Abstract
Zebrafish dead end (dnd) mRNA is specifically expressed in primordial germ cells (PGCs) and is required for PGC migration and survival. Previous studies have shown that zebrafish Dnd functions by protecting the 3′UTRs of nanos1 and TDRD7 from miR-430b-mediated RNA deadenylation. In this work, we demonstrate that zebrafish Dnd protein possesses Mg2+-dependent ATPase activity that is required for PGC formation. Michaelis-Menten analysis revealed that the ATPase has a kcat of 0.632 ± 0.036/min under optimal conditions, and mapping studies using Dnd truncates showed that ATPase resides in the last 91 aa of the Dnd C terminus. Internal deletion and point mutagenesis analysis of this region were used to identify key amino acids required for ATPase activity. Rescue experiments conducted by injecting mRNAs encoding the Dnd ATPase mutants into embryos in which the endogenous dnd expression was inhibited demonstrated that the ATPase activity is required for normal zebrafish PGC survival. Real-time PCR analysis showed that the expression of PGC markers nanos1 and TDRD7 but not vasa were down-regulated when dnd mutant proteins lacking ATPase were expressed in the rescued embryos, indicating that the Dnd ATPase is involved in protecting nanos1 and TDRD7 transcripts.—Liu, W., Collodi, P. Zebrafish dead end possesses ATPase activity that is required for primordial germ cell development.
Keywords: rescue, vasa, nanos1, TDRD7, miRNA
Primordial germ cells (PGCs) are the progenitors of the germ cell lineage able to develop into either spermatogonia in the male or oogonia in the female (1). In many species (i.e., Danio rerio, Drosophila melanogaster, Xenopus laevis, and Caenorhabditis elegans), PGC specification is determined by maternally derived germ plasm that is present in the oocyte and composed of fibrils, mitochondria, a nuclear envelope, and a mix of proteins and RNA molecules, including vasa, nanos1, and dnd(2,3,4,5). Vasa is an ATP-dependent RNA helicase that has been shown to be required for PGC formation in Drosophila(6, 7), Xenopus(8), C. elegans(9, 10), and Mus musculus(11). Inhibition of vasa expression in each of these species results in sterility. In zebrafish, although vasa is specifically expressed in the germ cell lineage, transient inhibition of its expression by an antisense morpholino (MO) did not block PGC formation presumably due to the stability of maternally deposited protein (12, 13). Nanos1 possesses a putative zinc-finger domain for RNA binding and plays a key role in PGC migration and survival during development, as well as in germ stem cell self renewal in the adult (14,15,16,17).
dnd mRNA was first identified in zebrafish embryos and shown to specifically localize in the germ cell lineage and be essential for normal PGC development (5). Inhibition of Dnd expression prevents zebrafish PGCs from migrating properly, resulting in the cells undergoing apoptosis. In 129 strain mice, the Ter mutation of Dnd1 leads to germ cell deficiency and causes testicular germ cell tumor formation (18, 19), while humans with testicular germ cell tumors rarely have mutations in the Dnd1 gene (20). In vitro and in vivo experiments have shown that mouse Dnd protein can interact with apolipoprotein B editing complex 3 (APOBEC3) (21), a multifunctional protein physically interacting with Argonaute1 and Argonaute2, leading to inhibition of miRNA-mediated mRNA repression (22, 23). Experiments with human germ cell lines have shown that Dnd1 protein can bind to p27-3′UTR and protect it from miR-221-mediated repression. Similarly, zebrafish Dnd can also bind to the 3′UTRs of nanos1 and TDRD7 to prevent miR-430b-mediated RNA repression (24, 25).
In zebrafish, Dnd protein localizes to the perinuclear germ granules along with the other components of the germ plasm (5). The specific subcellular localization of Dnd is attributed to its canonical RNA recognition motif (RRM) located in the N-terminal region (position 61–134) of the protein. Single amino acid substitutions (DndY72D, DndF89S, and DndN94K) in the RRM result in Dnd accumulating in the nucleus (26). In addition to the RRM, deletion experiments have shown that the C terminus of Dnd is also important for its function. A 126-aa deletion in the C-terminal region of Dnd resulted in an 85% reduction in the number of PGCs that were rescued in embryos treated with Dnd MO (26). In this study, we further examine the role of the Dnd C terminus and demonstrate that this region of the protein possesses ATPase activity that is necessary for its proper function. Deletion mapping reveals that the ATPase resides in the final 91 aa of the C terminus, and rescue experiments using embryos in which endogenous Dnd expression is blocked demonstrate that functional Dnd ATPase activity is required for normal zebrafish PGC development and survival.
MATERIALS AND METHODS
Fish stocks
vasa:EGFP transgenic zebrafish that exhibit germ cell-specific expression of GFP (27) and the AB wildtype fish were maintained according to the guidelines established by the Purdue University Animal Care and Use Committee.
Cloning, mutagenesis, and plasmid construction
vasa, dnd, and nanos1 cDNAs were synthesized and cloned from RNA isolated from early zebrafish embryos and verified by sequencing (primers listed in Table 1). Truncated dnd fragments were amplified (primers listed in Table 1), and dnd point mutations were made using the Stratagene Quickchange site-directed mutagenesis kit (Stratagene, La Jolla, CA, USA). For protein synthesis, after confirming each mutation by sequencing, the cDNAs were ligated into the EcoRI and XhoI sites of the pGEX-6p-1 plasmid (GE Healthcare, New York, NY, USA), and the correct reading frame was confirmed. For PGC rescue experiments, the dnd mutant cDNAs were cloned into the KpnI and XhoI sites of pBSK that contains sequence encoding the dnd 3′UTR and used to generate the rescue mRNAs.
Table 1.
