Abstract
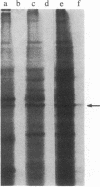
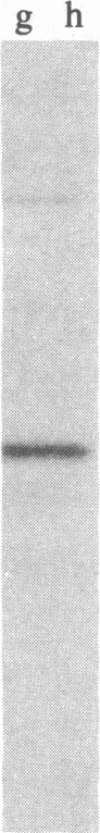
The isolation and characterization of a cDNA clone for the mRNA of human ornithine aminotransferase (OATase; ornithine-oxo-acid aminotransferase; L-ornithine:2-oxo-acid aminotransferase, EC 2.6.1.13), a nonabundant mitochondrial matrix enzyme that is severely deficient in a hereditary chorioretinal degenerative disease (gyrate atrophy), is described. Human liver, retina, and retinoblastoma (Y79) mRNAs were prepared and tested for the OATase mRNA content by in vitro translation, immunoprecipitation, and NaDodSO4/PAGE. The retinoblastoma cells were found to be expressing this enzyme at a relatively high level. The primary translation product of the OATase mRNA is larger than the pure OATase protein on NaDodSO4/PAGE by approximately equal to 4 kDa, suggesting a precursor protein. lambda gt11 cDNA libraries were prepared from the human mRNAs, and the recombinant clones were immunoscreened as plaques with two different preparations of rabbit anti-human OATase antibodies. A clone (lambda gtRB315) was isolated from the retinoblastoma library that reacts with both of the antibody preparations, and the DNA sequence of its 2.1-kilobase-pair cDNA insert was obtained. An open reading frame consisting of 1371 nucleotides is present in the sequence, and a putative translational initiation methionine codon is identified at position 55. A putative leader sequence consisting of 32 amino acid residues is identified, resulting in a precursor protein of 439 amino acid residues and a molecular mass of 48,534 Da and a mature protein of 407 residues and 45,136 Da. The amino acid sequences of seven tryptic peptides (115 amino acid residues) of the pure human OATase were obtained by microsequencing. When the tryptic peptide and cDNA-derived amino acid sequences were compared, homologies in 111 of 115 residues, including a match of 20 consecutive residues, were observed. An RNA blot hybridization of 32P-labeled OATase cDNA to normal human retina and retinoblastoma mRNAs demonstrated an OATase mRNA species of approximately equal to 2.2 kilobases. The level of OATase mRNA in the normal human retina is approximately equal to 1/100th the level of rhodopsin mRNA and 1/5th to 1/10th the level present in the retinoblastoma cells.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baralle F. E., Brownlee G. G. AUG is the only recognisable signal sequence in the 5' non-coding regions of eukaryotic mRNA. Nature. 1978 Jul 6;274(5666):84–87. doi: 10.1038/274084a0. [DOI] [PubMed] [Google Scholar]
- Biggin M. D., Gibson T. J., Hong G. F. Buffer gradient gels and 35S label as an aid to rapid DNA sequence determination. Proc Natl Acad Sci U S A. 1983 Jul;80(13):3963–3965. doi: 10.1073/pnas.80.13.3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boernke W. E., Stevens F. J., Peraino C. Effects of self-association of ornithine aminotransferase on its physicochemical characteristics. Biochemistry. 1981 Jan 6;20(1):115–121. doi: 10.1021/bi00504a020. [DOI] [PubMed] [Google Scholar]
- Chirgwin J. M., Przybyla A. E., MacDonald R. J., Rutter W. J. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry. 1979 Nov 27;18(24):5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- Dale R. M., McClure B. A., Houchins J. P. A rapid single-stranded cloning strategy for producing a sequential series of overlapping clones for use in DNA sequencing: application to sequencing the corn mitochondrial 18 S rDNA. Plasmid. 1985 Jan;13(1):31–40. doi: 10.1016/0147-619x(85)90053-8. [DOI] [PubMed] [Google Scholar]
