Abstract
Environmental influence on developmental plasticity impacts a wide diversity of animal life from insects to humans. We now understand the epigenetic basis for many of these altered phenotypes. The five environmental factors of nutrition, behavior, stress, toxins and stochasticity work individually and in concert to affect the developing epigenome. During early embryogenesis, epigenetic marks, such as DNA methylation, are reset at specific times. Two waves of global demethylation and reestablishment of methylation frame the sensitive times for early environmental influences and will be the focus of this review. Gene transcription, translation and post-translational modification of chromatin remodeling complexes are three mechanisms affected by developmental exposure to environmental factors. To illustrate how changes in the early environment profoundly affect these mechanisms, we provide examples throughout the animal kingdom. Herein we review the history, time points and mechanisms of epigenetic gene-environment interaction.
Key words: epigenetics, development, environment, DNA methylation, plasticity, environmental epigenomics
Background
The early environment during development is emerging as a strong predictor of phenotype and disease in later life. When and how does the environment alter these later life outcomes? The “developmental origins of adult disease” hypothesis posits gene-environment interactions that result in long-lasting effects and suggests epigenetic inheritance as a prime mechanism.1 These early environmental influences can be dietary (total caloric intake, specific nutrient level, phytochemicals), physical (behavior, temperature, species density, stress), chemical (toxins, endocrine disruptors, pharmaceuticals) or unknown (stochastic, random effects). Traditionally, epigenetics has been defined as changes in gene expression in the absence of underlying changes in genetic information. More recent, refinement in the usage of the term specifies that epigenetic changes must be heritable from cell to cell, hence through cell lineage development or even transgenerationally from parent to offspring to grand-offspring.2 Thus, the convergence of evolutionary developmental biology, environmental toxicology and epigenetics is particularly important at the earliest stages of development when epigenetic modifications, such as DNA methylation, are the most sensitive to perturbation resulting in lifelong and possibly transgenerational effects.
The importance of early environment influences in modifying developmental trajectory has a long and colorful history leading from Jean-Baptiste Lamarck's idea that use of a body part would cause a heritable increase in the size of that body part.3 His proposed mechanism was that organisms have a “tendency to progression” and that offspring can inherit traits acquired by the habits of the parents.4 Early environmental manipulations were tried by discredited Soviet biologist Trofim Lysenko in his claims that crops could be adapted to cold climates by exposing seeds to cold temperatures.5 This culminated in his attempts at feeding special diets to gestating cattle hybrids to produce offspring with greater milk productivity.6 Such misconceptions persisted despite early refutations such as August Weismann's experiment in cutting the tails off of rats over five generations while never observing the birth of a tailless rat.7 He explicitly refuted the idea of soft inheritance by proposing what is now called the “Weismann barrier,” stating that germ cells cannot inherit modifications acquired by the body. More recent studies, however, have shown that with an understanding of molecular mechanisms, we can better establish the link between the early environment and adult disease. Indeed, epigenetic changes resulting from early environmental exposure are being newly discovered at a rapid pace. The goal of this review is to examine the timing of DNA methylation reprogramming and the molecular mechanisms by which this and other epigenetic marks can be modified. DNA methylation is primarily a stable repressive mark; however, its regulation is more dynamic than previously believed, and it can be actively removed at specific loci and genome-wide at several stages during development.8 The early environmental time points we focus on are the post-fertilization and germ cell differentiation stages in male and female offspring. Here we showcase examples in animals from insect to man where the environment influences the epigenome through early developmental exposures.
The Five Early Developmental Influences Resulting in Lifelong Phenotypic Change
Nutrition.
Nutrition in early life is never guaranteed or consistent. In bees (Apis mellifera), early life nutritionally induced changes are the underlying cause of queen and worker honeybee differentiation. Bee larva fed royal jelly, a diet specially enhanced with royalactin proteins, shifts development to the queen phenotype and shows similar effects in the fruit fly (Drosophila melanogaster).9 Furthermore, recent work supports the notion that DNA methylation is a primary mechanism by which royal jelly acts on the genome.10 The methylation of dynactin p62 is decreased in worker bee heads as compared to bodies and averages 10% lower methylation in the queen bee's complementary tissues, an effect that has been experimentally induced by siRNA-mediated silencing of Dnmt3. For mice, in utero supplementation with methyl donor rich diets increases methylation and suppresses transcription of the Runx3 gene in lung tissue11 and permanently shifts the coat color pattern of mice carrying the Agouti viable yellow allele.12 Dietary phytoestrogens interact with the methyl donor pathway to similarly shift the coat color distribution of Agouti viable yellow mice.13 Whole genome hypomethylation is also seen in offspring from dams receiving folate deficient diets during pregnancy and lactation.14 An early post-natal methyl donor deficient diet also reduces methylation at the imprinted gene, Igf2.15 In fact, in utero malnutrition in rodents not only directly affects the expression and methylation of several genes, such as glucocorticoid receptor, Nr3c1 and Pparα, but also the neonatal response to leptin; moreover, these epigenetic effects persist later in life and affect the ultimate adult phenotype, adiposity.16 Humans are also affected by early life nutritional status as shown in the DNA methylation changes at the IGF2 locus in whole blood from individuals subject to the Dutch hunger winter.17 Human longevity also appears correlated to food abundance available to our grandparents during their prepubertal growth, an effect hypothesized to be epigenetic in origin, though the direct epigenetic mechanism remains unknown.18
Behavior.