Primers used in the study
| Primers for cDNA clone | |
|---|---|
| vasa | |
| vasa-f | 5′-TTT-GAA-TTC-AAT-ATG-GAT-GAC-TGG-GAG-GAA-3′ |
| vasa-r | 5′-TTT-CTC-GAG-TTA-TTC-CCA-TTC-CTC-ATC-GTC-3′ |
| Dnd | |
| dnd-f | 5′-TTT-GAA-TTC-CAG-ATG-GTC-GGA-GAC-ATG-GAT-3′ |
| dnd-r | 5′-TTT-CTC-GAG-TTA-GAA-AGG-CCG-TAA-ATT-TGA-GAC-T-3′ |
| nanos1 | |
| nanos1-f | 5′-TTT-GAA-TTC-AGC-ATG-GCT-TTT-TCT-CTT-CTC-3′ |
| nanos1-r | 5′-TTT-CTC-GAG-TCA-CCA-TGT-TGA-TTT-GGC-GTA-3′ |
| Primers for truncated dnd fragments | |
| truncate 1–141 | |
| dnd-f | 5′-TTT-GAA-TTC-CAG-ATG-GTC-GGA-GAC-ATG-GAT-3′ |
| dnd-r1 | 5′-TTT-CTC-GAG-CTG-GCG-CTT-CTC-GGT-GCT-CCT-3′ |
| truncate 1–258 | |
| dnd-f | 5′-TTT-GAA-TTC-CAG-ATG-GTC-GGA-GAC-ATG-GAT-3′ |
| dnd-r2 | 5′-TTT-CTC-GAG-TTA-CTG-GTG-AGC-TGG-GAC-GTC-ATA-3′ |
| truncate 63–258 | |
| dnd-f2 | 5′-TTT-GAA-TTC-ATC-AGT-CAG-ATC-CCG-AAC-GAC-3′ |
| dnd-r2 | 5′-TTT-CTC-GAG-TTA-CTG-GTG-AGC-TGG-GAC-GTC-ATA-3′ |
| truncate 63–411 | |
| dnd-f2 | 5′-TTT-GAA-TTC-ATC-AGT-CAG-ATC-CCG-AAC-GAC-3′ |
| dnd-r | 5′-TTT-CTC-GAG-TTA-GAA-AGG-CCG-TAA-ATT-TGA-GAC-T-3′ |
| truncate 135–411 | |
| dnd-f1 | 5′-TTT-GAA-TTC-AGG-AGC-ACC-GAG-AAG-CGC-CAG-3′ |
| dnd-r | 5′-TTT-CTC-GAG-TTA-GAA-AGG-CCG-TAA-ATT-TGA-GAC-T-3′ |
| truncate 230–411 | |
| dnd-f3 | 5′-TTT-GAA-TTC-ACT-CCC-CAG-GAA-GAC-AGC-TGC-GTA-3′ |
| dnd-r | 5′-TTT-CTC-GAG-TTA-GAA-AGG-CCG-TAA-ATT-TGA-GAC-T-3′ |
| truncate 230–351 | |
| dnd-f3 | 5′-TTT-GAA-TTC-ACT-CCC-CAG-GAA-GAC-AGC-TGC-GTA-3′ |
| dnd-r3 | 5′-TTT-CTC-GAG-TTA-CGC-GTG-ATG-GAA-GTG-GAC-TTC-3′ |
| truncate 322–411 | |
| dnd-f4 | 5′-TTT-GAA-TTC-ATT-AGC-CTC-GAC-GCC-GTG-GTG-TCT-CAT-3′ |
| dnd-r | 5′-TTT-CTC-GAG-TTA-GAA-AGG-CCG-TAA-ATT-TGA-GAC-T-3′ |
| Primers for modified dnd | |
| dnd-M2f | 5′-GCT-CAA-GCC-GCC-AGG-TCC-GAA-AGG-GAA-AGA-GG-3′ |
| dnd-M2r | 5′-CCT-CTT-TCC-CTT-TCG-GAC-CTG-GCG-GCT-TGA-GC-3′ |
| dnd-M3f | 5′-TTT-GAA-TTC-GGT-ACC-ACC-ATG-GAG-CTT-CAG-CAG-ATT-CTG-AAC-3′ |
| Primers for internal deletions | |
| dnd-Δ1 (Δ344–359) | |
| dnd-Δ1f | 5′-GGC-TCT-CCG-CAA-TAT-TTC-AAA-GTG-CTG-ATC-3′ |
| dnd-Δ1r | 5′-GAT-CAG-CAC-TTT-GAA-ATA-TTG-CGG-AGA-GCC-3′ |
| dnd-Δ2 (Δ360–375) | |
| dnd-Δ2f | 5′-ATT-CCT-CTA-CTT-CGC-CCA-GAT-CCT-GCC-AGG-CAC-3′ |
| dnd-Δ2r | 5′-GTG-CCT-GGC-AGG-ATC-TGG-GCG-AAG-TAG-AGG-AAT-3′ |
| dnd-Δ3 (Δ376–391) | |
| dnd-Δ3f | 5′-CTG-TAT-GGG-TTC-GTC-CGG-GCC-GCC-GCT-G-3′ |
| dnd-Δ3r | 5′-CAG-CGG-CGG-CCC-GGA-CGA-ACC-CAT-ACA-G-3′ |
| dnd-Δ3 (Δ392–406) | |
| dnd-Δ4f | 5′-GAG-TGA-AGT-TTA-CTT-ACG-GCC-TTT-CTA-AC-3′ |
| dnd-Δ4r | 5′-GTT-AGA-AAG-GCC-GTA-AGT-AAA-CTT-CAC-TC-3′ |
| dnd Δ4-1 (Δ392–396) | |
| dnd Δ4-1-f | 5′-aag-agt-gaa-gtt-tac-cag-gtg-atc-caa-acc-3′ |
| dnd Δ4-1-r | 5′-ggt-ttg-gat-cac-ctg-gta-aac-ttc-act-ctt-3′ |
| dnd Δ4-2 (Δ397–400) | |
| dnd Δ4-2-f | 5′-cgg-gcc-gcc-gct-gag-acc-ttg-tgc-cga-gtc-3′ |
| dnd Δ4-2-r | 5′-gac-tcg-gca-caa-ggt-ctc-agc-ggc-ggc-ccg-3′ |
| dnd Δ4-1 (Δ401–404) | |
| dnd Δ4-3-f | 5′-gag-cag-gtg-atc-caa-gtc-tca-aat-tta-cgg-3′ |
| dnd Δ4-3-r | 5′-ccg-taa-att-tga-gac-ttg-gat-cac-ctg-ctc-3′ |
| Primers for cDNA clone | |
| dnd Δ4-4 (Δ405–407) | |
| dnd Δ4-4-f | 5′-caa-acc-ttg-tgc-cga-tta-cgg-cct-ttc-taa-3′ |
| dnd Δ4-4-r | 5′-tta-gaa-agg-ccg-taa-tcg-gca-caa-ggt-ttg-3′ |
| Primers for single amino acid mutation | |
| dndR392E-f | 5′-gaa-gag-tga-agt-tta-cga-ggc-cgc-cgc-tga-gc-3′ |
| dndR392E-r | 5′-gct-cag-cgg-cgg-cct-cgt-aaa-ctt-cac-tct-tc-3′ |
| dndR392K-f | 5′-gaa-gag-tga-agt-tta-caa-ggc-cgc-cgc-tga-gc-3′ |
| dndR392K-r | 5′-gct-cag-cgg-cgg-cct-tgt-aaa-ctt-cac-tct-tc-3′ |
| dndA393T-f | 5′-gtg-aag-ttt-acc-gga-ccg-ccg-ctg-agc-3′ |
| dndA393T-r | 5′-gct-cag-cgg-cgg-tcc-ggt-aaa-ctt-cac-3′ |
| dndA394S-f | 5′-gtt-tac-cgg-gcc-tcc-gct-gag-cag-gtg-3′ |
| dndA394S-r | 5′-cac-ctg-ctc-agc-gga-ggc-ccg-gta-aac-3′ |
| dndA395S-f | 5′-gtt-tac-cgg-gcc-gcc-tct-gag-cag-gtg-atc-3′ |
| dndA395S-r | 5′-gat-cac-ctg-ctc-aga-ggc-ggc-ccg-gta-aac-3′ |
| dndE396D-f | 5′-cgg-gcc-gcc-gct-gac-cag-gtg-atc-caa-acc-3′ |
| dndE396D-r | 5′-ggt-ttg-gat-cac-ctg-gtc-agc-ggc-ggc-ccg-3′ |
| dndE396R-f | 5′-cgg-gcc-gcc-gct-cgg-cag-gtg-atc-caa-acc-3′ |
| dndE396R-r | 5′-ggt-ttg-gat-cac-ctg-ccg-agc-ggc-ggc-ccg-3′ |
| dndY104C-f | 5′-cgc-ggc-ttc-gcc-tgt-gct-aag-tac-ggt-gac-3′ |
| dndY104C-r | 5′-gtc-acc-gta-ctt-agc-aca-ggc-gaa-gcc-gcg-3′ |
| Primers for real time PCR | |
| odc-f | 5′-AGA-TGC-CTT-CTA-TGT-GGC-GGA-TCT-3′ |
| odc-r | 5′-TGC-TAT-CGT-TGC-ACT-TCA-CTG-CGT-3′ |
| vasa-f1 | 5′-CCC-AAT-ATG-GAT-GAC-TGG-GAG-3′ |
| vasa-r1 | 5′-GTC-ATT-TTC-CAT-GAG-CTA-CC-3′ |
| nanos1-f1 | 5′-GAG-AGC-AGC-ATG-GCT-TTT-TC-3′ |
| nanos1-r1 | 5′-TTC-CAA-GGC-TGA-AAG-TCC-TG-3′ |
| TDRD7-f2 | 5′-GAA-GTT-GCG-GCA-GCT-GCT-CAA-TAA-3′ |
| TDRD7-r2 | 5′-GAA-ATG-CAC-AGC-CAA-ATG-CCT-CCT-3′ |
Protein expression and purification