- Deshmukh D. R., Srivastava S. K. Purification and properties of ornithine aminotransferase from rat brain. Experientia. 1984 Apr 15;40(4):357–359. doi: 10.1007/BF01952550. [DOI] [PubMed] [Google Scholar]
- Erlich H. A., Cohen S. N., McDevitt H. O. Immunological detection and characterization of products translated from cloned DNA fragments. Methods Enzymol. 1979;68:443–453. doi: 10.1016/0076-6879(79)68034-5. [DOI] [PubMed] [Google Scholar]
- Ford G. C., Eichele G., Jansonius J. N. Three-dimensional structure of a pyridoxal-phosphate-dependent enzyme, mitochondrial aspartate aminotransferase. Proc Natl Acad Sci U S A. 1980 May;77(5):2559–2563. doi: 10.1073/pnas.77.5.2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto T., Wang J. C. Yeast DNA topoisomerase II is encoded by a single-copy, essential gene. Cell. 1984 Apr;36(4):1073–1080. doi: 10.1016/0092-8674(84)90057-6. [DOI] [PubMed] [Google Scholar]
- Graf-Hausner U., Wilson K. J., Christen P. The covalent structure of mitochondrial aspartate aminotransferase from chicken. Identification of segments of the polypeptide chain invariant specifically in the mitochondrial isoenzyme. J Biol Chem. 1983 Jul 25;258(14):8813–8826. [PubMed] [Google Scholar]
- Hayasaka S., Shiono T., Takaku Y., Mizuno K. Ornithine ketoacid aminotransferase in the bovine eye. Invest Ophthalmol Vis Sci. 1980 Dec;19(12):1457–1460. [PubMed] [Google Scholar]
- Herzfeld A., Knox W. E. The properties, developmental formation, and estrogen induction of ornithine aminotransferase in rat tissues. J Biol Chem. 1968 Jun 25;243(12):3327–3332. [PubMed] [Google Scholar]
- Hewick R. M., Hunkapiller M. W., Hood L. E., Dreyer W. J. A gas-liquid solid phase peptide and protein sequenator. J Biol Chem. 1981 Aug 10;256(15):7990–7997. [PubMed] [Google Scholar]
- Himeno M., Mueckler M. M., Gonzalez F. J., Pitot H. C. Cloning of DNA complementary to ornithine aminotransferase mRNA. J Biol Chem. 1982 May 10;257(9):4669–4672. [PubMed] [Google Scholar]
- Horwich A. L., Fenton W. A., Williams K. R., Kalousek F., Kraus J. P., Doolittle R. F., Konigsberg W., Rosenberg L. E. Structure and expression of a complementary DNA for the nuclear coded precursor of human mitochondrial ornithine transcarbamylase. Science. 1984 Jun 8;224(4653):1068–1074. doi: 10.1126/science.6372096. [DOI] [PubMed] [Google Scholar]
- Hunkapiller M. W., Hood L. E. Analysis of phenylthiohydantoins by ultrasensitive gradient high-performance liquid chromatography. Methods Enzymol. 1983;91:486–493. doi: 10.1016/s0076-6879(83)91045-5. [DOI] [PubMed] [Google Scholar]
- Kaiser-Kupfer M. I., Valle D., Del Valle L. A. A specific enzyme defect in gyrate atrophy. Am J Ophthalmol. 1978 Feb;85(2):200–204. doi: 10.1016/s0002-9394(14)75948-3. [DOI] [PubMed] [Google Scholar]
- Kemp D. J., Coppel R. L., Cowman A. F., Saint R. B., Brown G. V., Anders R. F. Expression of Plasmodium falciparum blood-stage antigens in Escherichia coli: detection with antibodies from immune humans. Proc Natl Acad Sci U S A. 1983 Jun;80(12):3787–3791. doi: 10.1073/pnas.80.12.3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemp D. J., Cowman A. F. Direct immunoassay for detecting Escherichia coli colonies that contain polypeptides encoded by cloned DNA segments. Proc Natl Acad Sci U S A. 1981 Jul;78(7):4520–4524. doi: 10.1073/pnas.78.7.4520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler S. W. Rapid isolation of antigens from cells with a staphylococcal protein A-antibody adsorbent: parameters of the interaction of antibody-antigen complexes with protein A. J Immunol. 1975 Dec;115(6):1617–1624. [PubMed] [Google Scholar]
