Abstract
The molecular phenotype of tumor vasculature is different from normal vasculature, offering new opportunities for diagnosis and therapy of cancer, but the identification of tumor-restricted targets remains a challenge. We investigated 13 tumor vascular markers (TVMs) from 50 candidates identified through expression profiling of ovarian cancer vascular cells and selected to be either transmembrane or secreted, and to be either absent or expressed at low levels in normal tissues while overexpressed in tumors, based on analysis of 1,110 normal and tumor tissues from publicly available Affymetrix microarray data. Tumor-specific expression of each TVM was confirmed at the protein level in tumor tissue and/or in serum. Among the 13 TVMs, 11 were expressed on tumor vascular endothelium; the remaining 2 TVMs were expressed by tumor leukocytes. Our results demonstrate that certain transmembrane TVMs such as ADAM12 and CDCP1 are selectively expressed in tumor vasculature and represent promising targets for vascular imaging or anti-vascular therapy of epithelial ovarian cancer, while secreted or shed molecules such as TNFRSF21/DR6 can function as serum biomarkers. We have identified novel tumor-specific vasculature markers which appear promising for cancer serum diagnostics, molecular imaging and/or therapeutic targeting applications and warrant further clinical development.
Key words: ovarian, cancer, vascular, biomarkers, diagnostics, serum, expression, profiling
Introduction
Tumor vasculature is different from normal vasculature at the molecular level.1–6 Tumor vessels are enmeshed in a complex tissue, which includes tumor cells, stromal cells and leukocytes. Interactions with these cells and their products (growth factors, cytokines etc.) alter the protein repertoire of endothelial cells, conferring a tumor-specific phenotype. The unique molecular features of the tumor vasculature offer novel and largely unexploited opportunities for cancer diagnosis and therapy.7 In fact, molecules secreted or cleaved off the tumor vasculature could provide biomarkers for serum tumor detection. In addition, vascular surface molecules could be targeted for molecular imaging. Because the angiogenic switch occurs when solid tumors reach few mm in diameter,8,9 it is expected that many of these phenotypic changes occur at the vasculature of tumors at the occult stage. Provided sensitive and specific affinity probes are developed, one can envision these targets serving as markers for early tumor serum detection or visualization of clinically occult tumors. Similarly, surface vascular molecules can be targeted therapeutically to achieve destruction of established tumors or to prevent solid tumor growth by disrupting the tumor vasculature. Recent encouraging results with VEGF blockade have demonstrated the feasibility and efficacy of targeting tumor angiogenesis,10–13 while vascular disrupting agents have produced dramatic results in preclinical models.14–16 Thus, vascular markers can serve important functions in tumor detection and therapy.
Important discovery work in the area of tumor vascular targeting has already been done6,17,18 and selective markers for tumor vasculature have been identified, including the tumor endothelial marker 1 (TEM1) and other TEMs; the extracellular domain B isoform of fibronectin; exposed phosphatidylserine; the prostate specific membrane antigen (PSMA); and integrin αvβ3.19 Some of these targets are being tested clinically.17,20–22 However, the heterogeneous expression of endothelial markers in tumors and the lack of suitable imaging beacons and effector molecules have delayed harnessing the potential of these targets. Thus, identifying additional markers of tumor vasculature and developing affinity reagents is a high priority.
We previously performed genome-wide high throughput discovery to characterize the signature of epithelial ovarian cancer (EOC) vasculature1,23 using gene expression profiling of CD31+ CD146+ ovarian tumor vascular cells (TVCs) isolated by immunohistochemistry-assisted laser capture microdissection (immuno-LCM).23 In our previous study, we validated our approach by demonstrating that 12 randomly selected genes were indeed tumor vascular specific. However, the protein products of many of these genes are intracellular and thus not useful for tumor diagnosis or surface targeting. To serve as imaging or therapy targets, candidate tumor vascular marker (TVM) molecules need to be expressed on the cell surface and be uniquely expressed or overexpressed by tumor vessels, while not expressed or present at significantly lower levels in normal tissues. Additionally, to serve as serum biomarkers, molecules need to be shed in the bloodstream by virtue of secretion or cleavage. Thus, careful selection of candidates and detailed validation is necessary for the identification of suitable candidates for further clinical development.
In this study we present a more focused investigation in order to identify TVMs that fulfil the above criteria. In particular, from our previous gene expression profiles of vascular cells micro-dissected from ovarian tumors and normal ovaries we identified 162 tumor vasculature-associated genes with transmembrane or secreted protein products. We used public Affymetrix data obtained from 1,110 normal and cancer tissues followed by quantitative RT-PCR validation to further select 50 genes with low or no expression in normal tissues. Expression and localization of 13 candidates was then validated at the mRNA and protein level in ovarian cancer tissue or serum. Receiver operating characteristic (ROC) curves were used to assess the potential clinical utility of 3 TVMs using serum samples from 19 cancer-free women with normal ovaries and 61 patients with EOC. We identified a number of novel tumor vascular biomarkers, which are suitable targets for serum diagnostics, vascular imaging or antivascular therapy of ovarian cancer.
Results
Identification of tumor vascular marker candidates.
We identified a more comprehensive list of genes using gene expression data from our previous study1,23 using two criteria for differential expression. The first used a nonparametric analysis in which the ranks for gene expression values were computed for each gene. Genes were included in the list if the sum of the ranks for the normal tissues was less than 20. For example, if the four normal specimens showed the lowest expression compared with all tumor samples, then they would rank in position 1, 2, 3 and 4, and the sum of their ranks would be equal to 10 (= 1 + 2 + 3 + 4) and this gene was selected for further characterization. The second used a parametric analysis that compared the ratio of gene expression in tumor cells compared with normal vascular cells for each gene. Genes were included in the list when a gene was at least 2-fold overexpressed. There were 17,920 genes included in this analysis after elimination of genes where the difference between tumor and normal mean expression level was less than its standard error. We identified 230 genes and 50 Expressed sequence tags (ESTs) upregulated in ovarian tumor vasculature as compared with normal ovarian vasculature, which were then considered the putative pool of genes from which tumor vascular biomarkers could be identified, a significantly higher number than the 70 genes in our previous study.1
We performed comprehensive review of the literature on PubMed for the above genes. For genes without publicly available data, we used Biology Workbench-UCSD Bioinformatics and Systems Biology Group (www.workbench.sdsc.edu/) to predict the presence of transmembrane domains. We thus classified the genes in the following groups, based on known or putative location of their protein products: (a) transmembrane (n = 98); (b) secreted/unknown (n = 64); and (c) intracellular (n = 71) (Table S1). The 162 genes with transmembrane or secreted/unknown protein products were candidates for further validation.