Escherichia coli BL21 (DE3) transformed with pGEX carrying each dnd mutant was grown (37°C) to A600 of 0.6, induced with isopropyl-β-d-thiogalactoside (0.6 mM, 6 h at 30°C) and collected by centrifugation. After sonication, the GST fusion proteins were purified on Glutathione-Sepharose 4B resin (Amersham Biosciences, Piscataway, NJ, USA), dialyzed (pH 7.5, 10 mM imidazole-Cl, 75 mM KCl, 0.2 mM EDTA, 3 mM Mg2+, 5% glycerol, 1 mM DTT, and protease inhibitor cocktail), and the concentration was measured using the Bradford assay (28). In some experiments, following affinity chromatography, the GST fusion proteins were further purified by gel filtration chromatography using Bio-Rad P-100 medium (Bio-Rad, Hercules, CA, USA) with a Pharmacia FRAC100 fraction collector and LKB Uvicord SII detector (Amersham Pharmacia, Piscataway, NJ, USA). Blue dextran was used to measure the void volume. ATPase activity in the fractions was measured as described below using 34 μl of each fraction, 5 mM Mg2+ and a 3 h incubation. Proteins contained in each fraction were visualized by 10% SDS-polyacrylamide-gel electrophoresis (PAGE) with silver staining.
Phosphate release assay
The phosphate release assay was performed using the molybdate-malachite green reaction (29,30,31,32). Recombinant Dnd proteins (final 0.25 μM) and ATP or NTP/dNTP (final 100 μM) were added to imidazole-Cl buffer (10 mM imidazole-Cl, 75 mM KCl, 0.2 mM EDTA, 3 mM MgCl2, pH 7.5) in a 96-well plate and incubated for 90 min or as indicated. HCl/Mo (35 μl; 4 vol of 2M HCl with 3 vol of 0.1M Na2MoO4) was added and followed 2 min later by 0.042% malachite green (25 μl) and then H2SO4 (50 μl 15%). The color reaction was complete in 30 min. Absorbance was measured (650 nm) and compared to a standard curve generated using KH2PO4. The data were corrected by subtracting absorbance from free orthophosphate determined from parallel reactions conducted by adding recombinant proteins to the reaction mix just before HCl/Mo. pH dependence was assessed using imidazole-Cl buffer at pH 4.5 to 9.5. Bivalent cation dependence was determined using solutions of MgCl2, CaCl2, FeCl2, CuSO4, MnCl2 and CoCl2 at 3 mM. To remove RNA contamination from the purified GST-Dnd, 50 μl of GST-Dnd (500 ng/μl) was treated with 2 μl of RNaseA (10 mg/ml) at 37°C for 30 min. For the determination of KM and kcat, ATP concentrations ranging from 6.25 to 200 μM were used and Michaelis-Menten calculations were performed with KaleidaGraph software (Synergy Software, Reading, PA, USA).
MO and mRNA microinjection
Capped mRNAs were synthesized in vitro using the T7 Message Machine kit (Ambion, Austin, TX, USA). Dnd antisense MO (5) and control MO (5′-CCTCTTACCTCAGTTACAATTTATA-3′) were obtained from Gene Tools (Philomath, OR, USA). Capped mRNA (100 ng/μl mRNA) and MO (1 μg/μl) were coinjected into 1-cell-stage vasa:EGFP embryos, and the number of PGCs was evaluated by fluorescence microscopy (Nikon Y-FL Epi-fluorescence microscope; Nikon, Tokyo, Japan) at 30 hpf. The rescue efficiency was calculated by dividing the number of embryos harboring PGCs at 30 hpf by the total number of surviving embryos.
Quantitative real-time PCR (qRT-PCR) analysis
Total RNA was isolated (TRIzol reagent, Invitrogen) from zebrafish embryos at bud stage (10 hpf) and treated with rDNaseI. Following first-strand cDNA synthesis (M-MLV reverse transcriptase; Promega, Madison, WI, USA) qRT-PCR was performed using SYBR Green Supermix (Bio-Rad) with the primers listed in Table 1. Odc1 was used as the internal control. Relative expression of germ cell markers was calculated using the 2−ΔΔCt method with a correction for different amplification efficiencies (33).
Statistical analysis
qRT-PCR data were analyzed by 1-way ANOVA followed by the Tukey’s test using SAS software (SAS Institute, Cary, NC, USA). Other results were analyzed by Student’s t test, and data are presented as means ± sd with significance at P < 0.05.
RESULTS
Zebrafish Dnd protein possesses ATPase activity
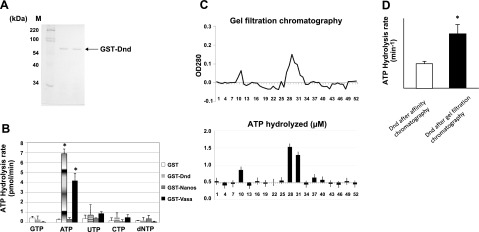
Results of molybdate-malachite green phosphate release assays demonstrated that zebrafish Dnd protein, synthesized in E. coli as a GST fusion protein and purified on glutathione resin (Fig. 1A), was able to hydrolyze ATP (Fig. 1B, second column). The ATPase activity comigrated with the Dnd protein during gel filtration chromatography following the initial purification on the glutathione resin (Fig. 1C). The specific activity of the ATPase increased 2-fold following the gel filtration step (Fig. 1D). A small amount of ATPase activity was detected in the gel filtration void volume, which was most likely due to dimerized GST-Dnd. Hydrolysis activity was specifically observed when ATP was the substrate, although a small amount of phosphate release was also detected with UTP (Fig. 1B). NTPase and dNTPase activity was not detected with GST alone or with GST-Nanos1 fusion protein; however, GST-Vasa did possess specific ATPase activity (Fig. 1B). The latter result is consistent with previous studies showing that Vasa has ATPase activity that can be stimulated by the presence of RNA (7). We were not able to detect a similar RNA stimulation of Dnd ATPase when the assay was conducted in the presence of yeast tRNA or total RNA, zebrafish embryo total RNA, or RNA encoding the zebrafish nanos1 3′UTR (data not shown). Treatment with RNaseA did not affect the Dnd ATPase activity (data not shown), indicating that the lack of stimulation by added RNA was not due to the presence of contaminating RNA that copurified with Dnd.