Behaviorally induced changes are likewise widespread from insects to mammals. The desert locust (Schistocerca gregaria) produces more offspring of the gregarious swarming phenotype when breeding in crowded conditions.19 Similarly, rats show persistent DNA methylation changes of the glucocorticoid receptor and many other loci in the hippocampus due to high versus low levels of maternal grooming in the first week of life.20 Falling under both behavior and stress, humans abused in early life also show increased DNA methylation at the NR3C1 glucocorticoid receptor promoter in the hippocampus.21
Stress.
Stress induces epigenetically controlled phenotypic changes in many animals. Recent work shows widely varying genomic methylation levels between insect species and that methylation level changes within a species during development suggesting a role for methylation in gene-environment interactions in insects.22 The pea aphid (Acyrthosiphon pisum) has a functional DNA methylation system and, under stress from crowded conditions or predators, will produce more winged offspring.23 The crowded mothers express more DNMT enzyme and the winged offspring show increased methylation at the apJHBP locus.24 Emerging evidence suggests that regulation of methylation is associated with stress and environmental response genes in the pacific oyster (Crassostrea gigas) and in the basal chordate Ciona intestinalis.25,26 After being in close proximity to cats, rats exhibit symptoms of post-traumatic stress syndrome concomitant with increases of methylation in the Bdnf gene in the hippocampus.27 Increased methylation of this gene is also seen in human suicide victims.28 In mice, stress in early life results in increased adult brain expression of arginine vasopressin (AVP) concomitant with increased methylation of the Avp gene in neurons.29 Stress from maternal separation in mice results in depressive behavior coupled with increased DNA methylation at Mecp2 and Cb1 and decreased methylation at Crfr2.30 Interestingly, these mice can transmit this phenotype and DNA methylation pattern transgenerationally through the male line with the shift of methylation in F1 sperm being mirrored by its pattern in F2 brain.30 Early life stress in humans is also linked with gene expression changes for a polymorphic form of serotonin receptor.31
Toxicants.
Toxins are widely dispersed and comprise the fourth category of early life epigenetic modifiers. Water fleas (Daphnia magna) show decreased DNA methylation when reared in the presence of vinclozolin, a fungicide and endocrine disruptor.32 Female rats exposed to vinclozolin during gestation produce male offspring with methylation changes in numerous genes in their sperm and female offspring with a greater incidence of tumor formation and pregnancy abnormalities.33–35 Because F1 and F2 germ cells are present in the exposed gestating female, these cells were therefore also exposed to vinclozolin during early development and indeed, these effects persist even transgenerationally. Bisphenol A (BPA), a widely studied endocrine disruptor, is ubiquitous in our environment and has been repeatedly shown to affect DNA methylation in multiple rodent tissues such as liver and brain.36,37 In primates, early exposure to lead (Pb) results in decreased DNA methyltransferase activity in the brain even 23 years later, suggestive of decreased DNA methylation, though this remains to be directly tested.38 Given the emerging weight of evidence linking developmental toxicant exposures to later disease states in animal models via methylation, unraveling these potential human health effects is particularly crucial.
Stochasticity.
Lastly, stochastically placed methylation marks laid down in early development have been observed at several loci. The well-studied viable yellow (Avy) mouse varies from brown, pseudoagouti, to yellow fur coloration due to randomly established levels of methylation at a recent contraoriently inserted intracisternal A-particle element (IAP) within the 5′ end of the Agouti gene. DNA methylation can vary by over 80% at several CpG sites within this IAP between animals.39 Similarly the CabpIAP locus in C57BL/6 mice also contains a contraoriented IAP element in intron 6, capable of stochastic DNA methylation.40 The Axin-fused (AxinFu) mouse has a dramatic kinky tail phenotype caused by another intronic metastable IAP element.41 These alleles are termed “metastable epialleles,” as they are variably expressed in genetically identical organisms due to epigenetic modifications that are established during early development.42 These elements are variable between individuals but consistent and stable in their patterns within a mouse throughout its life, implying that the level of methylation is set early in development and stable for life. The distribution of variable expressivity has been shifted at these metastable epialleles following maternal exposure to nutritional and environmental factors.12,43–47 It is likely that the underlying stochastic distribution of methylation at metastable epialleles may be affected by as yet uncharacterized environmental factors.
The five early developmental influences described here, nutrition, behavior, stress, toxins and stochasticity interact to influence methylation and other epigenetic marks that in turn affect life-stage phenotype and disease (Fig. 1). As indicated by the wide number of animal species discussed, it is likely that the capacity for epigenetic plasticity is evolutionarily selected and therefore likely that many more instances of environmental epigenetic influences remain to be elucidated.48 Of important note, however, not all animals use DNA methylation as a gene repression mechanism; for example the model organisms fruit fly (Drosophila melanogaster) and roundworm (Caenorhabditis elegans).
Figure 1.
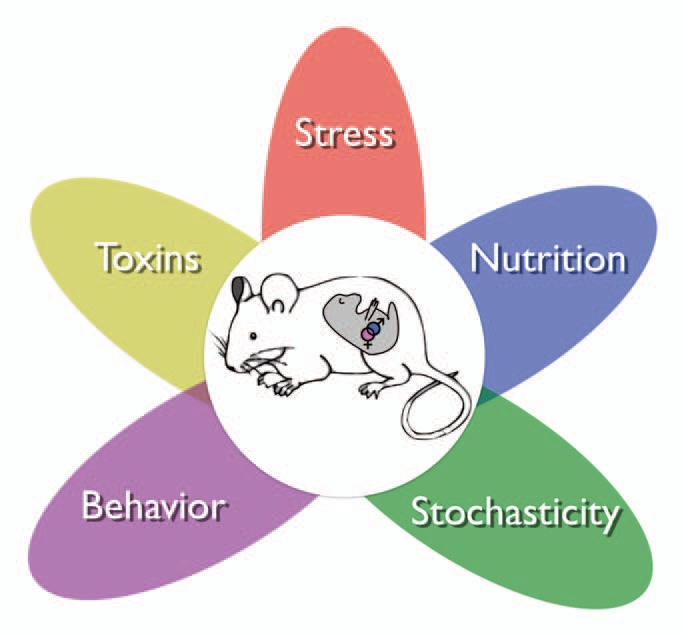
Environmental factors working individually and in concert. Five environmental influences that affect the developing embryo and its primordial germ cells (represented by the pink and blue dots). Each of these factors can act through a variety of mechanisms and result in an array of changes in epigenetic marks.