Identification of TVM with low expression in normal tissues.
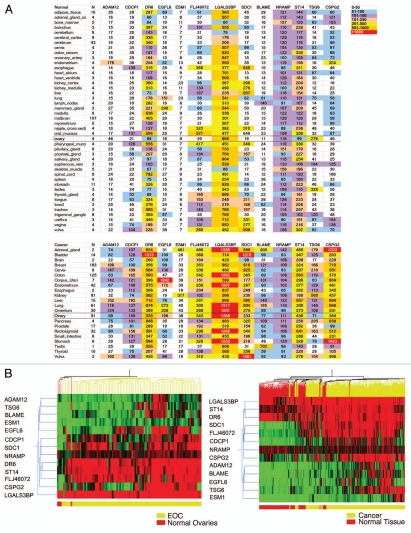
We used publicly available Affymetrix microarray data (Gene Expression Omnibus (GEO) Data sets) to evaluate the 162 putative transmembrane and/or secreted TVM candidates for expression in 355 normal human tissues and 755 human tumors. Figure 1A shows an example of this analysis for 13 genes. The average expression in tumors and in normal tissues was used to identify genes with low expression in normal tissues. Unsupervised hierarchical clustering further revealed the difference in gene expression between normal tissues and tumors (Fig. 1B). This high throughput approach led to the identification of 50 tumor vascular marker candidates with low or no expression in normal tissues and higher expression in tumors for further validation by qRT-PCR.
Figure 1.
(A) Heat-map summary of gene expression levels of 13 tumor vascular markers in normal human tissues and solid tumor types based on Affymetrix gene expression analysis. Rows represent different tissues or tumors. In the top part are shown normal tissues. Data represent 355 different tissue samples from five female and five male donors. The numbers of samples analyzed per each organ or tissue are shown in the second column. In the bottom part are shown tumors; data represent 755 tumor samples. The numbers of samples analyzed per each tumor type are shown in the second column. Columns represent genes. Each cell represents averaged Affymetrix gene expression level for each gene in each tissue or tumor. N indicates the number of different samples available for the specified tissue or tumor. For normal brain, the gene expression data set comprised multiple parts of cerebrum or mid brain, and these sites were collapsed. Color coding is arbitrary and based on scale provided at the top of list for quick data interpretation. (B) Unsupervised hierarchical clustering of 13 TVMs in four normal ovaries and 91 ovarian cancer samples derived from the above arrays (left), as well as in all 355 normal tissues and 755 tumors (right). All analyses were conducted using whole tissue Affymetrix expression data.
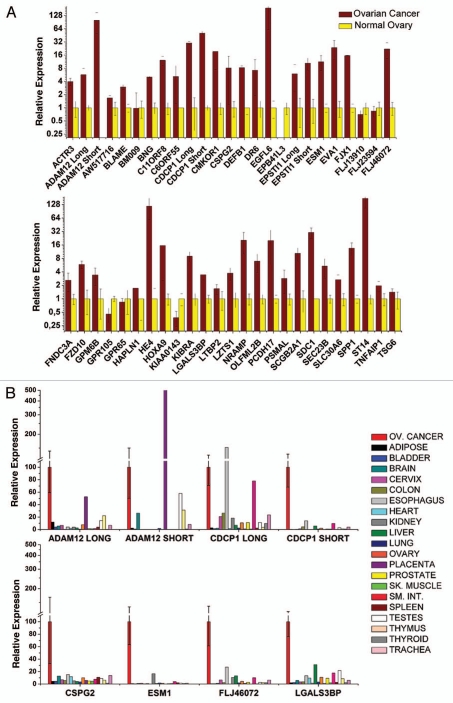
More than 30 genes were overexpressed in EOC tumors (n = 10) relative to postmenopausal normal ovary (n = 3) by qRT-PCR (Fig. 2A). Commercially available antibodies were screened and, based on the availability of reliable reagents, we selected 13 promising genes for further validation, most of which we had not validated in our previous study: ADAM metallopeptidase domain 12 (ADAM12), B-lymphocyte activator macrophage expressed (BLAME; SLAMF8), CUB domain containing protein 1 (CDCP1), chondroitin sulfate proteoglycan 2 (CSPG2; versican, VCAN), death receptor-6 (DR6; tumor necrosis factor receptor superfamily member 21, TNFRSF21), EGF-like-domain, multiple 6 (EGFL6), endothelial cell-specific molecule 1 (ESM1; endocan), FLJ46072 (FAM83H), lectin galactosidebinding soluble 3 binding protein (LGALS3BP), natural resistance-associated macrophage protein 1, (solute carrier family 11; NRAMP), syndecan 1 (SDC1), suppression of tumorigenicity 14 (ST14); and tumor necrosis factor α-induced protein 6 (TNFAIP6; TSG6). For ADAM12 and CDCP1 we used primers specific for two known splice variants. Importantly, EGFL6 and TNFRSF21/DR6 were previously validated in our previous study,1 and were used as positive controls. The 13 candidate genes were further validated with qRT-PCR in a part of 20 EOC specimens and 20 normal human tissues (Fig. 2B). Average expression was significantly higher in tumors than any other normal tissue tested for 9 out of 13 genes (ADAM12 long isoform, CDCP1 short isoform, CSPG2, EGFL6, ESM1/endocan, FLJ46072, LGALS3BP, ST14, TSG6/TNFAIP6).
Figure 2.
(A) qRT-PCR analysis of tumor vascular marker expression in ovarian cancer and normal ovaries. Expression for each cancer tissue was normalized against normal ovarian tissue, which was defined as 1 for each gene. Expression in all tissues was normalized to CD31. Means are presented with standard errors. (B) Quantitative real-time polymerase chain reaction analysis of tumor vascular marker expression in ovarian cancer and a part of normal human tissues. Expression for each normal tissue was normalized against ovarian cancer, which was defined as 100% for each gene. Expression was normalized to CD31. Means are presented with standard errors.
