Abstract
Cellular senescence is an irreversible growth arrest that is activated in normal cells upon shortening of telomere and other cellular stresses. Bypassing cellular senescence is a necessary step for cells to become immortal during oncogenic transformation. During the spontaneous immortalization of Li-Fraumeni Syndrome (LFS) fibroblasts, we found that CREG1 (Cellular Repressor of E1A-stimulated Genes 1) expression was decreased during immortalization and increased in senescence. Moreover, we found that repression of CREG1 expression occurs via an epigenetic mechanism, promoter DNA methylation. Ectopic expression of CREG1 in the immortal LFS cell lines decreases cell proliferation but does not directly induce senescence. We confirmed this in osteosarcoma and fibrosarcoma cancer cell lines, cancers commonly seen in Li-Fraumeni Syndrome. In addition, we found that p16INK4a is also downregulated in immortal cells and that coexpression of CREG1 and p16INK4a, an inhibitor of CDK4/6 and Rb phosphorylation, has a greater effect than either CREG1 and p16INK4a alone to reduce cell growth, induce cell cycle arrest and cellular senescence in immortal LFS fibroblasts, osteosarcoma and fibrosarcoma cell lines. Moreover, cooperation of CREG1 and p16INK4a inhibits the expression of cyclin A and cyclin B by inhibiting promoter activity, thereby decreasing mRNA and protein levels; these proteins are required for S-phase entry and G2/M transition. In conclusion, this is the first evidence to demonstrate that CREG1 enhances p16INK4a-induced senescence by transcriptional repression of cell cycle-regulated genes.
Key words: CREG1, p16INK4a, cellular immortalization, cellular senescence, Li-Fraumeni syndrome
Introduction
Cellular senescence was initially described by Leonard Hayflick who demonstrated that normal human cells had a limited ability to proliferate in culture.1 Senescent cells are characterized by (1) an irreversible growth arrest in G1 phase of the cell cycle, (2) changes of cell morphology (cell enlargement and flattening), (3) unresponsiveness to mitotic signals yet remain metabolically active, (4) alterations in gene expression patterns and (5) the expression of senescence-associated β-galactosidase (SA-β-gal).2,3 Cellular senescence appears to be one of the tumor suppressor mechanisms that is programmed to prevent cells from uncontrolled proliferation. Normal cells that bypass the pathways of senescence become immortal, which provides the opportunity to gain additional genetic and epigenetic events thus leading to tumor development.
Two major pathways, p53 and pRb, are responsible for activating senescence.4 p21INK1a, a p53 target gene and p16INK4a, a CDK4/6 inhibitor that can activate pRb, can induce the onset of cellular senescence.5–7 Induction of p16INK4a in some cancer cell lines and human diploid fibroblast is sufficient to induce senescence in pRb-dependent manner.8,9 Tumor suppressor genes involved in several pathways such as the cell cycle, DNA repair and apoptosis are hypermethylated and silenced in cancer, e.g., pRb, p16INK4a, p15INK4b and BRCA1.10 The inhibition of DNA methyltransferases (DNMTs) can restore gene expression silenced by promoter methylation. 5-aza-deoxycytidine (5-aza-dC) is a DNMT inhibitor widely used to demethylate DNA and restore silenced gene expression.11
Li-Fraumeni syndrome (LFS) is a rare inherited cancer syndrome characterized by early age of tumor onset with multiple types of cancers12,13 and 75% of LFS patients carry a germline mutation of a p53 allele.14 Fibroblasts from LFS patients spontaneously immortalize in culture and these cells can be transformed by an H-ras oncogene to form tumors in immunodeficient mice.15,16 Immortalization is a necessary but not sufficient for tumor development. Previous studies identified changes in gene expression profiles during immortalization that could be reversed by 5-aza-dC-induced senescence in four independent spontaneously immortalized LFS cell lines, MDAH041, MDAH0871, MDAH087-10 and MDAH087-N, derived from two LFS patients.17 CREG1, the cellular repressor of E1A-stimulated genes 1, is one of the genes whose expression is epigenetically downregulated in immortal LFS cells compared with preimmortal or precrisis cells from the same LFS patient and upregulated in 5-aza-dC-treated immortal LFS cells. Treatment of immortal LFS cells with 5-aza-dC induces senescence18,19 indicating that genes involved in the immortalization and senescence processes can be regulated epigenetically by DNA methylation.17
CREG1 is a secreted glycoprotein containing 220 amino acids with three consensus N-glycosylation sites. CREG1 was first identified by the yeast two-hybrid screen for proteins that interact with the Drosophila TATA-binding protein, TBP, and it shares sequence similarity with a region of the adenovirus E1A protein at its Rb binding sites.20 The human CREG1 protein binds to TBP and Rb-family pocket proteins, pRb/p105, RbL1/p107 and RbL2/p130 in vitro.20 CREG1 functions as transcriptional repressor of E2F transcriptional activation capable of antagonizing the ability of E1A and ras to transform primary cells.20,21 CREG1 enhances differentiation of embryonal carcinoma cells and its expression increases in differentiated cells.22 Overexpression of CREG1 in human teratocarcinoma cells inhibits cell cycle progression and cell proliferation.23 The secreted form of CREG1 interacts with its putative receptor, insulin-like growth factor2 receptor (IGF2R), which appears to affect its endogenous localization. Growth suppression by CREG1 is IGF2R-dependent.23,24 Moreover, Han et al. demonstrated that in vascular cells CREG1 promotes differentiation and growth arrest in this cell type.25 Adenovirus-mediated CREG1 expression in balloon-injured arteries reduces vascular hyperplasia.26 CREG1 overexpression was shown to attenuate cardiac hypertrophy in mice, involving downregulation of ERK signaling,27 inhibiting vascular smooth muscle cell migration as well as reducing the activity and expression of MMP-9 protein with increased protein expression of TIMP-1 and TIMP-2.28
In this study, we demonstrate a functional role for CREG1 in cellular senescence. We found that CREG1 expression was epigenetically silenced in immortal LFS cell lines. When overexpressed in immortalized LFS fibroblasts, or U2OS and HT1080 cancer cell lines, CREG1 decreased cell growth but did not directly induce senescence. Interestingly, we found that coexpression of CREG1 enhanced p16INK4a induced senescence and more strongly inhibited cell proliferation. This study provides the first evidence for the role of CREG1 in the mechanism of senescence related to the important p16INK4a-Rb pathway.
Results
CREG1 expression is epigenetically downregulated during immortalization and upregulated upon senescence in LFS fibroblasts.
