3 g yeast extract, 1 g NH4Cl, 1 g KH2PO4, 0.5 g CaCl2 × 2H2O, 0.5 g NaCl, 1 g MgSO4 × 7H2O, and 3 mg FeCl3 per litre) at 24°C for analysis of submitochondrial fractions (Sherman, 1991).
For growth analysis and mitochondrial preparations, strains were cultured as indicated at 24, 30 or 37°C in Lac medium, in YPD medium (1% yeast extract, 2% peptone, 2% glucose) (Sherman, 1991), or synthetic medium (1.7 g Yeast Nitrogen Base and 5 g (NH4)2SO4 per l) containing either 2% lactate (SLac) or 2% glucose (SD) (see Supplementary data for details).
Subfractionation of mitochondria
Mitochondria were swollen for 30 min on ice. Sucrose concentration was adjusted to 0.5 M followed by incubation for 15 min at 0°C and mild sonication. After a clarifying spin (20 000 g, 20 min, 4°C), vesicles were concentrated by centrifugation (120 000 g, 100 min, 4°C). Vesicles were resuspended and separated by a centrifugation of a continuous flotation sucrose gradient (200 000 g, 24 h, 4°C). For detailed protocol see Supplementary data.
Mass spectrometry analysis of proteins from submitochondrial fractions
For sample preparation and mass spectrometry, equal volumes of sucrose gradient fractions were mixed with SILAC standards and supplemented with twice the volume of denaturation buffer (9 M urea, 3 M thiourea, 100 mM Tris/HCl pH 8, 1.5 mM DTT). After alkylation with 2-iodoacetamide, proteins were digested overnight with endoproteinase LysC (Wako Bioproducts, Richmond, VA, USA) and peptides were desalted and concentrated via C18 StageTips (Rappsilber et al, 2003). LC–MS experiments with an Easy nLC nanoflow HPLC system coupled to an LTQ Orbitrap XL mass spectrometer (Thermo Fisher Scientific) were performed essentially as described previously (Olsen et al, 2004; Forner et al, 2006) with modifications. Columns of 40 cm length and an inner diameter of 75 μm, packed with 1.8 μm beads (Reprosil-AQ Pur, Dr Maisch, Entringen, Germany) (Thakur et al, 2011), were used and the gradient length was 5 h.
Raw data were analysed using the MaxQuant software environment (Cox and Mann, 2008). Peak lists were searched with Mascot (Perkins et al, 1999) against a database containing the translation of all 6809 gene models from the Saccharomyces Genome Database release from 12 December 2007 and 175 frequently observed contaminants as well as the reverse sequences of all entries. Both peptide and protein identification were accepted at a 1% false discovery rate, using a decoy database strategy. Protein quantification was exclusively based on peptides with unique sequences.
Electron microscopy
For standard EM, cells were fixed with glutaraldehyde, contrasted with osmium tetroxide, sectioned and subjected to EM (see Supplementary data for details).
For immuno-EM, cells were grown in lactate or glycerol medium to the exponential phase, chemically fixed, embedded in 12% gelatin and cryosectioned as described (Griffith et al, 2008). Sections were immunogold labelled using either anti-HA (a kind gift of Guojun Bu, Washington University) or anti-porin (Molecular Probes) antibodies and a protein A-gold 10 nm conjugate before being viewed in a JEOL 1010 electron microscope (JEOL, Tokyo, Japan). The quantitative evaluation of the gold-labelling experiments was performed as follows. A gold particle was assigned to the CJ if at a distance not >20 nm from this site. Likewise, and assigned to the mitochondrial envelope if at a distance not >20 nm from the OM/IM and not localizing to the CJ. The remaining particles present in the interior of the mitochondria were considered in the inner space, which comprises both the cristae and the matrix.
Miscellaneous
Proteolytic susceptibility assay. In all, 50 μg mitochondria were incubated with either SM buffer (0.6 M sorbitol, 20 mM MOPS, pH 7.4), swelling buffer (20 mM MOPS, pH 7.4) or lysis buffer (1% (v/v) Triton X-100, 20 mM MOPS, pH 7.4) for 30 min on ice. Proteinase K (final concentration of 0.2 mg/ml) was added and the suspension was incubated at 4°C for 15 min. Proteinase K was inhibited by the addition of phenylmethanesulfonyl fluoride (PMSF) to a final concentration of 4 mM and incubation for 10 min on ice. Samples were centrifuged for 20 min (20 000 g, 4°C), resuspended in SM buffer, and precipitated by addition of trichloroacetic acid (TCA; final concentration of 14%). The precipitate was resuspended in Laemmli buffer, subjected to SDS–PAGE and analysed by immunoblotting.