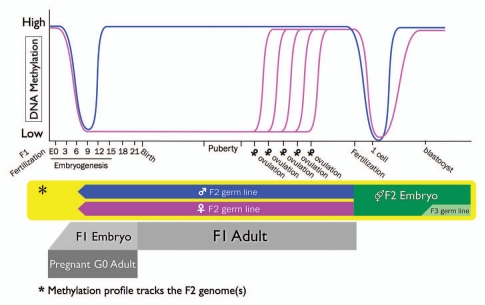
Time Points of DNA Methylation Lability
In the context of early environmentally modifiable epigenetic marks, it is important to determine the windows of greatest susceptibility. For example, the epigenome is most vulnerable to environmental factors during embryogenesis because the DNA synthetic rate is high, and the elaborate DNA methylation patterning required for normal tissue development is established during early development. The mammalian genome undergoes two waves of global DNA demethylation followed by de novo methylation, as illustrated in Figure 2 using the mouse as a representative mammalian animal model.49 In mammals, the mother, G0, hosts the development of the F1 offspring from zygote stage to birth. During the development of the F1 offspring, a separate lineage of cells within the F1, called the primordial germ cells (PGCs), migrate and differentiate into gamete precursor cells that will eventually become the F2 generation. By convention, the “first wave” of methylation resetting refers to the reprogramming of the epigenome within these PGCs, and the second wave refers to the reprogramming that happens shortly after zygote formation. Exposure of a pregnant mother can affect methylation status of both the first wave (in the F2 PGCs) and the second wave (in the post-fertilization F1 pluripotent somatic cells).
Figure 2.
Methylation reprogramming in a single genome occurs in two waves. Global demethylation events from the perspective of the mouse F2 genome from germ cells to newly combined somatic embryo. Within the pregnant G0 mouse, the F1 embryo generates a group of cells destined to become its gametes, which will form the F2 generation. These primordial germ cells (PGC) begin to migrate to the genital ridge around embryonic day 7.25 in the mouse, during which time they become demethylated in preparation to adopt the somatic methylation pattern and, for imprinted genes, the gender specific methylation pattern to match the genotype of the individual in which they now reside. In males, methylation is reestablished by E14. In females the PGC remain largely unmethylated until maturation in the F1 adult during each estrous cycle. During fertilization, the F2 gametes combine and undergo the second, more complete, wave of demethylation in preparation to establish somatic methylation patterns (with the exception of the F3 PGCs). Any environmental influences on the pregnant G0 adult can affect the development and adult disease susceptibility of both the F1 and F2 generations as their somatic and germline methylation patterns are being established, respectively.
Germ cell methylation reprogramming.
In mice, the first wave of reprogramming occurs in primordial germ cells (PGCs) during and after their migration to the genital ridge beginning at E7.25.50 This demethylation is largely complete in the mouse by E13.5 and allows resetting of imprinted genes in the PGCs to match the sex of the host in which they now find themselves.51,52 Many repetitive elements are also protected from demethylation to varying extents during this wave.53 Interestingly, this demethylation event appears to involve a base excision repair pathway.54 For humans, this wave of PGC reprogramming is especially important given our complement of imprinted genes and the high content of repetitive elements in our genomes. These genetic loci are more susceptible to loss of heterozygosity and increased methylation instability, respectively.55
In male mouse fetuses, PGCs differentiate into prospermatogonia, enter mitotic arrest and reestablish methylation starting at E15; this is the time point at which paternal sex-specific imprints are set.56,57 Consequently, this window is especially important for disruptions to loci that escape demethylation as well as resetting of global methylation in male offspring with any effects likely to be seen in the F2 generation.33,58 Non-mammalian animals, in general, do not have imprinted genes.53 Developmental exposures may affect the growth of the offspring by inheritance without necessarily having a lasting impact on the parent.
After sexual maturation in all male mammals, the prospermatogonia complete meiosis and differentiate into mature sperm and during this process, the chromosomes are almost entirely stripped of histones and repackaged with highly basic protamines. Because the protamines do not contain any modifiable tails, any epigenetic information carried on histones is unable to be passed through the male germ line.59 A small number of histones are retained in mammalian sperm; however, it is unknown whether they play a role in passing on any epigenetic information to the resulting zygote.57,60
In contrast to males, F1 female mammalian PGCs complete meiosis I while still in the developing embryo, followed by cell arrest until puberty.57 Thus, in human females, for example, the oocytes remain in a haploid demethylated state for years; therefore, the window of possible disruption to the establishment of methylation patterns in oocytes is much longer and repeatedly occurs during the maturation of each egg throughout fertility (Fig. 2).52
Zygotic methylation reprogramming.