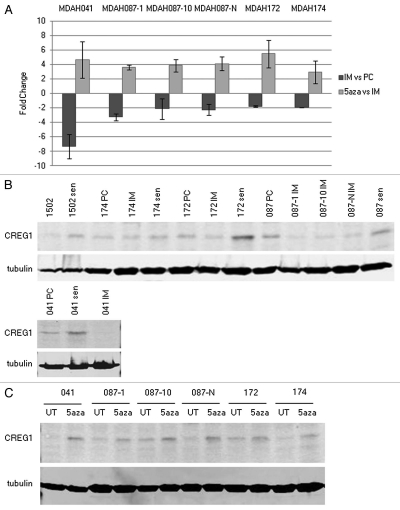
Gene expression profiling analysis in immortal LFS fibroblasts revealed that CREG1 was epigenetically downregulated during cellular immortalization.17 To confirm the microarray data of CREG1 expression, quantitative real-time PCR (Q-RT-PCR) was performed to measure CREG1 expression at mRNA levels in four immortal LFS cell lines used in microarray analysis, MDAH041, MDAH087-1, MDAH087-10 and MDAH087-N. Two additional immortal LFS cell lines, MDAH172 and MDAH174 that were not included in the original gene expression profiling study provided independent validation that CREG1 was frequently epigenetically downregulated in immortal cells. Using RNA from precrisis (population doubling [PD] <30) and immortal (PD >150) LFS fibroblasts as well as 5-aza-dC-treated and untreated immortal cells for Q-RT-PCR, we found the levels of CREG1 mRNA decreased in immortal cells compared to their precrisis counterparts and that treatment of immortal LFS cell lines with the DNA demethylating agent, 5-aza-dC, reactivated its expression (Fig. 1A).
Figure 1.
Expression of CREG1 is downregulated in immortal LFS cell lines and upregulated in 5-aza-dC-induced and replicative senescent fibroblasts. (A) The expression of CREG1 by Q-RT-PCR. CREG1 expression was examined by Q-RT-PCR analysis in six independent immortalized LFS fibroblasts. The dark bars show fold changes of CREG1 in immortal LFS (IM) cells compared with precrisis (PC) fibroblasts derived from the same patient. The light bars represent the fold changes of immortal cells treated with 5-aza-dC compared to untreated immortal cells. GAPDH mRNA was detected from the same sample for normalization. The bar graphs represent the mean fold changes from two different RNA preparations with three replicate for each reaction. (B) Western blot analysis of CREG1 in normal and LFS fibroblasts. CREG1 protein levels were examined in normally growing and in replicative senescent fibroblast CRL1502 and LFS precrisis (PC), immortal (IM) and senescent (sen) fibroblasts. The population doublings (PD) of examined fibroblasts are indicated as followed; 1502 (PD21), 1502 sen (PD46), 174 PC (PD13), 174 IM (PD132), 174 sen (PD52), 172 PC (PD14), 172 IM (PD172), 172 sen (PD51), 087 PC (PD26), 087-1 IM (PD298), 087-10 IM (PD258), 087-N IM (PD250), 087 sen (PD61), 041 PC (PD 19), 041 IM (PD351), 041 sen (PD 43). (C) Western blot analysis of CREG1 in 5-aza-dC treatment of immortal LFS cell lines. CREG1 proteins were detected in six independent immortal LFS cell lines untreated (UT) and treated (5aza) with 1 µM 5-aza-dC.
To expand on the Q-RT-PCR results, CREG1 protein levels were measured by western blotting protein extracts from precrisis and immortal LFS cell lines (Fig. 1B) as well as 5-aza-dC treated immortal LFS cells (Fig. 1C). Additionally, the protein levels of CREG1 were measured from normal and LFS cells at replicative senescence. The population doublings of replicatively senescent LFS fibroblasts are slightly varied among four independent cells lines; MDAH041 (PD43), MDAH087 (PD61), MDAH172 (PD51), MDAH174 (PD52) and normal fibroblast CRL-1502 (PD46). Interestingly, the protein levels of CREG1 were upregulated in replicative senescent LFS and normal CRL-1502 fibroblasts, as shown in Figure 1B. The expression level of CREG1 fits the criteria of senescence-associated gene, decreased during immortalization and increased in replicative and 5-aza-dC-induced senescence justifying further investigation of the role of CREG1 in cellular senescence.
Epigenetic mechanism by promoter hypermethylation regulates CREG1 expression.
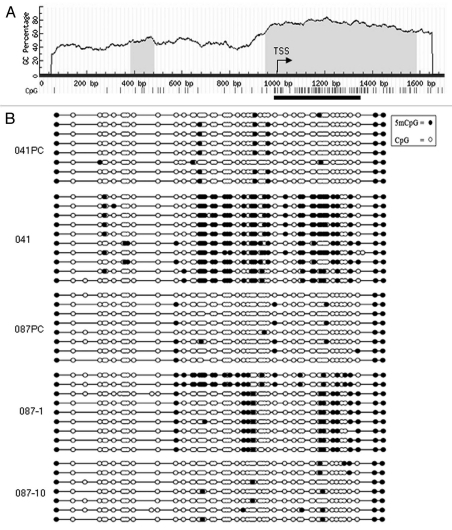
We found that treatment of immortal LFS cells with the DNA demethylating agent 5-aza-dC could reactivate CREG1 expression. DNA methylation at gene promoters has been shown to silence the gene expression, including many tumor suppressor genes in cancer cells.29 To investigate whether CREG1 expression is regulated by promoter hypermethylation, we analyzed the region of CREG1 promoter for CpG sites in the region of −1,000 to +800 bp relative to the transcription start site using MethPrimer software. We found that the CREG1 promoter contains CpG rich islands (Fig. 2A). Sodium bisulfite-modified genomic DNA samples from precrisis and immortal LFS fibroblasts were PCR-amplified followed by sequencing at a 383 bp CpG rich region. This region of CREG1 promoter analyzed is highly methylated in immortal MDAH041 cells (60%) and partially methylated in MDAH087-1 (30%) and MDAH087-10 (10%) compared to their precrisis counterparts (Fig. 2B). The degree of promoter methylation appeared to be related to the expression level of CREG1; lower CREG1 expression and higher methylation status was observed in MDAH041 than those in MDAH087, and this was also true when evaluating the degree of promoter methylation and expression levels in MDAH087-1 compared with MDAH087-10. However, CREG1 expression may not only be regulated by promoter methylation as we observed partial methylation in MDAH087-1 and low methylation in MDAH087-10. In this case, other transcriptional mechanisms may also be regulating CREG1 expression.
Figure 2.
CREG1 promoter contains CpG rich islands and is silenced by DNA methylation. (A) Schematic of CREG1 promoter from −1,000 to + 800 bp relevant to the transcription start site (TSS). The shaded area shows the CpG rich region and the vertical bars represent the CpG sites. The black line represents the region analyzed by bisulfite sequencing of sodium bisulfite treated genomic DNA. (B) Bisulfite sequencing of the CREG1 promoter from precrisis (PC) and immortal (IM) MDAH041, MDAH087-1 and MDAH087-10. The filled black circles represent the methylated CpG sites, whereas the open white ones represent the unmethylated CpG sites.
CREG1 overexpression decreases cell proliferation in immortal LFS and cancer cell lines.