Alkaline extraction. In all, 100 mg mitochondria were resuspended in 75 μl 20 mM HEPES and 75 μl 200 mM Na2CO3 were added. The suspension was mixed by vortexing for 15 s, incubated for 30 min on ice and centrifuged for 30 min (135 000 g, 4°C). The pellet was resuspended in Laemmli buffer. In all, 30% of the supernatant was TCA precipitated and resuspended in Laemmli buffer. Samples were subjected to SDS–PAGE and analysed by immunoblotting.
Co-isolation assays. For co-isolation of proteins with His-tagged Fcj1, Mcs10, Mcs19, Mcs27, Mcs29 or Mcs12, 1 mg mitochondria isolated from the respective strains were lysed with 1% (w/v) digitonin or 1% (v/v) Triton X-100 as indicated. After Ni-NTA affinity chromatography, fractions were analysed by SDS–PAGE followed by immunoblotting.
Size exclusion chromatography. Isolated mitochondria were incubated for solubilization in digitonin buffer (30 mM Hepes pH 7.4, 100 mM potassium acetate pH 7.4, 5 mM EDTA and 1 mM PMSF, 1% digitonin) at a protein/digitonin ratio of 1/1 for 30 min on ice. After centrifugation for 15 min at 60 000 g and 4°C, cleared lysates were subjected on Superose 6 size exclusion column (GE Healthcare; Elution buffer: 30 mM Hepes pH 7.4, 150 mM KAc pH 7.4, 5 mM EDTA, 1 mM PMSF, 0.1% digitonin). Fractions were analysed by SDS–PAGE and immunoblotting.
Blue native PAGE. Mitochondria (150 μg protein) were incubated with 20 μl solubilization buffer (50 mM NaCl, 50 mM imidazole, 2 mM 6-aminohexanoic acid, 1 mM EDTA, 1 mM PMSF, 3% digitonin, pH 7.0) for 15 min at 4°C followed by a clarifying spin for 20 min at 15 000 g and 4°C (Wittig et al, 2006). The supernatant was mixed with 2 μl Native PAGE™ 5% G-250 Sample Additive (Invitrogen) and subjected to BN-PAGE (Native PAGE 3–12% Bis-Tris, Invitrogen). After blotting on PVDF membranes (Roth), immunodecoration using the indicated antibodies was performed.
Supplementary Material
Acknowledgments
We thank Petra Heckmeyer, Sabine Tost, Christiane Kotthoff and Petra Robisch for excellent technical assistance. We are grateful to Kai Hell and Martin Ott for helpful discussions, the late Professor Fritz Miller for the EM micrographs of Neurospora mitochondria and Regina Rabl for providing the construct with N-terminally his-tagged Fcj1. This work was supported by the Deutsche Forschungsgemeinschaft, SFB 594. FR is supported by the Netherlands Organization for Health Research and Development (ZonMW-VIDI-917.76.329) and by the Netherlands Organization for Scientific Research (Chemical Sciences, ECHO Grant-700.59.003).
