Abstract
The present studies were initiated to determine in greater molecular detail the regulation of CHK1 inhibitor lethality in transfected and infected breast cancer cells and using genetic models of transformed fibrobalsts. Multiple MEK1/2 inhibitors (PD184352, AZD6244 [ARRY-142886]) interacted with multiple CHK1 inhibitors (UCN-01 [7-hydroxystaurosporine], AZD7762) to kill mammary carcinoma cells and transformed fibroblasts. In transformed cells, CHK1 inhibitor-induced activation of ERK1/2 was dependent upon activation of SRC family non-receptor tyrosine kinases as judged by use of multiple SRC kinase inhibitors (PP 2, Dasatinib; AZD0530), use of SRC/FYN/YES deleted transformed fibroblasts or by expression of dominant negative SRC. Cell killing by SRC family kinase inhibitors and CHK1 inhibitors was abolished in BAX/BAK−/− transformed fibroblasts and suppressed by overexpression of BCL-XL. Treatment of cells with BCL-2/BCL-XL antagonists promoted SRC inhibitor + CHK1 inhibitor-induced lethality in a BAX/BAK-dependent fashion. Treatment of cells with [SRC + CHK1] inhibitors radio-sensitized tumor cells. These findings argue that multiple inhibitors of the SRC-RAS-MEK pathway interact with multiple CHK1 inhibitors to kill transformed cells.
Key words: CHK1, SRC, apoptosis, breast cancer, kinase, therapeutics, intrinsic, caspase
Introduction
CHK1 inhibitors including UCN-01 (7-hydroxystaurosporine) and AZD7762 are currently being evaluated as anti-neoplastic agents in clinical trials, both alone and in combination with chemotherapeutic agents and ionizing radiation.1,2 These agents are postulated to enhance the cytotoxicity of established chemotherapeutic agents by inhibition of CHK1 and inappropriate cell cycle progression after DNA damage.3 Inhibition of CHK1 may directly promote activation of the protein phosphatase CDC25C and can also interfere with CDC25C elimination by blocking its binding to 14-3-3 proteins and subsequent degradation.3 Downregulation of CDC25C results in enhanced Cdc2 phosphorylation and inactivation of cyclin-dependent kinases (CDKs) such as p34, which are critically involved in G2/M cell cycle arrest following DNA damage.4 UCN-01 is known to have many other intracellular kinase targets including the downstream effector of PI3 kinase, PDK-1, as well as PKC isoforms.5
Based on initial phase I studies, the maximal free achievable concentration of UCN-01 in human plasma was thought to be at or below ∼100 nM with a long plasma half-life due to UCN-01 binding to human alpha1 acidic glycoprotein, which is a considerably lower concentration than that achievable in rodent plasma.6–8 The clinical utility of UCN-01 was limited due to its binding to α1-acidic glycoprotein and also to the possibility that off-target actions may be responsible for toxicity, such as hyperglycemia.9 A novel UCN-01 schedule has recently been developed in which UCN-01 is administered as a 3-h infusion (95 mg/m) q 3 weeks.10 This schedule, which appears to be well tolerated, results in free, salivary UCN-01 plasma concentrations of 800–1,400 nM that are sustained for up to 3 weeks following drug administration. Using the original infusion schedule for the kinase inhibitor, the combination of UCN-01 with topotecan or cisplatin has shown preliminary evidence of patient activity.11,12
Multiple intracellular signal transduction pathways e.g., ERK1/2 and PI3K/AKT are often highly activated in tumor cells, and have been proposed as therapeutic targets in preventing cancer cell growth in many malignancies.13–15 An alternative approach to killing tumor cells without the use of conventional cytotoxic agents is to exploit their reliance (i.e., addiction) to high levels of signaling pathway activity to maintain growth and viability. Previous studies by this group have demonstrated that, UCN-01, at clinically relevant concentrations in vitro causes activation of the ERK1/2 pathway in transformed cell types. Prevention of ERK1/2 pathway activation, by inhibition of either MEK1/2 or RAS function, rapidly promoted UCN-01-induced tumor cell death in a synergistic fashion.16–21 Indeed, UCN-01 was shown to promote RAS activation.19 Non-transformed cells from multiple tissues were noted in several studies to be insensitive to apoptosis-induction by this strategy.
The present studies are focused in mammary carcinoma cells, and also use additional genetic models of transformed fibroblasts and transfected HNSCC cells to determine the protein kinase(s) responsible upstream of RAS for the activation of the ERK1/2 pathway which were noted to be in the SRC non-receptor tyrosine kinase family. In addition, the present analyses solidified the overall concept of using multiple inhibitors of CHK1 combined with either multiple inhibitors of SRC kinases or multiple inhibitors of MEK1/2 to kill transformed cells.
Results
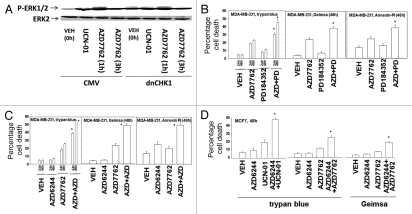
Prior studies have demonstrated that the MEK1/2 inhibitors PD184352 and AZD6244 interact with UCN-01 in a synergistic manner to kill mammary, prostate and pancreatic tumor cells.16,21 To prove or refute whether UCN-01 was mediating its ERK1/2 activating effects via inhibition of CHK1, we made use of a plasmid to express dominant negative CHK1. Expression of a dominant negative CHK1 protein in MCF7 cells, a cell line with a high level of transfection efficiency, enhanced basal levels of ERK1/2 phosphorylation within 24 h and blunted the ability of UCN-01 to stimulate ERK1/2 phosphorylation (Fig. 1A).21,22
Figure 1.
Inhibition of CHK1 enhances the toxicity of MEK1/2 inhibitors. (A) MCF7 cells were transfected with either an empty vector control plasmid or a plasmid to express dominant negative CHK1 (dnCHK1). Twenty-four hours after transfection, cells were treated with vehicle (VEH, DMSO), UCN-01 (100 nM) or AZD7762 (50 nM). Cells were isolated at the indicated time points and subjected to SDS PAGE followed by immunoblotting to determine the phosphorylation of ERK1/2 (P-ERK1/2) or the expression of GAPDH. Data are from a representative of two separate studies. (B) MDA-MB-231 cells plated in triplicate were treated with vehicle (DMSO), MEK1/2 inhibitor PD184352 (2 µM), CHK1 inhibitor AZD7762 (200 nM) or PD184352 and AZD7762. Cells were isolated at the indicated time points and subjected to the indicated various cell viability assays. Data for each assay is the mean of all data points from three studies ±SEM (*p < 0.05 greater than AZD7762 value). (C) MDA-MB-231 cells plated in triplicate were treated with either vehicle (DMSO), MEK1/2 inhibitor AZD6244 (400 nM), CHK1 inhibitor AZD7762 (200 nM) or AZD6244 and AZD7762. Cells were isolated at the indicated time points and subjected to the indicated various cell viability assays. Data for each assay is the mean of all data points from three studies ±SEM (*p < 0.05 greater than AZD7762 value). (D) MCF7 cells plated in triplicate were treated with either vehicle (DMSO), MEK1/2 inhibitor AZD6244 (400 nM), the CHK1 inhibitors AZD7762 (200 nM) or UCN-01 (100 nM), or AZD6244 and the indicated ChK1 inhibitor. Cells were isolated at the indicated time points and subjected to the indicated various cell viability assays. Data for each assay is the mean of all data points from three studies ±SEM (*p < 0.05 greater than CHK1 inhibitor value).
Recently, several additional novel MEK1/2 inhibitors and CHK1 inhibitors have become available for pre-clinical testing e.g., AZD6244, AZD7762. Thus, we next determined whether a variety of chemically dissimilar inhibitors of MEK1/2 and CHK1 interacted to kill breast cancer cells, as judged by various methods of analysis. AZD7762 and PD184352, and AZD7762 and AZD6244, interacted in a greater than additive fashion to kill “triple negative” MDA-MB-231 cells (Fig. 1B, C and Table 1). Similar data were obtained in estrogen dependent MCF7 cells (Fig. 1D).