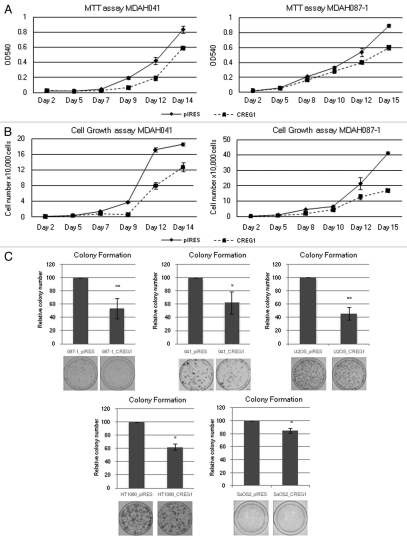
The upregulation of CREG1 in replicative and 5-aza-dC-induced senescence fibroblasts and its silencing by promoter methylation led us to further investigate whether CREG1 was functionally involved in cellular senescence. We overexpressed CREG1/pIRES and a control pIRES empty vector into two immortal LFS cell lines, MDAH041 and MDAH087-1, and studied the growth and senescence properties of the resulting cell lines. We found that CREG1 overexpression decreased cell growth in immortal LFS cells when compared with pIRES empty vector transfected cells demonstrated by both MTT and cell counting proliferation assays (Fig. 3A and 3B). To confirm the effects of CREG1 on cell proliferation, we determined the ability of CREG1 and control pIRES transfected cells to form colonies after transfection and drug selection. We found that CREG1-transfected immortal LFS cell lines displayed decreased colony formation (Fig. 3C). We also investigated the role of CREG1 on cell growth on cancer cell lines relevant to Li-Fraumeni Syndrome: osteosarcoma U2OS and SaOS2 and fibrosarcoma HT1080. The wild type or mutant status of pRb, p16INK4a and p53 of these cancer cell lines is shown in Table 1.30,31 Analysis of cell growth by colony formation assay after stable transfection of CREG1 and pIRES vectors into these cancer cell lines revealed that CREG1 transfected cells decreased colony formation in U2OS and HT1080 cell lines. However, CREG1 only slightly decreased the formation of colony in SaOS2 cell line (Fig. 3C).
Figure 3.
Ectopic expression of CREG1 decreases cell proliferation in immortal LFS and cancer cell lines. (A) MTT cell proliferation assay of MDAH041 and MDAH087-1 immortal cells. After stable transfection, 2,000 cells were seeded into 24-well plates in triplicate and the MTT assay was conducted every 2 days for 2 weeks. The data point with the standard deviation shows the result from a representative experiment of three independent experiments. (B) Cell proliferation assay by cell counting from MDAH041 and MDAH087-1 after stable transfection. After stable transfection, 1,000 cells were seeded into 24-well plates in triplicate and cell number was counted every 2 days. The data point with the standard deviation shows the result from a representative experiment of three independent experiments. (C) Colony formation assay. After 2 weeks of drug selection, 500–5,000 cells were seeded into 10 cm culture plates in triplicate and colonies were stained by Giemsa after 2 weeks in culture. The bar graphs represent the relative colonies number normalized to control pIRES vector that were set to 100% from at least two independent experiments. Overexpression of CREG1 in immortal LFS, U2OS and HT1080 cells decreases colony number compared with pIRES vector control and slightly decrease colony number in SaOS-2 cells (**p < 0.01; *p < 0.05).
Table 1.
Status of pRb, p16 and p53 in cell line used
| Cell line | pRb | p16 | p53 |
| Immortal LFS | Expressed | No expression | Gene mutation |
| U2OS | Expressed | No expression | Expressed |
| SaOS2 | No expression | Expressed | No expression |
| HT1080 | Expressed | No expression | Gene mutation |
CREG1 stable expression is not sufficient to induce senescence in LFS cells.
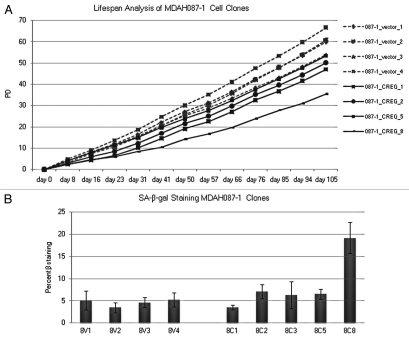
Using the SA-β-gal assay and cellular lifespan analysis, we tested whether CREG1 expression induced senescence after CREG1 transfection of immortal LFS cells. We transfected and isolated individual CREG1 stably transfected cell clones from MDAH087-1 immortal cells (PD 317), analyzed the lifespan by counting the population doubling every week over 105 days maintaining drug selection of the transgene and stained for SA-β-gal to determine the presence of senescent cells in the transfected cell population. At the end of the lifespan assay, the PD from three of the vector control clones exceeded 60 and the growth rate seemed to increase constantly, whereas three of CREG1 clones had PD ≤50 (Fig. 4A). CREG1 stably transfected clones appeared to slow growth compared with pIRES empty vector over this period of time but did not induce a senescence-like growth arrest of the cells as these cells maintained their initial cell morphology and growth rate. To determine whether CREG1-overexpressing cells developed a senescence-like phenotype, SA-β-galactosidase staining was examined in pIRES and CREG1-overexpressing MDAH087-1 clones from passage 6 (Fig. 4B). CREG1 expressing clones did not have a higher activity of SA-β-galactosidase staining than cells from vector control clones with the exception of clone 8 (8C8), which had high senescence-associated β-galactosidase activity. This might be due to clonal variation and a higher spontaneously induced senescence of MDAH087-1 clone 8 fibroblasts. This confirmed the negative growth function of CREG1 but did not indicate an independent role for the function of CREG1 in cellular senescence. We concluded that role of CREG1 in cellular senescence may function within the context of other genetic or epigenetic changes in gene expression.
Figure 4.
CREG1 stable expression is not sufficient to induce senescence in LFS cells. (A) Lifespan analysis of CREG1 overexpressing immortal cells. The population doublings of MDAH087-1 from pIRES and CREG1 expressing clones were calculated from total cell number counted every week over a period of 105 days in culture. (B) Senescence-associated β-galactosidate staining of MDAH087-1 cell clones stably transfected with pIRES empty vector and CREG1. MDAH087-1 cell clones stably expressed CREG1 (8C1, 8C2, 8C3, 8C5 and 8C8) and pIRES (8V1, 8V2, 8V3 and 8V4) and were stained at passage 6. Student's t-test of percent mean of SA-β-gal stained cells in the group of vector control (8V1, 8V2, 8V3 and 8V4) and CREG1 (8C1, 8C2, 8C3 and 8C5) shows p = 0.043.
Coexpression of CREG1 enhances p16INK4a induction of senescence.
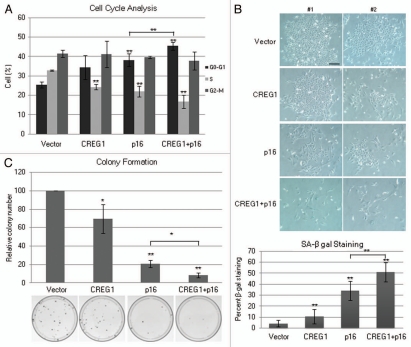
Because CREG1 was shown to interact with Rb-family pocket proteins in vitro and inhibited the transcription activity by E2F transcription factor,20 we hypothesized that CREG1 might affect senescence through the p16INK4a-pRb pathway, which regulates the E2F-family of transcription factors. We knew that p16INK4a expression was downregulated in immortal LFS fibroblasts and its expression was epigenetically regulated by promoter hypermethylation.17 p16INK4a is not expressed in U2OS and HT1080 cell lines. We next tested whether coexpression of CREG1 could enhance p16INK4a-induced senescence in U2OS and HT1080 cancer cell lines. CREG1/pIRESpuro and p16INK4a/pcDNA3.1neo were cotransfected into these cell lines, alone or in combination using the empty vectors to balance the amount of transfected DNA. We used double drug selection, puromycin and G418, to ensure similar transfection efficiencies and the expression of transfected genes was confirmed by western blotting. After 5 days of selection, U2OS cells were analyzed for growth and senescence. Cell cycle analysis showed an increase in the G0-G1 cell population and a decrease of S-phase population in cells cotransfected with CREG1 and p16INK4a compared to cells transfected with either CREG1 or p16INK4a alone (Fig. 5A). In p16-transfected cells and more so in cells cotransfected with CREG1 and p16INK4a, we observed cell characteristics indicative of senescence, a large and flat morphology with an increase of SA-β-galactosidase staining (Fig. 5B). To confirm the growth inhibition of CREG1 and p16INK4a coexpression, colony formation was assessed after stable transfection. Colony formation was inhibited in cells cotransfected with CREG1 and p16INK4a more than cells transfected with either p16INK4a or CREG1 alone (Fig. 5C). Although it was shown previously that p16INK4a overexpression alone induces cellular senescence in this cell line, U2OS, we showed here that coexpression of CREG1 with p16INK4a had an even stronger effect than p16INK4a expression alone.