The second wave of global demethylation occurs shortly after fertilization and before implantation. The male pronucleus is stripped of the protamines while DNA is actively demethylated and repackaged with newly synthesized histones in the zygote.61,62 The female complement of chromosomes becomes demethylated via a passive mechanism during replication.63 Not every gene is demethylated, since the oocyte contains egg-specific isoforms of DNMT1 and the early embryo synthesizes its own somatic DNMT1 isoform. The presence of these maintenance methyltransferases is required to ensure the preservation of gametically derived differential methylation for imprinted genes, particularly during global demethylation of most other regions of the genome.64 By blastocyst stage, E3.5 in mice and E5 in humans, DNMT1 is present in the nucleus and by E3.5 DNMT3b also relocates to the nucleus and the genome becomes fully remethylated.63,65 This round of demethylation is less comprehensive than the reprogramming in PGCs, with imprinting control regions retaining differential methylation depending on their parent of origin and some classes of repetitive elements retaining methylation.51 This wave of methylation cycling sets the pattern for all somatic cells in the resulting embryo and adult except for the PGCs, which will form the gametes for the next generation. The embryo (F1) would be most vulnerable during this window to environmental influences disrupting reestablishment of DNA methylation.
Somatic methylation lability.
The major global demethylation event that occurs in the somatic cells of adults is associated with aging and disease states.66 As mammals age, they undergo gradual DNA hypomethylation genome-wide concomitant with hypermethylation at the normally unmethylated CpG islands.67 Cancers are also widely hypomethylated as compared to normal tissue with notable hypermethylation of tumor suppressor genes.68 A complete review of methylation dynamics of aging and disease is outside the scope of this review but is particularly important in the understanding of environmental effects on the epigenome.
Mechanistic Targets of Environmental Exposure
Mounting evidence suggests that environmental pressures can exert effects on multiple levels of gene regulation. The weight of evidence supports three molecular targets, including gene transcription, protein translation and the post-translational modification of chromatin remodeling complexes. As indicated below, the ability of early environmental fluctuations to affect these molecular processes long-term is not necessarily uniform.
Gene transcription.
Gene transcription rates can be suppressed by DNA methylation of CpG islands and promoters as well as histone modification and nucleosome placement. For example the methyl group of the 5-methyl-cytosine extends into the major groove of DNA, inhibiting transcription by interfering with transcription factor binding proteins. In addition, DNMTs and methylated DNA interact with higher order chromatin proteins, such as the repressive Polycomb group (PcG) protein, Enhancer of Zeste homolog 2 (EZH2), to affect histone modifications and further compact chromatin. Thus, it is not only the modifications of DNA base pairs but also the higher order structure which controls access to transcription factors.
Any of these epigenetic marks can create a metastable epiallele, a locus of identical sequence harboring different epigenetic marks, most commonly DNA methylation, in different individuals. As introduced previously, the insertion of a metastable IAP in the Agouti gene in the Avy mouse strain provides a locus for variable DNA methylation to affect transcription. The CabpIAP, AxinFu and Run3x loci also show a correlation of increased methylation and decreased expression affected by environmental exposures. These loci provide a basis for the investigation of genome-wide changes due to methylation and a number of studies are finding novel metastable epialleles with variable methylation.69,70
Histone modifications are also targets of environmental exposure that vary in concert with DNA methylation and subsequently affect gene transcription.36,71 The numerous modifications of histone tails, collectively referred to as the histone code, comprise over 100 modifications, with many of these serving to modify transcriptional activity.72 The heritable propagation of histone marks such as the repressive H3K27me3, placed by the polycomb repressive complex 2 (PRC2), is beginning to be understood. For example, widespread trans effects are caused when the PRC2 subunit EED is mutated, normally transmitted H3K27me3 modifications are no longer transmitted and development is affected.73
Protein translation.
While environmental exposure can influence transitory effects on protein translation, it is unlikely to be a pathway of directly maintaining early environmentally induced changes over the life-course. This is due to the fact that mRNA is an inherently short-lived molecule serving as an intermediary to a translated protein. Similarly, methylation of transfer RNA (tRNA) by DNMT2 is unlikely to be a mediator of long-term changes but has been shown to be reduced in azacytidine exposed cells.74 The existence of paramutation in mice potentially provides a mechanism by which changes in small RNA levels could be maintained across many cell divisions thereby affecting translation levels long after the removal of the environmental stimulus. The most famous example of this phenomenon is the mutant phenotypic transmission to wild-type offspring resulting in white tail tips by transmission of mutant RNA or microRNA targeting the Kit locus in mice; however, these results are still quite controversial.75,76 Small, micro and anti-sense RNAs could and likely do alter the placement of other epigenetic marks, which are themselves stably maintained from early development to cause phenotypic changes long after the cessation of exposure that caused the initial shift in RNA-induced changes.
Chromatin remodeling complexes.
Post-translational modification of DNA-binding proteins is also a target of early environmental influences leading to changes in adult gene expression. Since chromatin modifying complexes act throughout the genome, relatively small changes in their activity can cause widespread trans effects. The methyltransferase EZH2 catalyzes H3K27 trimethlyation as part of the PRC2 and can be phosphorylated by the cyclin dependent kinases, CDK1 and CDK2. After phosphorylation it loses the ability to bind to non-coding RNAs and is thus unable to place the H3K27me3 mark in a sequence specific manner.77 The cell-to-cell inheritance of this mark is thereby disrupted and has implications for development. Any environmental influences that alter the activity of EZH2, or its upstream CDK enzymes, can thereby have widespread developmental effects lasting into adulthood. Placental mammals also have Rex1, a DNA-binding transcription factor thought to target chromatin remodeling complexes.78 In mouse Rex1 null mutants, the DNA methylation at the differentially methylated regions of several imprinted loci is disrupted in adult tissues despite the fact that Rex1 is only expressed very early in development.79
Moving Forward
It is increasingly recognized that environmental exposure to chemical, nutritional and behavioral factors alters gene expression and affects health and disease by not only mutating promoter and coding regions of genes, but also by modifying the epigenome. The investigation of early environmental effects can inform the fields of toxicology and environmental epidemiology by elucidating the mechanisms underlying developmental exposure and adult disease. Of the five environmental factors acting in development, nutrition is the best studied. Given the ubiquity of environmental toxins in our environment, and the proliferation of new compounds, their effects, singular and in combination, are in need of the most urgent study. These along with studies of stress and behavior will increase our understanding of the multiple factors governing stochastic changes that are commonly seen in model organisms.