Author contributions: MH, CK, DW, DM, JK and JG performed the experiments. UW, MM and FR supervised the study. WN was involved in conception of the project, supervision and writing of the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Andersen JS, Wilkinson CJ, Mayor T, Mortensen P, Nigg EA, Mann M (2003) Proteomic characterization of the human centrosome by protein correlation profiling. Nature 426: 570–574 [DOI] [PubMed] [Google Scholar]
- Bayer ME (1991) Zones of membrane adhesion in the cryofixed envelope of Escherichia coli. J Struct Biol 107: 268–280 [DOI] [PubMed] [Google Scholar]
- Boldogh IR, Pon LA (2006) Interactions of mitochondria with the actin cytoskeleton. Biochim Biophys Acta 1763: 450–462 [DOI] [PubMed] [Google Scholar]
- Brdiczka DG, Zorov DB, Sheu SS (2006) Mitochondrial contact sites: their role in energy metabolism and apoptosis. Biochim Biophys Acta 1762: 148–163 [DOI] [PubMed] [Google Scholar]
- Chan D, Frank S, Rojo M (2006) Mitochondrial dynamics in cell life and death. Cell Death Differ 13: 680–684 [DOI] [PubMed] [Google Scholar]
- Cox J, Mann M (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol 26: 1367–1372 [DOI] [PubMed] [Google Scholar]
- Darshi M, Mendiola VL, Mackey MR, Murphy AN, Koller A, Perkins GA, Ellisman MH, Taylor SS (2011) ChChd3, an inner mitochondrial membrane protein, is essential for maintaining crista integrity and mitochondrial function. J Biol Chem 286: 2918–2932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dengjel J, Jakobsen L, Andersen JS (2010) Organelle proteomics by label-free and SILAC-based protein correlation profiling. Methods Mol Biol 658: 255–265 [DOI] [PubMed] [Google Scholar]
- Dimmer KS, Fritz S, Fuchs F, Messerschmitt M, Weinbach N, Neupert W, Westermann B (2002) Genetic basis of mitochondrial function and morphology in Saccharomyces cerevisiae. Mol Biol Cell 13: 847–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donzeau M, Kaldi K, Adam A, Paschen S, Wanner G, Guiard B, Bauer MF, Neupert W, Brunner M (2000) Tim23 links the inner and outer mitochondrial membranes. Cell 101: 401–412 [DOI] [PubMed] [Google Scholar]
- Fawcett DW (1981) The Cell. Philadelphia: W.B. Saunders [Google Scholar]
- Forner F, Foster LJ, Campanaro S, Valle G, Mann M (2006) Quantitative proteomic comparison of rat mitochondria from muscle, heart, and liver. Mol Cell Proteomics 5: 608–619 [DOI] [PubMed] [Google Scholar]
- Foster LJ, de Hoog CL, Zhang Y, Xie X, Mootha VK, Mann M (2006) A mammalian organelle map by protein correlation profiling. Cell 125: 187–199 [DOI] [PubMed] [Google Scholar]
- Frey TG, Mannella CA (2000) The internal structure of mitochondria. Trends Biochem Sci 25: 319–324 [DOI] [PubMed] [Google Scholar]
- Fritz S, Rapaport D, Klanner E, Neupert W, Westermann B (2001) Connection of the mitochondrial outer and inner membranes by fzo1 is critical for organellar fusion. J Cell Biol 152: 683–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith J, Mari M, De Maziere A, Reggiori F (2008) A cryosectioning procedure for the ultrastructural analysis and the immunogold labelling of yeast Saccharomyces cerevisiae. Traffic 9: 1060–1072 [DOI] [PubMed] [Google Scholar]
- Griparic L, Head BP, Van der Bliek AM (2004) Mitochondrial division and fusion. Topics Curr Genet 8: 227–249 [Google Scholar]
- Hackenbrock CR (1968) Chemical and physical fixation of isolated mitochondria in low-energy and high-energy states. Proc Natl Acad Sci USA 61: 598–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harner M, Neupert W, Deponte M (2011) Lateral release of proteins from the TOM complex into the outer membrane of mitochondria. EMBO J 30: 3232–3241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Head BP, Zulaika M, Ryazantsev S, van der Bliek AM (2011) A novel mitochondrial outer membrane protein, MOMA-1, that affects cristae morphology in Caenorhabditis elegans. Mol Biol Cell 22: 831–841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hell K (2008) The Erv1-Mia40 disulfide relay system in the intermembrane space of mitochondria. Biochim Biophys Acta 1783: 601–609 [DOI] [PubMed] [Google Scholar]