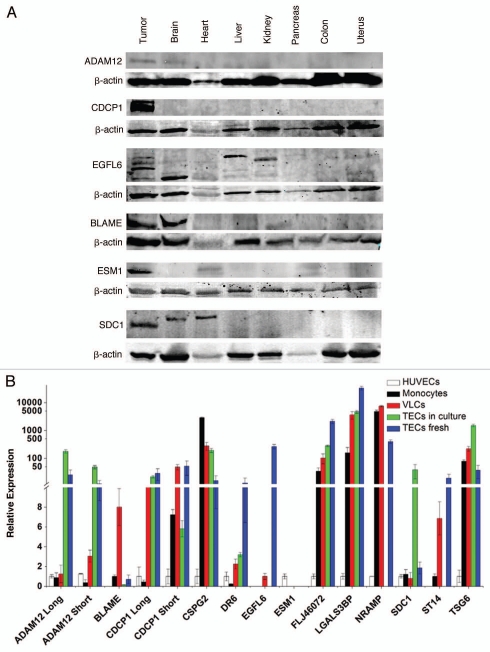
Next, the tissue expression of ADAM12, BLAME, CDCP1, EGFL6, ESM1/endocan and SDC1/syndecan proteins in ovarian cancers and normal human tissues were characterized by protein gel blot using commercially available antibodies (Fig. 3A). Validating mRNA findings, ADAM12 and CDCP1 proteins were not detected in any normal tissue checked, while a significant amount of protein was detected in EOC samples. Anti-EGFL6 antibody showed reactivity in normal brain, liver and heart, while anti-SDC1 reacted with normal brain and heart and BLAME showed reactivity in the brain. Interestingly, EGFL6 and SDC1/syndecan proteins migrated at a different molecular weight in normal tissues (Fig. 3A). This difference in molecular size may be due to differences in post-translational modification e.g., glycosylation, especially since both of these proteins have multiple glycosylation sites. Soluble factors released by tumor or inflammatory cells in the tumor microenvironment can profoundly alter the glycosylation pattern of surface proteins in endothelial cells, which is critical for their function in angiogenesis and metastasis.24,25 However, we cannot exclude the possibility of alternatively spliced isoforms expressed by tumors or the cross-reactivity of antibodies with other proteins.
Figure 3.
(A) Expression of TVM protein in ovarian cancer and normal tissues determined by protein gel blot. (B) Expression of TVMs by qRT-PCR in fresh or cultured immunopurified VE-cadherin+, CD45-TECs from cancer, VE-cadherin+ and CD45+ vascular leukocytes (VLCs) from ovarian cancer, HUVECs and monocytes from healthy donors were used for controls. Means are presented with standard errors.
TVM are expressed in tumor vasculature.
To validate that each of the newly identified TVMs is expressed by the tumor vasculature, we purified by flow sorting VE-cadherin+CD45− tumor endothelial cells and VE-cadherin+CD45+ tumor leukocytes from fresh tumor tissues and we analyzed TVM expression by qRT-PCR (Fig. 3B). With the exception of BLAME and NRAMP, we observed significantly higher expression of TVMs by the tumor endothelium compared with HUVEC, confirming that these are markers expressed by tumor endothelial cells. BLAME and NRAMP expression was absent in HUVEC and TEC, but expression was detected in tumor leukocytes as well as normal monocytes.
TVM are induced by inflammation or tumor cell hypoxia.
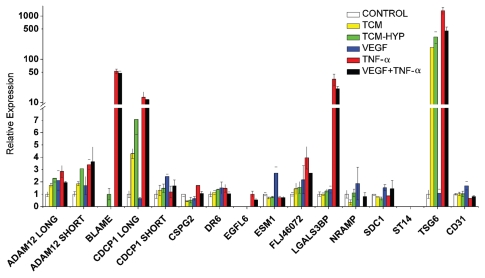
To further demonstrate that the identified genes are associated with tumor vasculature, we sought to determine whether their expression could be upregulated in quiescent endothelial cells upon exposure to conditions that mimic the tumor microenvironment. We cultured HUVEC in the presence of TNFα, a key inflammatory factor at the tumor microenvironment; VEGF, the key factor for angiogenesis; and tumor conditioned media (TCM) at normoxic conditions, to observe the impact of tumor derived growth factors at base line; or TCM generated under hypoxic conditions (tumor cells at 1.5% O2), to observe the effects of tumor cell hypoxia. Our results show that TNFα induced de novo expression of 7 out of 13 TVMs (ADAM12, BLAME, CDCP1, CSPG2/versican, EGFL6, LGALS3BP, TSG6) (Fig. 4). BLAME and EGFL6, although undetectable in untreated HUVEC, were detectable upon treatment with TNFα. CDCP1 (long isoform) and TSG6 were upregulated by TCM, and to a greater extent by media of hypoxic tumor cells. Interestingly, VEGF treatment led to only a modest increase in ESM1 and NRAMP levels. CD31 levels remained constant with all treatments. TNFα and VEGF did not show a positive interaction. On the contrary, in some cases (ADAM12 long, TSG6), TNFα treatment alone led to higher expression compared with TNFα and VEGF in combination. Thus, the identified TVMs are indeed markers of endothelium exposed to inflammatory conditions and/or factors released by hypoxic tumor cells, rather than VEGF alone.
Figure 4.
Expression of TVMs by qRT-PCR in HUVECs treated under inflammatory or tumor conditions. HUVECs were treated with Tumor Conditioned Media (TCM), Hypoxic TCM (TCM-Hyp), VEGF, TNFá or combination of VEGF and TNFá for 24 h. Means are presented with standard errors.
Validation of TVM that are suitable targets for therapy or imaging.
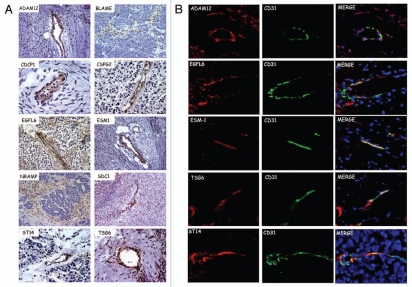
We examined whether predicted TVM targets are expressed in the tumor vasculature in vivo. Localization of the novel TVM proteins was determined by IHC (Fig. 5A). ADAM12, CDCP1, CSPG2, EGFL6, ESM1, SDC1, ST14, TSG6 and TNFRSF21/DR6 (as previously shown in ref. 1) localized to vascular-like structures in ovarian cancer but not in normal ovaries (Fig. S1). Expression of TVM proteins by tumor vessels varied between tumors, but overall stain was observed mostly in smaller, immature blood vessels. The only exceptions were BLAME and NRAMP, which were expressed in perivascular tumor stroma but not in the tumor islets. This observation is consistent with our qRT-PCR data demonstrating expression of these two genes by VLC and monocytes. Double immunofluorescent staining showed co-localization of ADAM12, ESM1/endocan, EGFL6, TSG6 and ST14 with the endothelial cell marker CD31 (Fig. 5B). Expression was not restricted to the tumor endothelium, but there was also diffuse staining in the extracellular matrix. This is consistent with the fact that EGFL6, ESM1 and TSG6 are secreted proteins, while ADAM12, CDCP1, SDC1, CSPG2 have both transmembrane and secreted forms. Thus, the tumor vasculature expresses unique protein products in vivo, which appear suitable candidates for direct targeting.