In order to translate epigenetic research to risk assessment or clinical practice we must first understand the most sensitive time points in the resetting of epigenetic marks. These vulnerable periods are likely distinct for males and females as well as for offspring and grand-offspring and may require specialized preventive and/or corrective actions. Additionally, the molecular mechanisms linking these sensitive time points to the adult presentation of disease need to be fully characterized. Environmentally induced disruptions in DNA methylation and histone modifications are currently best characterized while other mechanisms require further investigation. For example, posttranscriptional modification of chromatin modifying enzymes early in life can have profound consequences on the adult epigenome. Ultimately, researchers must integrate the layers of epigenetic changes with the times of sensitivity to understand and generate the best prescriptions for human health and disease.
Acknowledgments
We thank Dr. Craig Harris for critically reading this manuscript and Dr. Carl Anderson for assistance with figures. Research support was provided by NIH grants T32 ES007062 (C.F.), ES017524 (D.C.D.), and the University of Michigan NIEHS P30 Core Center P30ES017885 as well as NIH/EPA P20 grant ES018171/RD 83480001.
Abbreviations
- PGC
primordial germ cells
- GR
glucocorticoid receptor
- IAP
intracisternal A-particle
- PRC2
polycomb repressive complex 2
- tRNA
transfer RNA
- BPA
bisphenol A
References
- 1.Barker DJ, Eriksson JG, Forsen T, Osmond C. Fetal origins of adult disease: strength of effects and biological basis. Int J Epidemiol. 2002;31:1235–1239. doi: 10.1093/ije/31.6.1235. [DOI] [PubMed] [Google Scholar]
- 2.Youngson NA, Whitelaw E. Transgenerational epigenetic effects. Annu Rev Genomics Hum Genet. 2008;9:233–257. doi: 10.1146/annurev.genom.9.081307.164445. [DOI] [PubMed] [Google Scholar]
- 3.Lamarck JB. Philosophie Zoologique ou exposition des considérations relatives à l'histoire naturelle des animaux. Dentu et L'Auteur. 1809 (Fre). [Google Scholar]
- 4.Gould SJ. The Structure of Evolutionary Theory. Cambridge, MA: Belknap Press of Harvard University Press; 2002. [Google Scholar]
- 5.Soyfer VN. The consequences of political dictatorship for Russian science. Nat Rev Genet. 2001;2:723–729. doi: 10.1038/35088598. [DOI] [PubMed] [Google Scholar]
- 6.News of Science. Science. 1957;126:157–161. doi: 10.1126/science.126.3265.157. [DOI] [PubMed] [Google Scholar]
- 7.Weismann A. Essays Upon Heredity. London: Oxford at the Clarendon Press; 1891. [Google Scholar]
- 8.Wu SC, Zhang Y. Active DNA demethylation: many roads lead to Rome. Nat Rev Mol Cell Biol. 2010;11:607–620. doi: 10.1038/nrm2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kamakura M. Royalactin induces queen differentiation in honeybees. Nature. 2011;473:478–483. doi: 10.1038/nature10093. [DOI] [PubMed] [Google Scholar]
- 10.Kucharski R, Maleszka J, Foret S, Maleszka R. Nutritional control of reproductive status in honeybees via DNA methylation. Science. 2008;319:1827–1830. doi: 10.1126/science.1153069. [DOI] [PubMed] [Google Scholar]
- 11.Hollingsworth JW, Maruoka S, Boon K, Garantziotis S, Li Z, Tomfohr J, et al. In utero supplementation with methyl donors enhances allergic airway disease in mice. J Clin Invest. 2008;118:3462–3469. doi: 10.1172/JCI34378. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 12.Waterland R, Jirtle R. Transposable elements: targets for early nutritional effects on epigenetic gene regulation. Mol Cell Biol. 2003;23:5293–5300. doi: 10.1128/MCB.23.15.5293-5300.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dolinoy DC, Weidman JR, Waterland RA, Jirtle RL. Maternal genistein alters coat color and protects Avy mouse offspring from obesity by modifying the fetal epigenome. Environ Health Perspect. 2006;114:567–572. doi: 10.1289/ehp.8700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McKay JA, Waltham KJ, Williams EA, Mathers JC. Folate depletion during pregnancy and lactation reduces genomic DNA methylation in murine adult offspring. Genes Nutr. 2010;6:189–196. doi: 10.1007/s12263-010-0199-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Waterland RA, Lin JR, Smith CA, Jirtle RL. Post-weaning diet affects genomic imprinting at the insulin-like growth factor 2 (Igf2) locus. Hum Mol Genet. 2006;15:705–716. doi: 10.1093/hmg/ddi484. [DOI] [PubMed] [Google Scholar]
- 16.Gluckman PD, Lillycrop KA, Vickers MH, Pleasants AB, Phillips ES, Beedle AS, et al. Metabolic plasticity during mammalian development is directionally dependent on early nutritional status. Proc Natl Acad Sci USA. 2007;104:12796–12800. doi: 10.1073/pnas.0705667104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heijmans BT, Tobi EW, Stein AD, Putter H, Blauw GJ, Susser ES, et al. Persistent epigenetic differences associated with prenatal exposure to famine in humans. Proc Natl Acad Sci USA. 2008;105:17046–17049. doi: 10.1073/pnas.0806560105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaati G, Bygren LO, Pembrey M, Sjöström M. Transgenerational response to nutrition, early life circumstances and longevity. Eur J Hum Genet. 2007;15:784–790. doi: 10.1038/sj.ejhg.5201832. [DOI] [PubMed] [Google Scholar]