Table 1.
MEK1/2 inhibitors and CHK1 inhibitors synergize to kill tumor cells
| MDA-MB-231 | MCF7 | ||||||
| UCN-01 (nM) | PD184352 (µM) | Fraction affected | CI | UCN-01 (nM) | PD184352 (µM) | Fraction affected | CI |
| 25.0 | 0.30 | 0.39 | 0.44 | 25.0 | 0.30 | 0.12 | 0.51 |
| 33.3 | 0.40 | 0.47 | 0.34 | 33.3 | 0.40 | 0.17 | 0.56 |
| 41.6 | 0.50 | 0.57 | 0.27 | 41.6 | 0.50 | 0.32 | 0.41 |
| UCN-01 (nM) | AZD6244 (µM) | Fraction affected | CI | UCN-01 (nM) | AZD6244 (µM) | Fraction affected | CI |
| 25.0 | 0.20 | 0.28 | 0.41 | 20.0 | 0.20 | 0.15 | 0.52 |
| 50.0 | 0.40 | 0.35 | 0.41 | 25.0 | 0.25 | 0.24 | 0.50 |
| 75.0 | 0.60 | 0.47 | 0.47 | 30.0 | 0.30 | 0.38 | 0.60 |
| AZD7762 (nM) | AZD6244 (µM) | Fraction affected | CI | AZD7762 (nM) | AZD6244 (µM) | Fraction affected | CI |
| 50.0 | 0.10 | 0.26 | 0.63 | 50.0 | 0.20 | 0.28 | 0.71 |
| 100.0 | 0.20 | 0.39 | 0.58 | 100.0 | 0.25 | 0.39 | 0.78 |
| 150.0 | 0.30 | 0.49 | 0.44 | 150.0 | 0.30 | 0.53 | 0.79 |
In previous studies, we have demonstrated that inhibition of RAS protein function blocks the ability of UCN-01 to activate ERK1/2.16–24 A question thus arose as to what protein(s) upstream of RAS could be mediating the “activating” signal into the ERK1/2 pathway (due to inhibition of CHK1). Initial studies, examining a role for ERBB1, demonstrated that inhibition of this receptor antagonized UCN-01 lethality as judged by combination index (CI) values of greater than 1.00 suggesting that EGF receptor signaling was not part of the activation process (data not shown).
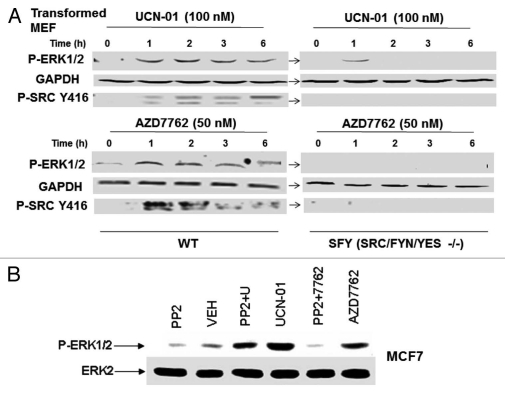
As ERBB1 was not a key factor in the ERK1/2 activation process by CHK1 inhibitors, we next determined whether SRC family protein kinases, molecules also associated with activation of the ERK1/2 pathway, were mechanistically involved. Treatment of wild type transformed mouse embryonic fibroblasts with UCN-01 or AZD7762 caused activation of ERK1/2 and phosphorylation of SRC kinases at Y416 (multiple bands), indicative of their activation (Fig. 2A). In matched transformed mouse embryonic fibroblasts genetically deleted for SRC/FYN/YES, CHK1 inhibitor-stimulated ERK1/2 activation was profoundly reduced and the phosphorylation of SRC kinases at Y416 was not detected by immunoblotting. However, we had great difficulty in observing a convincing similar activation of SRC in mammary carcinoma cells (data not shown); this was similar to contemporaneous data in blood cancer cells in Dai et al. (2010) wherein some myeloma cells exhibited increased SRC Y416 phosphorylation whereas other cell lines showed no induction effect.22 In MCF7 cells, treatment with the pan-SRC kinase inhibitor PP2 abolished AZD7762-induced activation of ERK1/2 and significantly suppressed UCN-01-induced ERK1/2 activation (Fig. 2B).
Figure 2.
SRC family kinases play a key role in the regulation of the ERK1/2 pathway by CHK1 inhibitors. (A) Transformed mouse embryonic fibroblasts, MEF (wild type, WT; deleted for SRC/FYN/YES, SYF) were treated with vehicle (VEH, DMSO), UCN-01 (100 nM) or AZD7762 (50 nM). Cells were isolated at the indicated time points and subjected to SDS PAGE followed by immunoblotting to determine the phosphorylation of ERK1/2 (P-ERK1/2), the phosphorylation of SRC Y461 (P-Y416 Src) or the expression of GAPDH. Data are from a representative of three separate studies. (B) MCF7 cells were treated with vehicle (VEH, DMSO), UCN-01 (100 nM), PP2 (10 µM), AZD7762 (50 nM) or UCN-01+PP2 or AZD7762+PP2. Cells were isolated 2 h after drug exposure and subjected to SDS PAGE followed by immunoblotting to determine the phosphorylation of ERK1/2 (P-ERK1/2) or the expression of ERK2 protein. Data are from a representative of two separate studies.
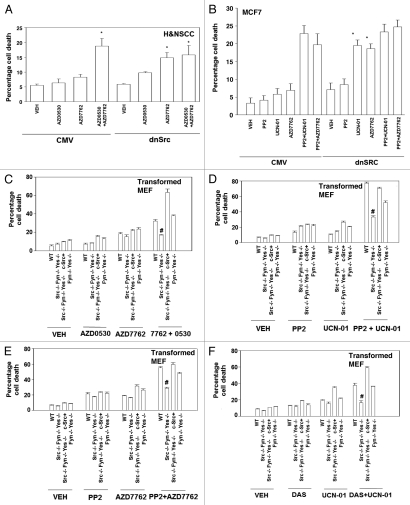
Based on these findings, we determined whether inhibition of SRC kinase function promoted the toxicity of CHK1 inhibitors in a SRC kinase-dependent fashion. Initial studies were performed in an extant human head and neck squamous carcinoma cell line, obtained by collaboration; a line that had been stably transfected to express dominant negative SRC. In these cells expression of dominant negative SRC enhanced the lethality of AZD7762 and abolished the toxic interaction between AZD0530 and AZD7762 (Fig. 3A). Furthermore, in subsequent studies using MCF7 cells transiently transfected with a plasmid to express dominant negative SRC we also noted that the lethality of AZD7762 and UCN-01 was enhanced and that the toxic interaction between these drugs and the SRC inhibitor PP2 was abolished (Fig. 3B).
Figure 3.