Figure 5.
(A) Immunohistochemical localization of TVMs in ovarian cancer. (B) Immunofluorescent colocalization of TVMs with CD31 in ovarian tumors. Merge includes 4,6-Diamidino-2-phenylindole (DAPI) nuclear staining.
Validation of TVM that are suitable biomarkers for serum detection.
Since tumor vascular proteins can be secreted or cleaved by vascular endothelium, we hypothesized that TVMs can be detected and informative in sera of patients with EOC. We have previously shown by protein gel blot that TNFRSF21/DR6 protein was detectable in sera of EOC patients and at higher levels compared with healthy donors.1 Availability of appropriate antibodies and peptides for protein gel blot allowed us to confirm that CDCP1 and LGALS3BP were also detectable in sera of ovarian carcinoma patients (n = 7) and at higher levels as compared with patients with benign tumors (n = 3) or healthy women (n = 3) (Fig. S2).
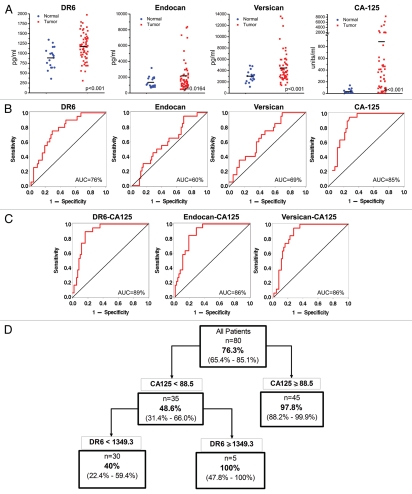
Based on commercially available reagents, we could develop bead-based quantitative immunoassays (Luminex) only for TNFRSF21/DR6, CSPG2/versican and ESM1/endocan. These TVMs were deemed suboptimal biomarkers because of expression in some normal tissues. Nevertheless, we screened blinded presurgical serum samples from 61 EOC patients (cases) and 19 healthy donors (controls) and evaluated these samples for levels of TNFRSF21/DR6, ESM1/endocan and CSPG2/versican as a proof of principle to test the notion that TVMs can be detected in serum by sensitive assays and can perform as predicted by molecular analyses of tumors. All three markers were significantly elevated in the EOC cases compared with the healthy controls (p < 0.01) (Fig. 6A). We also measured CA125 levels in these cases and controls using a commercially available Luminex assay for CA125. CA125 is a mucinous glycoprotein best known as a biomarker for ovarian cancer. It is clinically approved for following the response to treatment and predicting prognosis after treatment, being particularly useful for detecting the recurrence of ovarian cancer. As expected, CA125 levels were significantly elevated for the cases relative to normal controls (p < 0.001).
Figure 6.
DR6-ES M1-CSPG2 as serum biomarkers. Serum assays for DR6, ES M1 (endocan) and CSG2 (versican) were performed using bead-based immunoassays (Luminex assays). (A) Comparison of serum levels in the Torino cohort of normal controls (n = 19) and stage III/IV ovarian cancer cases (n = 61). A 2-tailed t-test with Welch's correction was performed to determine significance of difference of the biomarkers between the 2 groups. (B and C) Receiver operating characteristic (ROC) were computed for the indicated single (B) or composite marker (C) to compare the diagnostic accuracy as measured by the area under the curve (AUC). (D) Recursive partitioning was used to define a classification tree. A biomarker and a cut point is selected among all biomarkers and cut points to identify the homogeneous groups for the clinical outcome. Each subgroup is then split using the same algorithm until there is no further improvement in classification.
The ROC curves for the three TVM and CA125 were computed to assess their diagnostic accuracy to distinguish between normal and cancer samples. Compared with CA125, all three TVM individually performed suboptimally, with area under the ROC curve (AUC) values ranging from 60–76%, compared with the AUC value of 85% for CA125 (Fig. 6B). Logistic regression was used to develop predictive models, one for each TVM with CA125. The performance of the pairs of biomarkers was characterized by the ROC curve for the predicted probability of cancer using the predictive model. The AUCs for the predicted values of each model increased to 89, 86, 86%, for the composite markers of DR6-CA125, endocan-CA125 or versican-CA125 (Fig. 6C), respectively. However, the model fit improved significantly only when DR6 or endocan was paired with CA125.
A classification tree was developed to predict EOC cancer using TNFRSF21/DR6 and CA125 (Fig. 6D). This algorithm selected the best biomarkers from the four biomarkers, and a cut point for the selected marker at each decision point, to minimize misclassification errors. Based on this classification tree, a prediction rule that identified all samples when CA125 is greater than 88.5 units/mL or when CA125 is less than 88.5 units/mL but TNFRSF21/DR6 is greater than 1,349.3 pg/mL, identified 18 out of 19 of the normal cases (specificity = 95%; 95%-CI = 74–100%) and 50 out of 61 cancers (sensitivity = 82%; 95%-CI = 68–89%). This rule had improved specificity compared with the standard rule based on CA125 alone, where all samples with CA125 greater than or equal to 35 are classified as cancer.14 In this sample, the specificity and sensitivity of the prediction rule based on CA125 alone was 63% (95% CI = 38–84%) and 82% (95% CI = 70–91%), respectively.