Figure 5.
Coexpression of CREG1 and p16INK4a has an additive effect to inhibit cell growth and induce senescence. (A) Cell cycle analysis of U2OS cells transfected with control empty vectors, CREG1, p16INK4a and CREG1 + p16INK4a. Cells were analyzed at day 5 after drug selection. The bar graphs are average percent cell population in G0-G1, S and G2-M phases from three independent experiments. ANOVA analysis was performed to test the differences of each phases in all transfection conditions and showed significance differences of G0-G1 (p < 0.01) and S phases (p < 0.01) but not G2-M (p = 0.68). Student's t-test was performed to compare the effect of each transfection condition with control vector and p16INK4a with combination of CREG1 and p16INK4a in G0-G1 and S phase. **p < 0.01. (B) Cell morphology of U2OS cells cotransfected with CREG1 and p16INK4a, alone and in combination. Transfected cells were β-galactosidase stained and photographed at day 5 after selection. The pictures show two different fields on the culture plate from the representative experiment. Scale bar is 200 µm. The bar graphs show the percent of SA-β-gal stained (blue) cells from three independent experiments. Student's t-test was performed to compare the effect of each transfection condition with control vector and p16INK4a with combination of CREG1 and p16INK4a. **p < 0.01. (C) Colony number of U2OS cells transfected with CREG1 and p16INK4a. The bar graphs show the average relative colony number normalized to the vector control from three independent experiments. 2,000 cells were seeded into 10 cm cultured plate in triplicate at day 5 after double drug selection and the colonies were stained with Giemsa after 2 weeks. ANOVA analysis was performed to test the differences of relative colony number to vector control (p < 0.01). Student's t-test was performed to compare the effect of each transfection condition with control vector to p16INK4a in combination with CREG1, CREG1 alone or p16INK4a alone. **p < 0.01; *p < 0.05.
Coexpression of CREG1 and p16INK4a causes decreased of cyclin A and B expression.
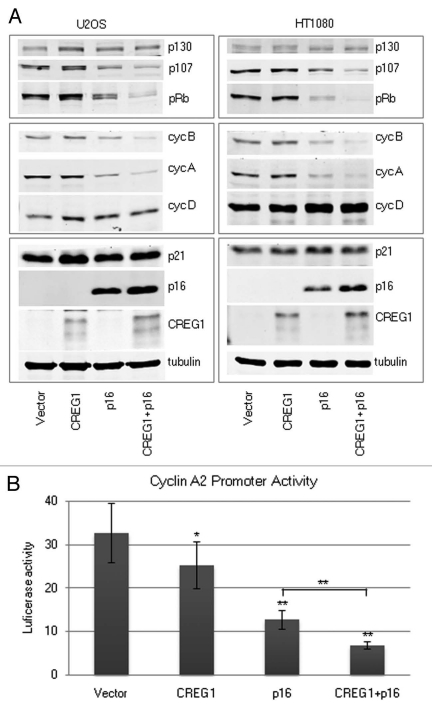
To investigate the additive effect of CREG1 and p16INK4a on cell cycle control, the expression of proteins involved in cell cycle progression was examined by western blotting. We found that cells coexpressing CREG1 and p16INK4a showed a marked decline in protein levels of cyclin A and B (Fig. 6A). We also showed that the decrease of mRNA level of cyclin A from CREG1 + p16INK4a cotransfected U2OS cells correlated with the decreased protein expression levels (data not shown). In addition, the protein levels of pRb (p105) and RbL1 (p107) were decreased whereas RbL2 (p130) expression was unchanged. Similar results were obtained from HT1080 cells cotransfected with CREG1 and p16INK4a (Fig. 6A).
Figure 6.
Cooperation between CREG1 and p16INK4a decreases cyclin A and B expression. (A) Western blot analysis of U2OS and HT1080 cell lines transfected with CREG1, p16INK4a alone or CREG1 and p16INK4a in combination at day 5 after selection. (B) Cyclin A2 reporter activity in U2OS cells transfected with CREG1 and p16INK4a. The bar graphs show the luciferase activity obtained from luciferase/renilla ratio from three independent experiments. ANOVA analysis was performed to test the differences of promoter activity relative to vector control (p < 0.01). Student's t-test was performed to compare the promoter activity of control vector with each transfection condition; CREG1, p16INK4a alone or CREG1 and p16INK4a in combination. **p < 0.01, *p < 0.05.
To further examine the effect of CREG1 with and without cotransfection of p16INK4a on cyclin A expression, we constructed a cyclin A reporter vector containing a cyclin A promoter driven luciferase gene. The cyclin A luciferase and TK-Renilla vectors were transfected into CREG1 and p16INK4a transfected U2OS cells 3 days after their transfection and double-drug selection. The luciferase activity was measured 40 h after reporter transfection. Consistent with the decrease in protein and mRNA levels of cyclin A, coexpression of CREG1 and p16INK4a resulted in a significant decrease of transcription activity directed by the cyclin A promoter (Fig. 6B).
Discussion
Cellular senescence results from the abrogation of gene expression and pathways regulating this process, which can cause cells to become immortal. This added lifespan leads to the additional genetic and epigenetic changes necessary for the development of cancer. Several studies were pursued to understand the mechanism of senescence. Previous analysis by gene expression profiling of four independent immortalized LFS fibroblasts, revealed changes of genes in cell cycle, interferon and cytoskeleton pathways.17 CREG1 is one of the 14 genes that was shown to be downregulated in all LFS cells during immortalization and upregulated after 5-aza-dC-induced senescence. Verification of CREG1 expression in six independent spontaneously immortalized LFS fibroblasts revealed that CREG1 levels decreased in immortal fibroblasts compared with each precrisis counterpart cell line and its expression was upregulated in replicative and 5-aza-dC-induced senescence fibroblasts. Analysis of the CREG1 promoter identified a CpG island rich region that was highly methylated in MDAH041, partially methylated in MDAH087-1 and weakly methylated in MDAH087-10 immortal fibroblasts. Possibly the two alleles of CREG1 in MDAH087-1 and MDAH087-10 are differentially methylated reflecting the intermediate expression level of CREG1 in these cells when compared with heavily methylated promoter from MDAH041. In addition, CREG1 is located at 1q24.1 region and this region of chromosome 1q has been shown to carry genes altered in cell lines assigned in complementation group C, which is involved in cellular senescence.32 We demonstrated for the first time that CREG1 expression is epigenetically silenced in immortal fibroblasts and this may contribute to cellular immortalization.