Loss of SRC/FYN/YES function abolishes the toxic interaction between CHK1 and SRC family kinase inhibitors in transformed fibroblasts. (A) 1483 H&N SC cells transfected with vector control or dominant negative Src (K296R) were plated in triplicate and treated with vehicle (VEH, DMSO), AZD0530 (125 nM), AZD7762 (50 nM) or AZD7762 and AZD0530. Cells were isolated 48 h after exposure and subjected to the indicated various cell viability assays. Data for each assay is the mean of all data points from two studies ±SEM (*p < 0.05 greater than corresponding empty vector value). (B) MCF7 cells were transfected with a plasmid to express dominant negative SRC. Twenty-four hours after transfection cells were treated with (VEH, DMSO), UCN-01 (100 nM), PP2 (10 µM), AZD7762 (50 nM) or UCN-01+PP2 or AZD7762+PP2. Cells were isolated 48 h after exposure and viability determined using a hemacytometer. Data for each assay is the mean of all data points from two studies ±SEM (*p < 0.05 greater than corresponding empty vector value). (C) Transformed mouse embryonic fibroblasts, MEF (wild type, WT; deleted for SRC/FYN/YES; deleted for SRC/FYN/YES with c-SRC knock in; deleted for FYN/YES) were plated in triplicate and treated with vehicle (VEH, DMSO), AZD0530 (125 nM), AZD7762 (50 nM) or AZD7762 and AZD0530. Cells were isolated 48 h after exposure and viability determined using a hemacytometer. Data for each assay is the mean of all data points from two studies ±SEM (#p < 0.05 less than WT cell value). (D) Transformed mouse embryonic fibroblasts, MEF (wild type, WT; deleted for SRC/FYN/YES; deleted for SRC/FYN/YES with c-SRC knock in; deleted for FYN/YES) were plated in triplicate and treated with vehicle (VEH, DMSO), PP2 (10 µM), UCN-01 (50 nM) or AZD7762 and AZD0530. Cells were isolated 48 h after exposure and viability determined using a hemacytometer. Data for each assay is the mean of all data points from two studies ±SEM (#p < 0.05 less than WT cell value). (E) Transformed mouse embryonic fibroblasts, MEF (wild type, WT; deleted for SRC/FYN/YES; deleted for SRC/FYN/YES with c-SRC knock in; deleted for FYN/YES) were plated in triplicate and treated with vehicle (VEH, DMSO), PP2 (10 µM), AZD7762 (50 nM) or AZD7762 and P 2. Cells were isolated 48 h after exposure and viability determined using a hemacytometer. Data for each assay is the mean of all data points from two studies ±SEM (#p < 0.05 less than WT cell value). (F) Transformed mouse embryonic fibroblasts, MEF (wild type, WT; deleted for SRC/FYN/YES; deleted for SRC/FYN/YES with c-SRC knock in; deleted for FYN/YES) were plated in triplicate and treated with vehicle (VEH, DMSO), dasatinib (DAS, 200 nM), UCN-01 (100 nM) or dasatinib and UCN-01. Cells were isolated 48 h after exposure and viability determined using a hemacytometer. Data for each assay is the mean of all data points from two studies ±SEM (#p < 0.05 less than WT cell value).
We next wished to further confirm our apoptosis findings using genetic models. Transformed mouse embryonic fibroblasts genetically deleted for SRC/FYN/YES were not, per se, profoundly more sensitive to exposure of either a SRC inhibitor (AZD0530; PP2; dasatinib) or to a CHK1 inhibitor (UCN-01, AZD7762) (Fig. 3C–F). However, in mouse embryonic fibroblasts genetically deleted for SRC/FYN/YES we noted that SRC inhibitors (AZD0530; PP2; dasatinib) did not interact with CHK1 inhibitors (UCN-01, AZD7762) to promote a greater than additive amount of short-term cell killing (Fig. 3). In transformed mouse embryonic fibroblasts genetically deleted for SRC/FYN/YES into which the gene for c-Src had been stably re-expressed back into the cells, we noted that the sensitivity of cells to SRC inhibitor + CHK1 inhibitor-induced lethality was restored to varying extents based on the kinase inhibitor regimens used (Fig. 3). In transformed mouse embryonic fibroblasts genetically deleted for only FYN/YES and not SRC, the lethality of SRC inhibitor + CHK1 inhibitor treatment was still competent to kill cells, arguing that SRC played a greater role than FYN and YES in this cell system to facilitate drug combination toxicity (Fig. 3).
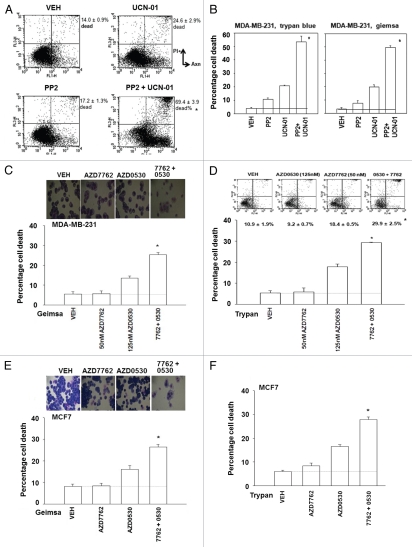
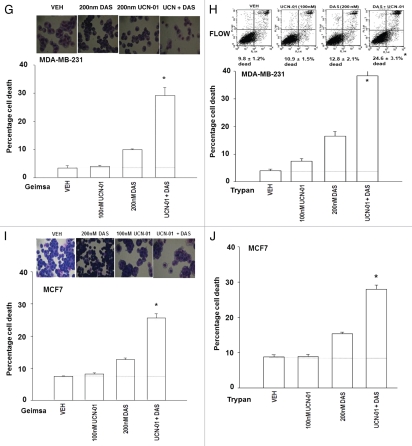
We next determined whether similar drug interaction/cell killing effects to those in genetically altered transformed fibroblasts were also observed in human tumor cell lines using a diverse range of SRC inhibitors and CHK1 inhibitors. SRC inhibitors (AZD0530; PP2; dasatinib) interacted with CHK1 inhibitors (UCN-01, AZD7762) to promote a greater than additive amount of short-term cell killing in MDA-MB-231 and MCF7 mammary carcinoma cells measured using a variety of cell viability assays/techniques (Fig. 4A–J).
Figure 4.
Loss of SRC function abolishes the toxic interaction between CHK1 and SRC family kinase inhibitors in breast cancer cells. (A and B) MDA-MB-231 cells were plated in triplicate and treated with vehicle (VEH, DMSO), PP2 (10 µM), UCN-01 (50 nM) or PP2 and UCN-01. Cells were isolated 48 h after exposure and subjected to the indicated various cell viability assays. Data for each assay is the mean of all data points from two studies ±seM (*p < 0.05 greater than CHK1 inhibitor value). (C and D) MDA-MB-231 cells were plated in triplicate and treated with vehicle (VEH, DMSO), AZD0530 (125 nM), AZD7762 (50 nM) or AZD7762 and AZD0530. Cells were isolated 48 h after exposure and subjected to the indicated various cell viability assays. Data for each assay is the mean of all data points from two studies ±SEM (*p < 0.05 greater than ChK1 inhibitor value). (E and F) MCF7 cells were plated in triplicate and treated with vehicle (VEH, DMSO), AZD0530 (125 nM), AZD7762 (50 nM) or AZD7762 and AZD0530. Cells were isolated 48 h after exposure and subjected to the indicated various cell viability assays. Data for each assay is the mean of all data points from two studies ±SEM (*p < 0.05 greater than CHK1 inhibitor value). (G and H) MDA-MB-231 cells were plated in triplicate and treated with vehicle (VEH, DMSO), dasatinib (DAS, 200 nM), UCN-01 (100 nM) or dasatinib and UCN-01. Cells were isolated 48 h after exposure and viability determined using the indicated method (Geimsa, Trypan blue exclusion, Annexin-pI flow cytometry). Data for each assay is the mean of all data points from two studies ±SEM (*p < 0.05 greater than CHK1 inhibitor value). (I and J) MCF7 cells were plated in triplicate and treated with vehicle (VEH, DMSO), dasatinib (DAS, 200 nM), UCN-01 (100 nM) or dasatinib and UCN-01. Cells were isolated 48 h after exposure and viability determined using the indicated method (Geimsa, Trypan blue exclusion). Data for each assay is the mean of all data points from two studies ±SEM (*p < 0.05 greater than CHK1 inhibitor value).