- 19.Maeno K, Tanaka S. Epigenetic transmission of phase in the desert locust, Schistocerca gregaria: determining the stage sensitive to crowding for the maternal determination of progeny characteristics. J Insect Physiol. 2010;56:1883–1888. doi: 10.1016/j.jinsphys.2010.08.010. [DOI] [PubMed] [Google Scholar]
- 20.McGowan PO, Suderman M, Sasaki A, Huang TCT, Hallett M, Meaney MJ, et al. Broad epigenetic signature of maternal care in the brain of adult rats. PLoS One. 2011;6:14739. doi: 10.1371/journal.pone.0014739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McGowan PO, Sasaki A, D'Alessio AC, Dymov S, Labonté B, Szyf M, et al. Epigenetic regulation of the glucocorticoid receptor in human brain associates with childhood abuse. Nat Neurosci. 2009;12:342–348. doi: 10.1038/nn.2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kronforst MR, Gilley DC, Strassmann JE, Queller DC. DNA methylation is widespread across social Hymenoptera. Curr Biol. 2008;18:R287–R288. doi: 10.1016/j.cub.2008.02.015. [DOI] [PubMed] [Google Scholar]
- 23.Weisser WW, Braendle C, Minoretti N. Predator-induced morphological shift in the pea aphid. Proceedings of the Royal Society of London Series B: Biological Sciences. 1999;266:1175–1181. [Google Scholar]
- 24.Walsh TK, Brisson JA, Robertson HM, Gordon K, Jaubert-Possamai S, Tagu D, et al. A functional DNA methylation system in the pea aphid, Acyrthosiphon pisum. Insect Mol Biol. 2010;19:215–228. doi: 10.1111/j.1365-2583.2009.00974.x. [DOI] [PubMed] [Google Scholar]
- 25.Gavery MR, Roberts SB. DNA methylation patterns provide insight into epigenetic regulation in the Pacific oyster (Crassostrea gigas) BMC Genomics. 2010;11:483. doi: 10.1186/1471-2164-11-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sasaki A, Satoh N. Effects of 5-aza-2′-deoxycytidine on the gene expression profile during embryogenesis of the Ascidian ciona intestinalis: a microarray analysis. Zoolog Sci. 2007;24:648–655. doi: 10.2108/zsj.24.648. [DOI] [PubMed] [Google Scholar]
- 27.Roth TL, Zoladz PR, Sweatt JD, Diamond DM. Epigenetic modification of hippocampal Bdnf DNA in adult rats in an animal model of post-traumatic stress disorder. J of Psychiatr Res. 2011 doi: 10.1016/j.jpsychires.2011.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Keller S, Sarchiapone M, Zarrilli F, Videtic A, Ferraro A, Carli V, et al. Increased BDNF promoter methylation in the Wernicke area of suicide subjects. Arch of Gen Psychiatry. 2010;67:258–267. doi: 10.1001/archgenpsychiatry.2010.9. [DOI] [PubMed] [Google Scholar]
- 29.Murgatroyd C, Patchev AV, Wu Y, Micale V, Bockmuhl Y, Fischer D, et al. Dynamic DNA methylation programs persistent adverse effects of early-life stress. Nat Neurosci. 2009;12:1559–1566. doi: 10.1038/nn.2436. [DOI] [PubMed] [Google Scholar]
- 30.Franklin TB, Russig H, Weiss IC, Graff J, Linder N, Michalon A, et al. Epigenetic transmission of the impact of early stress across generations. Biol Psychiatry. 2010;68:408–415. doi: 10.1016/j.biopsych.2010.05.036. [DOI] [PubMed] [Google Scholar]
- 31.Caspi A, Sugden K, Moffitt TE, Taylor A, Craig IW, Harrington H, et al. Influence of life stress on depression: moderation by a polymorphism in the 5-HTT gene. Science. 2003;301:386–389. doi: 10.1126/science.1083968. [DOI] [PubMed] [Google Scholar]
- 32.Vandegehuchte MB, Lemière F, Vanhaecke L, Vanden Berghe W, Janssen CR. Direct and transgenerational impact on Daphnia magna of chemicals with a known effect on DNA methylation. Comp Biochem Physiol C Toxicol Pharmacol. 2010;151:278–285. doi: 10.1016/j.cbpc.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 33.Guerrero-Bosagna C, Settles M, Lucker B, Skinner MK. Epigenetic transgenerational actions of vinclozolin on promoter regions of the sperm epigenome. PLoS ONE. 2010:5. doi: 10.1371/journal.pone.0013100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nilsson EE, Anway MD, Stanfield J, Skinner MK. Transgenerational epigenetic effects of the endocrine disruptor vinclozolin on pregnancies and female adult onset disease. Reproduction. 2008;135:713–721. doi: 10.1530/REP-07-0542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Anway MD, Cupp AS, Uzumcu M, Skinner MK. Epigenetic transgenerational actions of endocrine disruptors and male fertility. Science. 2005;308:1466–1469. doi: 10.1126/science.1108190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dolinoy DC, Weinhouse C, Jones TR, Rozek LS, Jirtle RL. Variable histone modifications at the A (vy) metastable epiallele. Epigenetics. 2010;5:637–644. doi: 10.4161/epi.5.7.12892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yaoi T, Itoh K, Nakamura K, Ogi H, Fujiwara Y, Fushiki S. Genome-wide analysis of epigenomic alterations in fetal mouse forebrain after exposure to low doses of bisphenol A. Biochem Biophys Res Commun. 2008;376:563–567. doi: 10.1016/j.bbrc.2008.09.028. [DOI] [PubMed] [Google Scholar]