CREG1 was initially identified as a protein that can bind to TBP and Rb family pocket proteins and was capable of transcriptionally repressing of oncoprotein E1A-activated genes as well as E2F target genes.20 We found that overexpression of CREG1 in immortal LFS cells and related cancer cell lines, osteosarcoma U2OS and fibrosarcoma HT1080, decreases cell proliferation, which supports the growth suppression function of CREG1 from previous studies. However, we did not observe the induction of cellular senescence in cells transfected with CREG1 alone. Interestingly, the growth suppression effect of CREG1 was lower in SaOS2 cell line, another osteosarcoma cell line that has no pRb expression, compared to two other cell lines studied that express pRb protein. In addition, the expression level of CREG1 in this cell line, SaOS2, seems to be higher than others (data not shown). This indicated that the growth suppressive function of CREG1 might depend on the pRb signaling pathway. Because the cells we used have lost the function of p16INK4a and that CREG1 was shown to interact with Rb family pocket proteins thus inhibiting the activity of E2F transcription factor, we hypothesized that CREG1 might be involved in p16INK4a-pRb-dependence senescence pathway. p16INK4a, the CDK inhibitor of pRb phosphorylation, is one the tumor suppressor genes that is commonly disrupted in immortal cells and some cancer cells. p16INK4a expression is also upregulated in senescent fibroblasts.5,7,33 Several studies showed that ectopic expression of p16INK4a caused cell cycle arrest and senescence in immortal LFS18 and human cancer cells including the U2OS cell line8,9,34,35 and these effects depended on the normal function of pRb. We showed that coexpression of CREG1 and p16INK4a in U2OS cells (pRb+/p16−/p53+) had a greater effect on inhibition cell growth, induction of cell cycle arrest, inhibition of colony formation and the induction of senescence than overexpression of p16INK4a alone. Consistent with its activity to induce cell cycle arrest, cells cotransfected with CREG1 and p16INK4a induced a marked decrease of cyclin A and B expression. The decreased expression of cyclin A was due to regulation at the transcription level. We also found that in p16INK4a or CREG1/p16INK4a transfected cells, the expression of pRb and p107 were decreased, but p130 expression was unchanged. The changes of Rb pocket proteins in our study are similar to those showed by Helmbold et al. that p130 accumulates and is important for the maintenance of senescence and that pRb and p107 decrease upon p16INK4a-induced senescence.36,37 Moreover, cyclin A expression was shown to be downregulated in cellular senescence by p130 recruitment to the cyclin A promoter to repress the transcription.36,38,39 Because we saw the same effect of exogenous expression of CREG1 and p16INK4a in cell lines with p53 wild-type (U2OS) and p53-deficient (HT1080), it appeared that CREG1 enhances p16INK4a to induce senescence through the p16INK4a-Rb pathway rather than the p53-p21INK1a pathway.
Here, we observed the additive effect of CREG1 and p16INK4a in cellular senescence. However, more mechanistic studies are needed to determine whether CREG1 directly participates in p16INK4a-pRb senescence pathway and if its role is limited to transcriptional repression of E2F target genes, such as cyclin A. It was shown by others that the growth inhibition function of CREG1 depends on its putative receptor, IGF2R, a multifunctional transmembrane protein that functions as scavenger receptor facilitating the clearance of extracellular insulin-like growth factor II (IGF-II).40 We too have observed this protein:protein interaction in cells (data not shown). Moreover, a recent study by Han et al. revealed that growth inhibition function of CREG1 involved in the regulation of IGF-II secretion28 which affects the biological function of IGF-II as a growth factor. However, no study has been clearly elucidated the localization of CREG1 in the nucleus as a transcriptional repressor. We have found that the CREG1 protein localizes to the cytoplasm (data not shown) which indicates a complex role in control of transcription. Nevertheless, we cannot exclude the possibilities that the additive effect of CREG1 and p16INK4a that was observed in our study may be due to the p16INK4a-pRb signaling pathway and that CREG1 is part of this mechanism, or that CREG1 functions through other pathway(s) such as its regulation of IGF-II signaling28 which may promote the p16INK4a-pRb signaling-induced senescence. Additional studies are needed to further investigate whether involvement of CREG1 in p16INK4a-induced senescence is due to its transcriptional repressor activity.
In summary, we demonstrated that CREG1 is in part regulated by epigenetic mechanism and its downregulation is consistently observed in spontaneous cellular immortalization of LFS cells. Ectopic expression of CREG1 in immortal LFS cells and cancer cells lacking endogenous p16INK4a results in decrease of cell proliferation but does not alone induce senescence. Analysis of how CREG1 enhances p16INK4a-induced senescence may provide a better understanding of cellular immortalization, an early step in human tumorigenesis, and could provide improved treatment or prevention of cancer at early stage.
Materials and Methods
Cell culture.
Immortal LFS fibroblasts, HT1080 fibrosarcoma and SaOs2 osteosarcoma cell lines were grown in Modified Eagles Medium (MEM, Invitrogen), and U2OS cell line were cultured in RPMI medium 1640 (Invitrogen). All cell lines were supplemented with 10% fetal bovine serum (Hyclone), 100 units/ml penicillin and 100 µg/ml streptomycin (Invitrogen) and were maintained at 37°C in 5% humidified CO2.
5-aza-dC treatment and gene expression analysis by Q-RT-PCR.
5-aza-dC (Sigma-Aldrich) treatment of immortal LFS fibroblasts was performed as previously described in reference 17. Total RNA was isolated from immortal and 5-aza-dC-treated immortal LFS cell lines using the QIAGEN RNeasy Kit (QIAGEN). cDNA was prepared from 2 µg of RNA using Superscript II (Invitrogen). Q-RT-PCR was performed using SYBR Green PCR Detection Kit (PE Biosystems) and analyzed on the ABI 5700 Sequence Detection System (Applied Biosystems). Primers for CREG1 Q-RT-PCR were F-5′CAG CTT CAG CCA GGG ACA AA and R-5′GGG CAG TTG AGG AAG CCT TAG, and GAPDH was used as an internal control. The relative fold change was calculated using CT method as follows:, 2−ΔΔCT, where, ΔΔCT = (CT Gene of interest − CT GAPDH) experiment − (CT Gene of interest − CT GAPDH) control.41 The calculated relative fold change between 0 and 1 was divided by −1.
Methylation analysis of the CREG1 promoter.
The presence of CpG islands in the promoter of CREG1 was identified using MethPrimer software (http://mail.ucsf.edu/urolab/methprimer/index1.html). The region of the gene, −1,000 to +800 relative to the transcription start was identified using UCSC Golden Path (http://genome.ucsc.edu). Genomic DNA prepared by Puregen DNA isolation kit (Gentra System) was modified with sodium bisulfite as previously described in reference 42. Briefly, 1 µg of genomic DNA was denatured and treated with 3.6 mol/L sodium bisulfite. The modified DNA was resuspended in water and amplified by PCR with two nested PCR reactions. The two sets of primers were, F1 5′-TTA AAA GGA GGG GGT GTG TG (−274), and R1 5′-CGA ACC CCA ACT CAC CTA CA (+1,297); F2 5′-TGC GGA GTT GTA GAG GGA TT (−65) and R2 5′-CCG CCT CCA ACG TAA AAA TA (+316). The numbers in parentheses indicate the position relative to the transcriptional start site of CREG1 (NM_003851). The PCR products were cloned into pCR2.1-TOPO vector (Invitrogen) using the TOPO-TA cloning kit (Invitrogen). DNA Sequencing of ten colonies was performed using M13-reverse and T7 primers. The sequencing results were analyzed by MethTools software (genome.imb-jena.de/methtools/).