Figure 1.
ATPase activity of recombinant zebrafish Dnd. A) SDS-PAGE gel analysis of zebrafish Dnd expressed in E. coli and purified to homogeneity by GST affinity chromatography. B) Phosphate release assay conducted with GST, GST-Dnd, GST-Vasa, and GST-Nanos1. A final concentration of 0.25 μM of each protein was added to the reaction mix with GTP, ATP, CTP, UTP, or dNTP (100 μM final concentration). Data are presented as means ± sd. C) Gel filtration chromatography of GST-Dnd that was initially purified by affinity chromatography. ATPase analysis was performed on the fractions eluted from the column as described in Materials and Methods. D) ATP hydrolysis rate of affinity chromatography-purified Dnd before and after additional purification by gel filtration chromatography. *P < 0.05.
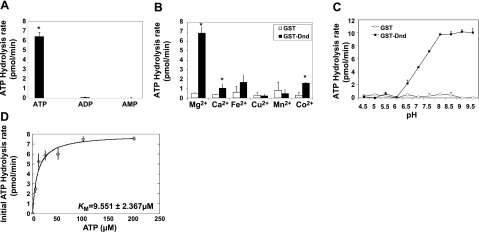
Although some ATPases, such as those of the apyrase and ecto-ATPase families (34, 35), can also hydrolyze ADP, we did not observe phosphate release when Dnd was assayed in the presence of either ADP or AMP substrates (Fig. 2A). The results indicate that Dnd can only cleave the β-γ phosphodiester bond of ATP. Competition experiments in which ATP was mixed with ADP or AMP in different ratios revealed that ADP but not AMP had some inhibitory effect on ATP hydrolysis (Supplemental Fig. 1A). Assays to determine bivalent cation dependence revealed that Dnd ATPase is dependent on Mg2+ concentration (Fig. 2B, Supplemental Fig. 1B). As a result, all of the subsequent ATPase experiments were performed in the presence of 3 mM MgCl2. Dnd ATPase activity was also pH dependent with no ATP hydrolysis detected at pH values < 6, while the activity increased sharply from pH 6 to 8. pH values > 8 did not increase ATP hydrolysis significantly (Fig. 2C).
Figure 2.
Biochemical characteristics of zebrafish Dnd ATPase. ATPase assays were performed with recombinant Dnd to determine: substrate specificity (A), optimal divalent metal ion (B), pH (C), and kinetic analysis of Dnd’s ATPase activity (D). ATPase assays were conducted as described in Materials and Methods; 3 mM of each metal ion was used in B. *P < 0.05.
To measure the kinetic parameters of Dnd ATPase, a time course of ATP hydrolysis was performed revealing that the reaction remained linear, at least during the first 45 min. The hydrolysis rate at 45 min was used as the initial rate of ATP hydrolysis for Michaelis-Menten analysis under the reaction condition described above. The results revealed a Vmax of 7.8 ± 0.45 pmol/min and Km of 9.551 ± 2.367 μM from the initial rate curve (Fig. 2D). On the basis of these results, Dnd ATPase has a kcat of 0.632 ± 0.036/min that did not increase in the presence of yeast tRNA or total RNA, zebrafish embryo total RNA, or nanos1 3′UTR, indicating that the ATPase activity is independent of RNA stimulation (data not shown).
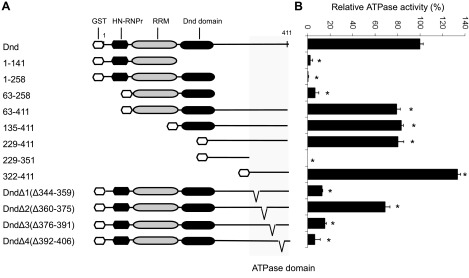
ATPase domain is located in the C terminus of Dnd
To determine the location of the domain responsible for ATPase activity, a series of truncated Dnd mutants were made, and the proteins were assayed for ATPase activity (Fig. 3A). The results revealed that truncates containing residues 1–141, 1–258, 63–258, and 229–351 completely lost ATPase activity, while truncates possessing residues 63–411, 135–411, 229–411, and 322–411 maintained 79, 83, 80, and 133% of the activity, respectively, compared to the intact Dnd protein (Fig. 3). These data indicate that the region responsible for ATPase activity resides within the last 91 aa of the C terminus (residues 322–411). Four deletions were made within this C-terminal 91-residue region, DndΔ1 (Δ344–359), DndΔ2 (Δ360–375), DndΔ3 (Δ376–391), and DndΔ4 (Δ392–406), and the ATPase activity of each mutant protein was measured (Fig. 3). The results revealed that DndΔ2 maintained 69% of the activity found in intact Dnd, while DndΔ1 and DndΔ3 each exhibited a dramatically lower level of activity (13 and 15%, respectively). Activity was totally abolished in DndΔ4 (7%), suggesting that residues 392–406 are essential for ATPase activity.
Figure 3.
Localization of the ATPase domain on zebrafish Dnd. A) Schematic showing the wild-type zebrafish Dnd protein along with truncates and deletion mutants used in this study. B) Relative ATPase activity of each Dnd mutant compared to wild-type Dnd. Data are presented as means ± sd. *P < 0.05.
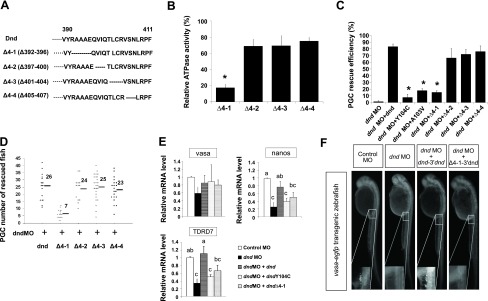
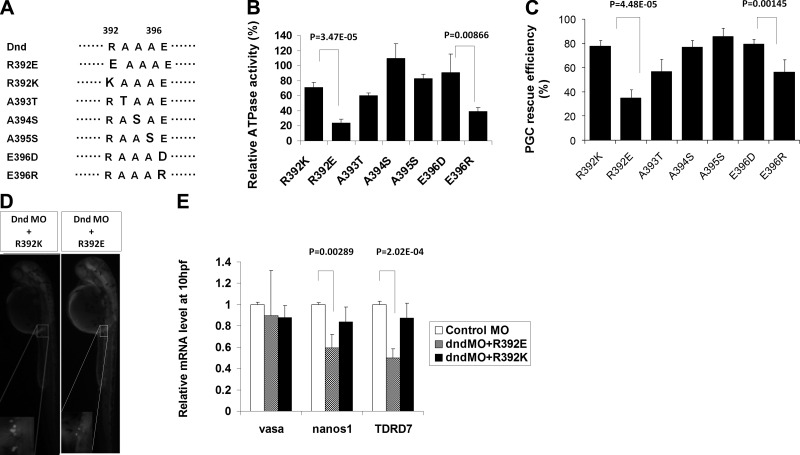
Since the reduction in ATPase activity was significantly greater (P<0.05) in DndΔ4 compared to DndΔ1 and DndΔ3, we examined this deletion in more detail. Four additional deletions were made within the 392–406 residues, DndΔ4-1 (Δ392–396), DndΔ4-2 (Δ397–400), DndΔ4-3 (Δ401–404), and DndΔ4-4 (Δ405–407) (Fig. 4A). ATPase assays performed on each of the mutants revealed that deletion of residues 392–396 (DndΔ4-1) abolished the activity, while the other deletions maintained most of the activity (69–75%) compared to intact Dnd (Fig. 4B). Single point mutations based on charge and size substitutions were made at each of the 5 residues 392–396 (Fig. 5A). Substitution of alanine-393 to threonine (dndA393T), or alanine-395 to serine (dndA395S) resulted in a loss of 40 and 17% of activity, respectively. Substituting alanine-394 to serine (dndA394S) actually increased ATPase activity by 9% compared to the intact Dnd protein. Interestingly, when arginine-392 was changed to glutamic acid (dndR392E), the ATPase activity decreased by ∼76%; however, when it was changed to lysine (dndR392K), the decline in activity was significantly less (∼29%) (Fig. 5B). Substitution of glutamic acid-396 with arginine (dndE396R) reduced the ATPase activity by 61%, while substituting with aspartic acid at this position (dndE396D) resulted in only a 9% loss of activity. Comparison of the wild-type and mutant forms of Dnd by nondenaturing PAGE and gel filtration chromatography revealed that the proteins possessed the same apparent molecular weight, indicating that the reduced ATPase activity associated with specific deletions and substitutions was not due to a conformational change of the protein (Supplemental Fig. 1C, D).