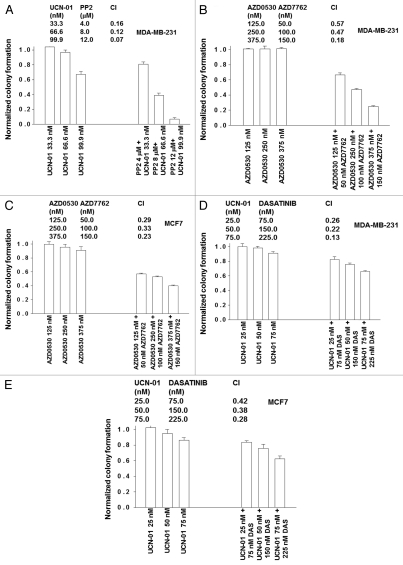
In median dose effect isobologram colony formation assays we noted that a 48 h exposure of either MDA-MB-231 or MCF7 cells to SRC inhibitors (AZD0530; PP2; dasatinib) with CHK1 inhibitors (UCN-01, AZD7762) resulted in a synergistic induction of cell killing as measured ∼14 days after drug removal in colony formation assays (Fig. 5A–E). A combination index (CI) value of less than 1.00 indicates a toxic synergy of interaction between the drugs when survival data are calculated within this computational assay (see Methods section).
Figure 5.
CHK1 and SRC family kinase inhibitors synergize to kill mammary carcinoma cells in colony formation assays. (A) MDA-MB-231 cells were plated as single cells in sextuplicate (250–2,000 cells per 60 mm dish) and 12 h after plating treated with increasing fixed dose concentrations of UCN-01 (33.3–99.9 nM) or PP2 (4–12 µM), or with the equivalent corresponding amount of vehicle control (DMSO) for 48 h. Media was then replaced and cells grown in the absence of drugs for 10–14 days to permit colonies of >50 cells to form. Cells were fixed, stained with crystal violet and colonies of >50 cells/colony counted. Colony formation data were entered into the Calcusyn program and combination index (CI) values determined. In the tabular portion of the part, a CI value of less than 1.00 indicates synergy. The data in lower bar graph are the means of data points from two separate experiments ±SEM. (B and C) MDA-MB-231 cells and MCF7 cells were plated as single cells in sextuplicate (250–2,000 cells per 60 mm dish) and 12 h after plating treated with increasing fixed dose concentrations of AZD0530 (125–375 nM) or AZD7762 (50–150 nM), or with the equivalent corresponding amount of vehicle control (DMSO) for 48 h. Media was then replaced and cells grown in the absence of drugs for 10–14 days to permit colonies of >50 cells to form. Cells were fixed, stained with crystal violet and colonies of >50 cells/colony counted. Colony formation data were entered into the Calcusyn program and combination index (CI) values determined. In the tabular portion of the part, a CI value of less than 1.00 indicates synergy. The data in lower bar graph are the means of data points from two separate experiments ±SEM. (D and E) MDA-MB-231 and MCF7 cells were plated in sextuplicate as single cells for colony formation assays, as described in the Methods. Cells were permitted to attach and 12 h after plating and each well individually treated for 48 h with the indicated concentrations of dasatinib and UCN-01. Following 48 h of drug treatment, media was carefully removed, the cells washed and fresh media lacking drugs added to the cultures. Cells were grown in the absence of drugs for 10–14 days to permit colonies of >50 cells to form. Cells were fixed, stained with crystal violet and colonies of >50 cells/colony counted. Colony formation data were entered into the Calcusyn program and combination index (CI) values determined. In the tabular portion of the part, a CI value of less than 1.00 indicates synergy. The data in lower bar graph are the means of data points from two separate experiments ±SEM.
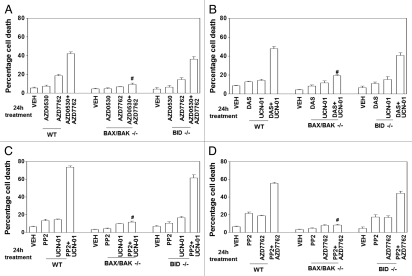
In previous studies combining MEK1/2 inhibitors or farnesyltransferase inhibitors with CHK1 inhibitors we published that cell killing by these drug combinations occurs via mitochondrial dysfunction and the release of cytochrome c into the cytosol (reviewed in ref. 21 and 24). In transformed embryonic fibroblasts genetically deleted for toxic BH3 domain proteins BAX and BAK, but not deleted for BID, the combination of a SRC inhibitor (AZD0530; PP2; dasatinib) with a CHK1 inhibitor (UCN-01, AZD7762) was unable to cause cell killing (Fig. 6A–D). In MDA-MB-231 or MCF7 cells, overexpression of BCL-XL or dominant negative caspase-9, but not the caspase-8 inhibitor CRM A, blocked the combination of a SRC inhibitor (AZD0530; PP2; dasatinib) with a CHK1 inhibitor (UCN-01, AZD7762) from causing death (data not shown).
Figure 6.
Loss of BAX and BAK expression abolishes the toxic interaction between CHK1 inhibitors and SRC family kinase inhibitors. Transformed mouse embryonic fibroblasts, MEF (wild type, WT; deleted for BAX and BAK, BAX/BAK−/−; and for BID, BID−/−) plated in triplicate were treated (as indicated in each graphical part) with: vehicle (VEH, DMSO), UCN-01 (50 nM), AZD7762 (50 nM), PP2 (10 µM); dasatinib (DAS, 200 nM); AZD0530 (50 nM) or the drugs in combination: (A) AZD0530+AZD7762; (B) DAS+UCN-01; (C) PP2+UCN-01; (D) PP2+AZD7762. Cells were isolated 24 h after exposure and subjected to trypan blue exclusion cell viability assays. Data for each assay is the mean of all data from two studies ±SEM (#p < 0.05 less than corresponding value in WT cells).
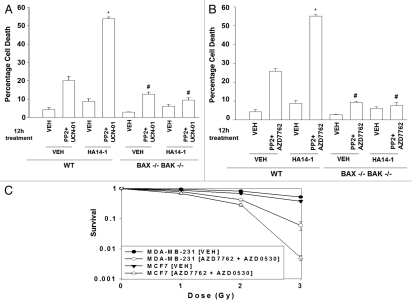
As the combination of a SRC inhibitor with a CHK1 inhibitor was promoting cell death via mitochondrial dysfunction, as previously shown for the combination of a MEK1/2 inhibitor with a CHK1 inhibitor, we determined whether the combination of these two agents with a third agent that inhibits BCL-2/BCL-XL function e.g., HA14-1, can further promote cell killing.22,25 Treatment of transformed mouse embryonic fibroblasts with HA14-1 rapidly promoted the toxicity of PP2 + UCN-01 and of PP2 + AZD7762 (Fig. 7A and B). In transformed mouse embryonic fibroblasts genetically deleted for toxic BH3 domain proteins BAX and BAK, HA14-1 was unable to promote SRC inhibitor + CHK1 inhibitor lethality, again arguing that the primary two drug combination kills transformed cells by initially causing mitochondrial dysfunction. Similar data were obtained with the clinically relevant BCL-2 inhibitor obatoclax, GX15-070 and in mammary carcinoma cells (data not shown).
Figure 7.