- Kozak M. How do eucaryotic ribosomes select initiation regions in messenger RNA? Cell. 1978 Dec;15(4):1109–1123. doi: 10.1016/0092-8674(78)90039-9. [DOI] [PubMed] [Google Scholar]
- Kozak M. Possible role of flanking nucleotides in recognition of the AUG initiator codon by eukaryotic ribosomes. Nucleic Acids Res. 1981 Oct 24;9(20):5233–5252. doi: 10.1093/nar/9.20.5233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law S. W., Dugaiczyk A. Homology between the primary structure of alpha-fetoprotein, deduced from a complete cDNA sequence, and serum albumin. Nature. 1981 May 21;291(5812):201–205. doi: 10.1038/291201a0. [DOI] [PubMed] [Google Scholar]
- Lehrach H., Diamond D., Wozney J. M., Boedtker H. RNA molecular weight determinations by gel electrophoresis under denaturing conditions, a critical reexamination. Biochemistry. 1977 Oct 18;16(21):4743–4751. doi: 10.1021/bi00640a033. [DOI] [PubMed] [Google Scholar]
- Leytus S. P., Chung D. W., Kisiel W., Kurachi K., Davie E. W. Characterization of a cDNA coding for human factor X. Proc Natl Acad Sci U S A. 1984 Jun;81(12):3699–3702. doi: 10.1073/pnas.81.12.3699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis T., Jeffrey A., Kleid D. G. Nucleotide sequence of the rightward operator of phage lambda. Proc Natl Acad Sci U S A. 1975 Mar;72(3):1184–1188. doi: 10.1073/pnas.72.3.1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzawa T., Katsunuma T., Katunuma N. Crystallization of ornithine transaminase and its properties. Biochem Biophys Res Commun. 1968 Jul 26;32(2):161–166. doi: 10.1016/0006-291x(68)90363-x. [DOI] [PubMed] [Google Scholar]
- Mueckler M. M., Himeno M., Pitot H. C. In vitro synthesis and processing of a precursor to ornithine aminotransferase. J Biol Chem. 1982 Jun 25;257(12):7178–7180. [PubMed] [Google Scholar]
- Mueckler M. M., Merrill M. J., Pitot H. C. Translational and pretranslational control of ornithine aminotransferase synthesis in rat liver. J Biol Chem. 1983 May 25;258(10):6109–6114. [PubMed] [Google Scholar]
- Mueckler M. M., Moran S., Pitot H. C. Transcriptional control of ornithine aminotransferase synthesis in rat kidney by estrogen and thyroid hormone. J Biol Chem. 1984 Feb 25;259(4):2302–2305. [PubMed] [Google Scholar]
- Mueckler M. M., Pitot H. C. Sequence of the precursor to rat ornithine aminotransferase deduced from a cDNA clone. J Biol Chem. 1985 Oct 25;260(24):12993–12997. [PubMed] [Google Scholar]
- Norrander J., Kempe T., Messing J. Construction of improved M13 vectors using oligodeoxynucleotide-directed mutagenesis. Gene. 1983 Dec;26(1):101–106. doi: 10.1016/0378-1119(83)90040-9. [DOI] [PubMed] [Google Scholar]
- Ohura T., Kominami E., Tada K., Katunuma N. Crystallization and properties of human liver ornithine aminotransferase. J Biochem. 1982 Dec;92(6):1785–1792. doi: 10.1093/oxfordjournals.jbchem.a134108. [DOI] [PubMed] [Google Scholar]
- Ohura T., Kominami E., Tada K., Katunuma N. Gyrate atrophy of the choroid and retina: decreased ornithine aminotransferase concentration in cultured skin fibroblasts from patients. Clin Chim Acta. 1984 Jan 16;136(1):29–37. doi: 10.1016/0009-8981(84)90244-4. [DOI] [PubMed] [Google Scholar]
- Okayama H., Berg P. High-efficiency cloning of full-length cDNA. Mol Cell Biol. 1982 Feb;2(2):161–170. doi: 10.1128/mcb.2.2.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelham H. R., Jackson R. J. An efficient mRNA-dependent translation system from reticulocyte lysates. Eur J Biochem. 1976 Aug 1;67(1):247–256. doi: 10.1111/j.1432-1033.1976.tb10656.x. [DOI] [PubMed] [Google Scholar]