- 38.Wu J, Basha MR, Brock B, Cox DP, Cardozo-Pelaez F, McPherson CA, et al. Alzheimer & aposis disease (AD)-like pathology in aged monkeys after infantile exposure to environmental metal lead (Pb): evidence for a developmental origin and environmental link for AD. J Neurosci. 2008;28:3–9. doi: 10.1523/JNEUROSCI.4405-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dolinoy DC, Weinhouse C, Jones TR, Rozek LS, Jirtle RL. Variable histone modifications at the A(vy) metastable epiallele. Epigenetics. 2010;5:637–644. doi: 10.4161/epi.5.7.12892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Druker R. Complex patterns of transcription at the insertion site of a retrotransposon in the mouse. Nucleic Acids Res. 2004;32:5800–5808. doi: 10.1093/nar/gkh914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vasicek TJ, Zeng L, Guan XJ, Zhang T, Costantini F, Tilghman SM. Two dominant mutations in the mouse fused gene are the result of transposon insertions. Genetics. 1997;147:777–786. doi: 10.1093/genetics/147.2.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rakyan VK, Blewitt ME, Druker R, Preis JI, Whitelaw E. Metastable epialleles in mammals. Trends Genet. 2002;18:348–351. doi: 10.1016/s0168-9525(02)02709-9. [DOI] [PubMed] [Google Scholar]
- 43.Cooney CA, Dave AA, Wolff GL. Maternal methyl supplements in mice affect epigenetic variation and DNA methylation of offspring. J Nutr. 2002;132:2393–2400. doi: 10.1093/jn/132.8.2393S. [DOI] [PubMed] [Google Scholar]
- 44.Dolinoy DC, Wiedman J, Waterland R, Jirtle RL. Maternal genistein alters coat color and protects Avy mouse offspring from obesity by modifying the fetal epigenome. Environ Health Perspect. 2006;114:567–572. doi: 10.1289/ehp.8700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dolinoy DC, Huang D, Jirtle RL. Maternal nutrient supplementation counteracts bisphenol A-induced DNA hypomethylation in early development. Proc Natl Acad Sci USA. 2007;104:13056–13061. doi: 10.1073/pnas.0703739104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kaminen-Ahola N, Ahola A, Maga M, Mallitt KA, Fahey P, Cox T, et al. Maternal Ethanol Consumption Alters the Epigenotype and the Phenotype of Offspring in a Mouse Model. PLoS Genet. 2010;6:1000811. doi: 10.1371/journal.pgen.1000811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Waterland RA, Dolinoy DC, Lin JR, Smith CA, Shi X, Tahiliani KG. Maternal methyl supplements increase offspring DNA methylation at Axin Fused. Genesis. 2006;44:401–406. doi: 10.1002/dvg.20230. [DOI] [PubMed] [Google Scholar]
- 48.Jablonka E, Raz G. Transgenerational epigenetic inheritance: prevalence, mechanisms and implications for the study of heredity and evolution. Q Rev Biol. 2009;84:131–176. doi: 10.1086/598822. [DOI] [PubMed] [Google Scholar]
- 49.Bernal AJ, Jirtle RL. Epigenomic disruption: the effects of early developmental exposures. Birth Defects Res A Clin Mol Teratol. 2010;88:938–944. doi: 10.1002/bdra.20685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yamazaki Y, Mann MRW, Lee SS, Marh J, McCarrey JR, Yanagimachi R, et al. Reprogramming of primordial germ cells begins before migration into the genital ridge, making these cells inadequate donors for reproductive cloning. Proc Natl Acad Sci USA. 2003;100:12207–12212. doi: 10.1073/pnas.2035119100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lees-Murdock DJ, Walsh CP. DNA methylation reprogramming in the germ line. Epigenetics. 2008;3:5–13. doi: 10.4161/epi.3.1.5553. [DOI] [PubMed] [Google Scholar]
- 52.Sasaki H, Matsui Y. Epigenetic events in mammalian germ-cell development: reprogramming and beyond. Nat Rev Genet. 2008;9:129–140. doi: 10.1038/nrg2295. [DOI] [PubMed] [Google Scholar]
- 53.Kaneda M, Okano M, Hata K, Sado T, Tsujimoto N, Li E, et al. Essential role for de novo DNA methyltransferase Dnmt3a in paternal and maternal imprinting. Nature. 2004;429:900–903. doi: 10.1038/nature02633. [DOI] [PubMed] [Google Scholar]
- 54.Hajkova P, Jeffries SJ, Lee C, Miller N, Jackson SP, Surani MA. Genome-wide reprogramming in the mouse germ line entails the base excision repair pathway. Science. 2010;329:78–82. doi: 10.1126/science.1187945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dolinoy DC, Jirtle RL. Environmental epigenomics in human health and disease. Environ Mol Mutagen. 2008;49:4–8. doi: 10.1002/em.20366. [DOI] [PubMed] [Google Scholar]
- 56.Reik W, Dean W, Walter J. Epigenetic reprogramming in mammalian development. Science. 2001;293:1089–1093. doi: 10.1126/science.1063443. [DOI] [PubMed] [Google Scholar]
- 57.Kota SK, Feil R. Epigenetic Transitions in Germ Cell Development and Meiosis. Developmental Cell. 2010;19:675–686. doi: 10.1016/j.devcel.2010.10.009. [DOI] [PubMed] [Google Scholar]