Loss of BAX/BAK function abolishes the toxic interaction between ChK1 inhibitors sRC family kinase inhibitors in transformed fibroblasts; cell killing is potentiated by inhibitors of BCL-2/BCL-XL function. (A) Transformed mouse embryonic fibroblasts, MEF (wild type, WT; deleted for BAX and BAK, BAX/BAK−/−) were plated in triplicate and treated with vehicle (VEH, DMSO), PP2+UCN-01 (10 µM + 50 nM), in the presence or absence of vehicle (DMSO) or HA14-1 (10 µM). Cells were isolated 12 h after exposure and viability determined using a hemacytometer and trypan blue exclusion staining. Data for each assay is the mean of all data points from two studies ±SEM. (B) Transformed mouse embryonic fibroblasts, MEF (wild type, WT; deleted for BAX and BAK, BAX/BAK−/−) were plated in triplicate and treated with vehicle (VEH, DMSO), PP2 + AZD7762 (10 µM + 50 nM), in the presence or absence of vehicle (DMSO) or HA14-1 (10 µM). Cells were isolated 12 h after exposure and viability determined using a hemacytometer and trypan blue exclusion staining. Data for each assay is the mean of all data points from two studies ±SEM (*p < 0.05 greater than CHK1 inhibitor value; **p < 0.05 greater than corresponding value in vehicle treated cells; #p < 0.05 less than corresponding value in WT cells). (C and D) MDA-MB-231 and MCF7 cells were plated in sextuplicate as single cells for colony formation assays, as described in the Methods. Cells were permitted to attach and 12 h after plating and each well individually treated for 48 h with AZD7762 (50 nM) and AZD0530 (125 nM). Six h after drug exposure cells are exposed to ionizing radiation (0–4 Gy). Following 48 h of drug treatment, media was carefully removed, the cells washed and fresh media lacking drugs added to the cultures. Cells were grown in the absence of drugs for 10–14 days to permit colonies of >50 cells to form. Cells were fixed, stained with crystal violet and colonies of >50 cells/colony counted. The data are the means of data points from two separate experiments ±SEM.
Radiotherapy is a primary modality for treating breast cancer patients. Treatment of MCF7 and MDA-MB-231 breast cancer cells with (AZD7762 + AZD0530) enhanced tumor cell radiosensitivity in colony formation assays (Fig. 7C and D). Collectively, our data demonstrate that SRC and CHK1 inhibitors interact to kill mammary carcinoma cells and to facilitate the lethal effects of established breast cancer therapies.
Discussion
Previous studies by this group have argued that MEK1/2 inhibitors or farnesyltransferase inhibitors interact with UCN-01 to promote tumor cell specific killing in a wide variety of malignancies including breast, prostate and multiple hematological cell types.16–24 The net output of the cytoprotective RAS-MEK1/2-ERK1/2 pathway has previously been shown to be a critical determinant of tumor cell survival. Furthermore, activation of this cascade has been observed as a compensatory response of tumor cells to various environmental stresses, including cytotoxic drugs. The present studies were initiated to determine in greater molecular detail than previously reported how CHK1 inhibitors activate the ERK1/2 pathway and whether multiple chemically dissimilar inhibitors of the CHK1 and ERK1/2 pathways can be utilized to achieve a similar cytotoxic effect in tumor cells.
Based on use of dominant negative CHK1, UCN-01 and AZD7762-induced activation of ERK1/2 was dependent upon inhibition of CHK1; furthermore, expression of dominant negative CHK1 enhanced basal levels of ERK1/2 phosphorylation arguing for a central regulatory role between CHK1 and the RAF-MEK-ERK1/2 pathway.22 Of note, ATM/checkpoint pathway signaling has previously been linked in our studies to regulation of the ERK1/2 pathway.26 Thus despite UCN-01 having the potential to inhibit several protein kinases e.g., PKC isoforms, PDK-1; our findings argue that inhibition of CHK1 is essential for activation of ERK1/2 to occur. Upstream of RAS proteins, we discovered that SRC family kinases, but not ERBB1, played a key role in the activation of ERK1/2 following CHK1 inhibitor exposure in breast cancer cells and transformed fibroblasts. Multiple CHK1 inhibitors (UCN-01, AZD7762) promoted ERK1/2 activation in a SRC kinase-dependent fashion. Inhibition of SRC kinase family function using multiple chemically distinct small molecule inhibitors (AZD0530, PP2, dasatinib) promoted the lethality of multiple CHK1 inhibitors (UCN-01, AZD7762), as we had previously noted for inhibitors of MEK1/2 and of RAS farnesylation.
Based on the use of BAX/BAK−/− cells the induction of mitochondrial dysfunction was shown to play a primary role in the synergistic induction of cell killing following treatment of cells with SRC inhibitors and CHK1 inhibitors. Of note, mammary carcinoma cells with very low basal levels of ERK1/2 activity and that are relatively non-invasive such as the estrogen-dependent line MCF7 were apparently as susceptible to being killed by SRC and CHK1 inhibitors as were carcinoma cells with very high basal levels of ERK1/2 activity and that are highly invasive such as the “triple-negative” line MDA-MB-231. SV40 large T-antigen transformed fibroblasts that are not tumorigenic in mice, but that are nonetheless transformed, were also sensitive to the drug schedule, although as previously noted, multiple non-established cell types are insensitive to CHK1 inhibitor + MEK1/2 inhibitor combined drug exposure regimen.16 Furthermore, we have previously noted that the lethal effects of combined inhibition of MEK1/2 and CHK1 does not require cells to be progressing through the cell cycle, as judged by the killing of non-proliferating primary multiple myeloma blasts.24 Our data suggests that CHK1 function plays a key role in maintaining cell viability in transformed cells and does so, in part, by regulating ERK1/2 pathway signaling.
In published studies treating animals with UCN-01 as an individual agent, noticeably higher drug concentrations than those used in prior in vivo studies by our group have been administered to show significant single agent anti-tumor effects.27 Based on initial phase I studies, the clinical utility of UCN-01 was thought to be limited due to its binding to α1-acidic glycoprotein in the plasma and also to the possibility that off-target actions may be responsible for dose-limiting toxicity in patients, such as hyperglycemia. A novel UCN-01 clinical schedule was developed which achieves much higher free plasma concentrations of the drug and that is also within the UCN-01 concentration range of our prior animal studies which showed a profound tumoricidal in vivo interaction between UCN-01 and MEK1/2 inhibitors. SRC inhibitors are already in clinical use, most notably, the FDA-approved drug dasatinib.28 Other inhibitors of SRC family kinases, such as AZD0530, are at present in clinical development.29 Whether the combination of CHK1 inhibitors and SRC kinase inhibitors will translate successfully into xenograft animal model systems of breast cancer, as was observed for the combination of CHK1 inhibitors and MEK1/2 inhibitors or of CHK1 inhibitors and RAS farnesylation inhibitors will need to be explored in a future study focused on animal pre-clinical translation.
Overexpression of mitochondrial BCL-2 family members has been shown in many tumor cell systems to raise the apoptotic threshold of tumor cells.21,22,30–33 As the potentiation of CHK1 inhibitor lethality by SRC/RAS/MEK inhibitors occurs primarily by promoting mitochondrial dysfunction, it would be assumed that over time, one of the mechanisms by which cells could survive this treatment will be a viability-selection based on increased expression of BCL-2 family members. With this general possibility in mind for multiple chemotherapeutic treatments, several drug companies have developed small molecule inhibitors of BCL-2, BCL-XL and MCL-1, including the drugs gossypol, ABT-737/ABT-263 and GX15-070 (Obatoclax).33–35 In the present studies we noted that a commercially available inhibitor of BCL-2 and BCL-XL, HA14-1, significantly enhanced the lethality of the two drug (CHK1 inhibitor + SRC inhibitor) regimen. Prior studies have also shown that HA14-1 can overcome the protective effect of BCL-XL in cells treated with UCN-01 and PD184352. Collectively, these findings demonstrate that the potentiation of CHK1 inhibitor lethality by SRC/MEK inhibitors can be profoundly enhanced by additional destabilization of mitochondrial function via inhibition of BCL-2 family member activity(ies).
Based on our present findings, an obvious question is posed, namely how does inhibition of CHK1 cause activation of SRC family kinases? In transformed fibroblasts lacking SRC/FYN/YES expression we observed a modest increase in SRC activity following CHK1 inhibitor administration. However, it is of note that activation of SRC family kinases was observed in some blood cancer cell lines but not in other cell lines, and not in our studies in mammary carcinoma cells, implying that different mechanisms may be induced in different cells types.24 SRC family kinases are auto-inhibited via phosphorylation at Y527 and trans-phosphorylate themselves for full catalytic activation at Y416.36 An overall reduction in the levels of phosphorylation of Y527 will promote SRC kinase activation. It is well established that CHK1 phosphorylation regulates the expression and function of the tyrosine phosphatases CDC25A and CDC25C.37 Phosphorylation of both enzymes by CHK1 would reduce the tyrosine phosphatase activity/expression of these enzymes; hence a CHK1 inhibitor will tend to promote tyrosine phosphatase activity/expression, in agreement with our observed activation of SRC kinases. It is also possible that CHK1 may directly regulate the kinases that control Y527 phosphorylation: Csk/CHK. Finally, it is possible that as inhibition of CHK1 causes a compensatory activation of ATM; that ATM can activate c-ABL; and that active c-ABL can signal to promote activation of SCR family kinases, it is conceivable that this pathway regulates SRC/ERK signaling levels.21,38,39 All of these complex studies will also need to be investigated in detailed future studies.