- Peterson R. C., Cheung L. C., Mattaliano R. J., Chang L. M., Bollum F. J. Molecular cloning of human terminal deoxynucleotidyltransferase. Proc Natl Acad Sci U S A. 1984 Jul;81(14):4363–4367. doi: 10.1073/pnas.81.14.4363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proudfoot N. J., Brownlee G. G. 3' non-coding region sequences in eukaryotic messenger RNA. Nature. 1976 Sep 16;263(5574):211–214. doi: 10.1038/263211a0. [DOI] [PubMed] [Google Scholar]
- Sanada Y., Suemori I., Katunuma N. Properties of ornithine aminotransferase from rat liver, kidney and small intestine. Biochim Biophys Acta. 1970 Oct 14;220(1):42–50. doi: 10.1016/0005-2744(70)90227-5. [DOI] [PubMed] [Google Scholar]
- Schatz G., Butow R. A. How are proteins imported into mitochondria? Cell. 1983 Feb;32(2):316–318. doi: 10.1016/0092-8674(83)90450-6. [DOI] [PubMed] [Google Scholar]
- Schwarzbauer J. E., Tamkun J. W., Lemischka I. R., Hynes R. O. Three different fibronectin mRNAs arise by alternative splicing within the coding region. Cell. 1983 Dec;35(2 Pt 1):421–431. doi: 10.1016/0092-8674(83)90175-7. [DOI] [PubMed] [Google Scholar]
- Shih V. E., Berson E. L., Mandell R., Schmidt S. Y. Ornithine ketoacid transaminase deficiency in gyrate atrophy of the choroid and retina. Am J Hum Genet. 1978 Mar;30(2):174–179. [PMC free article] [PubMed] [Google Scholar]
- Shih V. E. Regulation of ornithine metabolism. Enzyme. 1981;26(5):254–258. doi: 10.1159/000459187. [DOI] [PubMed] [Google Scholar]
- Shiono T., Hayasaka S., Mizuno K. Presence of ornithine ketoacid aminotransferase in human ocular tissues. Graefes Arch Clin Exp Ophthalmol. 1982;218(1):34–36. doi: 10.1007/BF02134098. [DOI] [PubMed] [Google Scholar]
- Steiner D. F., Quinn P. S., Chan S. J., Marsh J., Tager H. S. Processing mechanisms in the biosynthesis of proteins. Ann N Y Acad Sci. 1980;343:1–16. doi: 10.1111/j.1749-6632.1980.tb47238.x. [DOI] [PubMed] [Google Scholar]
- Trijbels J. M., Sengers R. C., Bakkeren J. A., De Kort A. F., Deutman A. F. L-Ornithine-ketoacid-transaminase deficiency in cultured fibroblasts of a patient with hyperornithinaemia and gyrate atrophy of the choroid and retina. Clin Chim Acta. 1977 Sep 1;79(2):371–377. doi: 10.1016/0009-8981(77)90431-4. [DOI] [PubMed] [Google Scholar]
- Valle D., Kaiser-Kupfer M. I., Del Valle L. A. Gyrate atrophy of the choroid and retina: deficiency of ornithine aminotransferase in transformed lymphocytes. Proc Natl Acad Sci U S A. 1977 Nov;74(11):5159–5161. doi: 10.1073/pnas.74.11.5159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volpe P., Sawamura R., Strecker H. J. Control of ornithin delta-transaminase in rat liver and kidney. J Biol Chem. 1969 Feb 25;244(4):719–726. [PubMed] [Google Scholar]
- Young R. A., Davis R. W. Efficient isolation of genes by using antibody probes. Proc Natl Acad Sci U S A. 1983 Mar;80(5):1194–1198. doi: 10.1073/pnas.80.5.1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young R. A., Davis R. W. Yeast RNA polymerase II genes: isolation with antibody probes. Science. 1983 Nov 18;222(4625):778–782. doi: 10.1126/science.6356359. [DOI] [PubMed] [Google Scholar]
- Zelenka P., Piatigorsky J. Isolation and in vitro translation of delta-crystallin mRNA from embryonic chick lens fibers. Proc Natl Acad Sci U S A. 1974 May;71(5):1896–1900. doi: 10.1073/pnas.71.5.1896. [DOI] [PMC free article] [PubMed] [Google Scholar]