Plasmids and DNA transfection.
CREG/pRC plasmid was kindly provided by Dr. Grace Gill from Tufts University. A 675-bp cDNA fragment of CREG1 was amplified from the CREG/pRC plasmid with primers flanking EcoRI and BamHI restriction sites and was cloned into pIRESpuro vector. p16INK4a was cloned into pcDNA3.1HisB vector.
Immortal LFS cell lines, MDAH041 and MDAH087-1, U2OS, HT1080 and SaOS2 cancer cell lines were transfected with 2 µg of CREG1/pIRES and pIRES empty vectors using lipofectamine transfection reagent and plus reagent (Invitrogen). For stable transfection, transfected cells were selected in standard culture media containing 0.5 µg/ml puromycin (Sigma-Aldrich) at 48 h after transfection. Stable transfected cells were used for cell growth assays including cell counting, MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) colorimetric and colony formation assays after 1 week of drug selection.
For coexpression of CREG1 and p16, U2OS and HT1080 cell lines were transfected with four expression conditions, (1) empty vector pIRES and pcDNA3.1, (2) CREG1/pIRES, (3) p16/pcDNA3.1, (4) CREG1/pIRES and p16/pcDNA3.1. Two µg of each vector was cotransfected using lipofectamine transfection reagent and plus reagent (Invitrogen). At 48 h posttransfection, the cells were selected in combination of 0.5 µg/ml puromycin (Sigma-Aldrich) and 200 µg/ml G418 (Gibco) double drug selection. Control untransfected cells died at day 3 after selection. At 5 days after selection, stable transfected cells were harvested for western blotting, flow cytrometry for cell cycle analysis, seeded for colony formation assay, fixed and SA-β-gal stained to detect senescence.
Cell cycle analysis.
Stably transfected cells were trypsinized, counted, and 106 cells were washed in 5 ml phosphate buffer saline (PBS) and centrifuged at 200x g. The resuspended cells (0.3 ml PBS) were fixed by adding 1 ml of cold 95% ethanol with mixing and incubated at −20°C overnight. The fixed cells were centrifuged at 200x g and the ethanol was removed by pipetting. The cells pellets were stained with 1 ml of propidium iodide (PI) staining solution containing 50 ug/ml PI (Invitrogen), 200 ug/ml RNaseA and 0.1% Triton X-100 in phosphate buffered saline (PBS) and incubated at room temperature for 30 minutes. The DNA content was measured at the Karmanos Cancer Institute Flow Cytometry Core using the Becton-Dickinson FACSCalibur™ Cytometer.
Western blotting.
To isolate protein, cells were washed twice with cold PBS and cold lysis buffer (50 mM Tris-HCl pH7.4, 150 mM NaCl, 1% NP40, 0.25% Na-deoxycholate, 1 mM EDTA) containing protease cocktail inhibitor, PMSF, NaPP, NaF and NaV, was added. The cell lysates were collected by scrapping from culture plates and centrifuged at 14,000 rpm for 15 minutes. Equal amounts of proteins were loaded onto 10 or 12% polyacrylamide gel, electrophoresed and transferred onto nitro-cellulose membrane. The membrane was blocked in 5% dry milk in Tris-buffered saline tween-20 (TBST). Proteins were detected by incubating overnight with primary antibodies diluted in 5% dry milk in TBST following by 1 h incubating with secondary antibodies. The images were detected using the Odyssey Infrared Imaging System (Li-Cor Biosciences). The following primary antibodies were used; anti-CREG1 (R&D system #AF2380), anti-p16INK4a (BD Biosciences #554079), anti-p21INK1a (Upstate #05-345), anti-cyclin A2 (BD Biosciences #611268), anti-cyclin B1 (Abcam #ab72), anti-cyclin D1 (Cell signaling #2926), anti-Rb (Cell signaling #9309), anti-p107 (Santa Cruz #sc-318), anti-p130 (Santa cruz #sc-317). The Alexa Fluor 680-labeled secondary antibodies were from Molecular Probes or the Li-Cor IRDye 800-labeled secondary antibodies were from Rockland Immunochemical.
Colony formation assay.
Stably transfected cells were seeded at low density (500–5,000 cells in 10 cm culture plates) in triplicate and cultured under drug selection, as described above, for 2 weeks. The colonies were washed twice with PBS and fixed with cold methanol, stained with diluted Giemsa (1:10 v/v) (Sigma-Aldrich) and incubated at room temperature for 30 minutes. The plates were then rinsed with tap water and air dried. The colonies were counted and mean percentages were calculated from independent experiments. The colonies (size ≥1 mm) were counted and mean percentages were calculated from at least three independent experiments.
MTT cell proliferation assay.
The MTT tetrazolium salt (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium) colorimetric assay was used to quantify metabolically active cells. Cells were seeded at a density of 2,000 cells per well in a 12-well plate. Stock 2 mg/ml filtered MTT (Sigma-Aldrich) solution was diluted 1:10 in culture media. One ml diluted MTT was added to each well and the plated were incubated 1.5 h at 37°C, 5% CO2. MTT was aspirated and 1 ml DMSO was added to each well. The plates were shaken at room temperature for 10 minutes. 100 µl of cell extract was pipette into a 96 well plate and the absorbance was measured at 540 nm on a Dynatech-MR-500 plate reader (Dynatech Laboratories). Absorbance is directly proportional to the number of live cells.
Senesncence-associated β-galactosidase assay.
Senescent cells express high β-galctosidase activity, which can be detected through 5-bromo-4-chloro-3-indolyl β-D-galactopyranoside (X-gal) hydrolysis at pH 6.2 A senescence detection kit (BioVision) was used for staining according to the manufacturer's instructions. Briefly, stably tranfected cells were stained for senescence-associated β-galactosidase activity after 5–7 days of drug selection. In order to count the senescence-associated β-galactosidase positive cells, stably transfected cells were seeded at equal amount (1 × 105 cells in 60-mm culture plate) after stable transfection and stained 24 h later. Cells were washed twice with PBS and fixed with fixative solution for 10–15 minutes. The fixed cells were stained with the staining solution containing X-gal and incubated at 37°C overnight.
Reporter gene assay.
The cyclin A2 promoter reporter was generated by cloning of a 347 bp fragment (−75 to +272) of cyclin A2 promoter into the KpnI and HindIII restriction sites of the pGL3-basic vector.43,44 U2OS cells (40,000) were transfected with 2 µg of each expression vector in a six-well cultured plate in triplicate. Selecting media was changed at 48 hours after transfection. Two µg of the cyclin A2 reporter construct and 0.1 µg of TK-renilla expression vector for normalizing the transfection efficiencies were transfected at day 3 after selection. The cells were then harvested for luciferase assay at 40 h after reporter vectors transfection. The luciferase activity was tested with the dual-luciferase reporter assay system (Promega) according to the manufacturer's instructions. The luciferase activity was calculated from three independent experiments.
Statistical analysis.