Discussion
Although the bulk of research on the development of biomarkers and therapeutic targets has focused on the tumor cell, the role of host cell populations in tumor growth has become quite apparent, and the investigation of such populations has the potential to identify additional biomarkers and therapeutic targets. Isolating and studying the small population of tumor vascular cells has been quite challenging. However, we have been able to characterize the gene expression signature of these cells in vivo with the use of immunohistochemistry-guided laser capture microdissection coupled with transcriptional profiling. We analyzed all genes we have identified to date from the ovarian cancer vasculature and applied a number of filters to identify tumor-specific candidates for diagnosis or therapy. First, we selected TVMs with known or putative transmembrane or secretory protein products. Then, we used publicly available Affymetrix expression data for over 1,000 normal and cancer tissues to select genes with low or no expression in normal tissues. Through this approach we identified 50 candidate genes, which were then validated by qRT-PCR. We chose 13 genes for detailed validation based on affinity reagents that could reliably identify the protein products. Most of these genes play important roles in processes that are in fact vital for tumor growth, such as regulation of endothelial permeability, leukocyte extravasation,26 cell adhesion,27,28 extracellular matrix degradation29–31 and metastasis.32
Nine of the genes (ADAM12, CDCP1, CSPG2, EGFL6, ESM1, FLJ46072, LGALS3BP, ST14 and TSG6) exhibited higher expression in ovarian cancer relative to normal tissues. It should be noted that these expression data are based on whole tissue analysis, but we normalized against endothelial gene CD31 to eliminate the influence of vascular density. The difference in expression between cancer and normal was quite pronounced when we compared purified endothelial cells. Thus, it is plausible that targeted imaging or therapy may reveal a higher tumor to normal ratio than analysis of whole tissue would suggest. After confirmation of our results at the protein level, we conclude that select transmembrane (ADAM12, CDCP1) as well as secreted (ESM1, EGFL6) proteins represent promising tumor vascular targets for imaging or therapy of EOC. These add to TVM genes identified in our previous work1 (adlican, COL11A1, F2RL1, FZD10 and OLFML2B), which also provide good candidates for therapeutic or imaging strategies. ESM1 showed low expression in freshly isolated TECs relative to HUVECs at the RNA level, but this may be due to sampling bias, as we show that ESM1 is expressed only in a subset of tumors, or to mRNA instability. The expression of ESM1 specifically in tumor endothelial cells (TECs) was established at the protein level in tumor tissue by immunohistochemistry. In addition, BLAME was not expressed at the RNA level in immunopiurified tumor endothelial cells, but was expressed by tumor-purified vascular related leukocytes (VLCs). These cells of monocyte-macrophage lineage associate structurally with tumor vasculature and perivascular stroma, and may play an important role in tumor vascular development.33 Pericyte-like expression of BLAME was confirmed by immunostaining. Because BLAME is not expressed by PBMC, it could be an interesting therapeutic target.
Tumor vascularization is a dynamic biological process regulated by complex microenvironment conditions that cannot be precisely recapitulated in vitro. Nevertheless, we attempted to subject HUVEC to known environmental/paracrine factors that are likely to play an important role in the tumor vasculature milieu: normoxic or hypoxic tumor cell supernatants; VEGF; and inflammatory mediators activating endothelial NFκB (TNFα). Under these controlled laboratory conditions that only in part reproduce the tumor microenvironment, 10 of 16 candidate genes were upregulated. For example, BLAME, EGFL6 and LGALS3BP showed strong expression after exposure to TNFα, while the expression was undetectable at baseline. CDCP1 (long) showed over 10-fold increase under tumor conditioned media as well as TNFα; TSG6 expression increased over 100-fold by exposure to tumor cell supernatants and almost 1,000-fold by TNFα. Other genes showed a more moderate or no increase. Interestingly, none of the gene candidates were upregulated by VEGF alone, suggesting that the signature we have uncovered does not pertain to physiologic angiogenesis, where VEGF is upregulated but inflammation or other tumor-derived paracrine factors are absent.
Our discovery rules required that vascular endothelial gene candidates be upregulated in many, not all, tumor vascular endothelial cell samples relative to normal endothelial cells. Thus, some of the identified genes were expressed at low levels in some tumors. This does not diminish their potential importance as tumor vascular markers or therapeutic targets in a subset of patients. Importantly, this is the case for tumor vascular markers that have already been validated and are currently being developed as therapeutic targets, e.g., TEM1,34 for which therapeutic antibodies are already under early phase clinical testing. Ideally, for the purpose of tumor detection, development of molecular imaging tools should focus on genes that are highly expressed in most EOC tumors. For example, as shown in Figure 1B, genes such as EGFL6, CDCP1 and FLJ46072 seem to be consistently elevated in most ovarian cancer samples. Thus, especially cell surface proteins may be suitable targets for tumor imaging. Of note, some markers were found to have high expression in some normal tissue. These could still serve as useful imaging targets, as long as one is aware of their expression in the normal adult. For example, BLAME shows its highest levels of expression in tonsil and lung tissue and therefore could be suitable for abdominal imaging. Furthermore, EGFL6 was only highly expressed in mammary gland, which again makes it a suitable target for abdominal imaging. Cell surface vascular proteins that are highly expressed in most tumors could be universal targets for therapy. In particular, the short CDCP1 isoform and EGFL6 appear promising candidates for therapy as normal tissues exhibit mostly undetectable expression (except for placenta in the case of EGFL6). On the other hand, ADAM12 is very highly expressed in some EOC samples compared with normal ovaries but a significant number of cancer samples exhibited low levels of ADAM12. Thus, ADAM12 may hold predictive value, and it could be used for therapy, especially the long isoform, for patients whose tumors express it. It is important to note that all of these molecules exhibit high levels of expression in many other solid tumors as well (Fig. 1), which underlines the possible application of these targeted approaches in cancers other than ovarian.