- Hess DC, Myers CL, Huttenhower C, Hibbs MA, Hayes AP, Paw J, Clore JJ, Mendoza RM, Luis BS, Nislow C, Giaever G, Costanzo M, Troyanskaya OG, Caudy AA (2009) Computationally driven, quantitative experiments discover genes required for mitochondrial biogenesis. PLoS Genet 5: e1000407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoppins S, Nunnari J (2009) The molecular mechanism of mitochondrial fusion. Biochim Biophys Acta 1793: 20–26 [DOI] [PubMed] [Google Scholar]
- John GB, Shang Y, Li L, Renken C, Mannella CA, Selker JM, Rangell L, Bennett MJ, Zha J (2005) The mitochondrial inner membrane protein mitofilin controls cristae morphology. Mol Biol Cell 16: 1543–1554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knop M, Siegers K, Pereira G, Zachariae W, Winsor B, Nasmyth K, Schiebel E (1999) Epitope tagging of yeast genes using a PCR-based strategy: more tags and improved practical routines. Yeast 15: 963–972 [DOI] [PubMed] [Google Scholar]
- Kozjak V, Wiedemann N, Milenkovic D, Lohaus C, Meyer HE, Guiard B, Meisinger C, Pfanner N (2003) An essential role of Sam50 in the protein sorting and assembly machinery of the mitochondrial outer membrane. J Biol Chem 278: 48520–48523 [DOI] [PubMed] [Google Scholar]
- Kunkele KP, Heins S, Dembowski M, Nargang FE, Benz R, Thieffry M, Walz J, Lill R, Nussberger S, Neupert W (1998) The preprotein translocation channel of the outer membrane of mitochondria. Cell 93: 1009–1019 [DOI] [PubMed] [Google Scholar]
- Larsson NG (2010) Somatic mitochondrial DNA mutations in mammalian aging. Annu Rev Biochem 79: 683–706 [DOI] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A III, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR (1998) Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14: 953–961 [DOI] [PubMed] [Google Scholar]
- Mannella CA (2006) The relevance of mitochondrial membrane topology to mitochondrial function. Biochim Biophys Acta 1762: 140–147 [DOI] [PubMed] [Google Scholar]
- Mannella CA (2008) Structural diversity of mitochondria: functional implications. Ann NY Acad Sci 1147: 171–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meeusen S, DeVay R, Block J, Cassidy-Stone A, Wayson S, McCaffery JM, Nunnari J (2006) Mitochondrial inner-membrane fusion and crista maintenance requires the dynamin-related GTPase Mgm1. Cell 127: 383–395 [DOI] [PubMed] [Google Scholar]
- Mulkidjanian AY, Cherepanov DA, Heberle J, Junge W (2005) Proton transfer dynamics at membrane/water interface and mechanism of biological energy conversion. Biochemistry (Mosc) 70: 251–256 [DOI] [PubMed] [Google Scholar]
- Odgren PR, Toukatly G, Bangs PL, Gilmore R, Fey EG (1996) Molecular characterization of mitofilin (HMP), a mitochondria-associated protein with predicted coiled coil and intermembrane space targeting domains. J Cell Sci 109(Part 9): 2253–2264 [DOI] [PubMed] [Google Scholar]
- Okamoto K, Shaw JM (2005) Mitochondrial morphology and dynamics in yeast and multicellular eukaryotes. Annu Rev Genet 39: 503–536 [DOI] [PubMed] [Google Scholar]
- Olsen JV, Ong SE, Mann M (2004) Trypsin cleaves exclusively C-terminal to arginine and lysine residues. Mol Cell Proteomics 3: 608–614 [DOI] [PubMed] [Google Scholar]
- Ong SE, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, Mann M (2002) Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics 1: 376–386 [DOI] [PubMed] [Google Scholar]
- Paschen SA, Waizenegger T, Stan T, Preuss M, Cyrklaff M, Hell K, Rapaport D, Neupert W (2003) Evolutionary conservation of biogenesis of beta-barrel membrane proteins. Nature 426: 862–866 [DOI] [PubMed] [Google Scholar]
- Pellegrini L, Scorrano L (2007) A cut short to death: Parl and Opa1 in the regulation of mitochondrial morphology and apoptosis. Cell Death Differ 14: 1275–1284 [DOI] [PubMed] [Google Scholar]
- Perkins DN, Pappin DJ, Creasy DM, Cottrell JS (1999) Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20: 3551–3567 [DOI] [PubMed] [Google Scholar]
- Perkins GA, Ellisman MH, Fox DA (2004) The structure-function correlates of mammalian rod and cone photoreceptor mitochondria: observations and unanswered questions. Mitochondrion 4: 695–703 [DOI] [PubMed] [Google Scholar]