The student's two-tailed t-test for independent samples (unpaired t-test) was used to test the statistic significant of the effect of CREG1 and control pIRES overexpression on cell growth by colony formation assay. In the coexpression of CREG1 and p16INK4a experiment, statistical significance was evaluated using analysis of variance analysis (Tukey's test), followed by Student's t-test. A p-value ≤0.05 was considered to be statistically significant.
Acknowledgements
This work was supported by the Barbara and Fred Erb Chair in Cancer Genetics to M.A.T. with support from the Genomics and Flow Cytometry Core laboratories of the KCI CCSG Grant P30-CA022453. B.M. was supported by a Predoctoral Fellowship from the Royal Thai Government. We thank Aviva Levine Fridman and Quanfang Li for the DNA samples used in analysis of CREG1 promoter and thank the Tainsky lab members for helpful comments and suggestions. We thank George Brush (Karmanos Cancer Institute) for expert comments on the manuscript, and Grace Gill (Tufts University) for her kind gift of CREG/pRC plasmid.
References
- 1.Hayflick L. The limited in vitro lifetime of human diploid cell strains. Exp Cell Res. 1965;37:614–636. doi: 10.1016/0014-4827(65)90211-9. [DOI] [PubMed] [Google Scholar]
- 2.Dimri GP, Lee X, Basile G, Acosta M, Scott G, Roskelley C, et al. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc Natl Acad Sci USA. 1995;92:9363–9367. doi: 10.1073/pnas.92.20.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Campisi J, d'Adda di Fagagna F. Cellular senescence: When bad things happen to good cells. Nat Rev Mol Cell Biol. 2007;8:729–740. doi: 10.1038/nrm2233. [DOI] [PubMed] [Google Scholar]
- 4.Shay JW, Pereira-Smith OM, Wright WE. A role for both RB and p53 in the regulation of human cellular senescence. Exp Cell Res. 1991;196:33–39. doi: 10.1016/0014-4827(91)90453-2. [DOI] [PubMed] [Google Scholar]
- 5.Alcorta DA, Xiong Y, Phelps D, Hannon G, Beach D, Barrett JC. Involvement of the cyclin-dependent kinase inhibitor p16 (INK4a) in replicative senescence of normal human fibroblasts. Proc Natl Acad Sci USA. 1996;93:13742–13747. doi: 10.1073/pnas.93.24.13742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hara E, Smith R, Parry D, Tahara H, Stone S, Peters G. Regulation of p16CDKN2 expression and its implications for cell immortalization and senescence. Mol Cell Biol. 1996;16:859–867. doi: 10.1128/mcb.16.3.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stein GH, Drullinger LF, Soulard A, Dulic V. Differential roles for cyclin-dependent kinase inhibitors p21 and p16 in the mechanisms of senescence and differentiation in human fibroblasts. Mol Cell Biol. 1999;19:2109–2117. doi: 10.1128/mcb.19.3.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Medema RH, Herrera RE, Lam F, Weinberg RA. Growth suppression by p16ink4 requires functional retinoblastoma protein. Proc Natl Acad Sci USA. 1995;92:6289–6293. doi: 10.1073/pnas.92.14.6289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dai CY, Enders GH. p16INK4a can initiate an autonomous senescence program. Oncogene. 2000;19:1613–1622. doi: 10.1038/sj.onc.1203438. [DOI] [PubMed] [Google Scholar]
- 10.Rountree MR, Bachman KE, Herman JG, Baylin SB. DNA methylation, chromatin inheritance and cancer. Oncogene. 2001;20:3156–3165. doi: 10.1038/sj.onc.1204339. [DOI] [PubMed] [Google Scholar]
- 11.Haaf T. The effects of 5-azacytidine and 5-azadeoxycytidine on chromosome structure and function: implications for methylation-associated cellular processes. Pharmacol Ther. 1995;65:19–46. doi: 10.1016/0163-7258(94)00053-6. [DOI] [PubMed] [Google Scholar]
- 12.Li FP, Fraumeni JF., Jr Soft-tissue sarcomas, breast cancer and other neoplasms. A familial syndrome? Ann Intern Med. 1969;71:747–752. doi: 10.7326/0003-4819-71-4-747. [DOI] [PubMed] [Google Scholar]
- 13.Chompret A. The Li-Fraumeni syndrome. Biochimie. 2002;84:75–82. doi: 10.1016/s0300-9084(01)01361-x. [DOI] [PubMed] [Google Scholar]
- 14.Malkin D, Li FP, Strong LC, Fraumeni JF, Jr, Nelson CE, Kim DH, Kassel J, et al. Germ line p53 mutations in a familial syndrome of breast cancer, sarcomas and other neoplasms. Science. 1990;250:1233–1238. doi: 10.1126/science.1978757. [DOI] [PubMed] [Google Scholar]
- 15.Bischoff FZ, Strong LC, Yim SO, Pratt DR, Siciliano MJ, Giovanella BC, Tainsky MA. Tumorigenic transformation of spontaneously immortalized fibroblasts from patients with a familial cancer syndrome. Oncogene. 1991;6:183–186. [PubMed] [Google Scholar]
- 16.Bischoff FZ, Yim SO, Pathak S, Grant G, Siciliano MJ, Giovanella BC, et al. Spontaneous abnormalities in normal fibroblasts from patients with Li-Fraumeni cancer syndrome: Aneuploidy and immortalization. Cancer Res. 1990;50:7979–7984. [PubMed] [Google Scholar]
- 17.Fridman AL, Tang L, Kulaeva OI, Ye B, Li Q, Nahhas F, et al. Expression profiling identifies three pathways altered in cellular immortalization: interferon, cell cycle and cytoskeleton. J Gerontol A Biol Sci Med Sci. 2006;61:879–889. doi: 10.1093/gerona/61.9.879. [DOI] [PubMed] [Google Scholar]
- 18.Vogt M, Haggblom C, Yeargin J, Christiansen-Weber T, Haas M. Independent induction of senescence by p16INK4a and p21CIP1 in spontaneously immortalized human fibroblasts. Cell Growth Differ. 1998;9:139–146. [PubMed] [Google Scholar]
- 19.Kulaeva OI, Draghici S, Tang L, Kraniak JM, Land SJ, Tainsky MA. Epigenetic silencing of multiple interferon pathway genes after cellular immortalization. Oncogene. 2003;22:4118–4127. doi: 10.1038/sj.onc.1206594. [DOI] [PubMed] [Google Scholar]
- 20.Veal E, Eisenstein M, Tseng ZH, Gill G. A cellular repressor of E1A-stimulated genes that inhibits activation by E2F. Mol Cell Biol. 1998;18:5032–5041. doi: 10.1128/mcb.18.9.5032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sacher M, Di Bacco A, Lunin VV, Ye Z, Wagner J, Gill G, et al. The crystal structure of CREG, a secreted glycoprotein involved in cellular growth and differentiation. Proc Natl Acad Sci USA. 2005;102:18326–18331. doi: 10.1073/pnas.0505071102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Veal E, Groisman R, Eisenstein M, Gill G. The secreted glycoprotein CREG enhances differentiation of NTERA-2 human embryonal carcinoma cells. Oncogene. 2000;19:2120–2128. doi: 10.1038/sj.onc.1203529. [DOI] [PubMed] [Google Scholar]