Cellular and molecular changes characterizing the angiogenic switch are essential for tumor growth beyond few millimeters.7 Interestingly, specialization of the vasculature may even precede tumor establishment at metastatic sites.35 We tested the notion that secreted or cleaved vascular proteins can function as serum biomarkers. We were restricted by the availability of antibodies for Luminex to the validation of three biomarkers: DR6, ESM1 and CSPG2. The discovery of DR6 as a tumor vascular marker was previous described.1 In the present manuscript we show the development of a new assay for detecting DR6 in serum using Luminex technology, followed by screening and evaluation of serum samples from patients and healthy individuals, and evaluation of DR6-based screening as well as a composite screening with both DR6 and CA-125 for increasing EOC detection specificity and sensitivity. DR6, ESM1 and CSPG2 were predicted to differentiate cancer from control cases but with suboptimal specificity, based on expression in normal tissues and suboptimal sensitivity based on lack of expression in many tumors. Screening sera from 61 EOC and 19 normal controls confirmed the predicted performance of these markers in cancer discrimination. All three markers could differentiate between normal controls and cancer patients. Furthermore, the bivariable logistic regression models that used two biomarkers, either CA125 and TNFRSF21/DR6 or CA125 and ESM1/endocan, fit the data better than the univariable model with CA125 alone. Classification using both CA125 and DR6 in patients with CA125 less than 89 IU/ml improved specificity with no change in sensitivity. Thus, DR6 appears promising for future biomarker validation in larger patient cohorts. It should be noted that these three markers were chosen as Luminex substrates merely based on feasibility, as proof of principle. It is possible that other TVM, predicted by our tissue-based analyses to have better discriminatory value, might in fact perform better as serum biomarkers and development of affinity reagents and assays then becomes a high priority.
In summary, our work emphasizes that the field of tumor vascular biology may provide applications in cancer therapeutics and diagnostics and has provided novel targets for further clinical development.
Materials & Methods
Gene expression array data.
The Affymetrix gene expression data comparing microdissected vascular endothelial cells from 21 stage III ovarian cancers and 4 normal ovaries has been reported earlier in reference 23. Data used for expression of the TVMs in normal and tumor tissue samples are available via the Gene Expression Omnibus (GEO; National Center for Biotechnology Information [NCBI], www.ncbi.nlm.nih.gov/geo) with series numbers GSE3526 (n = 355 including 4 normal ovaries) and GSE2109 (n = 755 including 91 ovarian cancers), respectively. All CEL files were downloaded and similarly processed using the rate monotonic algorithm (RMA).13
Tissues and serum samples.
Clinically annotated epithelial ovarian cancer (EOC) samples from patients were provided by the University of Turin (Turin, Italy). Stage III or IV EOC fresh specimens were obtained at the University of Pennsylvania. Tissue was snap frozen in liquid nitrogen and stored at −80°C until use. Normal human tissues were provided by the Cooperative Human Tissue which provides no clinical information. Human total RNA survey part was purchased from Ambion (Austin, TX). Serum samples from ovarian cancer cases and disease-free controls used to develop the serum assays were from the Biosample Repository at Fox Chase Cancer Center (FCCC). Pre-surgical serum samples from EOC patients were provided by the University of Turin, Turin, Italy (19 normal and 61 ovarian cancer samples, referred to as the Torino Cohort). All specimens were processed in compliance with institutional review board and Health Insurance Portability and Accountability Act (HIPAA) requirements.
Antibodies.
The following antibodies were used: for fluorescence-associated cell sorting (FACS), we used PE-conjugated anti-VE-cadherin (eBioscience, San Diego, CA) and APC-conjugated anti-CD45, (BD PharMingen, Franklin Lakes, NJ). For immunostaining and protein gel blot experiments we used anti-ESM1, anti-SDC-1, anti-TSG6, anti-BLAME, anti-LGALS3BP, and anti-CSPG2 (R&D Systems, Minneapolis, MN), anti-EGFL6 (Santa Cruz Biotechnology, Santa Cruz, CA), anti-NRAMP (Alpha Diagnostic, San Antonio, TX), anti-ST14 (Abnova, Taiwan), anti-CDCP1 (Novus Biologicals, Littleton, CO), anti-ADAM12 (Abcam, Cambridge, MA), mouse anti-β-actin (SIGMA, St. Louis, MO), Alexa 680-conjugated anti-rabbit (Invitrogen, Carlsbad, CA), and IRDye 800-conjugated anti-mouse (Rockland, Gilbertsville, PA). For the bead-based Luminex assays, we used capture and biotinylated detection antibodies against TNFRSF21/DR6, ESM1/endocan and CSPG2/versican from R&D Systems. For CSPG2/versican, biotinylation of the detection antibody was performed using the EZ-link biotinylation kit from Pierce (Rockford, IL) following the manufacturer's protocol.
Cells.
VE-cadherin+CD45− tumor endothelial cells (TEC) and VE-cadherin+CD45+ leukocytes were isolated from mechanically dispersed fresh human epithelial ovarian cancer specimens. Human umbilical-vein endothelial cells (HUVEC) were purchased from ATCC and maintained in EGM-2 media (Lonza, Walkersville, MD) containing 5% fetal bovine serum (FBS). Human ovarian cancer cell line A2008 was grown in RPMI containing 10% FBS. All cells were maintained at 37°C, 5% CO2 in humidified air. Tumor conditioned media (TCM) were generated from A2008 cells grown to 60% confluence in normal media, then serum starved for 4 h in 0.5% FBS media and incubated for 24 h in 0.5% FBS EBM-2 media (Cambrex, Charles City, IA) under normoxia or hypoxia (1.5% O2) conditions in a Hera Cell 240 (ThermoFisher, Asdhenville, NC). In some experiments, HUVEC were grown to confluence, serum starved overnight, and then incubated in hypoxic or normoxic TCM, or 0.5% FBS EBM-2 in the presence or absence of TNFα (25 ng/ml, Peprotech, Rocky Hill, NJ) or VEGF (100 ng/ml, Peprotech, Rocky Hill, NJ) for 24 h.
Immunostaining.
For validation studies, 8 µm sections of OCT embedded frozen tissues were immunostained as previously described in reference 36, using the VECTASTAIN ABC kit (Vector, Burlingame, CA). Immunoreaction was visualized with 3,3′-diaminobenzidine (Vector). Immunofluorescence was performed as previously described in reference 36. All staining steps were performed at room temperature.
RNA isolation and RT-PCR.
Fresh tissue or sections of tumors embedded in OCT were dissolved in Trizol reagent (Gibco, Carlsbad, CA). RNA was extracted as recommended by the manufacturer. RNA integrity and quantity were assayed using the Bioanalyzer (Agilent, Foster City, CA). Reverse transcription polymerase chain reaction (RT-PCR) was performed using Superscript first-strand synthesis system (Invitrogen, Carlsbad, CA). Quantitative real-time PCR (qPCR) was performed using primers to the 3′ end of transcripts spanning intron-exon boundaries whenever possible (Table S2). qPCR was performed for 40 cycles using SYBR Green (ABI, Foster City, CA) as recommended by the manufacturer, with primers at 100 nM concentrations. Expression of each gene in each tissue sample was normalized to CD31 expression levels in the same sample.