Figure 4.
Deletion analysis of zebrafish Dnd ATPase activity. A) Diagram showing the dndΔ4 deletions. B) Relative ATPase activity of each dnd Δ4 mutant compared to wild-type zebrafish Dnd. C) PGC rescue efficiency in dnd MO-treated embryos injected with mRNA encoding wild-type dnd that lacks the MO binding site, 2 of the dnd substitution mutants, dndY104C and dndA103V, and the dndΔ4 deletion mutants. D) Number of PGCs present in the rescued embryos at 30 hpf. E) qRT-PCR analysis showing the relative expression of vasa, nanos1, and TDRD7 in dnd MO-treated embryos injected with mRNA encoding wild-type dnd that lacked the MO binding site or mRNA encoding the dndY104C or dndΔ4-1 mutants. Data were analyzed with 1-way ANOVA followed by the Tukey test by SAS software. Vasa expression was not significantly different between the treated embryos. Bars that are labeled with the same letters are not significantly different (P>0.05). F) Representative 30 hpf embryos from each treatment. Inset: magnified view of GFP-positive PGCs. *P < 0.05.
Figure 5.
Amino acid substitution analysis to identify the residues required for Dnd ATPase activity. A) Diagram and sequence of amino acid substitutions within the Δ4-1 region of Dnd. B) Relative ATPase activity of R392K, R392E, A393T, A394S, A395S, E396D, and E396R compared to wild-type zebrafish Dnd. C) PGC rescue efficiency of each dnd substitution in dnd MO-treated embryos. D) Representative 30-hpf embryos rescued with each Dnd substitution. E) qRT-PCR analysis shows the the relative expression of vasa, nanos1, and TDRD7 in dnd MO-treated embryos rescued with mRNA encoding wild-type dnd or the R392E and R392K substitutions. P value was calculated by Student’s t test.
ATPase activity of Dnd is required for zebrafish PGC survival
Treatment of vasa:EGFP zebrafish embryos with a dnd MO that inhibits dnd expression resulted in a complete loss of PGC formation in 98 ± 2% of the treated embryos (ref. 5, Fig. 4C). PGC formation was rescued in 83 ± 4% of the MO-treated embryos that were coinjected with dnd mRNA that lacked the MO recognition sequence and contained the dnd 3′UTR to direct expression to the PGC lineage (Fig. 4C). Each rescued embryo contained an average of 26 PGCs at 30 hpf, which was approximately the same number found in nontreated vasa:EGFP embryos (Fig. 4D). Point mutations made in the RRM of the injected dnd mRNA greatly reduced the rescue activity (24). Substitutions dndY104C and dndA103V resulted in only 8 ± 5% and 18 ± 3% rescued embryos, respectively (Fig. 4C).
To test whether the ATPase activity of Dnd is also necessary for PGC rescue activity, the dnd MO-treated embryos were injected with dnd mRNA-containing deletions or substitutions in the ATPase domain. The 5-aa acid deletion dndΔ4-1 (Δ392–396) that abolished in vitro ATPase activity also reduced the in vivo rescue frequency to 15 ± 3% (Fig. 4C). The embryos rescued by dndΔ4-1 had an average of only 7 PGCs at 30 hpf compared to 26 PGCs in embryos rescued by intact dnd (Fig. 4D). The results indicate that the ATPase activity is essential for normal Dnd function during PGC development. Deletions dndΔ4-2, dndΔ4-3, and dndΔ4-4, that maintained most of the in vitro ATPase activities (Fig. 4B) were also able to rescue the majority of MO treated embryos (Fig. 4C). DndΔ4-2, dndΔ4-3, and dndΔ4-4 rescued 67 ± 14, 72 ± 7, and 76 ± 8% of the embryos with an average of 24, 25, and 23 PGCs/rescued embryo, respectively (Fig. 4C, D). Similarly, substitution dndR392E that resulted in a 76% loss of ATPase activity (Fig. 5B) also exhibited a significantly reduced rescue frequency (35±7%, P<0.001; Fig. 5C) and PGC number (15; P<0.005) compared to the control. Substitution dndR392K maintained 71% of the ATPase activity and rescued 78 ± 5% of the embryos with an average of 23 PGCs/embryo, which was not significantly different from the control (Fig. 5C). Other substitutions that did not significantly affect in vitro Dnd ATPase activity, including DndA394S and DndA395S, also did not affect the rescue efficiency or the total number of PGCs per rescued embryo. DndA394S had a rescue frequency of 77 ± 6% with an average of 25 PGCs, and DndA395S had a rescue frequency of 86 ± 7% with 23 PGCs/rescued embryos. Substitution DndA393T that exhibited 60 ± 4% of intact Dnd ATPase activity (Fig. 5B) had a reduced PGC rescue efficiency (57±10%, P<0.05; Fig. 5C), although the average number of PGCs per rescued embryo was 21, which was not significantly different than the control. Substitution dndE396D that exhibited 91% of the control ATPase activity was able to rescue 79 ± 4% of the embryos with 24 PGCs/rescued embryo. Interestingly, substitution dndE396R that maintained only 39% of ATPase activity was still able to rescue 56 ± 11% (P<0.005) of the embryos with 23 PGCs/embryo. Ectopic PGCs were not identified in any of the rescue experiments, indicating that the Dnd deletions, and substitutions used in this study did not affect normal migration of the PGCs (Fig. 4F, 5D).