Materials and Methods
Materials.
Phospho-/total-ERK1/2 antibodies were purchased from Cell Signaling Technologies (Worcester, MA). TUNEL kits were purchased from NEN Life Science Products (NEN Life Science Products, Boston, MA) and Boehringer Mannheim (Manheim, Germany), respectively. Trypsin-EDTA, RPMI, penicillin-streptomycin were purchased from GIBCOBRL (GIBCOBRL Life Technologies, Grand Island, NY). MDA-MB-231, MCF7 cells were purchased from the ATCC. SV40 Large T mouse embryonic fibroblasts lacking expression of various pro-apoptotic BH3 domain proteins were kindly provided by Dr. S. Korsmeyer (Harvard University, Boston, MA); the expression, or lack thereof of specific survival regulatory proteins was confirmed on the supplied cell lines (data not shown). SV40 Large T mouse embryonic fibroblasts lacking expression of SRC/FYN/YES or SRC/FYN/YES with c-SRC knock in were purchased from the ATCC (Bethesda, MD); the expression or lack thereof of SRC family kinase proteins was confirmed on the supplied cell lines (data not shown). PD98059, PP2 and AG1478 were purchased from Calbiochem/EMD sciences (San Diego, CA). The HNSCC cells (1,483 cells) transfected with vector control or dominant negative Src (K296R) were kindly supplied by Dr. J. Grandis (U. Pittsburgh, PA). UCN-01 was purchased from Sigma (St. Louis, MO). Dasatinib (BMS354825) was purchased from Eton Bioscience Inc. (San Diego, CA). AZD6244 (ARRY-142886), AZD7762 and AZD0530 were purchased from Selleck chemicals (Houston, TX). Other reagents and irradiation procedures using a Co., 60 source were as described in reference 16–24, 33 and 40–46.
Methods.
Culture and in vitro exposure of cells to drugs. Tumor cells for the studies in this manuscript were cultured at 37°C (5% v/v CO2) in vitro using RPMI supplemented with 10% (v/v) fetal calf serum. In vitro Vehicle/UCN-01/PD184352/AZD6244/PD98059/AZD0530/PP2/AZD7762/dasatinib/AG1478 treatment was from a 100 mM stock solution of each drug and the maximal concentration of Vehicle (DMSO) in media was 0.02% (v/v).
Cell treatments, SDS-PAGE and protein gel blot analysis.
For in vitro analyses of short-term apoptosis effects, cells were treated with Vehicle/UCN-01/PD184352/AZD6244/PD98059/AZD0530/PP2/AZD7762/dasatinib/AG1478 or their combination for the indicated times. Cells for colony formation assays were plated at 250–4,000 cells per well in sextupilcate and for in vitro assays 14 h after plating were treated with the individual or the drug combination(s) at a fixed increasing dose ratio according to the Method of T-C Chou and P Talalay, for 48 h followed by drug removal. Ten to fourteen days after exposure or tumor isolation, plates were washed in PBS, fixed with methanol and stained with a filtered solution of crystal violet (5% w/v). After washing with tap water, the colonies were counted both manually (by eye) and digitally using a ColCount plate reader (Oxford Optronics, Oxford, England). Data presented is the arithmetic mean (±SEM) from both counting methods from multiple studies. Colony formation was defined as a colony of 50 cells or greater.
For SDS PAGE and immunoblotting, cells were plated at 5 × 10 cells/cm and treated with therapeutic drugs at the indicated concentrations and after the indicated time of treatment, lysed with whole-cell lysis buffer (0.5 M Tris-HCl, pH 6.8, 2% SDS, 10% glycerol, 1% β-mercaptoethanol, 0.02% bromophenol blue), and the samples were boiled for 30 min. The boiled samples (10–100 µg based on the gel cassette being used) were loaded onto 10–14% SDS-PAGE and electrophoresis was run overnight. Proteins were electrophoretically transferred onto 0.22 µm nitrocellulose and immunoblotted with various primary antibodies against different proteins. All immunoblots were visualized by use of an Odyssey Infra Red Imaging System.
Short-term cell viability assays after drug exposure.
Cells were isolated at the indicated times in the figures by trypsinization, and either subjected to trypan blue cell viability assay by counting in a light microscope or fixed to slides, and stained using a commercially available Diff Quick (Geimsa) assay kit as per reference 16–24 and 40. The Annexin V/propidium iodide assay was carried to determine cell viability out as per the manufacturer's instructions (BD PharMingen) using a Becton Dickinson FACScan flow cytometer (Mansfield, MA). Media containing cells was collected. Cells were isolated by brief typsinization and this together with the growth media was centrifuged to isolate a cell pellet. The cell pellet was resuspended in PBS on ice containing an FITC conjugated anti-Annexin V antibody followed by addition to this suspension of propidium iodide. The suspension was then within ∼5 min subjected to flow cytometery to determine early apoptosis (Annexin+ cells); early necrosis (propidium+ cells); or later forms of cell death (Annexin+ propidium+). Unless otherwise stated, where the percentage of cell death was calculated using this method, the percentage cell death included all registering PI+ and all annexin+ cells.
Plasmid transfection.
Plasmid DNA (0.5 µg/total plasmid transfected) was diluted into 50 µl of growth media that lacked supplementation with FBS or with penicillin-streptomycin. Lipofectamine 2000 reagent (1 µl) (Invitrogen, Carlsbad, CA) was diluted into 50 µl growth media that lacked supplementation with FBS or with penicillin-streptomycin. The two solutions were then mixed together and incubated at room temperature for 30 min. The total mix was added to each well (4-well glass slide or 12-well plate) containing 200 µl growth media that lacked supplementation with FBS or with penicillin-streptomycin. The cells were incubated for 4 h at 37°C, after which time the media was replaced with growth media containing 5% (v/v) FBS and 1x pen-strep.16–24,40
Data analysis.
Comparison of the effects between various in vitro drug treatments was performed following ANOVA using the Student's t-test. Differences with a p value of < 0.05 were considered statistically significant. Experiments shown are the means of multiple individual points from multiple studies (±SEM). Median dose effect isobologram colony formation analyses to determine synergism of drug interaction were performed according to the Methods of T-C Chou and P Talalay using the Calcusyn program for Windows (BIOSOFT, Cambridge, UK). Cells were treated with agents at an escalating fixed concentration drug dose. A indicates synergy of interaction between the two drugs; a combination index of ∼1.00 indicates an additive interaction; a CI value of >1.00 indicates antagonism of action between the two agents.
Acknowledgments
This work was funded from PHS grants (R01-DK52825; P01-CA104177;R01-CA108325;R01-CA141703;R01-CA150214; R01-CA100866; R01-CA93738), Department of Defense Award (DAMD17-03-1-0262 and W81XWH-10-1-0009). The work was also funded by The Jim Valvano “Jimmy V” Foundation. P.D. is the holder of the Universal Inc. Professorship in Signal Transduction Research and S.G. is the holder of the Olsen Distinguished Professorship. “The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.”