Protein extraction and protein gel blot.
Fifteen 30 µm sections of OCT embedded tumors were cut and dispersed in cold PBS and centrifuged at 4°C. Pellets were dissolved in 100 µl RIPA buffer and incubated on ice for 20 min. Then samples were centrifuged and supernatants containing the protein were stored at −20°C. Total protein was quantified with the use of BCA protein assay kit (Pierce, Rockford, IL). Protein gel blots were scanned using the Odyssey Infra Red system (Li-Cor, Lincoln, NE). Band intensities were quantified using the Odyssey software v2.1 (Li-Cor).
Luminex serum assays.
Bead-based immunoassays for detection of TNFRSF21/DR6, ESM1/endocan and CSPG2/versican in serum samples were developed (detailed protocol in Sup. Materials).
Statistical analyses.
All means are presented with standard errors. Two-sample t-tests were used with Satterthwaitte's method when variances in the two groups were unequal. ROC curves and AUC were computed using PROC Logistic in SAS version 9.1. The significance of adding each TVM to CA125 was evaluated using the likelihood ratio statistic. Confidence intervals for sensitivity and specificity were obtained using the binomial distribution. Recursive partitioning was used to develop the classification tree in CART version 6.
Acknowledgments
This work was supported in part by the Ovarian Cancer SPORE at FCCC/UPenn P50 CA083638, the Department of Defense grant W81XWH-06-1-0143, the Ovarian Cancer Research Fund, the Honorable Tina Brozman Foundation, N01-CN-43309 and 5U01CA113916; and a grant by the Canary Foundation.
Supplementary Material
References
- 1.Buckanovich RJ, Sasaroli D, O'Brien-Jenkins A, Botbyl J, Hammond R, Katsaros D, et al. Tumor vascular proteins as biomarkers in ovarian cancer. J Clin Oncol. 2007;25:852–861. doi: 10.1200/JCO.2006.08.8583. [DOI] [PubMed] [Google Scholar]
- 2.Bhati R, Patterson C, Livasy CA, Fan C, Ketelsen D, Hu Z, et al. Molecular characterization of human breast tumor vascular cells. Am J Pathol. 2008;172:1381–1390. doi: 10.2353/ajpath.2008.070988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lu C, Bonome T, Li Y, Kamat AA, Han LY, Schmandt R, et al. Gene alterations identified by expression profiling in tumor-associated endothelial cells from invasive ovarian carcinoma. Cancer Res. 2007;67:1757–1768. doi: 10.1158/0008-5472.CAN-06-3700. [DOI] [PubMed] [Google Scholar]
- 4.Madden SL, Cook BP, Nacht M, Weber WD, Callahan MR, Jiang Y, et al. Vascular gene expression in non-neoplastic and malignant brain. Am J Pathol. 2004;165:601–608. doi: 10.1016/S0002-9440(10)63324-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Parker BS, Argani P, Cook BP, Liangfeng H, Chartrand SD, Zhang M, et al. Alterations in vascular gene expression in invasive breast carcinoma. Cancer Res. 2004;64:7857–7866. doi: 10.1158/0008-5472.CAN-04-1976.. [DOI] [PubMed] [Google Scholar]
- 6.St. Croix B, Rago C, Velculescu V, Traverso G, Romans KE, Montgomery E, et al. Genes expressed in human tumor endothelium. Science. 2000;289:1197–1202. doi: 10.1126/science.289.5482.1197. [DOI] [PubMed] [Google Scholar]
- 7.Folkman J. Role of angiogenesis in tumor growth and metastasis. Semin Oncol. 2002;29:15–18. doi: 10.1053/sonc.2002.37263. [DOI] [PubMed] [Google Scholar]
- 8.Folkman J. Angiogenesis. Annu Rev Med. 200;57:1–18. doi: 10.1146/annurev.med.57.121304.131306. [DOI] [PubMed] [Google Scholar]
- 9.Hanahan D, Folkman J. Patterns and emerging mechanisms of the angiogenic switch during tumorigenesis. Cell. 1996;86:353–364. doi: 10.1016/S0092-8674(00)80108-7. [DOI] [PubMed] [Google Scholar]
- 10.Van Cutsem E, Vervenne WL, Bennouna J, Humblet Y, Gill S, Van Laethem JL, et al. Phase III trial of bevacizumab in combination with gemcitabine and erlotinib in patients with metastatic pancreatic cancer. J Clin Oncol. 2009;27:2231–2237. doi: 10.1200/JCO.2008.20.0238. [DOI] [PubMed] [Google Scholar]
- 11.Hurwitz H, Fehrenbacher L, Novotny W, Cartwright T, Hainsworth J, Heim W, et al. Bevacizumab plus irinotecan, fluorouracil and leucovorin for metastatic colorectal cancer. N Engl J Med. 2004;350:2335–2342. doi: 10.1056/NEJMoa032691. [DOI] [PubMed] [Google Scholar]
- 12.Belani CP, Ramalingam S. Bevacizumab extends survival for patients with nonsquamous non-small-cell lung cancer. Clin Lung Cancer. 2005;6:267–268. [PubMed] [Google Scholar]
- 13.Miller JC, Pien HH, Sahani D, Sorensen AG, Thrall JH. Imaging angiogenesis: applications and potential for drug development. J Natl Cancer Inst. 2005;97:172–187. doi: 10.1093/jnci/dji023. [DOI] [PubMed] [Google Scholar]
- 14.Neri D, Bicknell R. Tumour vascular targeting. Nat Rev Cancer. 2005;5:436–446. doi: 10.1038/nrc1627. [DOI] [PubMed] [Google Scholar]
- 15.Siemann DW, Chaplin DJ, Horsman MR. Vascular-targeting therapies for treatment of malignant disease. Cancer. 2004;100:2491–2499. doi: 10.1002/cncr.20299. [DOI] [PubMed] [Google Scholar]
- 16.Tozer GM, Kanthou C, Baguley BC. Disrupting tumour blood vessels. Nat Rev Cancer. 2005;5:423–435. doi: 10.1038/nrc1628. [DOI] [PubMed] [Google Scholar]