- 23.Di Bacco A, Gill G. The secreted glycoprotein CREG inhibits cell growth dependent on the mannose-6-phosphate/insulin-like growth factor II receptor. Oncogene. 2003;22:5436–5445. doi: 10.1038/sj.onc.1206670. [DOI] [PubMed] [Google Scholar]
- 24.Han YL, Guo P, Sun MY, Guo L, Luan B, Kang J, et al. Secreted CREG inhibits cell proliferation mediated by mannose 6-phosphate/insulin-like growth factor II receptor in NIH3T3 fibroblasts. Genes Cells. 2008;13:977–986. doi: 10.1111/j.1365-2443.2008.01221.x. [DOI] [PubMed] [Google Scholar]
- 25.Han Y, Deng J, Guo L, Yan C, Liang M, Kang J, et al. CREG promotes a mature smooth muscle cell phenotype and reduces neointimal formation in balloon-injured rat carotid artery. Cardiovasc Res. 2008;78:597–604. doi: 10.1093/cvr/cvn036. [DOI] [PubMed] [Google Scholar]
- 26.Han Y, Guo L, Yan C, Guo P, Deng J, Mai X, et al. Adenovirus-mediated intra-arterial delivery of cellular repressor of E1A-stimulated genes inhibits neointima formation in rabbits after balloon injury. J Vasc Surg. 2008;48:201–209. doi: 10.1016/j.jvs.2008.01.061. [DOI] [PubMed] [Google Scholar]
- 27.Bian Z, Cai J, Shen DF, Chen L, Yan L, Tang Q, et al. Cellular repressor of E1A-stimulated genes attenuates cardiac hypertrophy and fibrosis. J Cell Mol Med. 2008;13:1302–1313. doi: 10.1111/j.1582-4934.2008.00633.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Han Y, Cui J, Tao J, Guo L, Guo P, Sun M, et al. CREG inhibits migration of human vascular smooth muscle cells by mediating IGF-II endocytosis. Exp Cell Res. 2009;315:3301–3311. doi: 10.1016/j.yexcr.2009.09.013. [DOI] [PubMed] [Google Scholar]
- 29.Baylin SB, Esteller M, Rountree MR, Bachman KE, Schuebel K, Herman JG. Aberrant patterns of DNA methylation, chromatin formation and gene expression in cancer. Hum Mol Genet. 2001;10:687–692. doi: 10.1093/hmg/10.7.687. [DOI] [PubMed] [Google Scholar]
- 30.Janderova-Rossmeislova L, Novakova Z, Vlasakova J, Philimonenko V, Hozak P, Hodny Z. PML protein association with specific nucleolar structures differs in normal, tumor and senescent human cells. J Struct Biol. 2007;159:56–70. doi: 10.1016/j.jsb.2007.02.008. [DOI] [PubMed] [Google Scholar]
- 31.Paulson TG, Almasan A, Brody LL, Wahl GM. Gene amplification in a p53-deficient cell line requires cell cycle progression under conditions that generate DNA breakage. Mol Cell Biol. 1998;18:3089–3100. doi: 10.1128/mcb.18.5.3089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hensler PJ, Annab LA, Barrett JC, Pereira-Smith OM. A gene involved in control of human cellular senescence on human chromosome 1q. Mol Cell Biol. 1994;14:2291–2297. doi: 10.1128/mcb.14.4.2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rogan EM, Bryan TM, Hukku B, Maclean K, Chang AC, Moy EL, et al. Alterations in p53 and p16INK4 expression and telomere length during spontaneous immortalization of Li-Fraumeni syndrome fibroblasts. Mol Cell Biol. 1995;15:4745–4753. doi: 10.1128/mcb.15.9.4745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Craig C, Kim M, Ohri E, Wersto R, Katayose D, Li Z, et al. Effects of adenovirus-mediated p16INK4a expression on cell cycle arrest are determined by endogenous p16 and Rb status in human cancer cells. Oncogene. 1998;16:265–272. doi: 10.1038/sj.onc.1201493. [DOI] [PubMed] [Google Scholar]
- 35.Mitra J, Dai CY, Somasundaram K, El-Deiry WS, Satyamoorthy K, Herlyn M, et al. Induction of p21(WAF1/CIP1) and inhibition of Cdk2 mediated by the tumor suppressor p16(INK4a) Mol Cell Biol. 1999;19:3916–3928. doi: 10.1128/mcb.19.5.3916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Helmbold H, Komm N, Deppert W, Bohn W. Rb2/p130 is the dominating pocket protein in the p53-p21 DNA damage response pathway leading to senescence. Oncogene. 2009;28:3456–3467. doi: 10.1038/onc.2009.222. [DOI] [PubMed] [Google Scholar]
- 37.Kapic A, Helmbold H, Reimer R, Klotzsche O, Deppert W, Bohn W. Cooperation between p53 and p130(Rb2) in induction of cellular senescence. Cell Death Differ. 2006;13:324–334. doi: 10.1038/sj.cdd.4401756. [DOI] [PubMed] [Google Scholar]
- 38.Jackson JG, Pereira-Smith OM. Primary and compensatory roles for RB family members at cell cycle gene promoters that are deacetylated and downregulated in doxorubicin-induced senescence of breast cancer cells. Mol Cell Biol. 2006;26:2501–2510. doi: 10.1128/MCB.26.7.2501-2510.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lacy S, Whyte P. Identification of a p130 domain mediating interactions with cyclin A/cdk 2 and cyclin E/cdk 2 complexes. Oncogene. 1997;14:2395–2406. doi: 10.1038/sj.onc.1201085. [DOI] [PubMed] [Google Scholar]
- 40.Kornfeld S. Structure and function of the mannose 6-phosphate/insulinlike growth factor II receptors. Annu Rev Biochem. 1992;61:307–330. doi: 10.1146/annurev.bi.61.070192.001515. [DOI] [PubMed] [Google Scholar]
- 41.Schmittgen TD, Zakrajsek BA, Mills AG, Gorn V, Singer MJ, Reed MW. Quantitative reverse transcription-polymerase chain reaction to study mRNA decay: comparison of endpoint and real-time methods. Anal Biochem. 2000;285:194–204. doi: 10.1006/abio.2000.4753. [DOI] [PubMed] [Google Scholar]
- 42.Li Q, Tang L, Roberts PC, Kraniak JM, Fridman AL, Kulaeva OI, et al. Interferon regulatory factors IRF5 and IRF7 inhibit growth and induce senescence in immortal Li-Fraumeni fibroblasts. Mol Cancer Res. 2008;6:770–784. doi: 10.1158/1541-7786.MCR-07-0114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ahmed-Choudhury J, Agathanggelou A, Fenton SL, Ricketts C, Clark GJ, Maher ER, Latif F. Transcriptional regulation of cyclin A2 by RASSF1A through the enhanced binding of p120E4F to the cyclin A2 promoter. Cancer Res. 2005;65:2690–2697. doi: 10.1158/0008-5472.CAN-04-3593. [DOI] [PubMed] [Google Scholar]
- 44.Doerksen LF, Bhattacharya A, Kannan P, Pratt D, Tainsky MA. Functional interaction between a RARE and an AP-2 binding site in the regulation of the human HOX A4 gene promoter. Nucleic Acids Res. 1996;24:2849–2856. doi: 10.1093/nar/24.14.2849. [DOI] [PMC free article] [PubMed] [Google Scholar]