To confirm the PGC rescue results, we used qRT-PCR to measure the expression of PGC-specific markers nanos1, vasa, and TDRD7 in embryos treated with the dnd-MO and each of the dnd mRNAs. Injection of the dnd-MO alone resulted in a significant decline in nanos1 (74.3±11.5%) and TDRD7 (66.1±12.3%) mRNA levels, while vasa expression was not significantly reduced from the control level (Fig. 4E). Coinjection of intact dnd mRNA together with the MO restored the expression of nanos1 and TDRD7 to a level not statistically different from the control. Coinjection of the MO and dndY104C mRNA encoding a substitution in the RRM that destroyed the PGC rescue activity resulted in embryos with reduced expression of nanos1 and TDRD7 (nanos1, 60.2±8.8%; TDRD7, 49.7±6.0%) (Fig. 4E). Similarly, embryos had reduced levels of nanos1 and TDRD7 expression (48.9±13.2 and 38.9±14.1%, respectively; Fig. 4E) when coinjected with MO and mRNA encoding the dndΔ4-1 deletion mutant that lacked ATPase (Fig. 4B) and rescue activities. The same result was obtained with embryos that were coinjected with MO and mRNA encoding the dndR392E point mutation that possessed reduced ATPase activity and did not rescue the PGCs. In contrast, coinjection of MO with mRNA encoding the dndR392K point mutation that did not affect ATPase or rescue activities restored nanos1 and TDRD7 expression to control levels (Fig. 5E).
DISCUSSION
In this study, we demonstrate that zebrafish Dnd possesses ATPase activity that resides in the C terminus of the protein. Comparison of Dnd orthologs from several fish species reveals that the C terminus is highly conserved (Supplemental Fig.1E) (36). Within this region, the amino acids spanning the DndΔ4-1 deletion that resulted in 82% loss of ATPase activity, share 80% homology between fish species with 4 of the 5 aa either identical or similar (Supplemental Fig. 1E). Although the C terminus of Dnd is conserved among fish, the core of the zebrafish Dnd ATPase, located in the final 91 residues of the C terminus, does not share homology with other ATPases, making it difficult to assign Dnd to a specific ATPase family. ATPases associated with kinases, helicases, and transportases are classified into different families, such as P-loop and GHL, based on conserved sequences within but not necessarily between each family (37, 38). Many ATPases, however, have been identified that share no conserved sequences such as phosphofructokinase, ATP/ADP translocase, transcarboxylase biotin subunit, and phospholipase A (39, 40). Therefore, it is not unusual that zebrafish Dnd does not share sequence with other known ATPase families.
On the basis of the deduced amino acid sequence of zebrafish Dnd, the protein has one conserved RRM (residue 62–125). Although occasionally found in single-strand DNA binding proteins, the RRM is usually identified in RNA binding proteins that have various functions, including heterogeneous nuclear ribonucleoproteins (hnRNPs), involved in alternative splicing (SR, U2AF, Sxl), small nuclear ribonucleoproteins (U1 and U2 snRNPs), and proteins that regulate RNA stability and translation (PABP, LA, Hu) (41, 42).
The presence of both an RRM and ATPase domain raises the possibility that zebrafish Dnd has RNA helicase activity. RNA helicases include a large family of enzymes that utilize the energy of NTP hydrolysis to remodel RNA or RNA-protein complexes, resulting in RNA duplex strand separation and/or protein displacement from the RNA molecules (43). A helicase can also serve as an RNA chaperone that functions to unwind misfolded RNAs to generate the correct active tertiary structure (44, 45). The germ plasm component Vasa is known to be an ATP-dependent RNA helicase that can unwind a poly U oligonucleotide in vitro(7). Interestingly, the catalytic rate for Dnd (0.632±0.036/min) is similar to other helicases such as eIF4A (kcat=0.3/min), UAP56 (kcat=0.25/min) (46, 47) and zebrafish vasa from our study. The possibility that Dnd also possesses helicase activity deserves further study.
Characterization of the catalytic properties of zebrafish Dnd ATPase has revealed that, unlike the ATPases associated with DEXD/H-box proteins such as Vasa, eIF4A, Mtr4p, and UAP56 (7, 46, 47), the Dnd ATPase is not stimulated by the presence of RNA in the reaction mix. Activity of the eIF4A ATPase can increase 10- to 14-fold in the presence of polyU, poly(A), or polyC (46). UAP56 ATPase can increase as much as 20-fold in the presence of short arbitrary polyU sequences or double-strand RNA (47). Dnd may associate in vivo with one or more partner proteins that can stimulate the ATPase activity in a manner similar to the ATPase associated with some RNA helicases. The eIF4A ATPase activity increases 3-fold in the presence of the eIF4B and eIF4H partner proteins (48), the weak ATPase activity of Dbp5 is stimulated by the Gle1 protein and the ATPase of Dbp8 is stimulated by Esf2 (49, 50). Further research is needed to determine whether partner proteins exist that might enhance zebrafish Dnd ATPase activity.
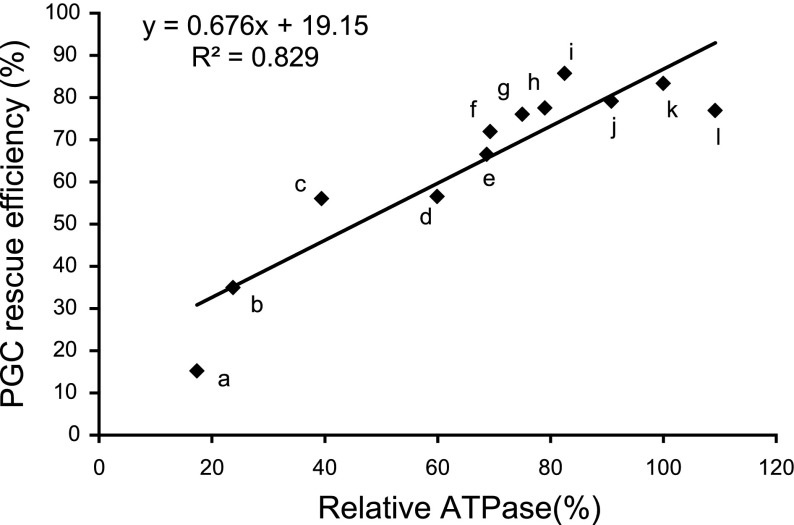
The results of our study show that Dnd ATPase activity is important for PGC development and survival. Introduction of mRNA encoding intact Dnd completely rescued PGC formation in MO-treated embryos. The capacity of the mutated Dnds to rescue the PGCs correlated with the level of ATPase activity exhibited by the mutant protein (r2=0.83, Fig. 6). The Dnd deletions and substitutions that retained most of the ATPase activity (DndΔ4-2, DndΔ4-3, DndΔ4-4, DndR392K, DndA394S, DndA395S, and DndE396D) rescued the majority of the PGCs in the MO-treated embryos while the mutants, DndΔ4-1and DndR392E, that completely lacked ATPase activity did not rescue. Mutations that resulted in a reduced level of ATPase activity such as DndE396R and DndA393T were able to partially rescue the PGCs.
Figure 6.
Linear regression analysis for Dnd ATPase and PGC rescue efficiency. Relative ATPase activities of dnd mutants used in the study and their PGC rescue efficiencies were fit into a linear regression equation. a, dnd Δ4-1; b, dndR392E; c, dndE396R; d, dndA393T; e, dnd Δ4-2; f, dnd Δ4-3; g, dnd Δ4-4; h, dndR392K; i, dndA395S; j, dndE396D; k, dnd; l, dndA394S. Correlation coefficient (R2) is 0.83.
A proposed function of zebrafish Dnd is to protect transcripts such as nanos1 and TDRD7 from microRNA-mediated degradation in the PGCs (24, 51). We are currently working to determine what role the Dnd ATPase might have in this function.
Supplementary Material
Acknowledgments
The authors thank Dr. Lisbeth Olsen (University of Bergen, Bergen, Norway) for the vasa:EGFP transgenic zebrafish and Ten-tsao Wong (Purdue University) for the pBSK-dnd3′UTR. This work was supported by a grant from the National Institutes of Health (R01GM069384).