- 17.Jennewein M, Lewis MA, Zhao D, Tsyganov E, Slavine N, He J, et al. Vascular imaging of solid tumors in rats with a radioactive arsenic-labeled antibody that binds exposed phosphatidylserine. Clin Cancer Res. 2008;14:1377–1385. doi: 10.1158/1078-0432.CCR-07-1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pasqualini R, Moeller BJ, Arap W. Leveraging molecular heterogeneity of the vascular endothelium for targeted drug delivery and imaging. Semin Thromb Hemost. 2010;36:343–351. doi: 10.1055/s-0030-1253456. [DOI] [PubMed] [Google Scholar]
- 19.Bieker R, Kessler T, Schwoppe C, Padro T, Persigehl T, Bremer C, et al. Infarction of tumor vessels by NGR-peptide-directed targeting of tissue factor: experimental results and first-in-man experience. Blood. 2009;113:5019–5027. doi: 10.1182/blood-2008-04-150318. [DOI] [PubMed] [Google Scholar]
- 20.Carson-Walter EB, Watkins DN, Nanda A, Vogelstein B, Kinzler KW, St. Croix B. Cell surface tumor endothelial markers are conserved in mice and humans. Cancer Res. 2001;61:6649–6655. [PubMed] [Google Scholar]
- 21.Castellani P, Viale G, Dorcaratto A, Nicolo G, Kaczmarek J, Querze G, et al. The fibronectin isoform containing the ED-B oncofetal domain: a marker of angiogenesis. Int J Cancer. 1994;59:612–618. doi: 10.1002/ijc.2910590507. [DOI] [PubMed] [Google Scholar]
- 22.Reinmuth N, Liu W, Ahmad SA, Fan F, Stoeltzing O, Parikh AA, et al. Alphavbeta3 integrin antagonist S247 decreases colon cancer metastasis and angiogenesis and improves survival in mice. Cancer Res. 2003;63:2079–2087. [PubMed] [Google Scholar]
- 23.Buckanovich RJ, Sasaroli D, O'Brien-Jenkins A, Botbyl J, Conejo-Garcia JR, Benencia F, et al. Use of immuno-LCM to identify the in situ expression profile of cellular constituents of the tumor microenvironment. Cancer Biol Ther. 2006;5:635–642. doi: 10.4161/cbt.5.6.2676. [DOI] [PubMed] [Google Scholar]
- 24.García-Vallejo JJ, Van Dijk W, Van Het Hof B, Van Die I, Engelse MA, Van Hinsbergh VW, et al. Activation of human endothelial cells by tumor necrosis factor-alpha results in profound changes in the expression of glycosylation-related genes. J Cell Physiol. 2006;206:203–210. doi: 10.1002/jcp.20458. [DOI] [PubMed] [Google Scholar]
- 25.Peng Y, Li J, Geng M. The glycan profile of endothelial cells in the present of tumor-conditioned medium and potential roles of beta-1,6-GlcNAc branching on HUVEC conformation. Mol Cell Biochem. 2010;340:143–152. doi: 10.1007/s11010-010-0411-z. [DOI] [PubMed] [Google Scholar]
- 26.Béchard D, Scherpereel A, Hammad H, Gentina T, Tsicopoulos A, Aumercier M, et al. Human endothelial-cell specific molecule-1 binds directly to the integrin CD11a/CD18 (LFA-1) and blocks binding to intercellular adhesion molecule-1. J Immunol. 2001;167:3099–3106. doi: 10.4049/jimmunol.167.6.3099. [DOI] [PubMed] [Google Scholar]
- 27.Bhatt AS, Erdjument-Bromage H, Tempst P, Craik CS, Moasser MM. Adhesion signaling by a novel mitotic substrate of src kinases. Oncogene. 2005;24:5333–5343. doi: 10.1038/sj.onc.1208582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Osada A, Kiyozumi D, Tsutsui K, Ono Y, Weber CN, Sugimoto N, et al. Expression of MAEG, a novel basement membrane protein, in mouse hair follicle morphogenesis. Exp Cell Res. 2005;303:148–159. doi: 10.1016/j.yexcr.2004.04.053. [DOI] [PubMed] [Google Scholar]
- 29.Asakura M, Kitakaze M, Takashima S, Liao Y, Ishikura F, Yoshinaka T, et al. Cardiac hypertrophy is inhibited by antagonism of ADAM12 processing of HB-EGF: metalloproteinase inhibitors as a new therapy. Nat Med. 2002;8:35–40. doi: 10.1038/nm0102-35. [DOI] [PubMed] [Google Scholar]
- 30.Milner CM, Higman VA, Day AJ. TSG-6: a pluripotent inflammatory mediator? Biochem Soc Trans. 2006;34:446–450. doi: 10.1042/BST0340446. [DOI] [PubMed] [Google Scholar]
- 31.Uhland K. Matriptase and its putative role in cancer. Cell Mol Life Sci. 2006;63:2968–2978. doi: 10.1007/s00018-006-6298-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Uekita T, Tanaka M, Takigahira M, Miyazawa Y, Nakanishi Y, Kanai Y, et al. CUB-domain-containing protein 1 regulates peritoneal dissemination of gastric scirrhous carcinoma. Am J Pathol. 2008;172:1729–1739. doi: 10.2353/ajpath.2008.070981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Conejo-Garcia JR, Buckanovich RJ, Benencia F, Courreges MC, Rubin SC, Carroll RG, et al. Vascular leukocytes contribute to tumor vascularization. Blood. 2005;105:679–681. doi: 10.1182/blood-2004-05-1906. [DOI] [PubMed] [Google Scholar]
- 34.Davies G, Cunnick GH, Mansel RE, Mason MD, Jiang WG. Levels of expression of endothelial markers specific to tumour-associated endothelial cells and their correlation with prognosis in patients with breast cancer. Clin Exp Metastasis. 2004;21:31–37. doi: 10.1023/B:CLIN.0000017168.83616.d0. [DOI] [PubMed] [Google Scholar]
- 35.Kaplan RN, Riba RD, Zacharoulis S, Bramley AH, Vincent L, Costa C, et al. VEGFR1-positive haematopoietic bone marrow progenitors initiate the pre-metastatic niche. Nature. 2005;438:820–827. doi: 10.1038/nature04186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Conejo-Garcia JR, Benencia F, Courreges MC, Kang E, Mohamed-Hadley A, Buckanovich RJ, et al. Tumor-infiltrating dendritic cell precursors recruited by a beta-defensin contribute to vasculogenesis under the influence of Vegf-.A. Nat Med. 2004;10:950–958. doi: 10.1038/nm1097. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.