- Pon L, Moll T, Vestweber D, Marshallsay B, Schatz G (1989) Protein import into mitochondria: ATP-dependent protein translocation activity in a submitochondrial fraction enriched in membrane contact sites and specific proteins. J Cell Biol 109: 2603–2616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabl R, Soubannier V, Scholz R, Vogel F, Mendl N, Vasiljev-Neumeyer A, Korner C, Jagasia R, Keil T, Baumeister W, Cyrklaff M, Neupert W, Reichert AS (2009) Formation of cristae and crista junctions in mitochondria depends on antagonism between Fcj1 and Su e/g. J Cell Biol 185: 1047–1063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappsilber J, Ishihama Y, Mann M (2003) Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal Chem 75: 663–670 [DOI] [PubMed] [Google Scholar]
- Reichert AS, Neupert W (2002) Contact sites between the outer and inner membrane of mitochondria-role in protein transport. Biochim Biophys Acta 1592: 41–49 [DOI] [PubMed] [Google Scholar]
- Renken C, Siragusa G, Perkins G, Washington L, Nulton J, Salamon P, Frey TG (2002) A thermodynamic model describing the nature of the crista junction: a structural motif in the mitochondrion. J Struct Biol 138: 137–144 [DOI] [PubMed] [Google Scholar]
- Scheffler I (ed). (2011) Mitochondria, 2nd edn. New York: Wiley and Sons Ltd [Google Scholar]
- Scorrano L, Ashiya M, Buttle K, Weiler S, Oakes SA, Mannella CA, Korsmeyer SJ (2002) A distinct pathway remodels mitochondrial cristae and mobilizes cytochrome c during apoptosis. Dev Cell 2: 55–67 [DOI] [PubMed] [Google Scholar]
- Sesaki H, Jensen RE (2001) UGO1 encodes an outer membrane protein required for mitochondrial fusion. J Cell Biol 152: 1123–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sesaki H, Jensen RE (2004) Ugo1p links the Fzo1p and Mgm1p GTPases for mitochondrial fusion. J Biol Chem 279: 28298–28303 [DOI] [PubMed] [Google Scholar]
- Shaw JM, Nunnari J (2002) Mitochondrial dynamics and division in budding yeast. Trends Cell Biol 12: 178–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F (1991) Getting started with yeast. Methods Enzymol 194: 3–21 [DOI] [PubMed] [Google Scholar]
- Strauss M, Hofhaus G, Schroder RR, Kuhlbrandt W (2008) Dimer ribbons of ATP synthase shape the inner mitochondrial membrane. EMBO J 27: 1154–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamai S, Iida H, Yokota S, Sayano T, Kiguchiya S, Ishihara N, Hayashi J, Mihara K, Oka T (2008) Characterization of the mitochondrial protein LETM1, which maintains the mitochondrial tubular shapes and interacts with the AAA-ATPase BCS1L. J Cell Sci 121: 2588–2600 [DOI] [PubMed] [Google Scholar]
- Thakur SS, Geiger T, Chatterjee B, Bandilla P, Froehlich F, Cox J, Mann M (2011) Deep and highly sensitive proteome coverage by LC-MS/MS without pre-fractionation. Mol Cell Proteomics 10: M110.003699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel F, Bornhovd C, Neupert W, Reichert AS (2006) Dynamic subcompartmentalization of the mitochondrial inner membrane. J Cell Biol 175: 237–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace DC, Fan W (2009) The pathophysiology of mitochondrial disease as modeled in the mouse. Genes Dev 23: 1714–1736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner S, Neupert W (1972) Functional and biogenetical heterogeneity of the inner membrane of rat-liver mitochondria. Eur J Biochem 25: 379–396 [DOI] [PubMed] [Google Scholar]
- Westermann B (2010) Mitochondrial fusion and fission in cell life and death. Nat Rev Mol Cell Biol 11: 872–884 [DOI] [PubMed] [Google Scholar]
- Wittig I, Braun HP, Schägger H (2006) Blue native PAGE. Nat Protoc 1: 418–428 [DOI] [PubMed] [Google Scholar]
- Wong ED, Wagner JA, Scott SV, Okreglak V, Holewinske TJ, Cassidy-Stone A, Nunnari J (2003) The intramitochondrial dynamin-related GTPase, Mgm1p, is a component of a protein complex that mediates mitochondrial fusion. J Cell Biol 160: 303–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie J, Marusich MF, Souda P, Whitelegge J, Capaldi RA (2007) The mitochondrial inner membrane protein mitofilin exists as a complex with SAM50, metaxins 1 and 2, coiled-coil-helix coiled-coil-helix domain-containing protein 3 and 6 and DnaJC11. FEBS Lett 581: 3545–3549 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.