References
- Wylie C. Germ cells. Cell. 1999;96:165–174. doi: 10.1016/s0092-8674(00)80557-7. [DOI] [PubMed] [Google Scholar]
- Strome S., Lehmann R. Germ versus soma decisions: lessons from flies and worms. Science. 2007;316:392–393. doi: 10.1126/science.1140846. [DOI] [PubMed] [Google Scholar]
- Knaut H., Pelegri F., Bohmann Kerstin, Schwarz H., Nusslei-Volhard C. Zebrafish vasa RNA but not its protein is a component of the germ plasm and segregates asymmetrically before germline specification. J Cell Biol. 2000;149:875–888. doi: 10.1083/jcb.149.4.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Extavour C., Akam M. Mechanism of germ cell specification across the metazoans: epigenesist and preformation. Development. 2003;130:5869–5884. doi: 10.1242/dev.00804. [DOI] [PubMed] [Google Scholar]
- Weidinger G., Stebler J., Slanchev K., Dumstrei K., Wise C., Lovell-Badge R., Thisse C., Thisse B, Raz E. Dead end, a novel vertebrate germ plasm component, is required for zebrafish primordial germ cell migration and survival. Curr Biol. 2003;13:1429–1434. doi: 10.1016/s0960-9822(03)00537-2. [DOI] [PubMed] [Google Scholar]
- Lasko P. F., Ashburner M. The product of the Drosophila gene vasa is very similar to eukaryotic initiation factor-4A. Nature. 1988;335:611–617. doi: 10.1038/335611a0. [DOI] [PubMed] [Google Scholar]
- Sengoku T., Nureki O., Nakamura A., Kobayashi S., Yokoyama S. Structural basis for RNA unwinding by the DEAD-box protein Drosophila vasa. Cell. 2006;125:287–300. doi: 10.1016/j.cell.2006.01.054. [DOI] [PubMed] [Google Scholar]
- Ikenishi K., Tanaka T. S. Involvement of the protein of Xenopusvasa homologue (Xenopus vasa-like gene 1, XVLG1) in the differentiation of primordial germ cells. Dev Growth Differ. 1997;39:625–633. doi: 10.1046/j.1440-169x.1997.t01-4-00010.x. [DOI] [PubMed] [Google Scholar]
- Gruidl M. E., Smith P. A., Kuznicki K. A., McCrone J. S., Kirchner J., Roussell D. L., Bennett K. Multiple potential germ-line helicases are components of the germ-line specific P granules of Caenorhabditis elegans. Proc Natl Acad Sci U S A. 1996;93:13837–13842. doi: 10.1073/pnas.93.24.13837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuznicki K. A., Smith P. A., Leung-Chiu W. M. A., Estevez A. O., Scott H. C., Bennett K. L. Combinatorial RNA interference indicates GLH-4 can compensate for GLH-1; these two P granule components are critical for fertility in C-elegans. Development. 2000;127:2907–2916. doi: 10.1242/dev.127.13.2907. [DOI] [PubMed] [Google Scholar]
- Tanaka S. S., Toyooka Y., Akasu R., Katoh-Fukui Y., Nakahara Y., Suzuki R., Yokoyama M., Noce T. The mouse homolog of Drosophila Vasa is required for the development of male germ cells. Genes Dev. 2000;14:841–853. [PMC free article] [PubMed] [Google Scholar]
- Yoon C., Kawakami K., Hopkins N. Zebrafish vasa homologue RNA is localized to the cleavage planes of 2- and 4-cell stage embryos and is expressed in the primordial germ cells. Development. 1997;124:3157–3166. doi: 10.1242/dev.124.16.3157. [DOI] [PubMed] [Google Scholar]
- Braat A. K., Water S., Korving J., Zivkovic D. A zebrafish vasa morphant abolishes vasa protein but does not affect the establishment of the germline. Genesis. 2001;30:183–185. doi: 10.1002/gene.1060. [DOI] [PubMed] [Google Scholar]
- Wang Z., Lin H. Nanos maintains germline stem cell self-renewal by preventing differentiation. Science. 2004;303:2016–2019. doi: 10.1126/science.1093983. [DOI] [PubMed] [Google Scholar]
- Koprunner M., Thisse C., Thisse B., Raz E. A zebrafish nanos-related gene is essential for the development of primordial germ cells. Genes Dev. 2001;15:2877–2885. doi: 10.1101/gad.212401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draper B. W., McCallum C. M., Moens C. B. nanos1 is required to maintain oocyte production in adult zebrafish. Dev Biol. 2007;305:589–598. doi: 10.1016/j.ydbio.2007.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda M., Sasaoka Y., Kiso M., Abe K., Haraguchi S., Kobayashi S., Saga Y. Conserved role of nanos proteins in germ cell development. Science. 2003;301:1239–1241. doi: 10.1126/science.1085222. [DOI] [PubMed] [Google Scholar]
- Youngren K., Youngrenm K. K., Coveney D., Peng X., Bhattacharya C., Schmidt L. S., Nickerson M. L., Lamb B. T., Deng J. M., Behringer R. R., Capel B., Rubin E. M., Nadeau J. H., Matin A. The ter mutation in the dead end gene causes germ cell loss and testicular germ cell tumors. Nature. 2005;435:360–364. doi: 10.1038/nature03595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya C., Aggarwal S., Zhu R., Kumar M., Zhao M., Meistrich M. L., Matin A. The mouse dead-end gene isoform alpha is necessary for germ cell and embryonic viability. Biochem Biophys Res Commun. 2007;35:194–199. doi: 10.1016/j.bbrc.2007.01.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linger R., Dudakia D., Huddart R., Tucker K., Friedlander M., Phillips K. A., Hogg D., Jewett M. A., Lohynska R., Daugaard G., Richard S., Chompret A., Stoppa-Lyonnet D., Bonaïti-Pellié C., Heidenreich A., Albers P., Olah E., Geczi L., Bodrogi I., Daly P. A., Guilford P., Fosså S. D., Heimdal K., Tjulandin S. A., Liubchenko L., Stoll H., Weber W., Einhorn L., McMaster M., Korde L., Greene M. H., Nathanson K. L., Cortessis V., Easton D. F., Bishop D. T., Stratton M. R., Rapley E. A. Analysis of the DND1 gene in men with sporadic and familial testicular germ cell tumors. Gene Chromosome Cancer. 2008;47:247–252. doi: 10.1002/gcc.20526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya C., Aggarwal S., Kumar M., Ali A., Matin A. Mouse apolipoprotein B editing complex 3 (APOBEC3) is expressed in germ cells and interacts with dead-end (DND1) PLoS ONE. 2008;5:e2315. doi: 10.1371/journal.pone.0002315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J., Liang Z., Yang B., Tian H., Ma J., Zhang H. Derepression of microRNA-mediated protein translation inhibition by apolipoprotein B mRNA-editing enzyme catalytic polypeptide-like 3G (APOBEC3G) and its family members. J Biol Chem. 2007;282:33632–33640. doi: 10.1074/jbc.M705116200. [DOI] [PubMed] [Google Scholar]