- 58.Hanel ML, Wevrick R. Establishment and maintenance of DNA methylation patterns in mouse Ndn: implications for maintenance of imprinting in target genes of the imprinting center. Mol Cell Biol. 2001;21:2384–2392. doi: 10.1128/MCB.21.7.2384-2392.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Balhorn R. The protamine family of sperm nuclear proteins. Genome Biol. 2007;8:227. doi: 10.1186/gb-2007-8-9-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gaucher J, Reynoird N, Montellier E, Boussouar F, Rousseaux S, Khochbin S. From meiosis to postmeiotic events: the secrets of histone disappearance. FEBS J. 2010;277:599–604. doi: 10.1111/j.1742-4658.2009.07504.x. [DOI] [PubMed] [Google Scholar]
- 61.Santos F, Hendrich B, Reik W, Dean W. Dynamic reprogramming of DNA methylation in the early mouse embryo. Dev Biol. 2002;241:172–182. doi: 10.1006/dbio.2001.0501. [DOI] [PubMed] [Google Scholar]
- 62.Nonchev S, Tsanev R. Protamine-histone replacement and DNA replication in the male mouse pronucleus. Mol Reprod Dev. 1990;25:72–76. doi: 10.1002/mrd.1080250113. [DOI] [PubMed] [Google Scholar]
- 63.Hemberger M, Dean W, Reik W. Epigenetic dynamics of stem cells and cell lineage commitment: digging Waddington's canal. Nat Rev Mol Cell Biol. 2009;10:526–537. doi: 10.1038/nrm2727. [DOI] [PubMed] [Google Scholar]
- 64.Cirio MC, Ratnam S, Ding F, Reinhart B, Navara C, Chaillet JR. Preimplantation expression of the somatic form of Dnmt1 suggests a role in the inheritance of genomic imprints. BMC Dev Biol. 2008;8:9. doi: 10.1186/1471-213X-8-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hirasawa R, Sasaki H. Dynamic transition of Dnmt3b expression in mouse pre- and early post-implantation embryos. Gene expression patterns: GEP. 2009;9:27–30. doi: 10.1016/j.gep.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 66.Calvanese V, Lara E, Kahn A, Fraga MF. The role of epigenetics in aging and age-related diseases. Ageing Res Rev. 2009;8:268–276. doi: 10.1016/j.arr.2009.03.004. [DOI] [PubMed] [Google Scholar]
- 67.Murgatroyd C, Wu Y, Bockmühl Y, Spengler D. The Janus face of DNA methylation in aging. Aging. 2010;2:107–110. doi: 10.18632/aging.100124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Feinberg AP. Phenotypic plasticity and the epigenetics of human disease. Nature. 2007;447:433–440. doi: 10.1038/nature05919. [DOI] [PubMed] [Google Scholar]
- 69.Luedi P, Hartemink A, Jirtle R. Genome-wide prediction of imprinted murine genes. Genome Res. 2005;15:875–884. doi: 10.1101/gr.3303505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Waterland RA, Kellermayer R, Laritsky E, Rayco-Solon P, Harris RA, Travisano M, et al. Season of conception in rural gambia affects DNA methylation at putative human metastable epialleles. PLoS Genet. 2010;6:1001252. doi: 10.1371/journal.pgen.1001252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Fernandez-Gonzalez R, Ramirez MA, Pericuesta E, Calle A, Gutierrez-Adan A. Histone modifications at the blastocyst Axin1(Fu) locus mark the heritability of in vitro culture-induced epigenetic alterations in mice. Biology of reproduction. 2010;83:720–727. doi: 10.1095/biolreprod.110.084715. [DOI] [PubMed] [Google Scholar]
- 72.Bannister AJ, Kouzarides T. Regulation of chromatin by histone modifications. Cell research. 2011;21:381–395. doi: 10.1038/cr.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Margueron R, Justin N, Ohno K, Sharpe ML, Son J, Drury WJ, et al. Role of the polycomb protein EED in the propagation of repressive histone marks. Nature. 2009;461:762–767. doi: 10.1038/nature08398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Schaefer M, Hagemann S, Hanna K, Lyko F. Azacytidine inhibits RNA methylation at DNMT2 target sites in human cancer cell lines. Cancer Res. 2009;69:8127–8132. doi: 10.1158/0008-5472.CAN-09-0458. [DOI] [PubMed] [Google Scholar]
- 75.Rassoulzadegan M, Grandjean V, Gounon P, Vincent S, Gillot I, Cuzin F. RNA-mediated non-mendelian inheritance of an epigenetic change in the mouse. Nature. 2006;441:469–474. doi: 10.1038/nature04674. [DOI] [PubMed] [Google Scholar]
- 76.Arnheiter H. Mammalian paramutation: a tail's tale? Pigment Cell Research. 2007;20:36–40. doi: 10.1111/j.1600-0749.2006.00351.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zeng X, Chen S, Huang H. Phosphorylation of EZH2 by CDK1 and CDK2: a possible regulatory mechanism of transmission of the H3K27me3 epigenetic mark through cell divisions. Cell Cycle. 2011;10:579–583. doi: 10.4161/cc.10.4.14722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kim JD, Faulk C, Kim J. Retroposition and evolution of the DNA-binding motifs of YY1, YY2 and REX1. Nucleic Acids Res. 2007;35:3442–3452. doi: 10.1093/nar/gkm235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kim JD, Kim H, Ekram MB, Yu S, Faulk C, Kim J. Rex1/Zfp42 as an epigenetic regulator for genomic imprinting. Hum Mol Genet. 2011;20:1353–1362. doi: 10.1093/hmg/ddr017. [DOI] [PMC free article] [PubMed] [Google Scholar]