Abbreviations
- ERK
extracellular regulated kinase
- MEK
mitogen activated extracellular regulated kinase
- JNK
c-Jun NH2-terminal kinase
- PI3K
phosphatidyl inositol-3-kinase
- MAPK
mitogen activated protein kinase
- CHK
checkpoint kinase
- ca
constitutively active
- dn
dominant negative
- EGFR
epidermal growth factor receptor
- PTEN
phosphatase and tensin homologue on chromosome ten
- ROS
reactive oxygen species
- CMV
empty vector plasmid or virus
- si
small interfering
- SCR
scrambled
- IP
immunoprecipitation
- Ad
adenovirus
- DISC
death-inducing signaling complex
- TUNEL
terminal deoxynucleotidyl transferase dUTP nick end labeling
- VEH
vehicle
Authorship's Contribution
P.D. designed the studies and directed C.M., H.A.H., N.C., Y.T., M.D.B., N.H. whom performed the studies. A.Y. and G.T. assisted P.D. in bench-side supervision of C.M., H.A.H., N.C., Y.T., M.D.B., N.H. S.G. and Y.D. assisted P.D. with the writing and proofing of the manuscript.
References
- 1.Mow BM, Blajeski AL, Chandra J, Kaufmann SH. Apoptosis and the response to anticancer therapy. Curr Opin Oncol. 2001;13:453–462. doi: 10.1097/00001622-200111000-00007. [DOI] [PubMed] [Google Scholar]
- 2.Prudhomme M. Novel checkpoint 1 inhibitors. Recent Patents Anticancer Drug Discov. 2006;1:55–68. doi: 10.2174/157489206775246520. [DOI] [PubMed] [Google Scholar]
- 3.Graves PR, Yu L, Schwarz JK, Gales J, Sausville EA, O'Connor PM, et al. The Chk1 protein kinase and the Cdc25C regulatory pathways are targets of the anticancer agent UCN-01. J Biol Chem. 2000;275:5600–5605. doi: 10.1074/jbc.275.8.5600. [DOI] [PubMed] [Google Scholar]
- 4.Peng CY, Graves PR, Thoma RS, Wu Z, Shaw AS, Piwnica-Worms H. Mitotic and G checkpoint control: regulation of 14-3-3 protein binding by phosphorylation of Cdc25C on serine-216. Science. 1997;277:1501–1505. doi: 10.1126/science.277.5331.1501. [DOI] [PubMed] [Google Scholar]
- 5.Komander D, Kular GS, Bain J, Elliott M, Alessi DR, Van Aalten DM. Structural basis for UCN01 (7-hydroxystaurosporine) specificity and PDK1 (3-phosphoinositide-dependent protein kinase-1) inhibition. Biochem J. 2003;375:255–262. doi: 10.1042/BJ20031119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sausville EA, Lush RD, Headlee D, Smith AC, Figg WD, Arbuck SG, et al. Clinical pharmacology of UCN-01: initial observations and comparison to pre-clinical models. Cancer Chemother Pharmacol. 1998;42:54–59. doi: 10.1007/s002800051080. [DOI] [PubMed] [Google Scholar]
- 7.Fuse E, Kuwabara T, Sparreboom A, Sausville EA, Figg WD. Review of UCN-01 development: a lesson in the importance of clinical pharmacology. J Clin Pharmacol. 2005;45:394–403. doi: 10.1177/0091270005274549. [DOI] [PubMed] [Google Scholar]
- 8.Hagenauer B, Maier-Salamon A, Thalhammer T, Zollner P, Senderowicz A, Jager W. Metabolism of UCN-01 in isolated perfused rat liver: role of Mrp2 in the biliary excretion of glucuronides. Oncol Rep. 2004;11:1069–1075. doi: 10.3892/or.11.5.1069. [DOI] [PubMed] [Google Scholar]
- 9.Fuse E, Tanii H, Kurata N, Kobayashi H, Shimada Y, Tamura T, et al. Unpredicted clinical pharmacology of UCN-01 caused by specific binding to human alpha1-acid glycoprotein. Cancer Res. 1998;58:3248–3253. [PubMed] [Google Scholar]
- 10.Dees EC, Baker SD, O'Reilly S, Rudek MA, Davidson SB, Aylesworth C, et al. A phase I and pharmacokinetic study of short infusions of UCN-01 in patients with refractory solid tumors. Clin Cancer Res. 2005;11:664–671. [PubMed] [Google Scholar]
- 11.Hotte SJ, Oza A, Winquist EW, Moore M, Chen EX, Brown S, et al. Phase I trial of UCN-01 in combination with topotecan in patients with advanced solid cancers: a Princess Margaret Hospital Phase II Consortium study. Ann Oncol. 2006;17:334–340. doi: 10.1093/annonc/mdj076. [DOI] [PubMed] [Google Scholar]
- 12.Perez RP, Lewis LD, Beelen AP, Olszanski AJ, Johnston N, Rhodes CH, et al. Modulation of cell cycle progression in human tumors: a pharmacokinetic and tumor molecular pharmacodynamic study of cisplatin plus the Chk1 inhibitor UCN-01 (NSC 638850) Clin Cancer Res. 2006;12:7079–7085. doi: 10.1158/1078-0432.CCR-06-0197. [DOI] [PubMed] [Google Scholar]
- 13.Wang S, Guo CY, Dent P, Grant S. Effect of Bcl-XL expression on taxol-induced apoptosis and cytotoxicity in human leukemia cells (U937) Leukemia. 1999;13:1564–1573. [Google Scholar]
- 14.Dent P, Yacoub A, Fisher PB, Hagan MP, Grant S. MAPK pathways in radiation responses. Oncogene. 2003;22:5885–5896. doi: 10.1038/sj.onc.1206701. [DOI] [PubMed] [Google Scholar]
- 15.Dent P, Qiao L, Grant S. Signaling by ErbB family receptors. Frontiers in Bioscience. 2002:376–389. doi: 10.2741/grant. [DOI] [PubMed] [Google Scholar]
- 16.McKinstry R, Qiao L, Yacoub A, Dai Y, Decker R, Holt S, et al. Pharmacologic inhibitors of the mitogen activated protein kinase cascade interact synergistically with UCN-01 to induce mitochondrial dysfunction and apoptosis in mammary and prostate carcinoma cells. Cancer Biol Ther. 2002;1:241–251. [Google Scholar]
- 17.Dai Y, Decker RH, McKinstry R, Dent P, Grant S. Pharmacologic inhibitors of the mitogen activated protein kinase cascade interact synergistically with UCN-01 to induce mitochondrial dysfunction and apoptosis in leukemia and lymphoma cells. Cancer Res. 2001;61:5106–1515. [PubMed] [Google Scholar]
- 18.Dai Y, Landowski TH, Rosen ST, Dent P, Grant S. Combined treatment with the checkpoint abrogator UCN-01 and MEK1/2 inhibitors potently induces apoptosis in drug-sensitive and -resistant myeloma cells through an IL-6-independent mechanism. Blood. 2002;100:3333–3343. doi: 10.1182/blood-2002-03-0940. [DOI] [PubMed] [Google Scholar]
- 19.Dai Y, Rahmani M, Pei XY, Khanna P, Han SI, Mitchell C, et al. Farnesyltransferase inhibitors interact synergistically with the Chk1 inhibitor UCN-01 to induce apoptosis in human leukemia cells through interruption of both Akt and MEK/ERK pathways and activation of SEK1/JNK. Blood. 2005;105:1706–1716. doi: 10.1182/blood-2004-07-2767. [DOI] [PubMed] [Google Scholar]
- 20.Hawkins W, Mitchell C, McKinstry R, Gilfor D, Starkey J, Dai Y, et al. Transient exposure of mammary tumors to PD184352 and UCN-01 causes tumor cell death in vivo and prolonged suppression of tumor regrowth. Cancer Biol Ther. 2005;4:1275–1284. doi: 10.4161/cbt.4.11.2286. [DOI] [PubMed] [Google Scholar]