- Gallois-Montbrun S., Kramer B., Swanson C. M., Byers H., Lynham S., Ward M., Malim M. H. Antiviral protein APOBEC3G localizes to ribonucleoprotein complexes found in P bodies and stress granules. J Virol. 2007;81:2165–2178. doi: 10.1128/JVI.02287-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedde M., Strasser M. J., Boldajipour B., Oude Vrielink J. A., Slanchev K., le Sage C., Nagel R., Voorhoeve P. M., van Duijse J., Orom U. A., Lund A. H, Perrakis A., Raz E., Agami R. RNA binding protein Dnd1 inhibits microRNA access to target mRNA. Cell. 2007;131:1273–1286. doi: 10.1016/j.cell.2007.11.034. [DOI] [PubMed] [Google Scholar]
- Ketting R. A dead end for microRNA. Cell. 2007;131:1226–1227. doi: 10.1016/j.cell.2007.12.004. [DOI] [PubMed] [Google Scholar]
- Slanchev K., Stebler J., Goudarzi M., Cojocaru V., Weidinger G., Raz E. Control of dead end localization and activity Implications for the function of the protein in antagonizing miRNA function. Mech Dev. 2008;126:270–277. doi: 10.1016/j.mod.2008.10.006. [DOI] [PubMed] [Google Scholar]
- Krovel A. V., Olsen L. C. Expression of a vas::EGFP transgene in primordial germ cells of the zebrafish. Mech Dev. 2002;116:141–150. doi: 10.1016/s0925-4773(02)00154-5. [DOI] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Carter S., Karl D. Inorganic phosphate assay with malachite green: an improvement and evaluation. J Biochem Biophys Methods. 1982;7:7–13. doi: 10.1016/0165-022x(82)90031-8. [DOI] [PubMed] [Google Scholar]
- Bernstein J., Patterson D., Wilson J., Toth E. Characterization of the essential activities of Saccharomyces cerevisiae Mtr4p, A 3′-5′ helicase partner of the nuclear exosome. J Biol Chem. 2008;283:4930–4942. doi: 10.1074/jbc.M706677200. [DOI] [PubMed] [Google Scholar]
- Sakowicz R., Berdelis S. M., Ray K., Blackburn C. L., Hopmann C., Faulkner D. J., Goldstein L. B. A marine natural product inhibitor of kinesin motors. Science. 1998;280:292–295. doi: 10.1126/science.280.5361.292. [DOI] [PubMed] [Google Scholar]
- Kimura Y., Kurzydlowski K., Tada M., MacLennan D. H. Phospholamban regulates the Ca2+-ATPase through intramembrane interactions. J Biol Chem. 1996;271:21726–21731. doi: 10.1074/jbc.271.36.21726. [DOI] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- LeBel D., Poirier G. G., Phaneuf S., St. Jean P., Laliberte J. F., Beaudoin A. R. Characterization and purification of a calcium-sensitive ATP diphosphohydrolase from pig pancreas. J Biol Chem. 1980;255:1227–1233. [PubMed] [Google Scholar]
- Tourneur L., Mistou S., Schmitt A., Chiocchia G. Adenosine receptors control a new pathway of Fas-associated death domain protein expression regulation by secretion. J Biol Chem. 2008;283:17929–17938. doi: 10.1074/jbc.M802263200. [DOI] [PubMed] [Google Scholar]
- Liu L., Hong N., Xu H., Li M., Yan Y., Purwanti Y., Yi M., Li Z., Wang L., Hong Y. Medaka dead end encodes a cytoplasmic protein and identifies embryonic and adult germ cells. Gene Expr Patterns. 2009;9:541–548. doi: 10.1016/j.gep.2009.06.008. [DOI] [PubMed] [Google Scholar]
- Walker J. E., Saraste M., Runswick M. J., Gay N. J. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1:945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarne A., Junop M. S., Yang W. Structure and function of the N-terminal 40-kDa fragment of human PMS2: a monomeric GHL ATPase. EMBO J. 2001;20:5521–5531. doi: 10.1093/emboj/20.19.5521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chene P. ATPase as drug targets: learning from their structure. Nat Rev Drug Discov. 2002;9:665–673. doi: 10.1038/nrd894. [DOI] [PubMed] [Google Scholar]
- Fry D. C., Kuby S. A., Mildvan A. S. ATP-binding site of adenylate kinase: mechanistic implications of its homology with ras-encoded p21, F1-ATPase, and other nucleotide-binding proteins. Proc Natl Acad Sci U S A. 1986;83:907–911. doi: 10.1073/pnas.83.4.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Query C. C., Bentley R.C., Keene J. D. A specific 31-nucleotide domain of U1 RNA directly interacts with the 70K small nuclear ribonucleoprotein component. Cell. 1989;57:89–101. doi: 10.1128/mcb.9.11.4872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandziulis R. J., Swanson M. S., Dreyfuss G. RNA binding proteins as developmental regulators. Genes Dev. 1989;3:431–437. doi: 10.1101/gad.3.4.431. [DOI] [PubMed] [Google Scholar]
- Bleichert F., Baserga S. The long unwinding road of RNA helicases. Mol Cell. 2007;27:339–352. doi: 10.1016/j.molcel.2007.07.014. [DOI] [PubMed] [Google Scholar]
- Lorsch J. RNA chaperones exist and DEAD box proteins get a life. Cell. 2002;109:797–800. doi: 10.1016/s0092-8674(02)00804-8. [DOI] [PubMed] [Google Scholar]
- Linder P. Dead-box proteins: a family affair—active and passive players in RNP-remodeling. Nucleic Acids Res. 2006;34:4168–4180. doi: 10.1093/nar/gkl468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abramson R. D., Dever T. E., Lawson T. G., Ray B. G., Thach R. E., Merrick W. C. J. Biol Chem. 1987;262:3826–3832. [PubMed] [Google Scholar]
- Sheng J., Zhang L., Zhao R. Biochemical characterization of the ATPase and helicase activity of UAP56, an essential pre-mRNA splicing and mRNA export factor. J Biochem Chem. 2007;282:22544–22550. doi: 10.1074/jbc.M702304200. [DOI] [PubMed] [Google Scholar]
- Rogers G. W., Richter N. J., Lima W. F., Merrick W. C. Modulation of the helicase activity of eIF4A by eIF4B, eIF4H and eIF4F. J Biol Chem. 2001;276:30914–30922. doi: 10.1074/jbc.M100157200. [DOI] [PubMed] [Google Scholar]
- Weirich C. S., Erzberger J. P., Flick J. S., Berger J. M., Thorner I., Weis W. Activation of the DExD/H-box protein Dbp5 by the nuclear-pore protein Gle1 and its coactivator InsP6 is required for mRNA export. Nat Cell Biol. 2006;8:668–676. doi: 10.1038/ncb1424. [DOI] [PubMed] [Google Scholar]
- Granneman S., Lin C. Y., Champion E. A., Nandineni M. R., Zorca C., Baserga S. J. The nucleolar protein Esf2 interacts directly with the DExD/H box RNA helicase, Dbp8, to stimulate ATP hydrolysis. Nucleic Acids Res. 2006;34:3189–3199. doi: 10.1093/nar/gkl419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishima Y., Giraldez A. J., Takeda Y., Fujiwara T., Sakamoto H., Schier A. F., Inoue K. Differential regulation of germline mRNAs in soma and germ cells by zebrafish miR-430. Curr Biol. 2006;16:2135–2142. doi: 10.1016/j.cub.2006.08.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.