- 21.Mitchell C, Park MA, Eulitt P, Yang C, Yacoub A, Dent P. PARP1 modulates the lethality CHK1 inhibitors in carcinoma cells. Mol Pharmacol. 2010;78:909–917. doi: 10.1124/mol.110.067199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dai Y, Chen S, Shah R, Pei XY, Wang L, Almenara JA, et al. Disruption of Src function potentiates Chk1 inhibitor-induced apoptosis in human multiple myeloma cells in vitro and in vivo. Blood. 2010;117:1947–1957. doi: 10.1182/blood-2010-06-291146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dai Y, Chen S, Pei XY, Almenara JA, Kramer LB, Venditti CA, et al. Interruption of the Ras/MEK/ERK signaling cascade enhances Chk1 inhibitor-induced DNA damage in vitro and in vivo in human multiple myeloma cells. Blood. 2008;112:2439–2449. doi: 10.1182/blood-2008-05-159392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dent P, Tang Y, Yacoub A, Dai Y, Fisher PB, Grant S. CHK1 Inhibitors in Combination Chemotherapy: Thinking Beyond the Cell Cycle. Mol Interv. 2011;11:133–140. doi: 10.1124/mi.11.2.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.O'Neill JW, Hockenbery DM. Bcl-2-related proteins as drug targets. Curr Med Chem. 2003;10:1553–1562. doi: 10.2174/0929867033457241. [DOI] [PubMed] [Google Scholar]
- 26.Golding SE, Rosenberg E, Neill S, Dent P, Povirk LF, Valerie K. Extracellular signal-related kinase positively regulates ataxia telangiectasia mutated, homologous recombination repair and the DNA damage response. Cancer Res. 2007;67:1046–1053. doi: 10.1158/0008-5472.CAN-06-2371. [DOI] [PubMed] [Google Scholar]
- 27.Patel V, Lahusen T, Leethanakul C, Igishi T, Kremer M, Quintanilla-Martinez L, et al. Antitumor activity of UCN-01 in carcinomas of the head and neck is associated with altered expression of cyclin D3 and p27(KIP1) Clin Cancer Res. 2002;8:3549–3560. [PubMed] [Google Scholar]
- 28.Jabbour E, Cortes J, Kantarjian H. Dasatinib for the treatment of Philadelphia chromosome-positive leukaemias. Expert Opin Investig Drugs. 2007;16:679–687. doi: 10.1517/13543784.16.5.679. [DOI] [PubMed] [Google Scholar]
- 29.Lee D, Gautschi O. Clinical development of SRC tyrosine kinase inhibitors in lung cancer. Clin Lung Cancer. 2006;7:381–384. doi: 10.3816/clc.2006.n.020. [DOI] [PubMed] [Google Scholar]
- 30.Dai Y, Grant S. Targeting multiple arms of the apoptotic regulatory machinery. Cancer Res. 2007;67:2908–2911. doi: 10.1158/0008-5472.CAN-07-0082. [DOI] [PubMed] [Google Scholar]
- 31.Cory S, Adams JM. Killing cancer cells by flipping the Bcl-2/Bax switch. Cancer Cell. 2005;8:5–6. doi: 10.1016/j.ccr.2005.06.012. [DOI] [PubMed] [Google Scholar]
- 32.Konopleva M, Watt J, Contractor R, Tsao T, Harris D, Estrov Z, et al. Mechanisms of antileukemic activity of the novel Bcl-2 homology domain-3 mimetic GX15-070 (obatoclax) Cancer Res. 2008;68:3413–3420. doi: 10.1158/0008-5472.CAN-07-1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mitchell C, Yacoub A, Hamed H, Martin AP, Bareford MD, Eulitt P, et al. Inhibition of MCL-1 in breast cancer cells promotes cell death in vitro and in vivo. Cancer Biol Ther. 2010;10:903–917. doi: 10.4161/cbt.10.9.13273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tse C, Shoemaker AR, Adickes J, Anderson MG, Chen J, Jin S, et al. ABT-263: a potent and orally bioavailable Bcl-2 family inhibitor. Cancer Res. 2008;68:3421–3428. doi: 10.1158/0008-5472.CAN-07-5836. [DOI] [PubMed] [Google Scholar]
- 35.Nguyen M, Marcellus RC, Roulston A, Watson M, Serfass L, Murthy Madiraju SR, et al. Small molecule obatoclax (GX15-070) antagonizes MCL-1 and overcomes MCL-1-mediated resistance to apoptosis. Proc Natl Acad Sci USA. 2007;104:19512–19517. doi: 10.1073/pnas.0709443104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ingley E. Src family kinases: regulation of their activities, levels and identification of new pathways. Biochim Biophys Acta. 2008;1784:56–65. doi: 10.1016/j.bbapap.2007.08.012. [DOI] [PubMed] [Google Scholar]
- 37.Boutros R, Lobjois V, Ducommun B. CDC25 phosphatases in cancer cells: key players? Good targets? Nat Rev Cancer. 2007;7:495–507. doi: 10.1038/nrc2169. [DOI] [PubMed] [Google Scholar]
- 38.Wang X, Zeng L, Wang J, Chau JF, Lai KP, Jia D, et al. A positive role for c-Abl in Atm and Atr activation in DNA damage response. Cell Death Differ. 2010;18:5–15. doi: 10.1038/cdd.2010.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Singh J, Aaronson SA, Mlodzik M. Drosophila Abelson kinase mediates cell invasion and proliferation through two distinct MAPK pathways. Oncogene. 2010;29:4033–4045. doi: 10.1038/onc.2010.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hamed H, Hawkins W, Mitchell C, Gilfor D, Zhang G, Pei XY, et al. Transient exposure of carcinoma cells to RAS/MEK inhibitors and UCN-01 causes cell death in vitro and in vivo. Mol Cancer Ther. 2008;7:616–629. doi: 10.1158/1535-7163.MCT-07-2376. [DOI] [PubMed] [Google Scholar]
- 41.Carter S, Auer KL, Reardon DB, Birrer M, Fisher PB, Valerie K, et al. Potentiation of Ionizing Radiation induced cell killing by inhibition of the Mitogen Activated Protein (MAP) kinase cascade in A431 human squamous carcinoma cells. Oncogene. 1998;16:2787–2796. doi: 10.1038/sj.onc.1201802. [DOI] [PubMed] [Google Scholar]
- 42.Mitchell JB, Choudhuri R, Fabre K, Sowers AL, Citrin D, Zabludoff SD. In vitro and in vivo radiation sensitization of human tumor cells by a novel checkpoint kinase inhibitor, AZD7762. Clin Cancer Res. 2010;16:2076–2084. doi: 10.1158/1078-0432.CCR-09-3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Park MA, Yacoub A, Rahmani M, Zhang G, Hart L, Hagan MP, et al. OSU-03012 stimulates PKR-like endoplasmic reticulum-dependent increases in 70-kDa heat shock protein expression, attenuating its lethal actions in transformed cells. Mol Pharmacol. 2008;73:1168–1184. doi: 10.1124/mol.107.042697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yacoub A, Park MA, Gupta P, Rahmani M, Zhang G, Hamed H, et al. Caspase-, cathepsin- and PERK-dependent regulation of MDA-7/IL-24-induced cell killing in primary human glioma cells. Mol Cancer Ther. 2008;7:297–313. doi: 10.1158/1535-7163.MCT-07-2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Certo M, Del Gaizo Moore V, Nishino M, Wei G, Korsmeyer S, Armstrong SA, et al. Mitochondria primed by death signals determine cellular addiction to antiapoptotic BCL-2 family members. Cancer Cell. 2006;9:351–365. doi: 10.1016/j.ccr.2006.03.027. [DOI] [PubMed] [Google Scholar]
- 46.Klinghoffer RA, Sachsenmaier C, Cooper JA, Soriano P. Src family kinases are required for integrin but not PDGFR signal transduction. EMBO J. 1999;18:2459–2471. doi: 10.1093/emboj/18.9.2459. [DOI] [PMC free article] [PubMed] [Google Scholar]